DOI:
10.1039/C9RA02225B
(Review Article)
RSC Adv., 2019,
9, 24539-24559
Metal nanoparticles fabricated by green chemistry using natural extracts: biosynthesis, mechanisms, and applications
Received
22nd March 2019
, Accepted 5th July 2019
First published on 8th August 2019
Abstract
Nanoparticles (NPs) are new inspiring clinical targets that have emerged from persistent efforts with unique properties and diverse applications. However, the main methods currently utilized in their production are not environmentally friendly. With the aim of promoting a green approach for the synthesis of NPs, this review describes eco-friendly methods for the preparation of biogenic NPs and the known mechanisms for their biosynthesis. Natural plant extracts contain many different secondary metabolites and biomolecules, including flavonoids, alkaloids, terpenoids, phenolic compounds and enzymes. Secondary metabolites can enable the reduction of metal ions to NPs in eco-friendly one-step synthetic processes. Moreover, the green synthesis of NPs using plant extracts often obviates the need for stabilizing and capping agents and yields biologically active shape- and size-dependent products. Herein, we review the formation of metallic NPs induced by natural extracts and list the plant extracts used in the synthesis of NPs. In addition, the use of bacterial and fungal extracts in the synthesis of NPs is highlighted, and the parameters that influence the rate of particle production, size, and morphology are discussed. Finally, the importance and uniqueness of NP-based products are illustrated, and their commercial applications in various fields are briefly featured.
Introduction
The past decade has witnessed the vast development and involvement of nanomaterials in many different areas of research due to their unique optoelectronic and physicochemical properties.1,2 Nanoparticles (NPs) are a particularly important class of nanomaterials with applications in diverse fields including electronics,3,4 catalysis,5 sensing, water treatment,6,7 oil recovery,8 corrosion inhibition,9 and drug delivery. Although NPs are usually defined as particles with diameters in the range of 1–100 nm,10 this term has been applied to particles with diameters of up to 500 nm in the context of biotechnology.11 The importance of NPs stems from their size, which profoundly affects their physicochemical properties.10 Particles with diameters below 100 nm often have properties that differ markedly from their corresponding bulk materials.12 Metallic NPs often exhibit surface plasmon resonance, leading to absorption in the UV-Vis region and distinctive optoelectronic properties.10 Importantly, changing the size and shape of NPs often changes their absorption spectra and interparticle properties.13 Because of these unique features, metallic NPs have reached the stage of preclinical and clinical trials.
Nanoscience-related development has been spurred by the major advances in nanoscale material research and thus, currently, a range of various nanomaterials are commercially available.14 In the coming years, it is expected that a great deal of nano-products will be translated into every day tools. They interact in many different ways with a wide range of biomolecules on the cell surfaces and inside cells, meaning that they can direct diverse cellular, physiochemical, and biochemical properties.15 The small size of NPs allows them to be taken up rapidly into cells and organelles. Moreover, they are readily transported across the placenta and the blood–brain barrier.14,16,17 As a result, they have been incorporated in more than 43 approved drug formulations, as enlisted in the discussion in nanopharmaceuticals by Weissig et al.18 Titanium dioxide and zinc oxide NPs are used in different types of sunscreens because they can block ultraviolet light while being transparent to visible light when applied to the skin.19 In addition, products based on single-walled carbon nanotubes were recently introduced in the Russian market.20 Food packaging and preservation techniques currently employ various nanosized materials to extend the shelf life of products and prevent the spoilage of edible items.21
In 2015, Vance et al.22 reviewed the entry of nanotechnology powered-products into the market, showing that there were at least 1814 nanomaterial-containing products consumed by 622 companies over 32 countries. Silver was found to be the most frequently used nanomaterial (435 products).22 Silver-based nanomaterials are used in restorative veils, shampoos, shirts, clothes, toothpastes, detergents, towels, toys, and humidifiers at concentrations ranging from 1.4 to 270![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 μg Ag per gram.23 Silver-containing products were washed in 500 mL of tap water to assess the potential release of silver into water-based natural networks such as surface water, saliva, and wastewater. Ag was discharged at levels of up to 45 μg Ag per gram, in particles of varying sizes including some with diameters above 100 nm and some with diameters below this level.22
000 μg Ag per gram.23 Silver-containing products were washed in 500 mL of tap water to assess the potential release of silver into water-based natural networks such as surface water, saliva, and wastewater. Ag was discharged at levels of up to 45 μg Ag per gram, in particles of varying sizes including some with diameters above 100 nm and some with diameters below this level.22
Currently, NPs are typically synthesized via chemical and physical methods, which are usually costly and environmentally hazardous.24 Therefore, the eco-friendly biosynthesis of NPs has attracted increasing attention over the last decade, in parallel with the increased interest in green chemistry and sustainability.25,26 Conventional physical and chemical methods for the synthesis of NPs often require harsh conditions, whereas biosynthetic processes involve simple, nontoxic, and environment-friendly protocols at ambient temperature and pressure. These processes can be classified as either intra- or extra-cellular syntheses depending on the location the NPs are formed. Due to the greater ease of product recovery, extracellular methods are generally preferred. Many different biological resources have been evaluated for use in intra- and extracellular NP synthesis, including plants, fungi, algae, viruses, bacteria, and yeast.27 Although there have been extensive studies on the biosynthesis of NPs, the exact mechanisms involved in these processes remain unclear.28 However, it is well established that biosynthetic methods for the preparation of NPs can be more cost-effective and have less environmental impact than conventional physicochemical syntheses. These methods use renewable microorganisms and plants (or their extracts) as both bio-reductants for capping and stabilizing agents, obviating the need for additional reagents. Furthermore, the addition of proteins and peptides as biomatrices help to form NPs with a defined size and shape.24 Similarly, biosynthetic methods can control the shape, and size of the formed NPs due to the highly specific interactions between the biomolecular templates and inorganic materials.29,30
The fundamentals of metallic NP biosynthesis
Although the mechanistic details of the biosynthesis processes are currently unclear,31,32 a range of mechanisms have been proposed to explain the formation of metallic NPs via bio-reduction, especially for Ag, Au, and Cd NPs.32,33 Most studies have focused on bio-reduction derived from bacteria, fungi, and plants.34
Polyphenol-mediated nanofabrication
Generally, phenolic compounds inactivate ions via chelation.35 The high nucleophilic nature of the phenolic aromatic rings are probably responsible for their chelating ability.36 Natural sources respond to heavy metal stress by metal complexation procedures such as the formation of phytochelations or by other metal-chelating peptides.19 Metal ions are captured and immobilized by biological elements and subsequently undergo reduction, sintering, and smelting processes, leading to the formation of NPs. The size and shape of the resulting NPs depend on the metal ion content and variation in the site of ion penetration.24,37 The morphology, dispersion, and yield of these biosynthetic NPs can be modified by controlling the reaction conditions.38 In the presence of high levels of polyphenols, protection against coalescence and aggregation was observed, which was ascribed to the production of a protective coating around the nascent NPs (Fig. 1).27
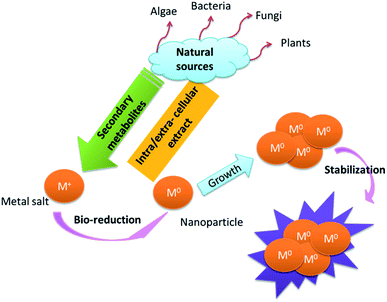 |
| | Fig. 1 Suggested general mechanisms for the synthesis of NPs.27 | |
Plants
Plants are rich in secondary metabolites such as alkaloids, flavonoids, saponins, steroids, tannins, and phenolic acids. Several studies have shown that many of these metabolites act as both reducing and stabilizing agents and inhibit the aggregation and agglomeration of the novel metallic NPs by non-hazardous means.39–41 The bio-reduction of metal NPs using plant extracts is divided into three main phases. The first is the activation step, in which the reduction and nucleation of metal ions occur. Secondly, the small adjacent NPs come together to form particles of a larger size, accompanied by an increase in the thermodynamic stability of the NPs, which is identified as the growth phase. Finally, the shape of the NPs is formed through the termination phase.24 Following the reduction of metal ions by the active metabolites, the NPs are centrifuged with the resultant precipitates and washed with a suitable solvent to remove impurities, where separation takes place through column chromatography prior to their further use.42 The type of plant extract and its concentration influences the morphology of the NPs formed, while the temperature and pH of the extract medium controls the growth and size of the NPs.43 The control of metal salt concentration has also been argued to play an important role in the morphology of the synthesized NPs.44 A list of some the plants used for the synthesis of NPs between 2014 and 2017 is presented in Table 1. The main functional groups involved in the reduction of metal ions are carbonyl, hydroxyl, amino and methoxide groups, which bind to the metal ions by electrostatic interaction, leading to their reduction.45 The leaf extract of Tabebuia berteroi is rich in polyphenols,46 and Withania coagulans is rich in flavonoids, tannins, phenolics, etc., which has been reported to be responsible for the reduction of Fe and Pd ions to their respective metal NPs together with graphene oxide to form a nanocomposite.47 Quercetin from the leaf extract in G biloba was used to reduce Cu(II) ions following a two-step process to CuNPs without the addition of any capping agents.48 Similarly, the flower extract of Anthemis xylopoda was attributed to the reduction of Au(III) to AuNPs, which were used as an effective catalyst.49 Euphorbia peplus is rich in flavonoids and rare disaccharides, which was reported to reduce AgNO3 in the presence of Fe3O4, leading to the immobilization of AgNPs with enhanced catalytic activity.50 Similarly, Pd/CuO NPs were successfully biosynthesized in vitro using Theobroma cocoa L. seed extracts, which contain phenolic antioxidants.51 Euphorbia heteradena Jaub contributed to the biosynthesis of very stable TiO2 NPs from titanyl hydroxide in the presence of potent phenolics, which were not only responsible for the reduction of the metal precursor but also capping the ligands onto the surface of the NPs, as evidenced via FTIR.52 The H-donating ability of polyphenols such as flavonoids and quercetin mostly involves the metal salts of nitrates, sulphates and chlorides as potent antioxidants in the reduction process of metal precursors. Here, the OH group in the reduced form of polyphenolics is converted into a carbonyl group with subsequent reduction of metal ions due to a redox reaction. These C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O groups on the oxidized form of polyphenol electrostatically stabilize the metal NPs. Hence, the mechanism for the biosynthesis of nM0 as zero-valent metal atoms by plant extract polyphenolics APOH when reacting with a metal halogen precursor is as follows:
O groups on the oxidized form of polyphenol electrostatically stabilize the metal NPs. Hence, the mechanism for the biosynthesis of nM0 as zero-valent metal atoms by plant extract polyphenolics APOH when reacting with a metal halogen precursor is as follows:
| nAPOH + nMn+ → nAPX + nM0 |
Table 1 Plant species used for the green biosynthesis of metallic NPs
| Plant species |
Plant origin |
Mode of synthesis |
NPs |
NPs size “nm” |
UV absorption “nm” |
Effect |
Reference |
| Andrographis paniculata |
Leaves |
Extracellular |
Ag |
40, 60 |
432 |
Antibacterial |
58 |
| Azadirachta indica |
Leaves |
Extracellular |
Ag |
100 |
430–435 |
— |
59 |
| Boerhaavia diffusa |
— |
Extracellular |
Ag |
25 |
418 |
Antibacterial |
60 |
| Brassica oleracea |
Aerial parts |
Extracellular |
Ag |
36 |
420 |
Antimicrobial |
61 |
| Brassica oleracea capitata |
42 |
428 |
| Calendula officinalis |
Leaves |
Extracellular |
Ag |
30–50 |
435 |
— |
62 |
| Callistemon viminalis |
Leaves |
Extracellular |
Ag |
|
|
Antibacterial |
63 |
| Camellia sinensis |
Leaves |
Extracellular |
ZnO |
9–17.5 |
450 |
Catalytic activity |
74 |
| Chenopodium ambrosioides |
— |
Extracellular |
Ag |
38.6 ± 16.4 (HFPM) |
414–432 |
Antioxidant |
34 |
| 25.0 ± 9.9 (MDPM) |
| 15.9 ± 4.6 (FFPM) |
| 11.2 ± 3.7 (DDPM) |
| 7.0 ± 2.9 (MFPM) |
| 21.8 ± 9.0 (HDPM) |
| Cucumis anguria |
Leaves |
Extracellular |
Ag |
11–27 |
420 |
Antibacterial |
65 |
| Dimocarpus longan Lour |
— |
Extracellular |
Ag |
9–32 |
380–450 |
Antibacterial and anticancer |
66 |
| Ipomoea pes-caprae |
Roots |
|
Ag |
10–50 |
430 |
Antimicrobial |
67 |
| Lansium domesticum |
Fruits |
Extracellular |
Au |
20–40 |
420, 540 |
Anti-microbial |
68 |
| Ag |
10–30 |
| Au–Ag alloy |
150–300 |
| Myrmecodia pendans |
— |
Extracellular |
Ag |
10–20 |
448 |
— |
69 |
| Oxalis corniculata |
Leaves and stem |
Extracellular |
Ag |
20–80 |
|
— |
70 |
| Phyllanthus emblica |
Fruits |
Extracellular |
Au |
|
|
Antimicrobial |
71 |
| Psychotria nilgiriensis |
Leaves |
Extracellular |
Ag |
40–60 |
422 |
Insecticidal |
72 |
| Quercus brantii Lindl. |
Leaves |
Extracellular |
Ag |
6 |
455 |
— |
73 |
| Origanum majorana |
Leaves |
Extracellular |
Ag |
35 |
440 |
— |
74 |
| Rhodomyrtus tomentosa |
— |
Extracellular |
Ag |
30 |
420 |
Anti-microbial |
75 |
| Rhynchotechum ellipticum |
Leaves |
Extracellular |
Ag |
510–730 |
459 |
— |
62 |
| Sapium sebiferum |
Leaves |
Extracellular |
Pd |
2–14 |
274 |
Bacterial and photocatalytic activities |
76 |
| Spinacia oleracea |
Leaves |
Extracellular |
ZnO |
40.9 |
375.4 |
Early seedling growth and growth characteristics of green gram |
77 |
| Tabernaemontana divaricata |
Leaves |
Extracellular |
Ag |
22.85 |
|
Cytotoxic activity against MCF-7 cell line |
78 |
| Ventilago maderaspatana |
Leaves |
|
Ag |
|
|
Anti-malaria |
79 |
| Ziziphora tenuior |
— |
Extracellular |
Ag |
8–40 |
|
Antioxidant |
80 |
| Ziziphus jujuba |
Leaves |
Extracellular |
Ag |
20–30 |
434 |
— |
81 |
| Ziziphus oenoplia |
Leaves |
Extracellular |
Ag |
10 |
436 |
Antibacterial |
82 |
| Ziziphus spina-christi |
Leaves |
Extracellular |
Ag |
30–70 |
442 |
Antibacterial |
83 |
The growth commences the assembly of metal atoms prior to the further reduction of metal ions with improved stability,
and the formation of (M
2n2n+)
n by collision, fusion of
nM
nn+ and ripening leads to the formation of metal NPs.
