Open Access Article
Open Access ArticleRecent progress in the imaging detection of enzyme activities in vivo
Chunjie Yang
ab,
Qian Wang
b and
Wu Ding
 *b
*b
aCollege of Health Science, Yuncheng Polytechnic College, Yuncheng, Shanxi 044000, PR China
bCollege of Food Science and Engineering, Northwest A&F University, Yangling, Shaanxi 712100, PR China. E-mail: dingwu10142000@hotmail.com
First published on 13th August 2019
Abstract
Enzymatic activities are important for normal physiological processes and are also critical regulatory mechanisms for many pathologies. Identifying the enzyme activities in vivo has considerable importance in disease diagnoses and monitoring of the physiological metabolism. In the past few years, great strides have been made towards the imaging detection of enzyme activity in vivo based on optical modality, MRI modality, nuclear modality, photoacoustic modality and multifunctional modality. This review summarizes the latest advances in the imaging detection of enzyme activities in vivo reported within the past years, mainly concentrating on the probe design, imaging strategies and demonstration of enzyme activities in vivo. This review also highlights the potential challenges and the further directions of this field.
1 Introduction
Enzymes play an important role in many physiological and biological processes, and can serve as critical biomarkers for the pathology of many diseases as abnormal enzyme activities are closely related to the pathology of many diseases.1–6 Thus, a large number of different methods of measuring the enzyme activity for a range of enzyme families have been developed over the past several decades. The traditional detection methods for performing the enzyme activity analysis mainly include colorimetric assays,7–10 electrochemical assays11–13 and luminescence assays.14–17 Although these technologies for determining enzymatic activities have great value, most researchers are not capable of measuring specific enzyme activity in biological systems in real time, and a non-destructive testing of the activity in vivo is still a challenging technical problem.18 Typically, these technologies are faced with two challenges: avoiding artificial aspects that affect the enzyme activity within the context of the natural biological environment in living subjects and accomplishing the rapid, sensitive and high-resolution detection in vivo, particularly, in relatively deep tissues. Recent developments in optical imaging, magnetic resonance imaging (MRI) and nuclear imaging have made it possible to visualize enzyme activities in deep tissues.19,20 Several pivotal reviews have discussed the advances in enzyme activity detection, with most reviews focusing on specific subcategories, such as small-molecule substrate reporters and protein-based reporters,21 labelled and unmodified substrates,22 gold nanoparticle-based biosensors,23 activatable probes capable of imaging enzyme activities,20 fluorescent probes24 and methods of measuring enzyme activities in vivo.25 We refer the reader to an earlier review for a more comprehensive coverage of the field.19 This critical review examined the approaches in the earlier literature towards the in vivo imaging of the activity, with an emphasis on the chemical perspective of the probe design, structure and function. Our review is not intended to be a comprehensive discussion of all aspects of enzyme activity detection in vivo, but instead, we aim to give a brief summary of the recent advances in this topic. This review summarizes recent advances in enzyme activity detection that have actually been used in vivo (i.e. that have actually been used in living mice subjects), including optical imaging, MRI, nuclear imaging and photoacoustic imaging-based methods. A systematic discussion is provided on the probe design, imaging strategies and imaging demonstrations of enzyme activities in vivo. The potential challenges and the further directions of this field have been discussed as well.2 Optical modality
Optical modality is a method that detects the photons emitted from bioluminescent or fluorescent contrast agents to generate a two- or three-dimensional image.19 The optical modality techniques used for enzyme activity detection in vivo mainly include fluorescence and bioluminescence imaging.2.1 Fluorescence imaging
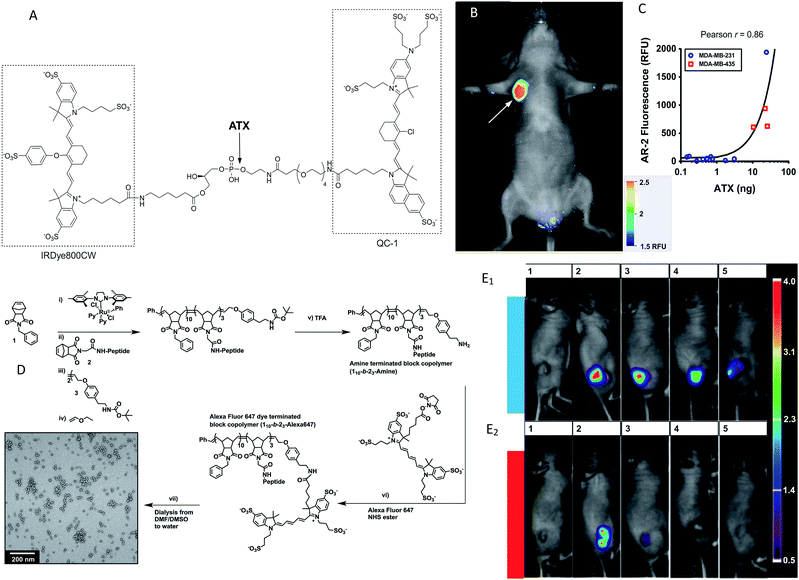
Although a few formal studies have shown that in vivo enzyme activities can be evaluated via visible fluorescence imaging in mouse models,26 one of the key limitations, particularly for larger animals (i.e. humans), is the limited penetration depth. Thick animal tissue absorbs and scatters photons and generates autofluorescence, all of which obscure an accurate collection and quantification of the signal.27 The penetration depth of macroscopic fluorescence imaging is limited to only 1–2 mm.28 Even the high-resolution visible-light optical imaging fails when the subcutaneous tissue depth is greater than 500 microns. As a result, in vivo optical imaging has traditionally been limited to endoscopically accessible areas.29 However, several strategies can overcome these limitations, such as varying the light's wavelength and applying new sophisticated optical approaches (e.g., optical coherence tomography).30 One of the major reasons that this technology has become feasible is the development of fluorescence probes emitting in the near-infrared (NIR) spectrum where tissues exhibit low absorption.31 It has been predicted that NIR fluorescent light can penetrate several centimetres,30,32,33 and possibly even more than ten centimetres.21 In addition, autofluorescence and light scattering are decreased in the NIR spectrum.34 In general, NIR imaging probes include two groups: small-molecule probes and nanoparticle (NP)-based probes. A number of approaches have been utilized to improve the activation or targeting of the NIR probes. Often these schemes are based on a quenching–dequenching effect, where in the native state, the probe is quenched and then following a specific molecular structure change with their intended enzyme, it becomes highly fluorescent. In the past years, these techniques have been extensively validated and used in various studies on enzyme activity detection in vivo (Table 1). Pre-quenched probes are widely used for imaging enzymes, and they are made up of three components: a fluorophore, a quencher dye and an enzyme-cleavable linker. The initial fluorescence of the fluorophore is quenched due to self-quenching, Förster resonance energy transfer (FRET) or non-radiative energy transfer. When the linker is cleaved by a target enzyme, the quencher is released to limit the quenching effect, resulting in fluorescence upon release.19,20 For an effective analysis of autotoxin (ATX) activity, Madan et al. developed and validated a fluorogenic analog of lysophosphatidylcholine (LPC), which enabled the visualization of ATX activity in vivo.56 This is a typical research for enzyme activity detection based on pre-quenched probes. The probe (AR-2) was designed as an analog of LPC, in which a quencher (IRDyeH QC-1) was linked to a mimic of the choline head group and a fluorescent dye (IRDyeH 800CW) was appended to the sn-1 acyl group (Fig. 1A). The fluorescence of AR-2 was quenched until it was activated by ATX, and this could be directly correlated with ATX activity in mice (Fig. 1B and C). In recent decades, similar works have been done on the detection of various enzyme activities, such as that of cysteine cathepsins,48 leucine aminopeptidase,52 nitroreductases50 and β-glucuronidase.60| Target analyte | Fluorescence donor | Emission wavelength (nm) | Experimental![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) subject subject |
Reference |
|---|---|---|---|---|
| Matrix metalloproteinases (MMPs) | Cy5.5 | 695 | MDA-MB-435 tumor-bearing mice | 35 |
| Alexa750 | 830 | 4T1luc tumors mice | 36 | |
| Alexa Fluor 647 | 670 | Tumor-bearing mice | 37 | |
| Alexa750 | 770 | Adult male BALB/cByJ mice | 38 | |
| Alexa680 | 700 | C57BL/6 mice | 39 | |
| Alexa680 | 700 | C57BL/6 mice | 40 | |
| Alexa680 | 700 | C57BL/6 mice | 41 | |
| Alexa680 | 700 | Tumor-bearing mice | 42 | |
| Cy5.5 | 692 | C57BL/6 mice | 43 | |
| Alexa680 | 700 | DBA1/J mice, C57Bl6/J mice | 44 | |
| II transmembrane ecto-enzyme (CD38) | Alexa680 | 702 | Mice | 45 |
| Renin | VivoTag-S680 | 690 | C57BL/6 mice | 46 |
| Cysteine cathepsin | Cy7 | 767 | Male CD-1 mice | 47 |
| Cy5 | 700 | C57BL/6J mice | 48 | |
| Cy5.5 | 720 | Tumor-bearing mice | 49 | |
| Nitroreductase | HXPI | 705 | Zebrafish | 50 |
| CytoCy5S | 660 | Mice | 51 | |
| Leucine aminopeptidase | Hemicyanine (HC) | 705 | Mice | 52 |
| β-Galactosidase | DCDHF | 615/665 | Mice | 53 |
| Dicyanomethylene-4H-pyran (DCM) | 500/685 | Mice | 54 | |
| Thrombin | Cy5 | 700 | Mice | 55 |
| Autotaxin (ATX) | IRDyeH800CW | 800 | Tumor-bearing mice | 56 |
| Protease | Alexa680 | 700 | C57BL/6 mice | 39 |
| Alexa750 | 800 | H292 xenograft mouse | 57 | |
| Cathepsin | Alexa680 | 693 | C57BL/6 mice | 39 |
| Alexa680 | 700 | DBA1/J mice | 44 | |
| Cy5.5 | 670–900 | Tumor-bearing mice | 58 | |
| Cy5 | 670 | Tumor-bearing mice | 59 | |
| β-Glucuronidase | Hemicyanine (HC) | 718 | Mice, zebrafish | 60 |
| IR-820 | 835 | Tumor-bearing mice | 61 | |
| 4-Hydroxy-1,8-naphthalimide (NH) | 445/556 | Zebrafish | 62 | |
| Carboxylesterase | 1,3-Dichloro-9,9-dimethyl-9H-acridin-2(7)-one (DDAO) | 700 | Mice | 63 |
| Prolyl endopeptidases | HiLyte fluo 647 | 700 | Tumor-bearing mice | 64 |
Besides, diverse fluorescent nanoprobes have proven useful as effective enzyme activity detection tools due to such nanoprobes presenting several advantages over conventional fluorescent probes (e.g. longer monitoring ability, higher signal-to-background ratio and higher intensity).65 Chien et al. designed a conjugated peptide polymeric nanoparticle to respond to the MMP enzyme.37 The preparation of enzyme-responsive nanoparticles is shown in Fig. 1D. Most importantly, the fluorescent aggregates were retained for at least 1 week (Fig. 1E). Similar work related to a transmembrane ectoenzyme (CD38) discrimination was demonstrated by Fumey et al., having generated human CD38-specific nanobodies from immunized llamas.45 These nanobodies were highly specific for modulating the enzymatic activity of CD38.
Recently, some self-assembly-based fluorescent probes for imaging enzyme activities were also developed. Ye et al. employed an optimized cyclization reaction to control the self-assembly of a fluorescent molecule, and applied it to image caspase-3/7 activity in vivo (Fig. 2A).58 The probe offered the following advantages: (1) a more-extensive biodistribution, (2) deeper tissue penetration and (3) longer residence time in the target tissue of interest. The same self-assembly strategy may be widely used for the non-invasive imaging of enzyme activities in vivo. Moreover, a self-assembled fluorescent nanoprobe was developed for the detection and imaging of legumain.66 The probe was constructed by Cys(StBu)–Ala–Ala–Asn–Lys(Cy5.5)–CBT, which was first reduced and subjected to a condensation reaction to self-assemble with self-quenched fluorescence by an aggregation-caused quenching (ACQ). The nanoparticles disassembled and the NIR fluorescence was released when the Ala–Ala–Asn peptide substrate was recognized and digested by legumain (Fig. 2B).
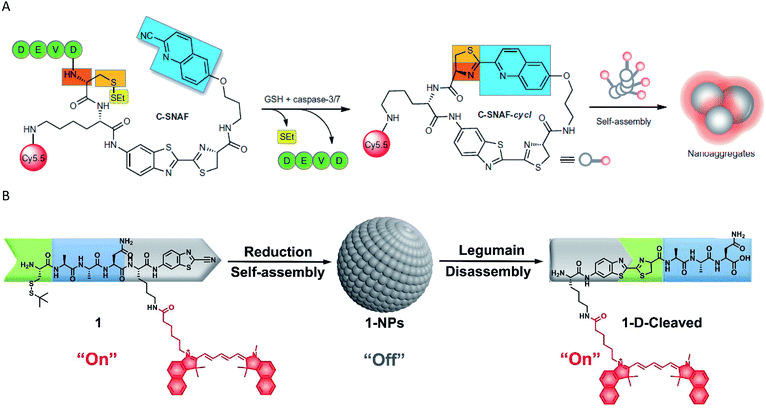 | ||
| Fig. 2 (A) Schematic representation of the mechanism of in vivo imaging by caspase-3/7 activity. Reproduced from ref. 58. Copyright 2014 Springer Nature. (B) Schematic representation of the mechanism of in vivo imaging by legumain. Reproduced from ref. 66. Copyright 2018 American Chemical Society. | ||
Although the emergence of various NIR fluorescence probes has led to significant advances in the field of enzyme activity detection in vivo, absolute intensity-dependent NIR fluorescence probes can fail to accurately quantify the enzyme activity due to the fact that the probes may be influenced by target concentration-independent experimental or physiological factors (such as cross-talk between the emission and excitation spectra, weakening the ability to capture valid signals).67 In response to this challenge, some strategies have been designed and proposed to mitigate the effects of a nonspecific background. Among these methods, one simple way to minimize nonspecific effects is to utilize a ratiometric fluorescence strategy. A detectable ratio signal was obtained from two independent signal fluctuations, avoiding the influence of various confounding factors; therefore, ratiometric fluorescence probes can facilitate more accurate and reliable quantitation. For example, a ratiometric fluorescent probe NH-Glu was developed for the selective sensing of β-glucuronidase (GLU) by Huo and colleagues.68 The probe was successfully used for the real-time visualization of GLU in zebrafish without the interference of the biological matrix. However, the probe is not very suitable for in vivo experiments due to the emission wavelength being in the visible light region. Gu and co-workers developed an enzyme-activatable ratiometric NIR probe (DCM-βgal) for real-time fluorescent β-gal activity quantification.54 The probe was designed by grafting a β-gal activatable unit onto a DCM-OH moiety. An obvious ratiometric fluorescent signal (I685nm/I500nm) was observed when excited at the isosbestic point of 450 nm (Fig. 3A). The probes share many advantages such as high photostability, large Stokes-shift and pH independence. More importantly, this probe was successfully applied for the in vivo imaging of mice bearing subcutaneously implanted tumors. In addition, the in vivo real-time capture of β-gal activity with a high-resolution 3D view was performed (Fig. 3B). Subsequently, a similar interesting work was presented by Kim's group, who developed a ratiometric fluorescent probe (DCDHF-βgal) for the in vivo visualization of β-gal.53 In this study, β-gal induced an obvious red-shift from 615 to 665 nm in the emission peaks with maximum intensity of the DCDHF-βgal (Fig. 3C). In vivo imaging experiments for ratiometric detection were conducted with an intravenous injection of DCDHF-βgal into mice bearing HepG2 xenograft tumors. The results shown in Fig. 3D suggest that the tumor could be clearly visualized by the in vivo fluorescence imaging with an emission at 600–700 nm. All these findings indicated that the ratiometric NIR fluorescent probes could be a promising imaging tool for the quantification of enzyme activity in vivo.
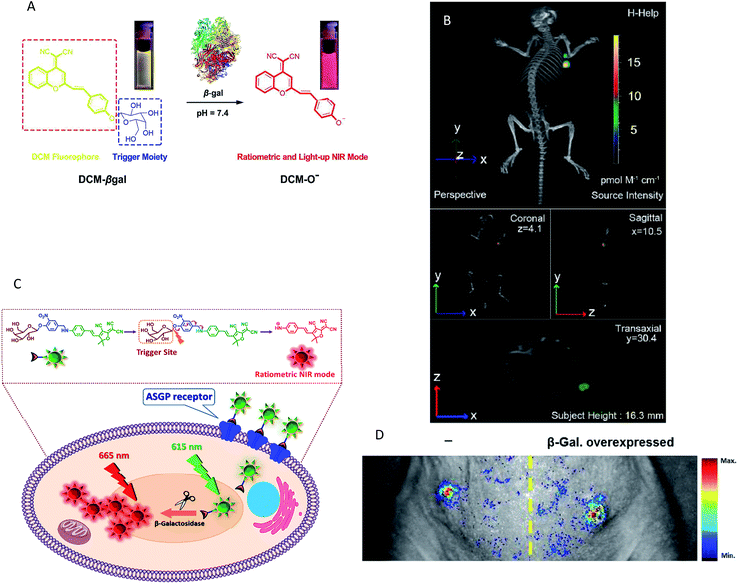 | ||
| Fig. 3 (A) Conceptual scheme of β-Gal enzymatic activation of DCM-βgal. (B) Three-dimensional in vivo imaging of DCM-βgal. (C) Conceptual scheme of DCDHF-βgal when encountering β-Gal and the following fluorescence change. (D) In vivo fluorescence imaging of DCDHF-βgal. (A and B) Reproduced from ref. 54. Copyright 2016 American Chemical Society. (C and D) Reproduced from ref. 53. Copyright 2017 Elsevier. | ||
However, in most studies, the penetrability of the NIR fluorescent probes was insufficient to detect the enzyme activity in organs deep within the body and imaging can only be done of the subcutaneous tumor of the mouse model. In recent years, several well-designed NIR fluorescent probes have been designed to monitor enzyme activities in deeper tissues, such as the GI tract,64 intestinal tract60,63 and liver.52,61,63 The recognition mechanism and deep tissue images of target enzymes using the special probes are shown in Fig. 4. It is noteworthy that all the probes were activatable and were designed to have fluorescence emission in the near-infrared (NIR) range to guarantee a high tissue-penetration capacity among these five studies (Fig. 4a–e). For example, Jin and co-workers designed an activatable NIR fluorescent probe (HC-glu) for the fluorescent evaluation of endogenous β-glu activity in the intestinal tracts of mice.60 The probe was designed by grafting a D-glucuronic acid residue onto a hemicyanine skeleton via a glycosidic bond (Fig. 4b). The initial maximum intensity of the absorption peak was approximately 670 nm. When HC was released by GLU from the reaction system, a remarkable fluorescence increase was observed at 718 nm (approximately a 40-fold increase). Moreover, a number of commercial NIR fluorescent probes have been developed for enzyme imaging in vivo.38–41,44 In addition, new generations of NIR detectors (fluorescence molecular tomography, FMT) allow the tomographic imaging of enzyme activities with high positional accuracy.31
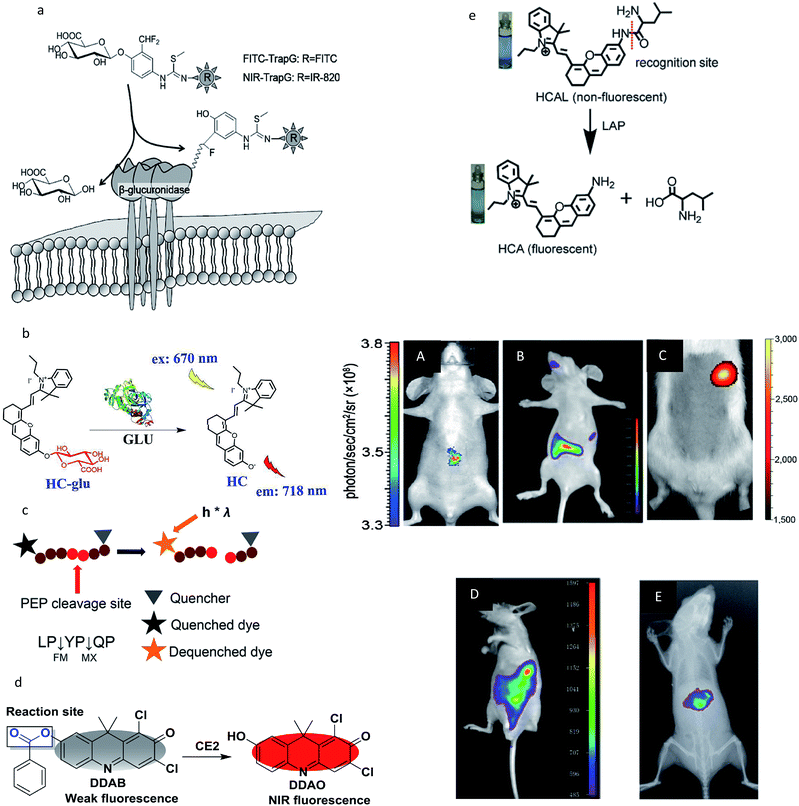 | ||
| Fig. 4 (a–e) Recognition mechanism of the probes. (A–E) Real-time activity measurement in deep tissues of a mouse. (a and A) Reproduced from ref. 61. Copyright 2012 American Chemical Society. (b and B) Reproduced from ref. 60. Copyright 2018 American Chemical Society. (c and C) Reproduced from ref. 64. Copyright 2011 National Academy of Sciences. (d and D) Reproduced from ref. 63. Copyright 2016 Elsevier. (e and E) Reproduced from ref. 52. Copyright 2017 Royal Society of Chemistry. | ||
However, the sensitive and selective detection of enzyme activities in deeper tissues by NIR fluorescent probes remains a formidable challenge because of the poor penetration and high scattering. To the best of our knowledge, there have been no successful studies on the non-invasive NIR fluorescent imaging of enzyme activities in deeper tissues. Fluorescence imaging in the NIR-II region (wavelength 1000–1700 nm) may be more desirable over traditional NIR-I imaging owing to its deeper tissue penetration, reduced photon scattering and lower autofluorescence even though it has some problems, such as poor biocompatibility and low fluorescence quantum yields, which limit its application for in vivo imaging with enough resolution.69–71 Over the years, several classes of fluorescent NIR-II probes, such as carbon nanotubes, small molecules, Ag2S dots and polymers, have been developed for NIR-II fluorescence imaging for diversified biomedical applications.72–75 In general, the use of NIR fluorescent probes for improving the effectiveness of enzyme activity imaging in deeper tissues is only in its infancy, and future years should see an increase in the use of these types of fluorescent probes with newer optical approaches, such as optical coherence tomography (OCT) and diffuse optical tomography.