Moreover, the stem extract of Callicarpa maingayi was used for the biosynthesis of AgNPs. The extract contained several aldehydes that reduced silver ions to metallic AgNPs, yielding a product referred to as [Ag (Callicarpa maingayi)] + complex. Spectroscopic analysis indicated that this material contained C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O and C
O and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N functional groups, suggesting that the amide and polypeptide groups served as capping agents, which stabilized the nascent metallic NPs from aggregation.53 A methanolic leaf extract of Vitex negundo was also used in the bio-synthesis of Ag-NPs under ambient conditions with no additives, resulting in a template that shaped NPs or accelerants that spontaneously protected the NPs from aggregation.54 El-Kemary et al. investigated the bio-synthesis of AgNPs using a leaf extract of Ambrosia maritime, and found that various secondary metabolites (notably sesquiterpene lactones and flavonoids) strongly influenced the NP yield.55 In addition, Coleus aromaticus leaf extract was found to be a suitable reductant for the rapid synthesis of AgNPs from silver salts. Rosmarinic acid was the major polyphenol in the extract.56 Our recent paper reported the successful fabrication of Ag2ONPs from chili as well due to the presence of alkaloid compounds.57 As noted above, the focus of much work has been on Cu and Ag NPs. Taken together, it has been reported that the synthesis of NPs using plant extracts proceeds more quickly than alternative processes using microbial cultures, and also yields more stable products with a greater range of shapes and sizes.24,40 A study on metal bioaccumulation in plants showed that the metals are usually deposited in the form of NPs.24
N functional groups, suggesting that the amide and polypeptide groups served as capping agents, which stabilized the nascent metallic NPs from aggregation.53 A methanolic leaf extract of Vitex negundo was also used in the bio-synthesis of Ag-NPs under ambient conditions with no additives, resulting in a template that shaped NPs or accelerants that spontaneously protected the NPs from aggregation.54 El-Kemary et al. investigated the bio-synthesis of AgNPs using a leaf extract of Ambrosia maritime, and found that various secondary metabolites (notably sesquiterpene lactones and flavonoids) strongly influenced the NP yield.55 In addition, Coleus aromaticus leaf extract was found to be a suitable reductant for the rapid synthesis of AgNPs from silver salts. Rosmarinic acid was the major polyphenol in the extract.56 Our recent paper reported the successful fabrication of Ag2ONPs from chili as well due to the presence of alkaloid compounds.57 As noted above, the focus of much work has been on Cu and Ag NPs. Taken together, it has been reported that the synthesis of NPs using plant extracts proceeds more quickly than alternative processes using microbial cultures, and also yields more stable products with a greater range of shapes and sizes.24,40 A study on metal bioaccumulation in plants showed that the metals are usually deposited in the form of NPs.24
Nano-synthesis and mechanisms using fungi, bacteria strains and algae
Extensive efforts are being made to understand the mechanisms of NP biosynthesis in fungi and bacteria. Per unit of biomass, fungi generally have a greater capacity for the bio-accumulation of metal ions than bacteria due to their exceptional biosorption of cations. However, the precise level of accumulation achieved by both types of microorganisms depends on the homogeneity of the experimental medium, temperature, initial metal ion concentration, and pH. In the presence of heavy metals, some fungi secrete large quantities of redox-active extracellular proteins that can bio-reduce metal ions, leading to the formation of insoluble metal–protein nanoconjugates, which eventually form nanocrystals. Fungi support large biologically diverse hotspots, which have not been well explored and may contain novel fungal proteins and enzymes capable of metal bio-reduction and detoxification.84
Even though the isolation and purification of NPs can be challenging, multiple studies have investigated the mechanisms of metal bioreduction by microorganisms.85 Two mechanisms have been proposed to explain the reduction of metal ions by fungi, extracellular and intracellular mechanisms. The extracellular mechanism is believed to involve nicotinamide adenine dinucleotide (NADH)-dependent enzymes, notably nitrate reductases, which are secreted into the reaction medium together with electron shuttles such as hydroxyquinoline.86 The structures of NADH and NAD+, and the reduction of Ag+ to AgNPs is mediated by electron transfer from NADPH, as shown in Fig. 2.87,88 Das and Thiagarajan86 examined the occurrence of the bio-synthesis of AgNPs, and suggested that the reduction of silver ions (Ag+) to AgNPs (Ag0) is mediated by an electron shuttle and an NADPH-dependent reductase specific to Fusarium oxysporum species.86,89 The reduction of the metal ions to form NPs is accompanied by the oxidation of NADPH to NADP+.86 Kalishwaralal et al.90 reported a similar process, in which silver ions were reduced by NADPH-dependent nitrate reductases secreted into the extracellular environment by non-pathogenic bacteria.90 Another study found that some bacteria reduce Fe3+ by forming and secreting small diffusible redox-active compounds that shuttle electrons between the microbes and the insoluble iron substrate.91 Microbial sources exhibit their action via the precipitation of NPs due to their metabolic activity. The bacterium Nocardia farcinica was demonstrated to induce the rapid extracellular reduction of gold ions into highly stable NPs, demonstrating that these processes are not restricted to Fe and Ag.92 The formation of the AuNPs was tentatively attributed to an electron shuttle and a nitrate reductase, which were found in the exudates of the bacterial cell culture.31 Nitrate reductases transfer electrons between nitrate ions and metal centers. A study on Fusarium oxysporum showed that silver and chloroaurate ions are reduced by a nitrate-dependent reducing enzyme and/or a quinone-based shuttle, supporting the extracellular mechanism theory.93 However, the precise mechanism of AgNP formation in this case is unknown.94
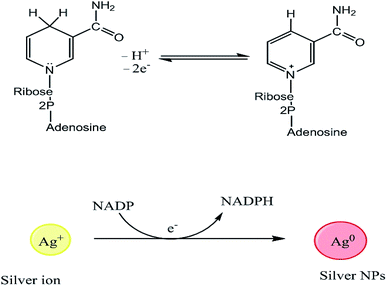 |
| | Fig. 2 Structures of NADH and NAD+, and the mechanism of Ag+ reduction by NADPH-dependent reductases to form AgNPs.74 | |
Several studies have also suggested that electron transfer is mediated by the constituents of Shewanella sp., such as flavins related to external electron acceptors,95 or direct electron transfer by redox proteins and membrane-bound cytochromes (direct electron transfer).96 An alternative extracellular mechanism was explained by the confinement at the cell wall by electrostatic interactions with positively charged functional groups (cell wall enzymes) during the formation of AuNPs from AuCl4− ions. The confined AuCl4− ions are then reduced to Au0 by proteins/enzymes in the cell wall, conferring extra stability to the biogenic NPs (Fig. 3).84,97,98 Alternatively, the NPs or metal ions may diffuse through the cell membrane and be reduced by redox mediators in the cytoplasmic matrix.99 The presence of particle aggregates in the cell wall, cytoplasmic membrane and cytoplasm was confirmed by microscopy studies.87 Another study on fungi (Verticillium sp.) ruled out the possibility that sugars in the cell wall are required for the reduction of Au3+ ions because NP formation was found to predominantly occur within the cytoplasmic membrane.100 The second proposed process for the bio-fabrication of NPs is the intracellular mechanism (Fig. 4), in which the fungal cell wall and various proteins play central roles in the bioreduction of metals. The primary fungal cell wall is an extracellular structural unit with a composition that varies over its life cycle.101 However, generally it consists of the polysaccharides chitin and glucan, which are bonded by 1–4 linkages. Inter- and intra-molecular H-bonding between the chitin and glucan units gives the cell wall its rigidity.101 It is worth mentioning that one of the precursors of Au reduction is HAuCl4, which dissociates into Au3+ ions, whereas AuCl dissociates into Au+.102,103 Since Au+ is much less soluble than Au3+, it has not been investigated as thoroughly. However, its solubility can be increased by the formation of coordination complexes with alkenes, alkanethiols, alkylamines, alkylphosphines and various anions.103 Investigations have shown that the Au+ concentration initially increases in the early stages of the biofabrication of AuNPs, but then falls as Au0 starts to appear, suggesting that Au3+ is initially reduced to Au+ and then finally to Au0. It should be noted that the reduction of Au+ to Au0 is a one-step, single-electron process, whereas that of Au3+ to Au0 involves several different chemical conversions.99
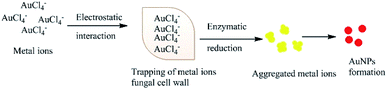 |
| | Fig. 3 Hypothetical mechanism for the extracellular synthesis of NPs. | |
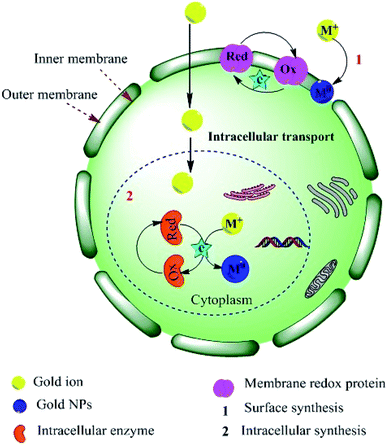 |
| | Fig. 4 Schematic illustration of the proposed mechanism for the intracellular synthesis of AuNPs.84,99 Adapted with permission from ref. 98; Copyright 2012, American Chemical Society. | |
Fungi
Biomass and extracellular materials from microorganisms such as fungi are widely used for the biotransformation of metal ions to NPs. Studies that investigated the bioreduction of metal ions using fungi are listed in Table 2. Fungi produce very large quantities of secreted proteins, which may increase the rate of NP formation.104 Many fungi have mycelia, which provide a larger surface area than that achievable with bacteria, supporting the interactions between metal ions and fungal biomass containing reducing agents. In principle, this should enhance the conversion of ions into metallic NPs. Fungi also have the advantage of straightforward downstream processing after extracellular NPs formation, enabling efficient NP extraction.104 Filamentous fungi are preferable to bacteria and unicellular organisms because they are easy to handle, highly tolerant of metals, and exhibit good wall binding capacity and the ability to synthesize NPs extracellularly.90,105 NPs synthesized by fungi were found to be more stable and monodisperse than that synthesized by other microorganisms.
Table 2 Fungi, bacteria and algae used in the synthesis of metal NPs
| Organism |
Mode of synthesis |
NPs |
NP Size |
Reference |
| Fungi |
| Aspergillus clavatus |
Extracellular |
Ag |
10–25 |
122 |
| Aspergillus fumigates |
Extracellular |
Ag |
5–25 |
123 |
| Aspergillus terreus |
Extracellular |
Se |
500 |
107 |
| Extracellular |
Ag |
1–20 |
88 |
| Bacillus megaterium |
Extracellular |
Ag |
46.9 |
122 |
| Bacillus licheniformis |
Extracellular |
Ag |
50 |
122 |
| Candida albicans |
Extracellular |
Au, |
20–40, |
106 and 124 |
| Ag |
60–80 |
| Cladosporium cladosporioides |
Extracellular |
Ag |
10–100 |
105 and 122 |
| Colletotrichum sp. |
Extracellular |
Au |
20–40 |
125 |
| Coriolus versicolor |
Extracellular |
Ag |
350–600 |
126 |
| Fusarium semitectum |
Extracellular |
Ag |
10–60 |
89 |
| Fusarium acuminatum |
Extracellular |
Ag |
5–40 |
122 |
| Fusarium oxysporum and Verticillium sp. |
Extracellular |
Magnetite |
20–50 |
125 |
| Fusarium oxysporum |
Extracellular |
Au |
20–40 |
125 |
| CdS |
5–20 |
| Zirconia |
3–11 |
| Fusarium oxysporum |
Extracellular |
Ag |
20–50 |
93 |
| Fusarium solani |
Extracellular |
Ag |
5–35 |
122 |
| Klebsiella pneumoniae |
Extracellular |
Ag |
50 |
122 |
| Lactobacillus strains |
Extracellular |
Ag |
500 |
122 |
| Neurospora crassa |
Extracellular |
Au |
32 |
127 |
| Penicillium fellutanum |
Extracellular |
Ag |
1–100 |
122 |
| Penicillium brevicompactum WA2315 |
Extracellular |
Ag |
23–105 |
122 |
| Phanerochaete chrysosporium |
Extracellular |
Ag |
50–200 |
127 and 128 |
| Phoma sp. 32883 |
Extracellular |
Ag |
75 |
126 |
| Pseudomonas stutzeri AG259 |
Extracellular |
Ag |
200 |
122 |
| Rhizopus stolonifer |
Extracellular |
Ag |
79.42 |
129 |
| Trichoderma asperellum |
Extracellular |
Ag |
13–18 |
122 |
| Trichoderma viride |
Extracellular |
Ag |
5–40 |
122 |
| Verticillium sp. |
Extracellular |
Ag |
25 |
126 |
| Yarrowia lipolytica |
Extracellular |
Au |
15 |
127 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
| Bacteria |
| Bacillus megaterium |
Extracellular |
Ag |
46.9 |
122 |
| Bacillus licheniformis |
Extracellular |
Ag |
50 |
122 |
| Bacillus cereus |
Extracellular |
Ag |
4–5 |
122 |
| Bacillus sp. |
Extracellular |
Ag |
5–15 |
122 |
| Bacillus subtilis |
Extracellular |
Ag |
5–60 |
122 |
| Bacillus safensis LAU 13 |
Extracellular |
Ag |
5–95 |
130 |
| Brevibacterium casei |
Extracellular |
Ag |
50 |
122 |
| Corynebacterium sp. |
Extracellular |
Ag |
10–15 |
122 |
| Escherichia coli |
Extracellular |
Ag |
50 |
131 |
| Extracellular |
Ag |
1–100 |
122 |
| Geobacter sulfurreducens |
Extracellular |
Ag |
200 |
122 |
| Klebsiella pneumoniae |
Extracellular |
Ag |
50 |
122 |
| Lactic acid bacteria |
Extracellular |
Ag |
11.2 |
122 |
| Lactobacillus strains |
Extracellular |
Ag |
500 |
122 |
| Morganella sp. |
Extracellular |
Ag |
20 ± 5 |
122 |
| Proteus mirabilis |
Extracellular |
Ag |
10–20 |
122 |
| Pseudomonas stutzeri AG259 |
Extracellular |
Ag |
200 |
122 |
| Pseudomonas aeruginosa KUPSB12 |
Extracellular |
Ag |
50–85 |
132 |
| Rhodopseudomonas capsulata |
Extracellular |
Au |
10–20 |
133 |
| Staphylococcus aureus |
Extracellular |
Ag |
1–100 |
122 |
| Vibrio alginolyticus |
Intracellular and extracellular |
Ag |
50–100 |
122 |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
| Algae |
| Chlorella vulgaris |
Intercellular |
Au |
75 |
115 |
| Chondrus crispus |
Extracellular |
Au, Ag |
— |
116 |
| Gracilaria edulis |
Extracellular |
Ag, ZnO |
55–99 |
134 |
| Kappaphycus alvarezii |
Extracellular |
Au |
|
121 |
| Sargassum wightii |
Extracellular |
Ag |
8–27 |
116 |
| Spirogyra insignis |
Extracellular |
Au, Ag |
— |
118 |
| Tetraselmis kochinensis |
Intercellular |
Au |
5–35 |
119 |
A cell-free extract of Candida albicans was used to prepare monodisperse and highly crystalline Au and AgNPs,106 and the marine fungus Aspergillus terreus was used to prepare selenium nanoparticles (SeNPs). The SeNPs had an average diameter of 500 nm, and FTIR analysis indicated that they may incorporate fungal protein groups, which facilitated their biosorption into the fungal cell wall.107 The A. terreus culture supernatant used to prepare SeNPs was also demonstrated to be competent in the reductive synthesis of AgNPs from aqueous Ag+ ions. The latter reaction was shown to be enzyme-mediated, which reached completion within a few hours at ambient temperature and yielded stable polydisperse spherical AgNPs with a smaller size (1–20 nm) (Fig. 2).88 Several Fusarium oxysporum strains were also investigated for the biosynthesis of AgNPs, which revealed that the extracellular reduction of the metal ions is mediated by a nitrate-dependent reductase and a shuttle quinine.32 The absence of an enzyme in the reaction medium validated the active enzymatic role in the complete process.87 Previous reports have shown that many active substances secreted by fungi can serve as reducing and capping agents in the biosynthesis of NPs. However, the precise processes are unclear, and further investigations may provide a deeper understanding of the molecular mechanisms involved in these processes.88
Bacteria
Bacteria are good candidates due to their environmental abundance and ability to adapt to extreme conditions.104 They are fast-growing, cheap to cultivate, and simple to manipulate.