2.2 Bioluminescence imaging
Bioluminescence generally refers to the emission of light that is generated by the enzymatic reaction of luciferase enzymes and its substrates from a living system.76 Bioluminescence imaging (BLI) has been broadly used as a powerful platform for in vivo animal imaging due to its favorable spectral properties and the lack of toxicity of its substrate.77 Bioluminescent light can usually be detected at a depth of few centimetres underneath the epidermis, although the exact limit is dependent on the sensitivity of the detector and the intensity of the light.78 The most widely used BLI method is based on the firefly luciferase (FLuc) and its substrate D-luciferin from Photinus pyralis, owing to its maximum emission being in the red and near-infrared wavelengths, making FLuc suitable for in vivo imaging. In general, bioluminogenic probes are designed as the “caged luciferin”, in which the specific position of luciferin (or aminoluciferin) is caged by some specific functional groups. These caged luciferins do not emit bioluminescence until the luciferins (or aminoluciferins) are generated or released via reacting with target enzymes and then oxidized by luciferase.79 On the basis of this strategy, highly responsive bioluminescent probes as these have been proven to be relatively cheap, animal friendly and to offer much more efficient penetrating power than fluorescence,80 and hence their use has been well established for many enzymes, such as monoamine oxidase,81 caspase,82,83 glutathione S-transferase,81,84 sulfatase,85 protein kinase,86 β-galactosidase,77,87 β-lactamase,88 CYP450,89 aminopeptidase N90 and nitroreductase.91–93 A typical example was given by Li and co-workers, who designed two bioluminogenic probes to detect the aminopeptidase N (APN) activity both in vitro and in vivo via grafting D-aminoluciferin onto the N-terminus of the APN recognition amino acid(s).90 In the presence of APN, the probes released free D-aminoluciferin due to the caging groups being removed by the enzymolysis reaction. The released D-aminoluciferin produced a photon via reaction with the firefly luciferase (Fig. 5A). Then, the two probes were used for the in vivo imaging of APN activity in nude mice. Further imaging analysis in Fig. 5B illustrated that the probes had the capacity to image APN activity in vivo. Subsequently, through similar strategies, several research groups have developed bioluminescent probes for the chemoselective visualization of nitroreductase (NTR)92 and tyrosinase94 in a tumor model. An excellent work was recently presented by Godinat's group, who demonstrated the in vivo applicability of this biocompatible reaction, a new method to visualize and quantify the activity of caspase 3/7 protease.95 This reaction was between D-cysteine and 6-hydroxy-2-cyanobenzothiazole (OHCBT). First, free D-cysteine was released from the caspase 3/7 peptide substrate (DEVD-(D-Cys)). Then, 6-amino-D-luciferin was formed with the progress of D-cysteine's reaction with 6-amino-2-cyanobenzothiazole (NH2-CBT). Finally, there was an emission of light that was generated by the enzymatic reaction of 6-amino-D-luciferin and luciferase (Fig. 5C). Importantly, the ratio between the signals produced from the d-GalN/lipopolysaccharide (LPS)-treated group and the control group was statistically significantly higher than that of the commercial substrate (DEVD-aminoluciferin) (Fig. 5D). All these findings showed that this strategy was superior to the commercially available substrate for the imaging of caspase 3/7 activity. Moreover, the split luciferin approach enables signal stability and a modular construction of bioluminescent probes. A further improvement in this approach was carried out by Bittner's group, who presented a general strategy for dual-analyte (H2O2 and caspase 8) detection in animals via utilizing the in situ formation of firefly luciferin from two complementary caged precursors.82 This strategy constituted an AND-type molecular logic gate. To establish this approach, the probe Peroxy Caged Luciferin-2 (PCL-2) was designed to generate 6-hydroxy-2-cyanobenzothiazole (HCBT) by reacting with H2O2. The probe z-Ile-Glu-ThrAsp-D-Cys (IETDC) was designed to release D-cysteine catalyzed by caspase 8. If and only if both chemicals were released, a bioluminescent signal was generated.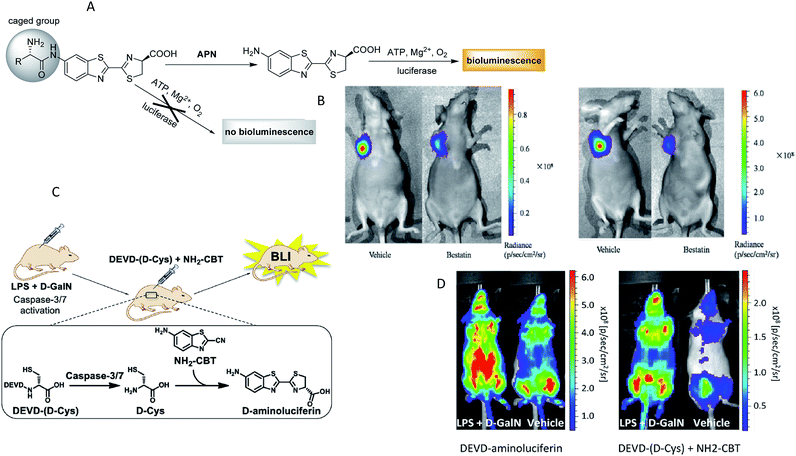 | ||
| Fig. 5 (A) Schematic of bioluminogenic probes for detecting APN activity. (B) Bioluminescence imaging of APN activity in the implanted ES-2-luc cells for probe 1(left), probe 2(right). (C) Schematic of caspase-3/7 activity imaging with DEVD-(D-Cys) peptide and NH2-CBT. (D) Representative image of mice treated with LPS and D-GalN or vehicle, injection of DEVD-aminoluciferin or a combination of DEVD-(D-Cys) and NH2-CBT reagents. (A and B): Reproduced from ref. 90. Copyright 2014 American Chemical Society. (C and D) Reproduced from ref. 95. Copyright 2013 American Chemical Society. | ||
Although great progress has been made over the past decade in the detection of enzyme activities in vivo by bioluminescence imaging, there are still many challenges ahead. First, though the enzyme–substrate specificity is crucial, many luciferases are sensitive to their environment, such as pH and reactive oxygen species, which can result in an increase in the number of false positives. Second, although many types of bioluminescence probes have been developed to target enzyme activity, the in vivo application of these probes is still unsatisfactory; for instance, the emitted light of all luciferases is inherently weak, and only a fraction of the light typically reaches the detector due to the majority of bioluminescent photons being scattered or absorbed by endogenous chromophores in biological tissues.78,96 Although these have been largely solved by engineering luciferases to produce near-infrared (NIR) bioluminescent photons, the improvement is often limited because the light intensity reduces with a red-shift in the peak emission.97 Finally, since the caged luciferins reaction require two co-existing enzymes (luciferase and target enzyme), caged luciferins are normally dedicated to transgenic animals for expressing luciferase. Owing to the requirement of genetic modification, this is limited in clinical applications. However, with the continued development of new luciferases and luciferins, these studies will enhance our ability to study enzyme activities in living organisms.
3 MRI modality
Magnetic resonance imaging (MRI) is a noninvasive imaging method that provides physiological and pathological information of living tissues.98 Although MRI is capable of high spatial resolution and unlimited tissue penetration, the technology generally requires exogenous contrast agents because of its low sensitivity.99,100 MR contrast agents can be separated into four categories: T1 agents, T2 agents, hyperpolarization agents and chemical exchange saturation transfer (CEST) agents.30,99Some effective strategies have been reported to design MR probes for enzyme activity analysis, such as oligomerization of the probe, the disassembly of nanoparticles, modulation of the hydration number, the switching of solubility, relaxivity enhancement via macromolecular binding and the unmasking of CEST-active protons.19 To date, 1H MRI has been broadly used in this area due to its high sensitivity. As a recent example for T1/T2 agents, several 1H MR probes have been designed for β-galactosidase activity imaging by Li's group.101 They designed a series of β-D-galactosides conjugated with various chelators for assessing β-gal activity in solution by 1H-MRI T1 and T2 relaxation mapping. However, 1H-MRI often has the disadvantage of a low signal-to-noise ratio because of large background signals,102 such that most of this type of contrast agents cannot be used for in vivo imaging.
For target-specific detection, heteronuclear MRI techniques have attracted more and more attention for alternative methodologies to the 1H-based MR imaging of enzymes. 19F MRI is considered a promising method mainly because 19F has higher selectivity than 1H MRI,103 and there is no interference from the background signals in vivo because fluorine is essentially absent in mammalian tissues. However, unlike the mechanism of 1H-MRI, 19F MRI usually offers ambiguous localization and poor sensitivity104,105 and few 19F MRI probes can be applied to detect enzymatic activity in vivo. Several promising enzyme in vivo sensing 19F MRI probes have been reported in the past few years.106–108 A representative study was reported by Yuan et al., who demonstrated a dual-functional probe for the specific detection of legumain (Lgmn) based on the self-assembly and disassembly of a probe conferring 19F MR signals “off” and “on”, respectively.103 The 19F nanoparticles were constructed by the self-assembly of a 19F probe (1-NPs) upon intracellular glutathione (GSH) reduction. When the nanoparticles were disassembled by Lgmn, the 19F MRI signal was output for Lgmn detection (Fig. 6A). In addition, a control compound probe (2-NPs), whose 19F MRI signal could be turned “off” in cells, was designed and studied. As shown in Fig. 6B, strong 19F MRI signals were detected from (or near) the tumors in zebrafish while healthy zebrafish did not show detectable 19F MRI signal when given an injection of the same dose of 1-NPs. In addition, 19F MRI signals could be detected from the nontumor sites of the tumor-bearing zebrafish and the healthy zebrafish after the injection of large-dose 2-NPs. These results illustrated that the 19F MRI probe 1-NPs could successfully be used to detect Lgmn activities in zebrafish at low doses. Most recently, Akazawa's group reported a similar nanoprobe FLAME-DEVD2 for detecting caspase-3/7 activity in mice spleen.107 In this study, a tandemly repeated substrate peptide sequence was incorporated into a 19F MRI signal activation strategy to improve the cleavage efficiency of peptides (Fig. 6C). After the injection of FLAME-DEVD 2 and an apoptosis-inducing reagent, the 19F MRI signals in the spleen were clearly enhanced, and the S/N ratios were 6.2-fold (Case 1) and 4.2-fold (Case 2) higher than before the clodronate liposome (CL) injection (Fig. 6D). These results demonstrate that the nanoprobe FLAME-DEVD2 could successfully be used for activity imaging of caspase-3/7 in the spleen of a living mouse.
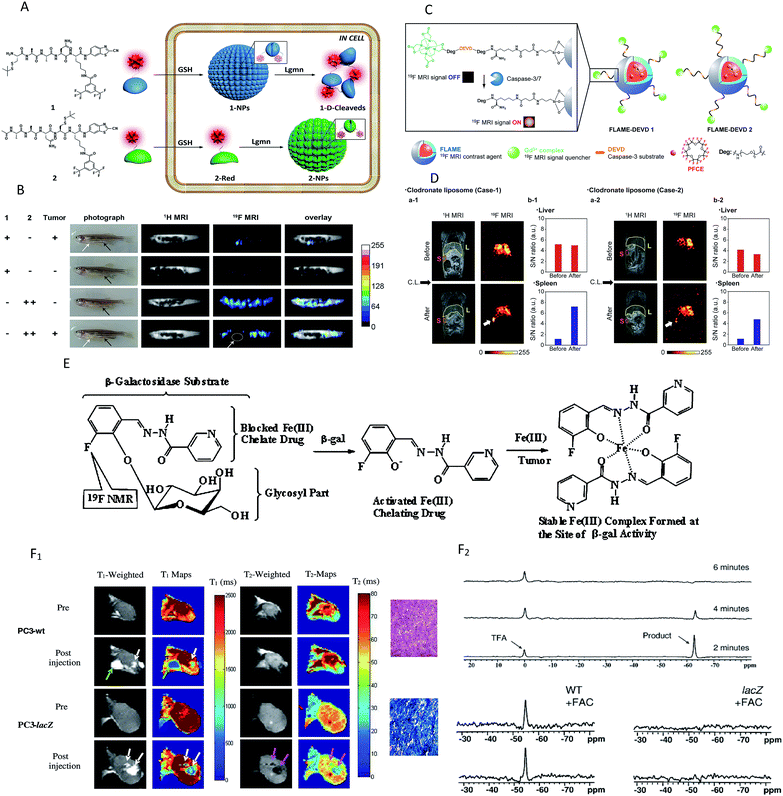 | ||
| Fig. 6 (A) Schematic illustration of the self-assembly followed by disassembly of 1-NPs, and the self-assembly of 2-NPs. (B) Representative image of tumour-bearing or healthy zebrafish after being injected with the probes. White arrows indicate the tumour sites, and black arrows indicate the injection sites. White circle indicates the tumour location. (C) Schematic of FLAME-DEVD X (X = 1, 2), enzyme-responsive 19F MRI nanoprobes for detecting caspase-3/7 activity. (D) 19F MRI of caspase-3/7 activity in a mouse. FLAME-DEVD was intravenously injected into a mouse (before). Then, clodronate liposome was intravenously injected (after). L: liver. S: spleen. (E) Schematic of MRI contrast for detecting β-gal activity. (F) F1: imaging β-gal activity in vivo by 1H MRI. Green arrow indicates anomalous injection outside the tumour. White arrows indicate injection inside th tumour. Pink arrows indicate the change in T-weighted images and T2 values was obvious in the lacZ-transfected tumours post-injection of the agent and FAC. F2: Detection of β-gal activity in vivo by 19F NMR. NaTFA served as a chemical shift reference at 0 ppm and remained quite constant. (A and B) Reproduced from ref. 103. Copyright 2015 American Chemical Society. (C and D) Reproduced from ref. 107. Copyright 2018 American Chemical Society. (E and F) Reproduced from ref. 109. Copyright 2012 American Chemical Society. | ||
The 19F and 1H MRI modalities both have their own advantages and disadvantages. Hence, several dual-function probes for a synergistic combination of 1H and 19F MRI to overcome the restraints of both imaging techniques have been designed to monitor the β-galactosidase activity, in which 19F-MRI recognizes substrate accumulation and conversion, whereas 1H-MRI reveals its location and magnitude.102,105,109 A typical example was found in the study by Yu and co-workers, who reported a dual-function MRI probe to detect the β-galactosidase activity in human tumor xenografts growing in mice.109 It is worth noting that the probe was introduced as a fluorine atom into iron chelating aroylhydrazone aglycones of β-D-galactopyranosides (Fig. 6E). When activated by β-gal, the fluoroaroylhydrazones were released and then could be trapped by Fe3+ at the site of enzyme activity to form highly relaxing complexes in situ. Meanwhile, the complexes caused strong T2 relaxation, providing clear 1H MRI to reveal β-gal activity. While the agents and ferric ammonium citrate (FAC) were injected into wild-type (WT) (no β-gal expression) and PC3 tumor (β-gal expression) xenografts growing in mice, T1 and T2 decreased significantly in lacZ tumors, but not in WT tumors (Fig. 6F1). Besides, no product or substrate 19F NMR signal was detected in the spectrum of lacZ tumors, but the agent was decreased slowly in the WT tumors (Fig. 6F2). In brief, the above results showed that the dual-modality approach can detect both the substrate and product and thus improves the reliability of enzyme detection.
Chemical exchange saturation transfer (CEST) MRI is a relatively new approach that allows the monitoring of protein properties in vivo.110 In this method, the CEST agents can reduce the MRI signal from water by the exchange of protons with bulk water. The CEST spectrum is a plot of the MR signal amplitude of bulk water versus a range of saturation frequencies.111 In the past few years, CEST MRI has been applied to detect the activity of the urokinase plasminogen activator, alkaline phosphatases, sulfatase enzyme, cathepsin, carboxypeptidase, cytosine deaminase, glutamyl transferase, kallikrein and so on.112–119 These studies, mostly from Pagel's group, utilized different types of diamagnetic or paramagnetic CEST agents to assess enzyme activities. For a typical example, this group detected the in vivo enzyme activity of g-glutamyl transferase (GGT) within mouse models using CEST MRI.118 As shown in Fig. 7A, the agent was synthesized by binding a glutamyl moiety to 4-amino salicylic acid. This agent generated two independent CEST signals, a signal at 4.8 ppm based on the aryl amide moiety and a signal at 9.2 ppm based on the salicylic acid moiety. When the glutamyl moiety of the CEST agent was cleaved by the GGT enzyme, the CEST signal at 4.8 ppm disappeared, whereas the CEST signal at 9.2 ppm remained approximately constant. This result showed that the ratio of the two signals could be used to detect the GGT enzyme activity. Further experiments in vivo were performed with mouse models of OVCAR-8 and OVCAR-3 human ovarian cancer as well as muscle, which had high, low, and no GGT activity, respectively. The parametric maps of the % CEST demonstrated high activity in the OVCAR-8 tumor, low activity in the OVCAR-3 tumor, and no activity in the muscle tissue (Fig. 7B). As a consequence, CatalyCEST MRI was shown to be an effective method for detecting GGT enzyme activity within in vivo tumor mice models. In a similar way, this group also successfully detected the activity of kallikrein 6 in an in vivo tumor model.119
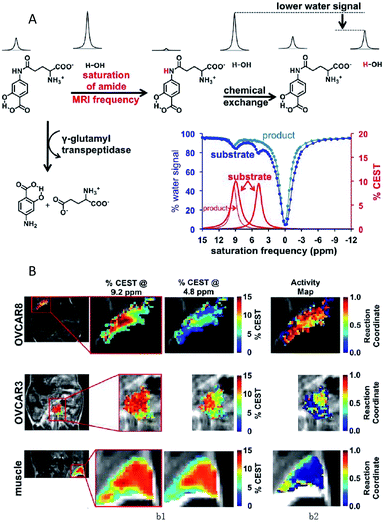 | ||
| Fig. 7 (A) The CEST-FISP MRI protocol of the agent. (B) CatalyCEST MRI of GGT activity in vivo. Reproduced from ref. 118. Copyright 2017 International Society for Magnetic Resonance in Medicine. | ||
Moreover, recent in vivo studies have demonstrated that MRI has a strong potential for the non-invasive, selective monitoring of enzymatic activity. However, only a few related studies have been performed in tumor mouse models, and there is hardly any monograph on MR imaging of enzymes activity in human body. The main drawbacks of MRI are represented by the limited sensitivity and potentially toxic effects of its probes. Thus, it is vital to design MRI contrast agents with lower toxicity and higher sensitivity. CEST and hyperpolarized probes appear to be promising potential tools in MR-based enzyme activity imaging applications. In addition, particular attention should be given to design “smart” MRI agents by combining MRI and nanotechnology.95 However, to achieve this aim, much more basic work should be done and related studies need to be performed.