Many bacteria have been isolated and used for the biosynthesis of NPs, both extracellularly by the reduction of metal ions on the surface of the microbial cells and intracellularly by the encapsulation of metal ions in the microbial cells inhabiting different pH, oxygenation incubation time and temperature conditions,104 thus offering high catalytic activity and surface area for the interaction of enzymes with metal salts.107,108 Enzymes play an important role in the biosynthesis of NPs, and hence optimized parameters are critical for maximum activity and enzymatic interaction.108 This property finds wide application in detoxification and remediation as well as in drug delivery applications by inducing the defence mechanism of microorganisms.109–111 There are important links between the ways NPs are synthesized and their potential uses. Johnston et al.112 showed that the formation of pure AuNPs by the bacterium Delftia acidovorans was mediated by a small non-ribosomal peptide, delftibactin, which was responsible for the resistance to otherwise toxic concentrations of gold ions.112 Delftibactin removes gold ions from solution by converting them into inert AuNPs, which are non-toxic to bacterial cells.112 The interesting study by Sintubin et al. examined the production of AgNPs by lactic acid bacteria. Many bacterial species were tested, but only four were found to synthesize AgNPs: Lactobacillus spp., Pediococcus pentosaceus, Enterococcus faecium and Lactococcus garvieae.113 NPs obtained through bacteria are precipitated within the cells after incubating the cultured supernatant with Ag+ or Au3+. A two-step mechanism for the biosynthesis of AgNP was proposed, in which the Ag ions initially accumulate at the cell wall via biosorption and then reduced to form metallic NPs.113 Sintubin et al. also suggested that the cell wall may act as a capping agent for the NPs, stabilizing them by preventing aggregation. These authors also showed that increasing the pH of the culture medium accelerated the reduction process and the formation of NPs.113
Algae
Algae are aquatic microorganisms that collect heavy metals and have been used in the biological synthesis of NPs (Table 2).114 Chlorella vulgaris was used to convert AuCl4− ions into algal-bound gold, which was subsequently reduced to tetrahedral, decahedral and icosahedral AuNPs that accumulated near the cell surface.115 Other groups have reported the extracellular synthesis of AgNPs using brown seaweed, Sargassum wightii,116 which enabled the rapid extracellular synthesis of AuNP, yielding highly stable mono-dispersed NPs with a size of 8 to 12 nm. The uniform biosynthesized particles were completely dispersed in the solution without aggregation.117 The biosynthesis of Au and AgNPs using red and green algae (Chondrus crispus and Spirogyra insignis) was also recently reported.118 Moreover, the alga Tetraselmis kochinensis has been used for the intracellular synthesis of AuNPs with dimensions of 5–35 nm and mostly were spherical in shape with occasional aggregations. The NPs formed in this way were observed to be more in the cell wall than on the cytoplasmic membrane, possibly indicating that the metal ions were reduced by enzymes in the cell wall rather than elsewhere.119 In addition, the bioreduction of Au using the biomass of the brown alga Fucus vesiculosus has been reported.120 The optimum pH range for the recovery and reduction process in this case was 4–9, with maximum uptake at pH 7. The hydroxyl groups present in the algal polysaccharides were involved in the gold bioreduction. Finally, Rajasulochana et al. have investigated the synthesis of extracellular AuNPs using Kappaphycus alvarezii.121
Natural products for metallic NP synthesis
As noted in the preceding section, the cultures and extracts used in the biosynthesis of metallic NPs contain a wide range of secondary metabolites, which contribute to the reduction of metal ions and stabilization of the NPs.135,136 The bioactive molecules include polysaccharides, enzymes, amino acids, vitamins, proteins, and organic acids, as shown in Table 3.137 This table also shows that most of the secondary metabolites known to be found in plant extracts, and relatively few derived from microbes.
Table 3 Natural metabolites used in the synthesis of metallic NPs
| Chemical name |
Class |
Source |
Reference |
| 21-Hydroxyonocera-8(26),14-diene-3-one |
Terpenoids |
Plants |
171 |
| 3β-Hydroxyonocera-8(26),14-dien-21-one |
Terpenoids |
Plants |
171 |
| A reducing hexose with the open chain form |
Sugar |
Plants |
24 |
| Diosgenin |
Steroids |
Plants |
145 |
| Eugenol |
Terpenoids |
Plants |
24 |
| Glutathione |
Tripeptide |
Yeast & fungi |
172 |
| Lansionic acid |
Terpenoids |
Plants |
171 |
| Lansic acid |
Terpenoids |
Plants |
171 |
| Lansiosides A |
Terpenoids |
Plants |
171 |
| Lansiosides B |
Terpenoids |
Plants |
171 |
| Lansiosides C |
Terpenoids |
Plants |
171 |
| Luteolin |
Flavonoids |
Plants |
24 |
| Quercetin |
Flavonoids |
Plants |
24 |
| Rosmarinic acid |
Acids |
Plants |
35 |
| Tryptophan |
Amino acids |
Plants |
24 |
| Tyrosine |
Amino acids |
Plants |
24 |
| Vitamin B2 |
Alkaloids |
|
173 |
| Vitamin C (ascorbic acid) |
Acids |
— |
170 |
Flavonoids
Flavonoids are a large group of polyphenolic compounds that can actively chelate metal ions and reduce them to form NPs.24 This ability to form NPs is attributed to their richness in key functional groups, such as multiple hydroxyl groups and the carbonyl moiety.138 Thus, the high flavonoid and phenolic content of an aqueous Rumex dentatus water extract enabled the facile bioreduction of Ag+ to Ag0.139 It has been postulated that the keto–enol tautomeric transformation of flavonoids may enable the release of reactive hydrogen atoms, which drive the reduction of metal ions.24 For example, the keto–enol form of the flavonoids luteolin and rosmarinic acid from Ocimum basilicum (sweet basil) extract was shown to play a key role in the formation of NPs (Fig. 5).35 In addition, the internal conversion of ketones to carboxylic acids in flavonoids is likely to be involved in the reduction of Au3+ ions. Interestingly, some flavonoids can chelate metal ions with their carbonyl groups or π-electrons. For example, apiin (apigenin glycoside) was extracted from Lawsonia inermis (lawsonite thornless, henna) and utilized for the synthesis of anisotropic AuNPs and quasi-spherical AgNPs with average sizes ranging from 21 to 39 nm.140 The size could be controlled by changing the ratio of metal salts to apiin in the reaction. FTIR analysis revealed that a carbonyl group within apiin was involved in the NP-forming process.140 Moreover, hesperetin isolated from citrus extract was used for initial complexation, which led to significant interactions with Au3+ ions to produce a net charge transfer and reduce gold to Au0.141 On the other hand, quercetin is a flavonoid with very strong chelating activity via its carboxyl group, hydroxyls at the C3 and C5 positions, and the C3′–C4′ catechol group. Flavonoids chelate metal ions such as Fe2+, Fe3+, Cu2+, Zn2+, Al3+, Cr3+, Pb2+, and Co2+, which explains why flavonoids are readily adsorbed onto the surfaces of nascent NPs. Thus, they can influence the early stages of NP formation (nucleation), restrict aggregation, and mediate bioreduction.24
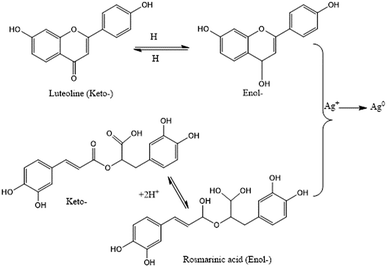 |
| | Fig. 5 Keto–enol tautomerism and its effects on the synthesis of Ag NPs.35 Adapted with permission from ref. 35; Copyright 2010, Elsevier. | |
Alkaloids
Ergoline alkaloids, indolizidine alkaloids, benzenoids and phenolic compounds in extracts of I. pes-caprae roots were shown to reduce and stabilize AgNPs.67 FTIR studies on mixtures of this extract with silver salts revealed peaks at 1660, 1043, and 635 cm−1, which were attributed to the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching vibrations of the amide bonds in proteins, the C–OH stretch of a cyclic alcohol, and aromatic C–H vibrations (suggesting the presence of free quinones derived from polyphenolic compounds), respectively.142 However, only a few publications have discussed the role of alkaloids in the formation of metallic NPs, and thus further investigations in this area are needed.
O stretching vibrations of the amide bonds in proteins, the C–OH stretch of a cyclic alcohol, and aromatic C–H vibrations (suggesting the presence of free quinones derived from polyphenolic compounds), respectively.142 However, only a few publications have discussed the role of alkaloids in the formation of metallic NPs, and thus further investigations in this area are needed.
Terpenoids
FTIR studies have shown that terpenoids are frequently associated with biosynthetic metallic NPs.143 The formation of AuNPs upon the reaction of chloroaurate ions with geranium leaves was attributed primarily to the content of terpenoids in the leaves.97 Similarly, the bio-reduction of HAuCl4 and AgNO3 by Cinnamomum zeylanisum (cinnamon) extracts was largely attributed to the high content of the basic terpenoid eugenol in the extracts.97 Previous studies have suggested that the deprotonation of the hydroxyl group in eugenol yields an anion, which can undergo further oxidation by metal ions, leading to their reduction and formation of NPs.144 In addition, the steroidal saponin diosgenin was found to act as a reducing and capping agent in the synthesis of AgNPs, and the mechanism proposed is shown in Fig. 6.145
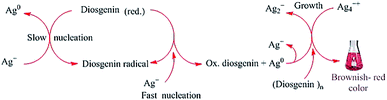 |
| | Fig. 6 Proposed mechanism for the formation of AgNPs via the reduction of Ag+ ions by the reducing steroidal saponin diosgenin. The OH groups are deprotonated to yield reducing anions, which convert Ag+ into Ag0. The neutral Ag0 then reacts with an Ag+ ion to form the comparatively stable Ag2+ cation. Ag2+ ions then dimerize to yield Ag42+.145 Adapted with permission from ref. 145; Copyright 2014, Elsevier. | |
Enzymes
Several studies have demonstrated the involvement of different enzymes in the biosynthesis of various metallic NPs. Enzymes are natural proteins produced in large quantities by living organisms.146 Enzymes can promote the formation of NPs with a wide variety of shapes and sizes, but they also vary widely in their catalytic activity, making it essential to carefully select the optimal enzyme for a given synthesis.147 For example, during the bio-synthesis of CdS NPs via the reaction of an aqueous CdSO4 solution with the fungus Fusarium oxysporum, NP formation was mediated by the secreted sulfate reductase enzymes. These enzymes reduced the sulphate ions to sulfides, which then reacted with aqueous Cd2+ ions to yield highly stable CdS NPs.148 The ubiquitous coenzyme NADH can also act as a reducing agent in the biosynthesis of NPs.88
The growing number of established pathways for the biosynthesis of NPs using fungal enzymes raises the exciting prospect of developing a rational unified biosynthetic strategy for nanomaterials with tailored chemical compositions and morphologies.148
Proteins
Proteins can be involved in both the bio-synthesis and stabilization of metallic NPs.149 A protein is a large biomolecule or macromolecule consisting of one or more long chains of amino acid residues. FTIR spectra clearly show the presence of various carbonyl groups on the surface of NPs, and the carbonyl groups of amino acid residues tend to act as capping ligands for NPs, thereby preventing their agglomeration and stabilizing them in aqueous media.106 However, proteins and peptides can also bind to AgNPs via their free amino groups.150 Naik et al.151 demonstrated the biosynthesis of AgNPs using peptides that can bind to the surface of nascent silver particles. The resulting metal clusters exhibited diverse structures, some of which resembled that of mature crystals. During the biogenesis of NPs, a plethora of biomolecules including proteins and amino acids with exposed disulphide bridges and thiol groups acted as non-enzymatic capping and reducing agents.152 The involvement of proteins as capping agents in the biosynthesis of AgNPs was demonstrated by Jain et al.,33 who identified two extracellular proteins with molecular weights of 32 kDa and 35 kDa. The two proteins were isolated from a fungal strain and were employed in the ecofriendly synthesis of stable AgNPs.33 Fig. 7 presents the suggested mechanism for the synthesis of AgNPs by these two proteins, in which the 32 kDa protein (presumed to be a reductase) reduces bulk Ag+ to Ag0, and the newly synthesized NPs are then capped and stabilized by the 35 kDa protein. Consistent with this suggested mechanism, proteins have been shown to bind immediately to the surfaces of NPs on contact.153 Gulsuner et al. designed of a multidomain peptide for the green, size-controlled, one-pot synthesis of biofunctionalized AuNPs (Fig. 8).154 The peptide incorporates the 3,4-dihydroxy-L-phenylalanine (L-DOPA) functional group, which has a catechol side chain, shown to be crucial for the reduction of the Au cations and the stabilization of the resulting NPs together with a lysine residue.