4 Nuclear modality
Enzyme activity imaging based on nuclear imaging mainly includes positron emission tomography (PET) and single-photon emission computed tomography (SPECT). PET is an imaging technique that relies on the detection of positrons that are produced by the decay of radioactive isotopes.120,121 The positron has the ability to penetrate the human body, so the probe can always be imaged, regardless of the location of the target.100,122 The major advantages of PET are its high sensitivity and precision spatial quantitation capabilities. SPECT is a similar technique based on the detection of gamma ray emission from radioactive decay.123 However, PET normally is an order of magnitude more sensitive than SPECT.124 The common isotopes that can be incorporated within the radioisotope-based agents mainly include 18F, 124I, 123I, 68Ga and 111In. Over the past years, nuclear imaging has been widely developed for the detection of several enzyme activities, such as β-glu, MMP, histone deacetylases, urokinase plasminogen activator (uPA) and drug metabolizing enzymes.125–131 Thus, in this section, several representative examples were selected to provide a brief illustration. Chuang and colleagues developed a PET imaging technology for detecting the MMP activity in vivo.126 In this report, the probe was synthesized in three steps: first, a polyethylene glycol (PEG5000) molecule was linked to the N-terminus of a MMP2/9 peptide substrate, then PEG-peptide was attached to a tetramethyl rhodamine group (TMR), and finally 18F was labelled to form the probe (PEGpeptide-18F-TMR). The test mechanism was as follows: first, the hydrophobic 18F-TMR was released by MMP and then preferentially accumulated at the active sites of the protease. Finally, MMP activity in vivo was determined through the detection of radioactivity from 18F-TMR by PET (Fig. 8A). As shown in Fig. 8B, radio signals were selectively accumulated in HT1080 tumors but not in MCF-7 tumors when mice bearing HT1080 (MMP-expressing) and MCF-7 tumors (MMP-no expressing) were injected with the same dosage of PEG-peptide-18F-TMR. These results showed that PEG-peptide-18F-TMR was effective for the detection of MMP activity in vivo. Using a similar design principle, PET imaging was also developed for the assessment of β-Glu activity in vivo.128 In addition, SPECT imaging has proved to be a feasible strategy to evaluate in vivo uPA activity.129 Here, the SPECT probe MICA-401 was appropriate to selectively display uPA activity in vivo since it can selectively bind to uPA by a covalent bond (Fig. 8C). In vivo SPECT images are shown in Fig. 8D. Both the tumors were easily detectable and a moderate tumor uptake was observed for the HT-29 (uPA-expressing) and MDA-MB-231 (uPA-expressing) models. These results showed that the imaging of uPA activity in vivo by SPECT was feasible.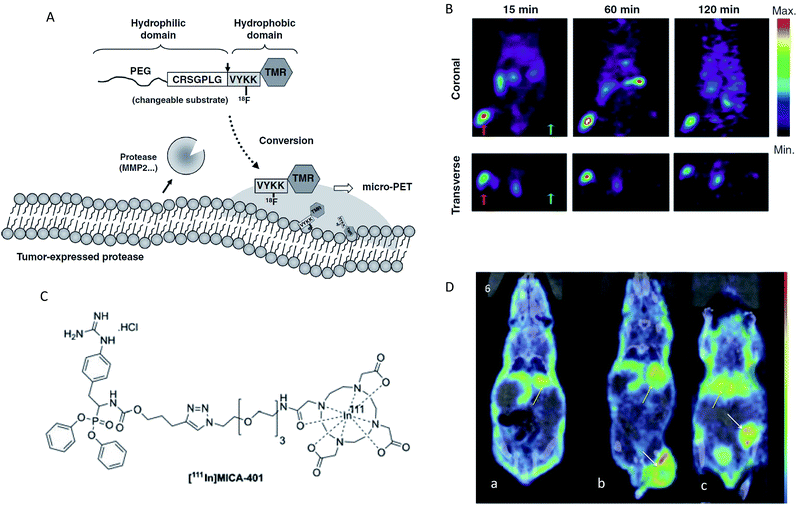 | ||
| Fig. 8 (A) Schematic representation of the MMP activity-based PET strategy. (B) Micro-PET imaging of MMP activity in vivo. Mice bearing established HT1080 (left hind legs) and MCF-7 (right hind legs) tumours were injected with PEG-peptide-18F-TMR. The red arrows indicate HT1080 human fibrosarcomas in left hind legs. The green arrows indicate MCF-7 human breast adenocarcinomas in right legs. (C) Chemical structures of fluor-18 and indium-111 labelled [111In]MICA-401. (D) Representative SPECT images at 95 h post injection of the uPA activity-based probe [111In]MICA-401 in the three models. (a) Healthy control mouse; (b) HT-29 tumour-bearing mouse; (c) MDA-MB-231 tumour-bearing mouse. The white arrows indicate tumours and yellow arrows indicate liver. (A and B) Reproduced from ref. 126. Copyright 2012 American Association for Cancer Research. (C and D) Reproduced from ref. 129. Copyright 2016 John Wiley & Sons, Ltd. | ||
However, nuclear imaging involves radiation exposure, a high cost of entry and high ongoing costs.132,133 The design of PET and SPECT tracers used for enzymatic target imaging is a challenging task because the agents need to have high specificity and affinity towards the target enzyme.134 In addition, storage and handling of these radioactive materials also involve a certain level of danger and may result in harmful consequences.100 Although these disadvantages have limited its broader practical application, it is still a potential technique for the detection of enzyme activities in a large animal model due to the penetration depth of this technique being almost unlimited.
5 Photoacoustic modality
The ultrasonic technique is a major clinical tool for soft-tissue imaging because of its low cost, high penetration depths and easy operation.135,136 However, ultrasound is limited by its inability to identify all abnormalities in tissues. Photoacoustic imaging (PAI) is able to complement and enhance ultrasound imaging. PAI is based on optical excitation and ultrasonic detection. First, a thermally induced pressure jump is caused by the light absorption of tissues, and then the pressure jump creates and launches ultrasonic waves. Finally, two-dimensional or three-dimensional images are formed by collecting and processing the ultrasonic signal.137–139 PAI integrates the advantage of the high penetration of ultrasound imaging and the high contrast of optical imaging.140,141In recent years, several studies have successfully shown the potential of PAI for the detection of enzyme activities in in vivo applications.142–149 An interesting study was recently presented by Liu's group, who used an activatable photoacoustic (PA) probe to estimate the distribution of MMP2 cleavage sites inside living tumor tissues.142 In this probe (GPD), a fluorescent dye molecule Dye680 (absorption peak at ∼680 nm) was used to couple with gold nanocages (GNCs) (absorption peak at ∼800 nm) via a specific enzymatic peptide substrate (SH-PEG-NH2). As shown in Fig. 9A, the Dye680 were released through the rupture of the peptide substrate by MMP-2. The PA signals changed significantly because of the change in the intrinsic absorption profile caused by the different retention speeds of GNCs and Dye680p. In vivo PAI images for MMP 2 activity are shown in Fig. 9B, where it can be seen that strong PA signals appeared immediately at both 680 and 770 nm after injection in a GPD injection group, while no obvious changes at either 680 or 770 nm were observed in a GPD + inhibitor injection group. In other words, the experiment showed that the probe has the ability to estimate the distribution of the protease activity in the tumor periphery region.
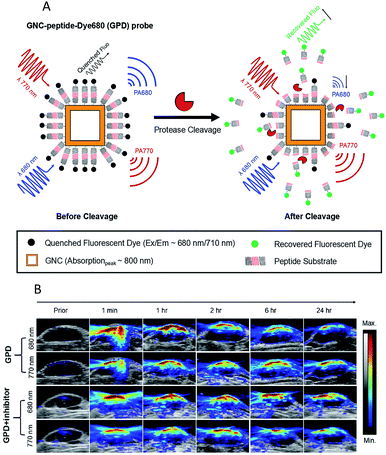 | ||
| Fig. 9 (A) Schematic design of the activatable probe GPD. The probe is expected to produce a strong PA signal from the GNCs and Dye680, conjugated by a cleavable peptide substrate. (B) PAI after intratumoral injections of GPD probe ± inhibitor. Reproduced from ref. 142. Copyright 2018 World Molecular Imaging Society. | ||
In addition, if the excitation light source is far-infrared light or microwaves, the imaging technique (usually called thermoacoustic imaging, TAI) can provide higher penetration than PAI.140,150 Despite all this, only a few PAI (TAI) examples exist so far for the detection of enzyme activities in vivo because the research on exogenous contrast agents is still at the entry level, and most contrast agents have been validated only at a pre-commercial, proof-of-concept stage without approval for clinical use. Much more work will need to be done to improve the PAI probes performance, such as solubility, stability and specific targeting capability.151 Nevertheless, with the compelling advantages of imaging performance characteristics, such as rivalling the speed of ultrasound imaging, the resolution of MRI and the specificity of nuclear imaging,152 imaging technologies based on PAI, therefore, are expected to have huge development potential and good application prospects in the detection of enzyme activities in vivo.
6 Multifunctional modality
Clearly to date, each method has both advantages and disadvantages. Therefore, the combination of two or more methods may prove more applicable. Multimodality imaging allows the integration of the advantages of a single modality and provides multi-parameter information in a composite imaging platform,153 so a growing number of multimodal probes have been designed to clearly delineate the localization and expression of biochemical markers.154 For the imaging of enzyme activities, the combination of different imaging modalities mainly include NIR-PET imaging,48,155 NIR-MR imaging,156 NIR-PA imaging157 and SPECT/CT.158,159 A recent significant study described a dual labelled (optical and PET/CT) probe BMV101, which could be used for the imaging of active cathepsins in fibrotic lesions of patients with idiopathic pulmonary fibrosis (IPF).48 This probe contained a near infrared fluorophore Cy5 for optical imaging and a NOTA chelator for labelling with 64Cu or 68Ga for PET/CT imaging (Fig. 10A).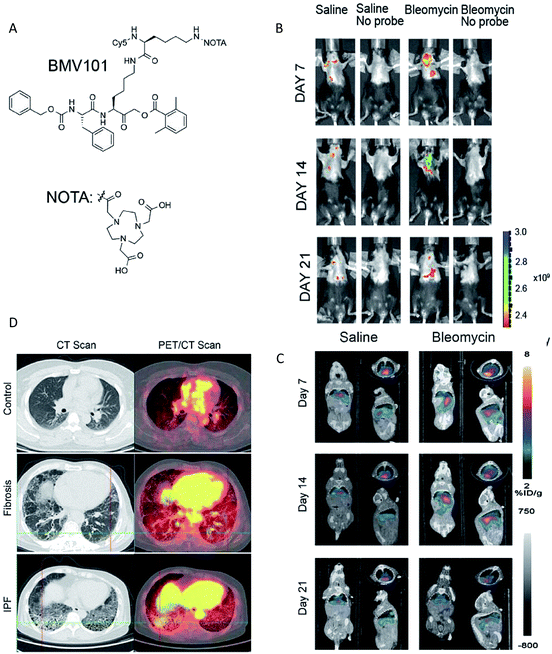 | ||
| Fig. 10 (A) Structures of the activity based probe BMV101. (B and C) Application of the 64Cu-BMV101 as a dual optical (B)/PET/CT (C) imaging probe. (D) First-in-human application of imaging probe 68Ga-BMV101. Representative scans of patients with idiopathic pulmonary fibrosis (IPF) and unclassifiable pulmonary fibrosis (fibrosis) compared to healthy controls. Reproduced from ref. 48. Copyright 2016 Springer Nature. | ||
Optical imaging and PET/CT of cathepsin activity by 64Cu-BMV101 in the bleomycin lung fibrosis model are shown in Fig. 10B and C. The PET-CT signal increased until day 14 and reduced on day 21 in the bleomycin-induced lung fibrosis mice model. In addition, the optical probe signal was consistent with the PET-CT results. These results confirmed that the intensity of signals observed by optical and PET/CT imaging correlated with the levels of cathepsin activity. More importantly, the non-invasive imaging of active cathepsins in human trials was demonstrated for the first time using 68Ga-BMV101 probe (Fig. 10D). In summary, although the overall signal-to-noise ratios were rather poor and more effective probe backbone structures need to be further identified, this research represents significant progress in human trials.
Typically, the multimodality imaging procedure is as follows: the test subject is scanned by different devices, and the images from each device are then overlaid, resulting in a fused image. This may lead to an imprecise co-registration of images and involves a time delay; so, simultaneous measurements by different techniques are critical to the measurement accuracy.160 Additionally, there are many challenges in the design of multimodal imaging probes, though efforts have been made in this field in recent years.161,162 Challenges for multimodality imaging probes are summarized nicely in a succinct review published in Chemical Reviews,163 and we refer the reader there for more information. However, in any case, the combination should draw on each other's merits to maximize the synergistic effect rather than choosing imaging modalities at random.
7 Conclusions and outlook
In the past few years, great progress has been made in the detection of enzyme activities in vivo based on optical modality, MRI modality, nuclear modality, photoacoustic modality and multifunctional modality. Overall, optical modality is the most thoroughly developed technique, though we anticipate that the other four techniques will all find more important applications in this area. Each imaging modality has its advantages along with limitations. For instance, despite the fact that the NIR optical modality exhibits unique optical properties (e.g. relatively low cost, high sensitivity, high biocompatibility and deep penetration) that make it widely used in the imaging of enzyme activities in animal models, the development of alternative strategies of imaging in deeper tissues while retaining a high resolution and signal intensity remains a big challenge. For another example, compared with optical imaging, MR imaging provides deeper tissue penetration, rendering it more appropriate for in vivo deep tissue imaging, but MRI provides relatively low sensitivity, and involves a high cost and long imaging time. Therefore, the use of multiple modalities in conjunction seems to be a powerful tool to implement each techniques' advantages, and more effort may be expected in the coming years in this regard. Furthermore, although the imaging of enzyme activity using these technologies has been demonstrated in small animal models, there are still several limitations. First, the type of enzymes that had been detected in vivo is far less than in vitro. The enzymes currently detected in vivo are mainly the hydrolyses used as tumor markers. Further studies should attempt to detect the other classes of enzymes in vivo, although it is a challenging task. Second, although some progress has been achieved in the research of the enzyme activity detection in vivo over the past few years, more preclinical and clinical trials should be encouraged to ascertain the clinical applicability of these techniques. Additionally, in order to make these technologies more suitable to clinical practice, several characteristics have to be taken into consideration as follows: (1) deep tissue penetration; (2) high sensitivity; (3) high specificity; (4) low toxicity; (5) easy removal and (6) low cost. Finally, advanced imaging equipment, such as multimodal instruments, should be developed for more effective use in in vivo applications.Over recent years, the number of contrast agents for diversified imaging applications is booming significantly. In clinical practice, small-molecule contrast agents are currently prevalent for use in imaging procedures because of their high biocompatibility and favorable excretion pharmacokinetics.164 However, small molecules cannot be easily multiplexed and cannot be easily designed.165 In recent decades, the design of various nanomaterial-based contrast agents has played a key role in improving imaging techniques. In general, most nanomaterial-based contrast agents have several promising advantages compared with their small-molecule counterparts, such as high sensitivity, good specificity and multimodal ability.67 Despite the advantages of using nanomaterial-based contrast agents in vivo, the pace of these contrast agents moving into clinical trials remains relatively gradual because of nanomaterial safety concerns. In brief, much work is required in the future to develop more advanced contrast agents for clinical needs.
Furthermore, it is worth noting that another promising area of future development is wireless sensor capsule for in situ measuring of biochemical compositions in a large animal or human models.166–171 This detection scheme using a capsule to bypass the hostile detection environment (e.g. deep tissues) offers challenges but potentially provides a new approach for the in vivo detection of enzyme activities. We believe that this technology based on optical or electrochemical wireless sensors will become a new trend and have quite high possibilities to become an important research tool, with applications across the detection of enzyme activity in deep tissues, such as in the gastrointestinal tract.
This review summarizes the latest advances in the imaging detection of enzyme activities in vivo within the past few years, mainly concentrating on the probe design, imaging strategies and the demonstration of enzyme activity in vivo. Although challenges are abound because current approaches are still difficult to achieve the quantitation of enzyme activity in deep tissues with satisfactory results, there is no doubt that the field of detection of enzyme activity in vivo will continue to evolve as more innovations in material chemistry, photophysics, biology and clinical medical examinations are achieved.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Key Research & Development Program, [Grant no. 2016 YFD 0401500, China], Key Disciplines Construction Foundation of the “1331” Project of Shanxi Province (098-091704) and the Fund for Shanxi Key Subject Construction (FSKSC). In addition, special thanks to Prof. Wei Zhao from the School of Food Science and Technology, Jiangnan University, who provided writing advice.References
- A. H. Palamakumbura and P. C. Trackman, Anal. Biochem., 2002, 300, 245–251 CrossRef CAS PubMed.
- R. H. Wijdeven, J. Neefjes and H. Ovaa, Trends Cell Biol., 2014, 24, 751–760 CrossRef CAS PubMed.
- M. B. Youdim, D. Edmondson and K. F. Tipton, Nat. Rev. Neurosci., 2006, 7, 295–309 CrossRef CAS PubMed.
- J. Yuan, Y. Q. Xu, N. N. Zhou, R. Wang, X. H. Qian and Y. F. Xu, RSC Adv., 2014, 4, 56207–56210 RSC.
- G. Leto, N. Gebbia, L. Rausa and F. M. Tumminello, Anticancer Res., 1992, 12, 235–240 CAS.
- Y. Urano, M. Sakabe, N. Kosaka, M. Ogawa, M. Mitsunaga, D. Asanuma, M. Kamiya, M. R. Young, T. Nagano, P. L. Choyke and H. Kobayashi, Sci. Transl. Med., 2011, 3, 110–119 Search PubMed.
- J. Wang, L. Wu, J. Ren and X. Qu, Small, 2012, 8, 259–264 CrossRef CAS PubMed.
- X. Xie, W. Xu and X. Liu, Acc. Chem. Res., 2012, 45, 1511–1520 CrossRef CAS PubMed.
- C. Jiang, C. Yan, J. Jiang and R. Yu, Anal. Chim. Acta, 2013, 766, 88–93 CrossRef CAS PubMed.
- A. Hayat, G. Bulbul and S. Andreescu, Biosens. Bioelectron., 2014, 56, 334–339 CrossRef CAS PubMed.
- V. Chikkaveeraiah, A. A. Bhirde, N. Y. Morgan, H. S. Eden and X. Chen, ACS Nano, 2012, 6, 6546–6561 CrossRef PubMed.
- L. Wu, J. Wang, L. Feng, J. Ren, W. Wei and X. Qu, Adv. Mater., 2012, 24, 2447–2452 CrossRef CAS PubMed.
- G. Wang, X. He, G. Xu, L. Chen, Y. Zhu, X. Zhang and L. Wang, Biosens. Bioelectron., 2013, 43, 125–130 CrossRef CAS PubMed.
- L. J. Kricka, J. C. Voyta and I. Bronstein, Methods Enzymol., 2000, 305, 370–390 CAS.
- A. Wang, S. Yan, R. Huang, S. Feng, B. Fu, X. Weng and X. Zhou, Analyst, 2013, 138, 2825–2828 RSC.
- H. Ao, Z. Qian, Y. Zhu, M. Zhao, C. Tang, Y. Huang, H. Feng and A. Wang, Biosens. Bioelectron., 2016, 86, 542–547 CrossRef CAS PubMed.
- Y. Su, S. F. Hickey, S. G. Keyser and M. C. Hammond, J. Am. Chem. Soc., 2016, 138, 7040–7047 CrossRef CAS PubMed.
- C. H. Tung, U. Mahmood, S. Bredow and R. Weissleder, Cancer Res., 2000, 60, 4953–4958 CAS.
- A. Razgulin, N. Ma and J. Rao, Chem. Soc. Rev., 2011, 40, 4186–4216 RSC.
- R. Yan and D. Ye, Sci. Bull., 2016, 61, 1672–1679 CrossRef CAS.
- A. Baruch, D. A. Jeffery and M. Bogyo, Trends Cell Biol., 2004, 14, 29–35 CrossRef CAS PubMed.
- J. P. Goddard and J. L. Reymond, Trends Biotechnol., 2004, 22, 363–370 CrossRef CAS PubMed.
- E. Hutter and D. Maysinger, Trends Pharmacol. Sci., 2013, 34, 497–507 CrossRef CAS PubMed.
- T. Wei, F. Wang, Z. Zhang, J. Qiang, J. Lv, T. Chen, J. Li and X. Chen, Curr. Med. Chem., 2018, 25, 1–33 CrossRef PubMed.
- Y. Ou, R. E. Wilson and S. G. Weber, Annu. Rev. Anal. Chem., 2018, 11, 509–533 CrossRef CAS PubMed.
- M. Chen, K. W. Cheng, Y. J. Chen, C. H. Wang, T. C. Cheng, K. C. Chang, A. P. Kao and K. H. Chuang, Sci. Rep., 2017, 7, 3142 CrossRef PubMed.
- W. F. Cheong, S. A. Prahl and A. J. Welch, IEEE J. Quantum Electron., 1990, 26, 2166–2185 CrossRef.
- M. Yang, E. Baranov, P. Jiang, F. X. Sun, X. M. Li, L. Li, S. Hasegawa, M. Bouvet, M. Al-Tuwaijri, T. Chishima, H. Shimada, A. R. Moossa, S. Penman and R. M. Hoffman, Proc. Natl. Acad. Sci. U. S. A., 2000, 97, 1206–1211 CrossRef CAS PubMed.