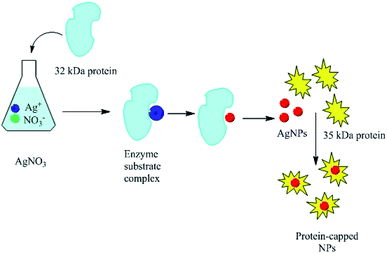 |
| | Fig. 7 Suggested mechanism explaining the involvement of two proteins (32 and 35 kDa) in the synthesis of AgNPs.33 Adapted with permission from ref. 33; Copyright 2011, Royal Society of Chemistry. | |
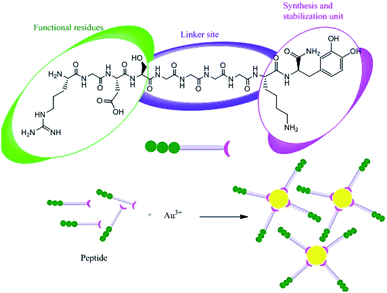 |
| | Fig. 8 Synthesis of biofunctionalized AuNPs using a designed peptide.153 Adapted with permission from ref. 153; Copyright 2015, American Chemical Society. | |
A linker domain was also incorporated to enable secondary functionalization and the connection of these reducing and stabilizing moieties to functional domains, which permitted effective biological targeting. Under appropriate experimental conditions, the oxidation of DOPA was coupled with the reduction of Au(III) ions, leading to the eco-friendly formation and stabilization of biofunctionalized AuNPs in a single-step process, which permits further functionalization and requires no additional reagents. The interaction of the peptide with the metal clusters provided a chemically reducing environment, accelerating the reduction of silver ions at the interface between the peptides and the metal (Fig. 9). In addition, the peptides adhere to the silver nuclei, reducing the surface energy of the crystal lattice.105,151,155 Recently, Paul and Sinha investigated the protein-mediated reduction of AgNO3 using a Pseudomonas aeruginosa culture supernatant.132 In addition, zein protein has been used as a reducing and capping agent for the biosynthesis of bio-conjugated AuNPs. It was found that a coating of zein protein on the surface of the NPs reduced their toxicity and hemolytic activity.156 Gruen observed that amino acids such as cysteine, arginine, methionine, and lysine can bind to silver ions.157
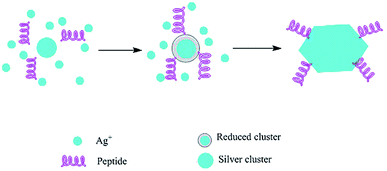 |
| | Fig. 9 Silver nanocrystal formation mediated by silver-binding peptides.150 Adapted with permission from ref. 150; Copyright 2002, Nature Publishing Group. | |
Complementary investigations established that Capsicum annuum L. extracts contain proteins with free amine groups that act as reducing and controlling agents during the solution-phase formation of AgNPs.158 Li et al.159 studied the extract of C. annuum L. and showed that the proteins and vitamin C present in the extract induced the rapid formation and precipitation of a-Se/protein NPs when combined with SeO32− ions at room temperature. The proteins stabilized the NPs by coating their surfaces and forming an a-Se/protein composite.159 The size and shell thickness of the a-Se/protein composite increased with an increase in the concentration of the extract but decreased at low pH values.159 Similar studies on the biosynthesis of AgNPs have shown that proteins can act as both reducing agents and protective surface coatings. These proteins prevent the agglomeration of AgNPs in the medium and are responsible for the formation of highly stable nanostructures.160,161 As discussed above, amino acids can bind to metal ions via the amino and carbonyl groups of their main chain or via side chains such as the carboxyl groups of aspartic and glutamic acid or the nitrogen atoms in the imidazole ring of histidine. Other side chains that can bind to metal ions include thiols (cysteine),89 thioethers (methionine), hydroxyl groups (serine, threonine, and tyrosine), and carbonyl groups (asparagine and glutamine).162 Tyrosine residues in proteins or peptides have been implicated in the production of AgNPs and AuNPs. The particles can be stabilized by the interactions between the metal surface and the free N terminus of the peptide. Bhattacharjee et al.163 produced AuNPs using a new tripeptide (H2N-Leu-Aib-Tyr-OMe) sequence containing a C-terminal tyrosine residue to reduce gold ions and a free N terminus that can bind to the nascent particles, forming stable AuNPs, as shown in Fig. 10. When amino acids are connected to a peptide chain, their individual capacity to bind and reduce metal ions may change. For example, the formation of the peptide backbone changes the functionality of the carboxyl group and the amino group. Capping proteins have important advantages for the synthesis of NPs compared to polymers and surfactants, including they are cost effective, safe, eco-friendly, and do not need special reaction conditions.129 They can also act as an anchoring layer for drug or genetic materials to be transported into human cells,164 and the presence of natural capping proteins increases the uptake and retention of NPs inside human cells.165 Moreover, the use of peptides that act as both reductants and capping agents eliminates the need for a separate capping step, which is very important for most medical applications of NPs.166
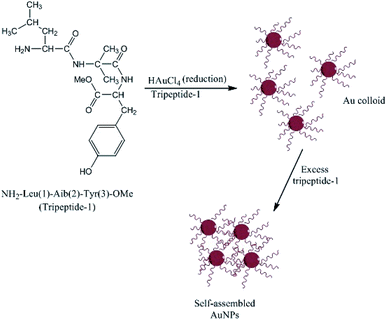 |
| | Fig. 10 Structure of tripeptide-1 and the self-assembly of AuNPs.162 | |
Sugars
Plant sugar extracts can affect the production and stability of metallic NPs.24 It was recently demonstrated that the capping properties of sugar extracts depend on their content of non-soluble carbohydrates such as starch.13 For example, β-D-glucose (monosaccharide) and soluble starch (polysaccharide) were used as structure-directing and stabilizing agents for the synthesis of spherical NPs and nanowires of silver and gold.167 The AgNPs formed in a plant extract (Rumex dentatus) were found to be very stable, possibly because the starch content in the extract prevented the agglomeration of the NPs.139 The synthesis of AuNPs was also attempted using selected glucose derivatives.168
In reactions with glycosides substituted at the anomeric carbon center, the C-6 position of the sugar was oxidized to a carboxylic acid with concomitant reduction of auric acid to yield AuNPs (Fig. 11).147 With glucose or glucuronic acid (an analogue of glucose with a carboxylic acid group at the C-6 position), auric acid reduction appeared to have been enabled by anomerisation and subsequent oxidation of the liberated aldehyde. This explains why the synthesis did not work using glucuronic acid derivatives substituted at the anomeric position, which cannot undergo oxidation at C-1 or C-6 (Fig. 11).147
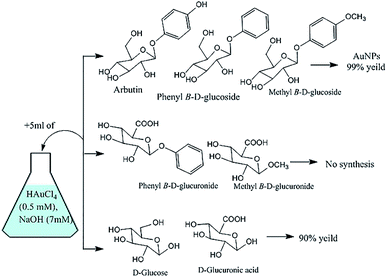 |
| | Fig. 11 Results obtained in the attempted synthesis of AuNPs using some glucose derivatives.147 | |
Thus, the choice of sugar derivative affected the final yield of AuNPs as well as their morphologies and sizes. Similar results were obtained in another study on the synthesis of AuNPs from auric acid using glycosides,169 where, 13C NMR experiments indicated that phenyl β-D-glucoside afforded the highest yield of mono-disperse round AuNPs.169
Vitamins
Various other purified secondary metabolites have also been used to prepare noble metal NPs.35 Vitamin B2 was employed in the synthesis of Au and PtNPs, yielding products with interesting morphologies,170 and vitamin C was used to prepare AuNPs in molecular gels and liquid crystal systems (Fig. 12).170 However, the use of vitamins in the biosynthesis of metallic NPs is currently under-explored.
 |
| | Fig. 12 (a), (b) and (c) Secondary metabolites used in the synthesis of metallic NPs. | |
Factors affecting the biosynthesis of metallic NPs
Various factors have been shown to affect the mechanism and rate of the bio-synthesis of metallic NPs, and the morphology of the resulting particles (Fig. 13).28,137 This section discusses each of these factors and their known effects on the synthesis of NPs.
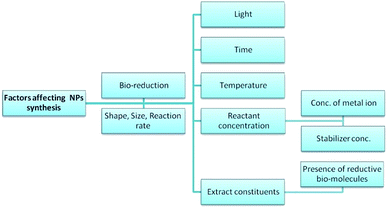 |
| | Fig. 13 Factors affecting the biosynthesis of NPs. | |
Light
Exposure to sunlight has repeatedly been shown to affect the rate of biosynthesis processes. Raut et al.137 observed that exposure to sunlight reduced the time required for the completion of the biosynthesis of AgNPs from 12 h to 5 min. This dramatic reduction was attributed to a photochemical process, which greatly increased the electron transfer rate to the metal ions.137 The effect of varying the wavelength of incident light on the biosynthesis of metallic NP was recently studied by wrapping reaction vessels with violet, red, green, and yellow filters made of cellophane paper. The violet filter was found to favor the formation of smaller spherical AgNPs with diameters of around 20 nm.174 In recent years, the light-assisted in situ preparation of AgNPs has emerged as a very promising strategy. It offers considerable versatility and selectivity together with opportunities to control the morphology of the resulting AgNPs without requiring severe reaction conditions.175 Photo-induced reaction protocols can be used to prepare nanostructures with different shapes and sizes, including larger spheres, nanorods, nanoplates, and dodecahedra (Fig. 14).175
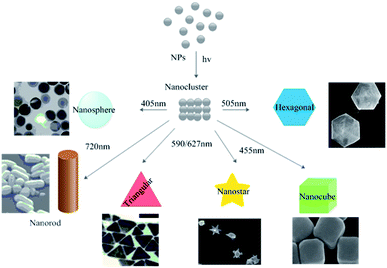 |
| | Fig. 14 Photo-induced production of AgNPs with various morphologies. Adapted with permission from ref. 175; copyright 2015, Springer. | |
Temperature and heating rate
The temperature can strongly affect the size and shape of biosynthetic NPs as well as the rate of their formation. Higher temperatures increase the reaction rate and the competence of some microorganisms with respect to the synthesis of NPs.8 For example, the synthesis of AgNPs from AgNO3 using cultures of the fungi Chrysosporium tropicum and Fusarium was studied at temperatures between 25 °C and 30 °C. The rate of AgNP formation increased with the incubation temperature and correlated with the absorbance of the reaction matrix. At the lowest temperature (25 °C), the majority of the formed AgNPs were very small, and incubation at the higher temperature of 30 °C reduced the proportion of smaller particles and promoted the formation of larger ones.176 A separate study showed that the size-selective synthesis of CuNPs could be achieved by adjusting the reaction temperature, yielding products with diameters ranging from 5 to 25 nm. The melting point of these CuNPs, and particularly the surface melting temperature, was believed to play a vital role in interparticle coalescence, causing them to grow more rapidly as the reaction temperature increased. In addition, the morphology of the CuNPs varied with the reaction temperature, enabling the formation of rod-shaped and cubical products.177 Temperature was also shown to affect the synthesis of AgNPs using the extract of Citrus sinensis peel, in which the particle size was found to depend on the relative rates of nucleation and particle growth. At the low temperature of 25 °C, the rate of growth exceeded that of nucleation, and thus relatively large particles were obtained, and at 60 °C, the rate of nucleation was higher, leading to smaller particles.178 Finally, Gericke and Pinches reported that higher temperatures increased the rate of AuNP formation when fungal cultures were incubated with AuCl4−, suggesting an increased rate of reduction. This in turn affected the particle morphology, where at lower temperatures, the main products were spherical AuNPs, whereas higher temperatures yielded rod-like and plate-like NPs.179 These results demonstrate that the reaction rate influences the rate of particle formation, which appears to increase with the reaction temperature. In addition, the reaction temperature has an important effect on particle size and morphology.114
pH
pH is well known to have important effects on the shapes and sizes of NPs. Sathishkumar et al.180 reported that the yield of synthesized AgNPs using Cinnamon zeylanicum bark extract increased with an increase in the extract concentration and pH (the pH was 5 or above). In addition, higher pH values favored the formation of more spherical NPs.180 Conversely, in other cases, acidic conditions favored the production of spherical monodispersed AgNPs.92,177 It has also been shown that lower pH values favor the formation of larger NPs.49,50 For example, rod-shaped AuNPs synthesized using Avena sativa biomass at pH 2 were larger (25 to 85 nm) than that formed at pH 3 or 4 (5 to 20 nm).181 This suggested that the number of functional groups in the extract acting as nucleation sites at pH 2 was lower than that at pH 3 and 4, leading to the formation of larger AuNPs.114 Conversely, a pear extract was shown to yield hexagonal and triangular gold nanoplates under alkaline conditions, but afforded no NPs at all under acidic conditions.182 Additionally, He et al. demonstrated that the reduction of gold ions using Rhodopseudomonas capsulate yielded spherical AuNPs with diameters in the range of 10–20 nm at pH 7, but gold nanoplates were obtained at pH 4.133
The pH also strongly affects the reduction of silver ions by the amino acid tyrosine, which functions as a reducing agent under alkaline conditions. Reduction occurs at high pH because the phenol group of tyrosine is ionized, allowing it to transfer electrons to silver ions, and thereby become oxidized to a semi-quinone (Fig. 15). The ability of tyrosine to reduce silver ions to prepare Au core–Ag shell nanostructures at high pH was exploited so that the amino groups of the tyrosine molecules coordinated to the NPs. The tyrosine-decorated NPs were then treated with an Ag salt under basic conditions, resulting in the formation of a reduced Ag shell around the Au core (Fig. 15).183
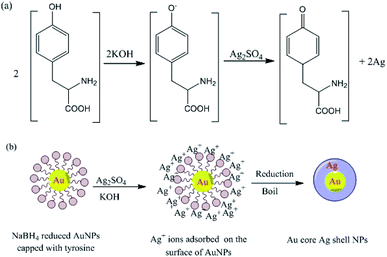 |
| | Fig. 15 (a) Tyrosine-catalyzed formation of AgNPs at high pH and (b) formation of Au-core–Ag-shell NPs under basic conditions.183 Adapted with permission from ref. 183. Copyright 2004, American Chemical Society. | |
Time
Reaction time is another key factor affecting the size and morphology of NPs. An increase in reaction rate has been observed to reduce the size of the synthesized NPs and the breadth of the particle size distribution.28,184 This can be due to the excitation of dissimilar multiple modes during the faceted and anisotropic growth of particles.145 The effect of reaction time on the synthesis of AgNPs was evaluated using the extract of Ananas comosus. Treatment of silver salts with this extract resulted in a pronounced color change within 2 min and the development of a broad UV-Vis absorption peak centered at around 430 nm, characteristic of Ag nanoclusters.61 The intensity of this peak increased over time, presumably due to the gradual increase in the number of NPs present in the suspension.185 The rate of particle growth was also time-dependent in this case, where the number of NPs increased over time, but their rate of growth decreased. There are several ways by which the number and morphology of NPs in a given suspension may change over time. For example, during prolonged storage, mixtures of NPs and plant extracts may undergo aggregation, and particles may shrink or grow.186 However, some AgNPs suspensions are stable for over 3 weeks. This can be due to the limited availability of extract molecules or a limited availability of Ag+ ions relative to each other's.174 The absorption of AgNPs increased linearly with time, i.e. A ∝ T where A and T denote absorbance and time, respectively.184
Reactant concentration
The initial concentration of the metal ions in the reaction mixture is another crucial factor. For example, when the leaf extract of Coleus aromaticus was used for the bioreduction of silver ions, it was found that NPs were efficiently synthesized when the concentration of silver ions was 1 mM or below, but higher concentrations caused aggregation of the newly formed NPs.56 Chauhan et al.124 reported the synthesis of AuNPs via the reaction of chloroaurate ions with a Candida albicans cytosolic extract. The shape of the NPs could be controlled by varying the concentration of the cytosolic extract in the reaction mixture, as shown in Fig. 16. The resulting NPs were non-spherical when the extract concentration was low, but spherical when it was high. Shervani and Yamamoto reported the synthesis of AuNPs and AgNPs, and found that treatment with excess NaBH4 caused the initially dark NP suspension to become clear, yielding a precipitate with a mesh-like or wire-like structure rather than monodisperse particles.167
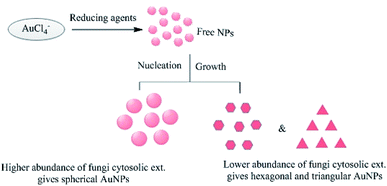 |
| | Fig. 16 Effects of varying the concentration of C. albicans cytosolic extract on the morphology of the AuNPs.64 | |
The concentration-dependent effect of added stabilizers on the biosynthesis of NPs has also been examined.63 Studies on AgNPs synthesis using the extract of Callistemon viminalis showed that adding cetyltrimethylammonium bromide at different concentrations yielded different morphological nano-couples. Imaging studies revealed that dimeric NPs with a range of different morphologies were obtained under these conditions.63
Industrial applications of metallic NP
In recent years, nanotechnology has emerged as a new field in fundamental science with broad applications.187 Nanotechnology plays an important role in many fields, especially health and life sciences, as shown in Fig. 17.188 It may be an answer for environmental and industrial challenges in the areas of solar energy, catalysis, medicine, water treatment and pollutant degradation.10,189–191 There are other fields that require the interaction of nanotechnology with other sciences to promote their bio-diverse applications in drug delivery, electronics, automobile, biosensors, semiconductors, cosmetics, the food industry and livestock feed additives.
 |
| | Fig. 17 Applications of metallic NPs. | |
Drug encapsulation
Pharmaceutical industries have become increasingly interested in nanotechnological advances, especially drug encapsulation.192 Although NPs provide great advantages in the pharmaceutical industry since they protect drugs from the harsh environment in vivo, improve their stability, increase and sustain their activity, improve their delivery efficiency and selectivity, extend their release and immobilize poorly soluble drugs, insufficient information is available on the long-term adverse effects of NPs on human health.193,194 The encapsulation is a vesicular immobilization system, where the conventional agents are confined or trapped inside the vesicle of the spherical structure. The biopolymer spherical structure can be constructed into a bilayer or a single layer sphere, depending on its properties and preparation techniques. Whereas, matrix-type sphere immobilization systems do not have a hollow cavity inside their structure, instead their whole structure is solidified and filled with the biomaterial. Modern drug encapsulation devices allow the efficient loading of drug molecules inside nanocarriers, thus reducing the systemic toxicity associated with drugs. Doxorubicin, which used in the treatment of different caner types, has been found to be associated with toxicities such as mucositis, gastrointestinal myelosuppression, and alopecia. This problem was solved by its encapsulation in liposomes, which reduced its toxicity and increased its retention time, whereas there was decrease in its systematic uptake with micelles compared to the free drug.195,196 Tretinoin is also used to treat various skin diseases, such as epidermotropism, psoriasis, acne, and T-cell lymphomas. However, it causes skin irritation, is highly unstable in the presence of air, light and heat and has low solubility, its use as a drug is limited. Therefore, tretinoin was encapsulated in liposomes. Negatively charged liposomes strongly improved the retention of tretinoin in the skin.197 On the other hand, encapsulating the drug together with a coating material is one of the few ways to load a drug on NPs. This provides better opportunity to attach several drugs for targeted delivery. For example, deferasirox (DFX), an anticancer drug, was loaded on Fe3O4NPs (Fe3O4@DFX), which displayed a higher apoptosis-inducing effect against different cancer cell lines, including MCF-7, HeLa, HT-29, K-562, Neuro-2a, and L-929 cells, with enhanced drug release.198 Methotrexate (MTX) was covalently bound with Fe3O4NPs for targeted drug delivery to cancer cells having overexpressed folate receptors on their surface.199 Treatment of MCF-7 and HeLa cells with the developed nanocomposite showed their internalization at a high level in the lysosome. In the lysosome, methotrexate was released at low pH by enzymatic cleavage of the peptide bond. These studies have documented that magnetic NPs can be applied as a positive carrier for controlled and targeted drug delivery.200 Furthermore, Paclitaxel (PTX) loaded on Fe3O4 NPs resulted in lower cell viability in ovary cancer cell lines, (A2780 and OVCAR-3) compared to doxorubicin (DOX)-loaded NPs with the same concentration. The result revealed a significant antineoplastic effect compared to the free drug.201
Livestock feed additives
Feed additives are food supplements added to livestock feed for animals that cannot get enough nutrients from regular meals. These additives include vitamins, amino acids, fatty acids, and minerals. Despite the advantages of higher quality of feed additives, most diets of livestock still consist of maize, wheat, and soybean to maintain cost-effectiveness. Currently, the major challenge is to increase the production of livestock, while maintaining the product quality, providing food security, and protecting the environment from hazardous elements.202 Nanotechnologies have the ability to improve the functionality of feed molecules, while delivering novel vehicles for nutrient supplementation. Minerals in the form of NPs can pass through the intestinal wall and cells more quickly due to their small size, thus enhancing their bioavailability.203 For example, the addition of ZnNPs to the livestock feed improved the growth, immunity, reproduction and increased the milk production in cows.204 Yang and Sun205 reported a significant drop in the diarrhea incidences by supplying graded doses of ZnONPs in the diet. In addition, nanosensors were introduced to study the causes of abortion in animals.205 Thus, the application of these techniques can possibly eliminate reproductive disturbances, and thus improve farming and the economy.206 Additionally, the addition of AgNPs (up to 100 nm) to animal feed was effective as an anti-microbial agent and selectively combated potential pathogens.207 Thus, NPs are a promising tool in animal feeding to improve the bioavailability of nutrients, production performance and immune status in livestock.
Future direction of the application of bio-derived nanomaterials
Currently, the various limitations in conventional therapy such as high cost and toxicity have made it necessary to design and develop novel drugs. Accordingly, opportunity to apply a wide range of nanosized materials in a greener way (eco-friendly, cost effective, easily available, and safe without the use of tough chemicals) opens a new door.208 Over the past decade, the rapid advancement of nanotechnology has promoted biomedical and industrial applications including detection, imaging, and drug delivery.209 From the relevant studies, it appears that bio-derived nanomaterials have been investigated in vitro, whereas there are no reports on their in vivo applications. Furthermore, there is insufficient information on the potential mechanisms of toxicity of metallic NPs.210 Therefore, in vivo trials should be carried out and tested thoroughly to generate data sets that depict nanomedicine behavior before initiating preclinical research.211 The applications of bio-derived nanomaterials will continue to grow, which is expected to shed light on their long-term effect on humans and animals and their impact on the environment.210 Additional studies are still required to understand their actual molecular mechanism and dynamics within the body tissues. In the coming years, biosynthesized metallic nanoparticles will strongly contribute to the nanodrug market and furnish other commercial applications.210
Limitations of biosynthesized NPs
Although the green biosynthesis of metallic NPs has gained tremendous interest in recent years, there are some limitations that should be considered prior to their wide industrial application. The size and morphology of NPs change dramatically depending on several factors. The localization of NPs can influence their movement and penetration after alterations in the tissue content of metal ions. This feature can increase the level of metal deposition around already existing NPs, and also the prospect of new nucleation events (initiation of NP formation).212 Moreover, the heterogeneity of the NPs formed may hamper their use in different applications, where specific and precise shapes and sizes are needed.24 Equally important is the difficulty related to the efficient extraction, isolation and purification process of NPs with a low recovery.24 The extraction of metal NPs for further applications is not well investigated. In the case of intracellular production of NPs, the cell wall around the plant and cells of other organisms is inflexible, which makes NP extraction difficult. Hence, organic solvents and surfactants are good candidates for the extraction and stabilization of NPs, whereas centrifugation can be used for extraction and purification, but aggregation may occur.40 For example, AuNPs were extracted from inactivated tissues of wheat and oat biomass.181 The extraction process was performed under sonication and required the use of a citrate and cetyltrimethylammonium bromide (CTAB) mixture for moving the NPs from the biomass into the aqueous solution. This study revealed that the extraction with citrate is more efficient than CTAB. The extracted solution was also examined under TEM, displaying the smaller radius of the AuNPs that appeared mainly at the beginning of the extraction, whereas the larger AuNPs were extracted later. A number of physicochemical approaches were applied to extract the formed NPs such as heating, freeze-thawing, and osmotic shock. However, these methods interfered with the structure of the NPs and caused some problems such as aggregation, precipitation, and sedimentation. Subsequently, the properties of the NPs changed as their size and the shape changed. Possibly, enzymatic lysis (e.g., cocktail of enzymes isolated from Trichoderma longibrachiatum) could be used, but this process is expensive and not applied on a large scale for industrial purposes.213 The other limitations of biosynthesized NPs are related to their storage conditions and long-term stability. Velgosová and Mražíková recently studied the long-term stability of AgNPs and discussed the influence of light and temperature on the storage of the AgNPs, where the stability loss was most significant in daylight at room temperature (21 °C). After 200 days, a large number of agglomerated particles appeared. AgNPs stored in the dark at room temperature showed better long-term stability, and agglomeration was not as evident. AgNPs stored in the dark and at a temperature of 5 °C showed the best long-term stability with a dominant spherical shape and no agglomeration even after more than six months.214
Conclusions
This review presented information about the eco-friendly biosynthesis of nanoparticles (NPs) with particular emphasis on the underlying mechanisms. Biomolecules of plant extracts are involved in the reduction of metal ions to nanoparticles in a one-step synthesis. A variety of plant species and plant parts have been successfully extracted and utilized for the synthesis of NPs since they contain a wide range of metabolites, including carbohydrates, alkaloids, terpenoids, phenolic compounds, and enzymes. Herein, a list of the plants involved in the green synthesis of nanoparticles in the period between 2014–2017 was tabulated. The mechanisms by which the biomolecules of other organisms such as fungi and algae are involved in the biosynthesis of NPs are still under debate; however, they showed shape and size-dependent biological activities and eliminated the need for stabilizing and capping agents. The current review also highlighted the factors affecting the biosynthesis of NPs as and some of their commercial and industrial applications. Considering the promise of nanotechnology applications in everyday life, more research is warranted to unravel the long-term chemical and physical properties of biosynthesized nanoparticles.
Conflicts of interest
There are no conflicts to declare.
Abbreviation
| NP | Nanoparticles |
| HFPM | Hexane–fresh plant material |
| HDPM | Hexane–dry plant material |
| FFPM | Dichloromethane–FPM |
| DDPM | Dichloromethane–DPM |
| MFPM | Methanol–FPM |
| MDPM | Methanol–DPM |
Acknowledgements
H. R. El-Seedi is very grateful to the Swedish Research links grant 2016-05885 (VR for the years 2017–2019) and to Al-Rayan Colleges, Medina, Saudi Arabia & Jiangsu University, China for generous financial support, consultant and Adjunct Professor fellowships, respectively.
Notes and references
- M. F. Abdelbar, H. S. El-Sheshtawy, K. R. Shoueir, I. El-Mehasseb, E. Z. M. Ebeid and M. El-Kemary, RSC Adv., 2018, 8, 24617 RSC.
- N. El-Shafai, M. E. El-Khouly, M. El-Kemary, M. Ramadan, I. Eldesoukey and M. Masoud, RSC Adv., 2019, 9, 3704 RSC.
- K. Shoueir, H. El-Sheshtawy, M. Misbah, H. El-hosainy, I. El-mehasseb and M. El-Kemary, Carbohydr. Polym., 2018, 197, 17–28 CrossRef CAS PubMed.
- H. El-Sheshtawy, H. El-hosainy, K. R. Shoueir, I. El-mehasseb and M. El-Kemary, Appl. Surf. Sci., 2019, 467, 268–276 CrossRef.
- A. Mezni, M. M. Ibrahim, M. El-Kemary, A. A. Shaltout, N. Y. Mostafa, J. Ryl and M. A. Amin, Electrochim. Acta, 2018, 290, 404 CrossRef CAS.