- A. H. Gandjbakhche, V. Chernomordik, D. Hattery and M. Hassan, Technol. Cancer Res. Treat., 2003, 2, 537 CrossRef PubMed.
- B. R. Smith and S. S. Gambhir, Chem. Rev., 2017, 117, 901–986 CrossRef CAS PubMed.
- V. Ntziachristos, C. Bremer and R. Weissleder, Eur. J. Radiol., 2003, 13, 195–208 Search PubMed.
- N. Won, S. Jeong, K. Kim, J. Kwag, J. Park, S. G. Kim and S. Kim, Mol. Imaging, 2012, 11, 338–352 CrossRef CAS PubMed.
- V. Ntziachristos, J. Ripoll and R. Weissleder, Opt. Lett., 2002, 27, 333–335 CrossRef PubMed.
- S. A. Hilderbrand and R. Weissleder, Curr. Opin. Chem. Biol., 2010, 14, 71–79 CrossRef CAS PubMed.
- L. Zhu, F. Zhang, Y. Ma, G. Liu, K. Kim, X. Fang, S. Lee and X. Chen, Mol. Pharm., 2011, 8, 2331–2338 CrossRef CAS PubMed.
- M. Solomon, K. Guo, G. P. Sudlow, M. Y. Berezin, W. B. Edwards, S. Achilefu and W. J. Akers, J. Biomed. Opt., 2011, 16, 066019 CrossRef PubMed.
- M. P. Chien, A. S. Carlini, D. Hu, C. V. Barback, A. M. Rush, D. J. Hall, G. Orr and N. C. Gianneschi, J. Am. Chem. Soc., 2013, 135, 18710–18713 CrossRef CAS PubMed.
- T. Fukui, E. Tenborg, J. H. Yik and D. R. Haudenschild, Biochem. Biophys. Res. Commun., 2015, 460, 741–746 CrossRef CAS PubMed.
- P. B. Satkunananthan, M. J. Anderson, N. M. De Jesus, D. R. Haudenschild, C. M. Ripplinger and B. A. Christiansen, Osteoarthritis Cartilage, 2014, 22, 1461–1469 CrossRef CAS PubMed.
- J. Schwenck, C. M. Griessinger, K. Fuchs, D. Bukala, N. Bauer, M. Eichner, M. Rocken, B. J. Pichler and M. Kneilling, Mol. Imaging, 2014, 13, 7290 Search PubMed.
- J. Schwenck, F. C. Maier, M. Kneilling, S. Wiehr and K. Fuchs, J. Visualized Exp., 2017, e55180 Search PubMed.
- R. A. Sheth, A. Kunin, L. Stangenberg, M. Sinnamon, K. E. Hung, R. Kucherlapati and U. J. M. I. Mahmood, Mol. Imaging, 2012, 11, 417–425 CrossRef CAS PubMed.
- A. K. Steingraber, S. Schelhaas, A. Faust, A. H. Jacobs, M. Schafers and T. Goerge, Exp. Dermatol., 2013, 22, 730–735 CrossRef PubMed.
- E. A. Vermeij, M. I. Koenders, A. B. Blom, O. J. Arntz, M. B. Bennink, V. D. B. Wb, P. L. van Lent and V. D. L. Fa, Mol. Imaging, 2014, 13, 1–10 CrossRef PubMed.
- W. Fumey, J. Koenigsdorf, V. Kunick, S. Menzel, K. Schutze, M. Unger, L. Schriewer, F. Haag, G. Adam, A. Oberle, M. Binder, R. Fliegert, A. Guse, Y. J. Zhao, H. Cheung Lee, F. Malavasi, F. Goldbaum, R. van Hegelsom, C. Stortelers, P. Bannas and F. Koch-Nolte, Sci. Rep., 2017, 7, 14289 CrossRef PubMed.
- J. Zhang, D. V. Preda, K. O. Vasquez, J. Morin, J. Delaney, B. Bao, M. D. Percival, D. Xu, D. McKay, M. Klimas, B. Bednar, C. Sur, D. Z. Gao, K. Madden, W. Yared, M. Rajopadhye and J. D. Peterson, Am. J. Physiol., 2012, 303, 593–603 CrossRef PubMed.
- D. Caglic, A. Globisch, M. Kindermann, N. H. Lim, V. Jeske, H. P. Juretschke, E. Bartnik, K. U. Weithmann, H. Nagase, B. Turk and K. U. Wendt, Bioorg. Med. Chem., 2011, 19, 1055–1061 CrossRef CAS PubMed.
- N. P. Withana, X. Ma, H. M. Mcguire, M. Verdoes, W. A. V. D. Linden, L. O. Ofori, R. Zhang, H. Li, L. E. Sanman, K. Wei, S. Yao, P. Wu, F. Li, H. Huang, Z. Xu, P. J. Wolters, G. D. Rosen, H. R. Collard, Z. Zhu, Z. Cheng and M. Bogyo, Sci. Rep., 2016, 6, 19755 CrossRef CAS PubMed.
- Y. Zhao, Z. Hai, H. Wang, L. Su and G. Liang, Anal. Chem., 2018, 90, 8732–8735 CrossRef CAS PubMed.
- Z. Li, X. He, Z. Wang, R. Yang, W. Shi and H. Ma, Biosens. Bioelectron., 2015, 63, 112–116 CrossRef CAS PubMed.
- S. Bhaumik, T. V. Sekar, J. Depuy, J. Klimash and R. Paulmurugan, Gene Ther., 2012, 19, 295–302 CrossRef CAS PubMed.
- X. He, L. Li, Y. Fang, W. Shi, X. Li and H. Ma, Chem. Sci., 2017, 8, 3479–3483 RSC.
- E. J. Kim, R. Kumar, A. Sharma, B. Yoon, H. M. Kim, H. Lee, K. S. Hong and J. S. Kim, Biomaterials, 2017, 122, 83–90 CrossRef CAS PubMed.
- K. Gu, Y. Xu, H. Li, Z. Guo, S. Zhu, S. Zhu, P. Shi, T. D. James, H. Tian and W. H. Zhu, J. Am. Chem. Soc., 2016, 138, 5334–5340 CrossRef CAS PubMed.
- E. S. Olson, M. A. Whitney, B. Friedman, T. A. Aguilera, J. L. Crisp, F. M. Baik, T. Jiang, S. M. Baird, S. Tsimikas, R. Y. Tsien and Q. T. Nguyen, Integr. Biol., 2012, 4, 595–605 CrossRef CAS PubMed.
- D. Madan, C. G. Ferguson, W. Y. Lee, G. D. Prestwich and C. A. Testa, PLoS One, 2013, 8, e79065 CrossRef PubMed.
- K. R. Wong, E. Menendez, C. S. Craik, W. M. Kavanaugh and O. Vasiljeva, Biochimie, 2016, 122, 62–67 CrossRef CAS PubMed.
- D. Ye, A. J. Shuhendler, L. Cui, L. Tong, S. S. Tee, G. Tikhomirov, D. W. Felsher and J. Rao, Nat. Chem., 2014, 6, 519–526 CrossRef CAS PubMed.
- M. Verdoes, K. Oresic Bender, E. Segal, W. A. van der Linden, S. Syed, N. P. Withana, L. E. Sanman and M. Bogyo, J. Am. Chem. Soc., 2013, 135, 14726–14730 CrossRef CAS PubMed.
- Y. Jin, X. Tian, L. Jin, Y. Cui, T. Liu, Z. Yu, X. Huo, J. N. Cui, C. P. Sun, C. Wang, J. Ning, B. Zhang, L. Feng and X. Ma, Anal. Chem., 2018, 90, 3276–3283 CrossRef CAS PubMed.
- T. C. Cheng, S. R. Roffler, S. C. Tzou, K. H. Chuang, Y. C. Su, C. H. Chuang, C. H. Kao, C. S. Chen, H. I-Hong, K. Y. Liu, L. T. Cheng and Y. L. Leu, J. Am. Chem. Soc., 2012, 134, 3103–3110 CrossRef CAS PubMed.
- X. Huo, X. Tian, Y. Li, L. Feng, Y. Cui, C. Wang, J. Cui, C. Sun, K. Liu and X. Ma, Sens. Actuators, B, 2018, 262, 508–515 CrossRef CAS.
- Q. Jin, L. Feng, D. D. Wang, J. J. Wu, J. Hou, Z. R. Dai, S. G. Sun, J. Y. Wang, G. B. Ge, J. N. Cui and L. Yang, Biosens. Bioelectron., 2016, 83, 193–199 CrossRef CAS PubMed.
- G. Fuhrmann and J. C. Leroux, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 9032–9037 CrossRef CAS.
- H. Lee and Y. P. J. B. R. Kim, BMB Rep., 2015, 48, 313–318 CrossRef CAS PubMed.
- Y. Zhao, Z. Hai, H. Wang, L. Su and G. Liang, Anal. Chem., 2018, 90, 8732–8735 CrossRef CAS PubMed.
- X. Huang, J. Song, B. C. Yung, X. Huang, Y. Xiong and X. Chen, Chem. Soc. Rev., 2018, 47, 2873–2920 RSC.
- X. Huo, X. Tian, Y. Li, L. Feng, Y. Cui, C. Wang, J. Cui, C. Sun, K. Y. Liu and X. Ma, Sens. Actuators, B, 2018, 262, 508–515 CrossRef CAS.
- A. M. Smith, M. C. Mancini and S. Nie, Nat. Nanotechnol., 2009, 4, 710–711 CrossRef CAS PubMed.
- S. Diao, G. Hong, A. L. Antaris, J. L. Blackburn, K. Cheng, Z. Cheng and H. Dai, Nano Res., 2015, 8, 3027–3034 CrossRef CAS.
- Y. Fan and F. Zhang, Adv. Opt. Mater., 2019, 7, 1801417 CrossRef.
- Y. Sun, F. Ding, Z. Zhou, C. Li, M. Pu, Y. Xu, Y. Zhan, X. Lu, H. Li, G. Yang, Y. Sun and P. J. Stang, Proc. Natl. Acad. Sci. U. S. A., 2019, 116, 1968–1973 CrossRef PubMed.
- F. Ding, Z. Chen, W. Y. Kim, A. Sharma, C. Li, Q. Ouyang, H. Zhu, G. Yang, Y. Sun and J. S. Kim, Chem. Sci., 2019, 10, 7023–7028 RSC.
- F. Ding, C. Li, Y. Xu, J. Li, H. Li, G. Yang and Y. Sun, Adv. Healthcare Mater., 2018, 7, e1800973 CrossRef PubMed.