- M. A. Akl, A. A. Sarhan, K. R. Shoueir and A. M. Atta, J. Dispersion Sci. Technol., 2013, 34, 1399 CrossRef CAS.
- A. Salama, K. R. Shoueir and H. A. Aljohani, Fibers Polym., 2017, 18, 1825 CrossRef CAS.
- R. R. Fouad, H. A. Aljohani and K. R. Shoueir, Mar. Pollut. Bull., 2016, 112, 46 CrossRef CAS PubMed.
- A. M. Atta, G. A. El-Mahdy, H. A. Al-Lohedan and K. R. Shoueir, Int. J. Electrochem. Sci., 2015, 10, 870 CAS.
- I. Khan, K. Saeed and I. Khan, Arabian J. Chem., 2017 DOI:10.1016/j.arabjc.2017.05.011.
- V. V. Mody, R. Siwale, A. Singh and H. R. Mody, J. Pharm. BioAllied Sci., 2010, 2, 282–289 CrossRef CAS PubMed.
- T. Yokoyama, H. Masuda, M. Suzuki, K. Ehara, K. Nogi, M. Fuji, T. Fukui, H. Suzuki, J. Tatami, K. Hayashi, and K. Toda, Nanoparticle technology handbook, 2008, pp. 3–48 Search PubMed.
- V. K. Sharma, R. A. Yngard and Y. Lin, Adv. Colloid Interface Sci., 2009, 145, 83–96 CrossRef CAS PubMed.
- R. Kessler, Environ. Health Perspect., 2011, 119, 120–125 Search PubMed.
- V. V. Mody, M. I. Nounou and M. Bikram, Adv. Drug Delivery Rev., 2009, 61, 795–807 CrossRef CAS PubMed.
- K. Takeda, K.-I. Suzuki, A. Ishihara, M. Kubo-Irie, R. Fujimoto, M. Tabata, S. Oshio, Y. Nihei, T. Ihara and M. Sugamata, J. Health Sci., 2009, 55, 95–102 Search PubMed.
- W. J. Trickler, S. M. Lantz, R. C. Murdock, A. M. Schrand, B. L. Robinson, G. D. Newport, J. J. Schlager, M. G. Paule, S. J. Oldenburg, W. Slikker Jr, S. M. Hussain and S. F. Ali, Toxicol. Sci., 2010, 118, 160–170 CrossRef CAS PubMed.
- V. Weissig, T. K. Pettinger and N. Murdock, Int. J. Nanomed., 2014, 9, 4357–4373 CrossRef CAS PubMed.
- B. Gulson, M. McCall, M. Korsch, L. Gomez, P. Casey, Y. Oytam, A. Taylor, M. Mc Culloch, J. Trotter, L. Kinsley and G. Greenoak, Toxicol. Sci., 2010, 118, 140–149 CrossRef CAS PubMed.
- A. V. Krestinin, N. N. Dremova, E. I. Knerel`man, L. N. Blinova, V. G. Zhigalina and N. A. Kiselev, Nanotechnol. Russ., 2015, 10, 537–548 CrossRef CAS.
- R. Ravichandran, Int. J. Green Nanotechnol., 2010, 1, 72–96 CrossRef.
- M. E. Vance, T. Kuiken, E. P. Vejerano, S. P. McGinnis, M. F. Hochella Jr, D. Rejeski and M. S. Hull, Beilstein J. Nanotechnol., 2015, 6, 1769–1780 CrossRef CAS PubMed.
- T. Benn, J. Environ. Qual., 2010, 39, 1875–1882 CrossRef CAS PubMed.
- V. V. Makarov, A. J. Love, O. V. Sinitsyna, S. S. Makarova, I. V. Yaminsky, M. E. Taliansky and N. O. Kalinina, Acta Nat., 2014, 6, 35–44 CrossRef CAS.
- L. Jia, Q. Zhang, Q. Li and H. Song, Nanotechnology, 2009, 20, 385601 CrossRef PubMed.
- J. Y. Song, H. K. Jang and B. S. Kim, Process Biochem., 2009, 44, 1133–1138 CrossRef CAS.
- P. Velusamy, G. V. Kumar, V. Jeyanthi, J. Das and R. Pachaiappan, Toxicol. Res., 2016, 32, 95 CrossRef CAS PubMed.
- K. Punjabi, P. Choudhary, L. Samant, S. Mukhejee, S. Vaidya and A. Chowdhary, Int. J. Pharm. Sci. Rev. Res., 2015, 30, 219 Search PubMed.
- H. colfen, Nat. Mater., 2010, 9, 960 CrossRef PubMed.
- S. K. Das, C. Dickinson, F. Lafir, D. F. Brougham and E. Marsili, Green Chem., 2012, 14, 1322–1334 RSC.
- K. N. Thakkar, S. S. Mhatre and R. Y. Parikh, Nanomedicine, 2010, 6, 257–262 CrossRef CAS PubMed.
- N. Duran, P. D. Marcato, M. Duran, A. Yadav, A. Gada and M. Rai, Appl. Microbiol. Biotechnol., 2011, 90, 1609–1624 CrossRef CAS PubMed.
- N. Jain, A. Bhargava, S. Majumdar, J. C. Tarafdar and J. Panwar, Nanoscale, 2011, 3, 635–641 RSC.
- L. M. Carrillo-López, H. A. Zavaleta-Mancera, A. Vilchis-Nestor, R. M. Soto-Hernández, J. Arenas-Alatorre, L. I. Trejo-Téllez and F. Gómez-Merino, J. Nanomater., 2014, 2014, 198 Search PubMed.
- N. Ahmad, S. Sharma, M. d. K. Alam, V. N. Singh, S. F. Shamsi, B. R. Mehta and A. Fatma, Colloids Surf., B, 2010, 81, 81–86 CrossRef CAS PubMed.
- J. F. Moran, R. V. Klucas, R. J. Grayer, J. Abian and M. Becana, Free Radical Biol. Med., 1997, 22, 861–870 CrossRef CAS PubMed.
- J. Huang, Q. Li, D. Sun, Y. Lu, Y. Su, X. Yang, H. Wang, Y. Wang, W. Shao, N. He, J. Hong and C. Chen, Nanotechnology, 2007, 18, 105104 CrossRef.
- R. Singh, U. U. Shedbalkar, S. A. Wadhwani and B. A. Chopade, Appl. Microbiol. Biotechnol., 2015, 99, 4579–4593 CrossRef CAS PubMed.
- P. Kuppusamy, M. M. Yusoff, G. P. Maniam and N. Govindan, Saudi Pharm. J., 2016, 24, 473 CrossRef PubMed.
- S. Iravani, Green Chem., 2011, 13, 2638 RSC.
- O. V. Kharissova, H. V. R. Dias, B. I. Kharisov, B. O. Perez, M. Victor and J. Perez, Trends Biotechnol., 2013, 31, 240 CrossRef CAS PubMed.
- M. Nasrollahzadeh and S. M. Sajadi, Ceram. Int., 2015, 41, 14435 CrossRef CAS.
- R. Sankar, P. Manikandan, V. Malarvizhi, T. Fathima, K. S. Shivashangari and V. Ravikumar, Spectrochim. Acta, Part A, 2014, 121, 746 CrossRef CAS PubMed.
- L. Castro, M. L. Blázquez, J. A. Muñoz, F. González, C. García-Balboa and A. Ballester, Process Biochem., 2011, 46, 1076 CrossRef CAS.
- S. Küünal, P. Rauwel and E. Rauwel, Plant extract mediated synthesis of nanoparticles, in Emerging Applications of Nanoparticles and Architecture Nanostructures, ed. A. Barhoum and A. S. H. Makhlouf, Elsevier, 2018, ch. 14, pp. 411–446 Search PubMed.
- B. Vellaichamy and P. Periakaruppan, RSC Adv., 2016, 6, 88837 RSC.
- M. Atarod, M. Nasrollahzadeh and S. M. Sajadi, J. Colloid Interface Sci., 2016, 465, 249 CrossRef CAS PubMed.
- M. Nasrollahzadeh and S. M. Sajadi, J. Colloid Interface Sci., 2015, 457, 141 CrossRef CAS PubMed.
- M. Nasrollahzadeh and S. M. Sajadi, RSC Adv., 2015, 5, 46240 RSC.
- M. Sajjadi, M. Nasrollahzadeh and S. M. Sajadi, J. Colloid Interface Sci., 2017, 497, 1 CrossRef CAS PubMed.
- M. Nasrollahzadeh and S. M. Sajadi, J. Colloid Interface Sci., 2015, 448, 106 CrossRef CAS PubMed.
- M. Nasrollahzadeh and S. M. Sajadi, Ceram. Int., 2015, 41, 14435 CrossRef CAS.
- K. Shameli, M. Ahmad, E. A. J. Al-Mulla, N. A. Ibrahim, P. Shabanzadeh, A. Rustaiyan and Y. Abdollahi, Molecules, 2012, 17, 8506 CrossRef CAS PubMed.
- M. Zargar, A. A. Hamid, F. A. Bakar, M. N. Shamsudin, K. Shameli, F. Jahanshiri and F. Farahani, Molecules, 2011, 16, 6667 CrossRef CAS PubMed.
- M. El-Kemary, M. Zahran, S. A. M. Khalifa and H. R. El-Seedi, Micro Nano Lett., 2016, 11, 311 CrossRef CAS.
- M. Vanaja, S. Rajeshkumar, K. Paulkumar, G. Gnanajobitha, C. Malarkodi and G. Annadurai, Adv. Appl. Sci. Res., 2013, 4, 50 CAS.
- R. El-Shabasy, N. Yosri, H. El-Seedi, K. Shoueir and M. El-Kemary, Optik- Int. J. Light Elect. Optics, 2019, 192, 162943 CrossRef CAS.
- S. N. Sinha and D. Paul, Spectrosc. Lett., 2015, 48, 600 CrossRef CAS.
- K. Vivehananthan, S. Lakshmi, Y. Sudha, W. H. De Silva and S. A. Kumar, World J. Pharm. Res., 2015, 4, 77 CAS.
- P. P. N. V. Kumar, S. V. N. Pammi, P. Kollu and U. Shameem, Ind. Crops Prod., 2014, 52, 562 CrossRef.
- R. Tamileswari, M. Haniff Nisha and S. S. Jesurani, Int. J. Eng. Res. Technol., 2015, 4, 1071 Search PubMed.
- M. El-Kemary, E. Ibrahim, M. F. Al-Ajmi, S. A. M. Khalifa, A. Alanazi and H. R. El-Seedi, Int. J. Electrochem. Sci., 2016, 11, 10795 CrossRef CAS.
- J. M. Rahisuddin and A. Akrema, Spectrosc. Lett., 2016, 49, 268 CrossRef.
- O. J. Nava, P. A. Luque, C. M. Gomez-Gutiérrez, A. R. Vilchis-Nestor, A. Castro-Beltran, M. L. Mota-Gonzalez and A. Olivas, J. Mol. Struct., 2017, 1134, 121 CrossRef CAS.
- M. M. Netai, N. Stephen and C. Musekiwa, Afr. J. Biotechnol., 2017, 16, 1911–1921 CrossRef CAS.
- H. Yan, D. Zhiyun, M. Shijing, C. Shupeng, J. Sen, L. Yue, L. Dongli, H. Huarong, Z. Kun and Z. Xi, Nanoscale Res. Lett., 2016, 11, 1 CrossRef PubMed.
- V. Subha, R. R. S. Ernest, P. Sruthi and S. Renganathan, Asian J. Pharm. Clin. Res., 2015, 8, 103 Search PubMed.
- S. Shankar, L. Jaiswal, R. S. L. Aparna and R. G. S. V. Prasad, Mater. Lett., 2014, 137, 75 CrossRef CAS.
- O. Zuas, N. Hamim and Y. Sampora, Mater. Lett., 2014, 123, 156 CrossRef CAS.
- P. Biparva, S. M. Abedirad, S. Y. Kazemi and M. Shanehsaz, Sens. Actuators, B, 2016, 234, 278 CrossRef CAS.
- A. Bashir, H. Nabia, B. Shumaila, U. Midrar, A. Sadiq, K. Ibrar and N. Sahar, Pak. J. Bot., 2015, 47, 139 Search PubMed.
- K. Kovendan, B. Chandramohan, D. Dinesh, D. Abirami, P. Vijayan, M. Govindarajan, S. Vincent and G. Benelli, J. Asia-Pac. Entomol., 2016, 19, 1001 CrossRef.
- H. Korbekandi, M. R. Chitsazi, G. Asghari, R. B. Najafi, A. Badii and S. Iravani, Pharm. Biol., 2015, 53, 807 CrossRef CAS PubMed.
- M. Zahran, M. El-Kemary, S. Khalifa and H. El-Seedi, Green Process. Synth., 2017, 7, 100–105 Search PubMed.
- S. Shankar, J. Chorachoo, L. Jaiswal and S. P. Voravuthikunchai, Mater. Lett., 2014, 137, 160 CrossRef CAS.
- K. Tahir, S. Nazir, B. Li, A. Ahmad, T. Nasir, A. U. Khan, S. A. A. Shah, Z. U. H. Khan, G. Yasin and M. U. Hameed, J. Photochem. Photobiol., B, 2016, 164, 164 CrossRef CAS PubMed.
- S. J. Lakshmi, R. R. Bai, H. Sharanagouda, C. T. Ramachandra, S. Nadagouda and S. R. Doddagoudar, Int. J. Curr. Microbiol. Appl. Sci., 2017, 6, 3376–3384 CrossRef.
- P. Devaraj, R. Arun, C. Aarti and P. Kumari, Int. J. Pharm. Pharm. Sci., 2014, 6, 86–90 CAS.
- T. Azarudeen, R. M. Sait, G. Marimuthu, A. M. Marzouq, A. F. Saeed, A. Abubucker and B. Giovanni, J. Cluster Sci., 2017, 28, 369 CrossRef.
- B. Sadeghi and F. Gholamhoseinpoor, Spectrochim. Acta, Part A, 2015, 134, 310 CrossRef CAS PubMed.
- N. L. Gavade, A. N. Kadam, M. B. Suwarnkar, V. P. Ghodake and K. M. Garadkar, Spectrochim. Acta, Part A, 2015, 136, 953 CrossRef CAS PubMed.
- S. Soman and J. G. Ray, J. Photochem. Photobiol., B, 2016, 163, 391 CrossRef CAS PubMed.
- E. A. Masoud, A. M. Al-Hajry and A. Al-Marrani, Int. J. Curr. Microbiol. Appl. Sci., 2016, 5, 226 CAS.
- M. Kitching, M. Ramani and E. Marsili, Mechanism and scale up, Microb. Biotechnol., 2015, 8, 904–917 CrossRef CAS PubMed.
- T. Klaus, R. Joerger, E. Olsson and C. G. Granqvist, Proc. Natl. Acad. Sci. U. S. A., 1999, 96, 13611–13614 CrossRef CAS PubMed.
- K. Das and P. Thiagarajan, Int. J. Nanotechnol., 2012, 32 Search PubMed.
- A. Ahmad, P. Mukherjee, S. Senapati, D. Mandal, M. I. Khan, R. Kumar and M. Sastry, Colloids Surf., B, 2003, 28, 313–318 CrossRef CAS.
- G. Li, D. He, Y. Qian, B. Guan, S. Gao, Y. Cui, K. Yokoyama and L. Wang, Int. J. Mol. Sci., 2012, 13, 466–476 CrossRef CAS PubMed.
- S. Basavaraja, S. D. Balaji, A. Lagashetty, A. H. Rajasab and A. Venkataraman, Mater. Res. Bull., 2008, 43, 1164–1170 CrossRef CAS.
- K. Kalishwaralal, V. Deepak, S. Ramkumarpandian, H. Nellaiah and G. Sangiliyandi, Mater. Lett., 2008, 62, 4411–4413 CrossRef CAS.
- D. K. Newman and R. Kolter, Nature, 2000, 405, 94 CrossRef CAS PubMed.
- G. Oza, S. Pandey, A. Gupta, R. Kesarkar and M. Sharon, J. Microbiol. Biotechnol. Res., 2012, 2, 511–515 CAS.
- N. Duran, P. D. Marcato, O. L. Alves, G. I. H. De Souza and E. Esposito, J. Nanobiotechnol., 2005, 13, 3–8 Search PubMed.
- P. Jeevan, K. Ramya and A. E. Rena, Indian J. Biotechnol., 201, 11, 72–76 Search PubMed.
- H. von Canstein, J. Ogawa, S. Shimizu and J. R. Lloyd, Appl. Environ. Microbiol., 2008, 74, 615–623 CrossRef CAS PubMed.
- C. Schwalb, S. K. Chapman and G. A. Reid, Biochem. Soc. Trans., 2002, 30, 658–662 CrossRef CAS PubMed.
- S. S. Shankar, A. Ahmad, R. Pasricha and M. Sastry, J. Mater. Chem., 2003, 13, 1822–1826 RSC.
- M. Rai, A. Yadav and A. Gade, Current Research Technology and Education Topics in Applied Microbiology and Microbial Biotechnology, ed. A. Mendez-Vilas, 2010 Search PubMed.
- S. K. Das, J. Liang, M. Schmidt, F. Laffir and E. Marsili, ACS Nano, 2012, 6, 6165 CrossRef CAS PubMed.
- P. Mukherjee, A. Ahmad, D. Mandal, S. Senapati, S. R. Sainkar, M. I. Khan, R. Ramani, R. Parischa, P. V. Ajayakumar, M. Alam, M. Sastry and R. Kumar, Angew. Chem., Int. Ed., 2001, 40, 3585 CrossRef CAS PubMed.
- J. F. Pebberdy, Fungal cell walls - A review, in Biochemistry of cell walls and membranes in fungi, ed. P. J. Kuhn, A. P. J. Trinci, M. J. Jung, M. W. Goosey and L. G. Coping, Springer-Verlag, Germany, 1990, pp. 5–30 Search PubMed.
- M. M. Khan, S. Kalathil, T. H. Han, J. Lee and M. H. Cho, J. Nanosci. Nanotechnol., 2013, 13, 6079 CrossRef CAS PubMed.
- J. Zeng, Y. Ma, U. Jeong and Y. Xia, J. Mater. Chem., 2010, 20, 2290 RSC.
- N. Pantidos and L. E. Horsfall, J. Nanomed. Nanotechnol., 2014, 5, 2 Search PubMed.
- M. A. Dias, I. C. Lacerda, P. F. Pimentel, H. F. de Castro and C. A. Rosa, Lett. Appl. Microbiol., 2002, 34, 46 CrossRef CAS.
- T. Ahmad, I. A. Wani, N. Manzoor, J. Ahmed and A. M. Asiri, Colloids Surf., B, 2013, 107, 227 CrossRef CAS PubMed.
- Y. Yin, X. Yang, L. Hu, Z. Tan, L. Zhao, Z. Zhang, J. Liu and G. Jiang, Environ. Sci. Technol. Lett., 2016, 3, 160–165 CrossRef CAS.
- J. Ali, N. Ali, L. Wang, H. Waseem and G. Pan, J. Microbiol. Methods, 2019, 159, 18–25 CrossRef CAS PubMed.
- B. Fadeel and A. E. Garcia-Bennett, Adv. Drug Delivery Rev., 2010, 62, 362–374 CrossRef CAS PubMed.
- H. Tang, M. Yan, H. Zhang, M. Xia and D. Yang, Mater. Lett., 2005, 59, 1024–1027 CrossRef CAS.
- M. Mergeay, S. Monchy, T. Vallaeys, V. Auquier, A. Benotmane, P. Bertin, S. Taghavi, J. Dunn, D. Van Der Lelie and R. Wattiez, FEMS Microbiol. Rev., 2003, 27, 385–410 CrossRef CAS PubMed.
- C. W. Johnston, M. A. Wyatt, X. Li, A. Ibrahim, J. G. Shuster Southam and N. A. Magarvey, Nat. Chem. Biol., 2013, 9, 241–243 CrossRef CAS PubMed.
- L. Sintubin, W. De Windt, J. Dick, J. Mast, D. van der Ha, W. Verstraete and N. Boon, Appl. Microbiol. Biotechnol., 2009, 84, 741–749 CrossRef CAS PubMed.
- M. Shah, D. Fawcett, S. Sharma, S. K. Tripathy and G. E. J. Poinern, Materials, 2015, 8, 7278–7308 CrossRef CAS PubMed.
- T. Luangpipat, I. R. Beattie, Y. Chisti and R. G. Haverkamp, J. Nanopart. Res., 2011, 13, 6439–6445 CrossRef CAS.
- K. Govindaraju, V. Kiruthiga, V. G. Kumar and G. Singaravelu, J. Nanosci. Nanotechnol., 2009, 9, 5497–5501 CrossRef CAS PubMed.
- G. Singaravelu, J. S. Arockiamary, V. G. Kumar and K. Govindaraju, Colloids Surf., B, 2007, 57, 97–101 CrossRef CAS PubMed.
- L. Castro, M. L. Blázquez, J. A. Muñoz, F. Gonzaález and A. Ballester, IET Nanobiotechnol., 2013, 7, 109–116 CrossRef CAS PubMed.
- S. Senapati, A. Syed, S. Moeez, A. Kumar and A. Ahmad, Mater. Lett., 2012, 79, 116–118 CrossRef CAS.
- Y. N. Mata, M. L. Blázquez, A. Ballester, F. González and J. A. Muñoz, J. Hazard. Mater., 2009, 166, 612–618 CrossRef CAS.
- P. Rajasulochana, R. Dhamotharan, P. Murugakoothan, S. Murugesan and P. Krishnamoorthy, Int. J. Nanosci., 2010, 9, 511–516 CrossRef CAS.
- D. Venkataraman, K. Kalimuthu, R. K. P. Sureshbabu, and G. Sangiliyandi. Metal Nanoparticles in Microbiology, ed. M. Rai and N. Duran, Springer, 2011, vol. XI, pp. 17–35 Search PubMed.
- K. C. Bhainsa and S. F. D'Souza, Colloids Surf., B, 2006, 47, 160–164 CrossRef CAS PubMed.
- A. Chauhan, S. Zubair, S. Tufail, A. Sherwani, M. Sajid, S. C. Raman, A. Azam and M. Owais, Int. J. Nanomed., 2011, 6, 2305 CAS.
- R. K. Abhilash and B. D. Pandey, Bull. Mater. Sci., 2011, 34, 191–198 CrossRef.
- P. Sukumaran and K. P. Eldho, Int. Nano Lett., 2012, 2, 2–10 CrossRef.
- L. Xiangqian, X. Huizhong, Z. Chen and G. Chen, J. Nanomater., 2011, 2011, 1–16 Search PubMed.
- N. Vigneshwaran, A. A. Kathe, P. V. Varadarajan, R. P. Nachane and R. H. Balasubramanya, Colloids Surf., B, 2006, 53, 55–59 CrossRef CAS PubMed.
- K. AbdelRahim, S. Y. Mahmoud, A. M. Ali, K. S. Almaary, A.-Z. M. A. Mustafa and S. M. Husseiny, Saudi J. Biol. Sci., 2017, 24, 208–216 CrossRef CAS PubMed.
- A. Lateef, S. A. Ojo and S. M. Oladejo, Process Biochem., 2016, 51, 1406–1412 CrossRef CAS.
- S. Gurunathan, K. Kalishwaralal, R. Vaidyanathan, V. Deepak, S. R. K. Pandian, J. Muniyandi, N. Hariharan and S. H. Eomb, Colloids Surf., B, 2009, 74, 328–335 CrossRef CAS PubMed.
- D. Paul and S. N. Sinha, Jordan J. Biol. Sci., 2014, 7, 245–250 CrossRef.
- S. He, Z. Guo, Y. Zhang, S. Zhang, J. Wang and N. Gu, Mater. Lett., 2007, 61, 3984 CrossRef CAS.
- R. K. Abhilash and B. D. Pandey, Bull. Mater. Sci., 2011, 34, 191–198 CrossRef.
- Z. A. Abdul Aziz, S. A. M. Ali, A. Ahmad and S. H. Mohd-Setapar, Der Pharm. Lett., 2016, 8, 161–167 CAS.
- M. Govindarajan, M. Rajeswary, U. Muthukumaran, S. L. Hoti, H. F. Khater and G. Benelli, J. Photochem. Photobiol., B, 2016, 161, 482–489 CrossRef CAS PubMed.
- R. W. Raut, V. D. Mendhulkar and S. B. Kashid, J. Photochem. Photobiol., B, 2014, 132, 45 CrossRef CAS PubMed.
- G. Marslin, K. Siram, Q. Maqbool, R. K. Selvakesavan, D. Kruska, P. Kachlicki and G. Franklin, Materials, 2018, 11, 940–965 CrossRef PubMed.
- O. El-Shahaby, M. El-Zayat, E. Salih, I. M. El-Sherbiny and F. M. Reicha, J. Nanomed. Nanotechnol., 2013, 4, 1–7 Search PubMed.
- J. Kasthuri, S. Veerapandian and N. Rajendiran, Colloids Surf., B, 2009, 68, 55–60 CrossRef CAS PubMed.
- J. A. Sierra, C. R. Vanoni, M. A. Tumelero, C. C. P. Cid, R. Faccio, D. F. Franceschini, T. B. Creczynski-Pasa and A. A. Pasa, New J. Chem., 2016, 40, 1420–1429 RSC.
- R. Krishanan and G. B. Maru, Food Chem., 2006, 94, 331–340 CrossRef.
- S. Kavitha, M. Dhamodaran, R. Prasad and M. Ganesan, Int. Nano Lett., 2017, 7, 141–147 CrossRef CAS.
- A. Singh, M. Talat, D. Singh and O. N. Srivastava, J. Nanopart. Res., 2010, 12, 1667–1675 CrossRef CAS.
- S. Hussain, O. Bashir, Z. Khan and S. A. Al-Thabaiti, J. Mol. Liq., 2014, 199, 489 CrossRef CAS.
- X. Zhang, S. Yan, R. D. Tyagi and R. Y. Surampalli, Chemosphere, 2011, 82, 489–494 CrossRef CAS PubMed.
- J. M. Palomo and M. Filice, Nanomaterials, 2016, 6, 84 CrossRef PubMed.
- A. Ahmad, P. Mukherjee, D. Mandal, S. Senapati, M. I. Khan, R. Kumar and M. Sastry, J. Am. Chem. Soc., 2002, 124, 12108–12109 CrossRef CAS PubMed.
- D. S. Balaji, S. Basavaraja, R. Deshpande, D. Mahesh, B. K. Prabhakar and A. Venkataraman, Colloids Surf., B, 2009, 68, 88–92 CrossRef CAS PubMed.
- A. Gole, C. Dash, V. Ramachandran, S. Sainkar, A. Mandale, M. Rao and M. Sastry, Langmuir, 2001, 17, 1674–1679 CrossRef CAS.
- R. R. Naik, S. J. Stringer, G. Agarwal, S. E. Jones and M. O. Stone, Nat. Mater., 2002, 1, 169–172 CrossRef CAS PubMed.
- M. Duran, C. Silveirs and N. Duran, IET Nanobiotechnol., 2015, 9, 314 CrossRef PubMed.
- J. L. Elechiguerra, J. L. Burt, J. R. Morones, A. Camacho-Bragado, X. Gao, H. H. Lara and M. J. Yacaman, J. Nanobiotechnol., 2005, 3, 6 CrossRef PubMed.
- H. U. Gulsuner, H. Ceylan, M. O. Guler and A. B. Tekinay, ACS Appl. Mater. Interfaces, 2015, 7, 10677 CrossRef PubMed.
- V. Gopinath, D. Mubarak, S. Priyadarshini, P. Meera, N. Thajuddin and P. Velusamy, Colloids Surf., B, 2012, 96, 69–74 CrossRef CAS PubMed.
- A. Mahal, P. Khullar, H. Kumar, G. Kaur, N. Singh, M. Jelokhani-Niaraki and M. S. Bakshi, Chem. Eng., 2013, 1, 627–639 CAS.
- L. C. Gruen, Biochim. Biophys. Acta, Protein Struct., 1975, 386, 270–274 CrossRef CAS.
- S. Li, Y. Shen, A. Xie, X. Yu, L. Qiu, L. Zhang and Q. Zhang, Green Chem., 2007, 9, 852–858 RSC.
- S. Li, Y. Shen, A. Xie, X. Yu, X. Zhang, L. Yang and C. Li, Nanotechnology, 2007, 18, 405101 CrossRef.
- C. G. Kumar and S. K. Mamidyala, Colloids Surf., B, 2011, 84, 462–466 CrossRef CAS PubMed.
- S. Sudha, K. Rajamanickam and J. Rengaramanujam, Indian J. Exp. Biol., 2013, 51, 393–399 CAS.
- J. Glusker, A. Katz and C. Bock, Rigaku J., 1999, 16, 8–16 CAS.
- R. R. Bhattacharjee, A. K. Das, D. Haldar, S. Si, A. Banerjee and T. K. Mandal, J. Nanosci. Nanotechnol., 2005, 5, 1141–1147 CrossRef CAS PubMed.
- J. Hu, L. Zhang, S. Aryal, C. Cheung, H. Fang and L. Zhang, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 10980–10985 CrossRef PubMed.
- L. Rodriguez, T. Harada, A. Christian, A. Pantano, K. Tsai and E. Discher, Science, 2013, 339, 971–975 CrossRef PubMed.
- S. Chowdhury, A. Basu and S. Kundu, Nanoscale Res. Lett., 2014, 9, 1–11 CrossRef CAS PubMed.
- Z. Shervani and Y. Yamamoto, Carbohydr. Res., 2011, 346, 651 CrossRef CAS PubMed.
- A. A. Sousa, S. A. Hassan, L. L. Knittel, A. Balbo, M. A. Aronova, P. H. Brown, P. Schuck and R. D. Leapman, Nanoscale, 2016, 8, 6577–6588 RSC.
- J. Jung, S. Park, S. Hong, M. W. Ha, H. G. Park, Y. Park, H. J. Lee and Y. Park, Carbohydr. Res., 2014, 386, 57–61 CrossRef CAS PubMed.
- M. N. Nadagouda and R. S. Varma, Green Chem., 2006, 8, 516–518 RSC.
- T. Tanaka, M. Ishibashi, H. Fujimoto, E. Okuyama, T. Koyano, T. Kowithayakorn, M. Hayashi and K. Komiyama, J. Nat. Prod., 2002, 65, 1709–1711 CrossRef CAS.
- Y. L. Chen, H. Y. Tuan, C. W. Tien, W. H. Lo, H. C. Liang and Y. C. Hu, Biotechnol. Prog., 2009, 25, 1260–1266 CrossRef CAS PubMed.
- P. K. Vemula, U. Aslam, V. A. Mallia and G. John, Chem. Mater., 2007, 19, 138–140 CrossRef CAS.
- S. K. Srikar, D. D. Giri, D. B. Pal, P. K. Mishra and S. N. Upadhyay, Green Sustainable Chem., 2016, 6, 26 CrossRef CAS.
- N. L. Pacioni, C. D. Borsarelli, V. Rey and A. V. Veglia, Silver Nanoparticle Applications, ed. E.I. Alarcon, Springer International Publishing Switzerland, 2015, pp. 13–46 Search PubMed.
- N. Soni and S. Prakash, Am. J. Nanotechnol., 2011, 2, 112 CAS.
- D. Mott, J. Galkowski, L. Wang, J. Luo and C. J. Zhong, Langmuir, 2007, 23, 5740 CrossRef CAS PubMed.
- S. Kaviya, J. Santhanalakshmi, B. Viswanathan, J. Muthumary and K. Srinivasan, Spectrochim. Acta, Part A, 2011, 79, 594 CrossRef CAS PubMed.
- M. Gericke and A. Pinches, Hydrometallurgy, 2006, 83, 132 CrossRef CAS.
- M. Sathishkumar, K. Sneha, S. W. Won, C. W. Cho, S. Kim and Y. S. Yun, Colloids Surf., B, 2009, 73, 332 CrossRef CAS PubMed.
- V. Armendariz, I. Herrera, J. R. Peralta-Videa, M. Jose-Yacaman, H. Troiani, P. Santiago and J. L. Gardea-Torresdey, J. Nanopart. Res., 2004, 6, 377 CrossRef CAS.
- G. S. Ghodake, N. G. Deshpande, Y. P. Lee and E. S. Jin, Colloids Surf., B, 2010, 75, 584 CrossRef CAS PubMed.
- P. R. Selvakannan, A. Swami, D. Srisathiyanarayanan, P. S. Shirude, R. Pasricha, A. B. Mandale and M. Sastry, Langmuir, 2004, 20, 7825 CrossRef CAS PubMed.
- M. Darroudi, M. B. Ahmad, R. Zamiri, A. K. Zak, A. H. Abdullah and N. A. Ibrahim, Int. J. Nanomed., 2011, 6, 677 CrossRef CAS PubMed.
- N. Ahmad and S. Sharma, Green Sustainable Chem., 2012, 2, 141 CrossRef.
- D. R. Baer, J. Surf. Anal., 2011, 17, 163 CrossRef CAS PubMed.
- M. El-Kemary, I. El-mehasseb, K. R. Shoueir, S. E. El-Shafey, El-O. I. Shafey, H. A. Aljohani and R. R. Fouad, J. Dispersion Sci. Technol., 2018, 39, 911–921 CrossRef CAS.
- S. Rajeshkumar, C. Malarkodi, K. Paulkumar, M. Vanaja, G. Gnanajobitha and G. Annadurai, Nanosci. Nanotechnol., 2013, 3, 21–25 Search PubMed.
- J. A. Dahl, B. L. S. Maddux and J. E. Hutchison, Chem. Rev., 2007, 107, 2228 CrossRef CAS PubMed.
- J. E. Hutchison, ACS Nano, 2008, 2, 395 CrossRef CAS PubMed.
- K. Shoueir, S. Kandil, H. El-hosainy and M. El-Kemary, J. Cleaner Prod., 2019, 230, 383 CrossRef CAS.
- J. Siepmann and A. T. Florence, Pharmaceutics: New Challenges, New Paradigms, in Modern Pharmaceutics, CRC Press, 2009, vol. 1, pp. 18–38 Search PubMed.
- N. Lubick, Environmental Science Technology, On Line News, February 20, 2008 Search PubMed.
- B. Bonifácio, P. da Silva, M. Ramos, K. Negri, T. Maria Bauab and M. Chorilli, Int. J. Nanomed., 2014, 9, 1–5 CrossRef PubMed.
- M. Kale, P. Suruse, R. Singh, G. Malhotra and P. Raut, Int. J. Biol. Pharm. Res., 2012, 3, 308–316 Search PubMed.
- A. Marin, H. Sun, G. A. Husseini, W. G. Pitt, D. A. Christensena and N. Y. Rapoport, J. Controlled Release, 2002, 84, 39–47 CrossRef CAS PubMed.
- C. Sinico, M. Manconi, M. Peppi, F. Lai, D. Valenti and A. M. Fadda, J. Controlled Release, 2005, 103, 123–136 CrossRef CAS PubMed.
- F. Taghavi, M. Gholizadeh and A. S. Saljooghi, New J. Chem., 2016, 40, 2696 RSC.
- N. Kohler, C. Sun, J. Wang and M. Zhang, Langmuir, 2005, 21, 8858–8864 CrossRef CAS PubMed.
- A. Kumari, R. Singla, A. Guliani and S. K. Yadav, Exp. Clin. Sci. J., 2014, 13, 265–286 Search PubMed.
- Y. P. Yew, K. Shameli, M. Miyake, N. B. B. A. Khairudin, S. E. B. Mohamad, T. Naiki and K. X. Lee, Arabian J. Chem., 2018 DOI:10.1016/j.arabjc.2018.04.013.
- A. Kamalzadeh, M. Rajabbaigy and A. Kiasat, J. Agric. Soc. Sci., 2008, 4, 183–188 Search PubMed.
- P. Singh, Int. J. Livest. Res., 2016, 6, 1–14 CrossRef.
- D. Rajendran, Res. J. Biotechnol., 2013, 8, 1–3 CAS.
- Z. P. Yang and L. P. Sun, J. Shanxi Agric. Sci., 2006, 3, 024 Search PubMed.
- P. S. Swain, D. Rajendran, S. B. N. Rao and G. Dominic, Vet. World, 2015, 8, 888 CrossRef CAS PubMed.
- M. Fondevilain, in Silver Nanoparticles, ed. D. P. Peres, Rijeka, Croatia: InTech, 2010, pp. 325–334 Search PubMed.
- A. Schröfel, G. Kratošová, I. Šafařík, M. Šafaříková, I. Raška and L. M. Shor, Acta Biomater., 2014, 10, 4023–4042 CrossRef PubMed.
- R. G. Saratale, I. Karuppusamy, G. D. Saratale, A. Pugazhendhi, G. Kumar, Y. Park and H. S. Shin, Colloids Surf., B, 2018, 170, 20 CrossRef CAS PubMed.
- T. M. Abdelghany, A. M. H. Al-Rajhi, M. A. Al Abboud, M. M. Alawlaqi, A. Ganash Magdah, E. A. M. Helmy and A. S. Mabrouk, BioNanoScience, 2018, 8, 5–16 CrossRef.
- J. I. Hare, T. Lammers, M. B. Ashford, S. Puri, G. Storm and S. T. Barry, Adv. Drug Delivery Rev., 2017, 108, 25–38 CrossRef CAS PubMed.
- J. L. Gardea-Torresdey, J. Parsons, E. Gomez, J. Peralta- Videa, H. Troiani, P. Santiago and M. Yacaman, Nano Lett., 2002, 2, 397–401 CrossRef CAS.
- J.-F. Manen, O. Sinitsyna, L. Aeschbach, A. V. Markov and A. Sinitsyn, BMC Plant Biol., 2005, 5, 23 CrossRef PubMed.
- O. Velgosová and A. Mražíková, AIP Conf. Proc., 2017, 1918, 020004 CrossRef.
|
| This journal is © The Royal Society of Chemistry 2019 |
Click here to see how this site uses Cookies. View our privacy policy here.  Open Access Article
Open Access Article *abcd,
Rehan M. El-Shabasyde,
Shaden A. M. Khalifafg,
Aamer Saeedh,
Afzal Shah
*abcd,
Rehan M. El-Shabasyde,
Shaden A. M. Khalifafg,
Aamer Saeedh,
Afzal Shah i,
Raza Shahj,
Faiza Jan Iftikharh,
Mohamed M. Abdel-Daim
i,
Raza Shahj,
Faiza Jan Iftikharh,
Mohamed M. Abdel-Daim k,
Abdelfatteh Omrilm,
Nahid H. Hajrahandlm,
Jamal S. M. Sabirlm,
Xiaobo Zou
k,
Abdelfatteh Omrilm,
Nahid H. Hajrahandlm,
Jamal S. M. Sabirlm,
Xiaobo Zou b,
Mohammed F. Halabic,
Wessam Sarhann and
Weisheng Guo*o
b,
Mohammed F. Halabic,
Wessam Sarhann and
Weisheng Guo*o
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 μg Ag per gram.23 Silver-containing products were washed in 500 mL of tap water to assess the potential release of silver into water-based natural networks such as surface water, saliva, and wastewater. Ag was discharged at levels of up to 45 μg Ag per gram, in particles of varying sizes including some with diameters above 100 nm and some with diameters below this level.22
000 μg Ag per gram.23 Silver-containing products were washed in 500 mL of tap water to assess the potential release of silver into water-based natural networks such as surface water, saliva, and wastewater. Ag was discharged at levels of up to 45 μg Ag per gram, in particles of varying sizes including some with diameters above 100 nm and some with diameters below this level.22
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O groups on the oxidized form of polyphenol electrostatically stabilize the metal NPs. Hence, the mechanism for the biosynthesis of nM0 as zero-valent metal atoms by plant extract polyphenolics APOH when reacting with a metal halogen precursor is as follows:
O groups on the oxidized form of polyphenol electrostatically stabilize the metal NPs. Hence, the mechanism for the biosynthesis of nM0 as zero-valent metal atoms by plant extract polyphenolics APOH when reacting with a metal halogen precursor is as follows:![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O and C
O and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N functional groups, suggesting that the amide and polypeptide groups served as capping agents, which stabilized the nascent metallic NPs from aggregation.53 A methanolic leaf extract of Vitex negundo was also used in the bio-synthesis of Ag-NPs under ambient conditions with no additives, resulting in a template that shaped NPs or accelerants that spontaneously protected the NPs from aggregation.54 El-Kemary et al. investigated the bio-synthesis of AgNPs using a leaf extract of Ambrosia maritime, and found that various secondary metabolites (notably sesquiterpene lactones and flavonoids) strongly influenced the NP yield.55 In addition, Coleus aromaticus leaf extract was found to be a suitable reductant for the rapid synthesis of AgNPs from silver salts. Rosmarinic acid was the major polyphenol in the extract.56 Our recent paper reported the successful fabrication of Ag2ONPs from chili as well due to the presence of alkaloid compounds.57 As noted above, the focus of much work has been on Cu and Ag NPs. Taken together, it has been reported that the synthesis of NPs using plant extracts proceeds more quickly than alternative processes using microbial cultures, and also yields more stable products with a greater range of shapes and sizes.24,40 A study on metal bioaccumulation in plants showed that the metals are usually deposited in the form of NPs.24
N functional groups, suggesting that the amide and polypeptide groups served as capping agents, which stabilized the nascent metallic NPs from aggregation.53 A methanolic leaf extract of Vitex negundo was also used in the bio-synthesis of Ag-NPs under ambient conditions with no additives, resulting in a template that shaped NPs or accelerants that spontaneously protected the NPs from aggregation.54 El-Kemary et al. investigated the bio-synthesis of AgNPs using a leaf extract of Ambrosia maritime, and found that various secondary metabolites (notably sesquiterpene lactones and flavonoids) strongly influenced the NP yield.55 In addition, Coleus aromaticus leaf extract was found to be a suitable reductant for the rapid synthesis of AgNPs from silver salts. Rosmarinic acid was the major polyphenol in the extract.56 Our recent paper reported the successful fabrication of Ag2ONPs from chili as well due to the presence of alkaloid compounds.57 As noted above, the focus of much work has been on Cu and Ag NPs. Taken together, it has been reported that the synthesis of NPs using plant extracts proceeds more quickly than alternative processes using microbial cultures, and also yields more stable products with a greater range of shapes and sizes.24,40 A study on metal bioaccumulation in plants showed that the metals are usually deposited in the form of NPs.24

![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif)
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif)

![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching vibrations of the amide bonds in proteins, the C–OH stretch of a cyclic alcohol, and aromatic C–H vibrations (suggesting the presence of free quinones derived from polyphenolic compounds), respectively.142 However, only a few publications have discussed the role of alkaloids in the formation of metallic NPs, and thus further investigations in this area are needed.
O stretching vibrations of the amide bonds in proteins, the C–OH stretch of a cyclic alcohol, and aromatic C–H vibrations (suggesting the presence of free quinones derived from polyphenolic compounds), respectively.142 However, only a few publications have discussed the role of alkaloids in the formation of metallic NPs, and thus further investigations in this area are needed.