- F. Ding, Y. Zhan, X. Lu and Y. Sun, Chem. Sci., 2018, 9, 4370–4380 RSC.
- S. Wu, E. Chang and Z. Cheng, Curr. Org. Synth., 2011, 8, 488–497 CrossRef CAS.
- T. S. Wehrman, G. V. Degenfeld, P. O. Krutzik, G. P. Nolan and H. M. Blau, Nat. Methods, 2006, 3, 295–301 CrossRef CAS PubMed.
- M. A. Paley and J. A. Prescher, MedChemComm, 2014, 5, 255–267 RSC.
- T. Zhang, L. Du and M. Li, Chin. Chem. Lett., 2015, 26, 919–921 CrossRef CAS.
- D. A. Dart and C. L. Bevan, Methods Mol. Biol., 2016, 1443, 203 CrossRef CAS PubMed.
- W. Zhou, M. P. Valley, J. Shultz, E. M. Hawkins, L. Bernad, T. Good, D. Good, T. L. Riss, D. H. Klaubert and K. V. Wood, J. Am. Chem. Soc., 2006, 128, 3122–3123 CrossRef CAS PubMed.
- G. C. Van de Bittner, C. R. Bertozzi and C. J. Chang, J. Am. Chem. Soc., 2013, 135, 1783–1795 CrossRef CAS.
- E. Adamová, M. Lišková, E. Matalová and K. Klepárník, Anal. Bioanal. Chem., 2014, 406, 5389–5394 CrossRef PubMed.
- W. Zhou, J. W. Shultz, N. Murphy, E. M. Hawkins, L. Bernad, T. Good, L. Moothart, S. Frackman, D. H. Klaubert, R. F. Bulleit and K. V. Wood, Chem. Commun., 2006, 38, 4620–4622 RSC.
- J. S. Rush, K. E. Beatty and C. R. Bertozzi, ChemBioChem, 2010, 11, 2096–2099 CrossRef CAS.
- J. Ø. Moskaug, H. Carlsen and R. Blomhoff, Mol. Imaging, 2015, 7, 35–41 Search PubMed.
- R. Geiger, E. Schneider, K. Wallenfels and W. Miska, Biol. Chem. Hoppe-Seyler, 1992, 373, 1187–1191 CrossRef CAS PubMed.
- H. Yao, M. K. So and J. Rao, Angew. Chem., Int. Ed., 2010, 46, 7161–7164 Search PubMed.
- Y. Gao, Y. Lin, T. Liu, H. Chen, X. Yang, C. Tian, L. Du and M. Li, Anal. Chem., 2017, 89, 12488–12493 CrossRef CAS PubMed.
- J. Li, L. Chen, W. Wu, W. Zhang, Z. Ma, Y. Cheng, L. Du and M. Li, Anal. Chem., 2014, 86, 2747–2751 CrossRef CAS PubMed.
- R. H. Wong, T. Kwong, K. H. Yau and H. Y. Au-Yeung, Chem. Commun., 2015, 51, 4440–4442 RSC.
- P. Feng, H. Zhang, Q. Deng, W. Liu, L. Yang, G. Li, G. Chen, L. Du, B. Ke and M. Li, Anal. Chem., 2016, 88, 5610–5614 CrossRef CAS PubMed.
- A. G. Vorobyeva, M. Stanton, A. Godinat, K. B. Lund, G. G. Karateev, K. P. Francis, E. Allen, J. G. Gelovani, E. McCormack, M. Tangney and E. A. Dubikovskaya, PLoS One, 2015, 10, e0131037 CrossRef PubMed.
- S. Li, R. Hu, S. Wang, X. Guo, Y. Zeng, Y. Li and G. Yang, Anal. Chem., 2018, 90, 9296–9300 CrossRef CAS PubMed.
- A. l. Godinat, H. M. Park, S. C. Miller, K. Cheng, D. Hanahan, L. E. Sanman, M. Bogyo, A. Yu, G. F. Nikitin and A. J. Stahl, ACS Chem. Biol., 2013, 8, 987–999 CrossRef CAS PubMed.
- A. M. Loening, A. M. Wu and S. S. Gambhir, Nat. Methods, 2007, 4, 641–643 CrossRef CAS PubMed.
- S. T. Adams Jr and S. C. Miller, Curr. Opin. Chem. Biol., 2014, 21, 112–120 CrossRef PubMed.
- M. A. Hahn, A. K. Singh, P. Sharma, S. C. Brown and B. M. Moudgil, Anal. Bioanal. Chem., 2011, 399, 3–27 CrossRef CAS PubMed.
- E. Terreno, D. D. Castelli, A. Viale and S. Aime, Chem. Rev., 2010, 110, 3019–3042 CrossRef CAS PubMed.
- M. Carril, J. Mater. Chem. B, 2017, 5, 4332–4347 RSC.
- X. Li, Z. Zhang, Z. Yu, J. Magnusson and J. X. Yu, Mol. Pharm., 2013, 10, 1360–1367 CrossRef CAS PubMed.
- J. X. Yu, V. D. Kodibagkar, L. Liu, Z. Zhang, J. Magnusson and Y. Liu, Chem. Sci., 2013, 4, 2132–2142 RSC.
- Y. Yuan, S. Ge, H. Sun, X. Dong, H. Zhao, L. An, J. Zhang, J. Wang, B. Hu and G. Liang, ACS Nano, 2015, 9, 5117–5124 CrossRef CAS PubMed.
- H. Matsushita, S. Mizukami, Y. Mori, F. Sugihara, M. Shirakawa, Y. Yoshioka and K. Kikuchi, ChemBioChem, 2012, 13, 1579–1583 CrossRef CAS PubMed.
- A. Keliris, I. Mamedov, G. E. Hagberg, N. K. Logothetis, K. Scheffler and J. Engelmann, Contrast Media Mol. Imaging, 2012, 7, 478–483 CrossRef CAS PubMed.
- Y. Yuan, H. Sun, S. Ge, M. Wang, H. Zhao, L. Wang, A. Linna, J. Zhang, H. Zhang, B. Hu, J. Wang and G. Liang, ACS Nano, 2015, 9, 761–768 CrossRef CAS PubMed.
- K. Akazawa, F. Sugihara, T. Nakamura, S. Mizukami and K. Kikuchi, Bioconjugate Chem., 2018, 29, 1720–1728 CrossRef CAS PubMed.
- M. Kazuya, K. Rui, M. Keigo, I. Hirohiko, T. Yuki, N. Michiko, M. Tetsuya, T. Yousuke and H. Itaru, Chem.–Eur. J., 2013, 19, 12875–12883 CrossRef PubMed.
- J. X. Yu, V. D. Kodibagkar, R. R. Hallac, L. Liu and R. P. Mason, Bioconjugate Chem., 2012, 23, 596–603 CrossRef CAS PubMed.
- G. Liu, Y. Liang, A. Bar-Shir, K. W. Chan, C. S. Galpoththawela, S. M. Bernard, T. Tse, N. N. Yadav, P. Walczak, M. T. McMahon, J. W. M. Bulte, P. C. M. van Zijl and A. A. Gilad, J. Am. Chem. Soc., 2011, 133, 16326–16329 CrossRef CAS PubMed.
- B. Yoo and M. D. Pagel, J. Am. Chem. Soc., 2006, 128, 14032–14033 CrossRef CAS PubMed.
- G. Liu, Y. Liang, A. Bar-Shir, K. W. Chan, C. S. Galpoththawela, S. M. Bernard, T. Tse, N. N. Yadav, P. Walczak, M. T. McMahon, J. W. Bulte, P. C. van Zijl and A. A. Gilad, J. Am. Chem. Soc., 2011, 133, 16326–16329 CrossRef CAS PubMed.
- M. Haris, A. Singh, I. Mohammed, R. Ittyerah, K. Nath, R. P. Nanga, C. Debrosse, F. Kogan, K. Cai, H. Poptani, D. Reddy, H. Hariharan and R. Reddy, Sci. Rep., 2014, 4, 6081 CrossRef CAS PubMed.
- Y. Jamin, T. R. Eykyn, E. Poon, C. J. Springer and S. P. Robinson, Mol. Imaging Biol., 2014, 16, 152–157 CrossRef PubMed.
- B. Yoo, V. R. Sheth, C. M. Howison, M. J. Douglas, C. T. Pineda, E. A. Maine, A. F. Baker and M. D. Pagel, Magn. Reson. Med., 2014, 71, 1221–1230 CrossRef CAS PubMed.
- I. Daryaei, M. M. Ghaffari, K. M. Jones and M. D. Pagel, ACS Sens., 2016, 1, 857–861 CrossRef CAS PubMed.
- S. Sinharay, G. Fernandez-Cuervo, J. P. Acfalle and M. D. Pagel, Chem.–Eur. J., 2016, 22, 6491–6495 CrossRef CAS PubMed.
- S. Sinharay, E. A. Randtke, K. M. Jones, C. M. Howison, S. K. Chambers, H. Kobayashi and M. D. Pagel, Magn. Reson. Med., 2017, 77, 2005–2014 CrossRef CAS PubMed.
- S. Sinharay, E. A. Randtke, C. M. Howison, N. A. Ignatenko and M. D. Pagel, Mol. Imaging Biol., 2018, 20, 240–248 CrossRef CAS PubMed.
- T. F. Massoud and S. S. Gambhir, Genes Dev., 2003, 17, 545–580 CrossRef CAS PubMed.
- A. Sundin, U. Garske and H. Orlefors, Best Pract. Res., Clin. Endocrinol. Metab., 2007, 21, 69–85 CrossRef CAS PubMed.
- A. Rahmim, J. Qi and V. Sossi, Med. Phys., 2013, 40, 064301 CrossRef PubMed.
- M. T. Madsen, J. Nucl. Med., 2007, 48, 661–673 CrossRef PubMed.
- A. Rahmim and H. Zaidi, Nucl. Med. Commun., 2008, 29, 193–207 CrossRef PubMed.
- I. F. Antunes, H. J. Haisma, P. H. Elsinga, J. W. Sijbesma, A. Waarde, A. T. Willemsen, R. A. Dierckx and E. F. de Vries, Nucl. Med. Biol., 2012, 39, 854–863 CrossRef CAS PubMed.
- C. H. Chuang, K. H. Chuang, H. E. Wang, S. R. Roffler, J. T. Shiea, S. C. Tzou, T. C. Cheng, C. H. Kao, S. Y. Wu, W. L. Tseng, C. M. Cheng, M. F. Hou, J. M. Wang and T. L. Cheng, Clin. Cancer Res., 2012, 18, 238–247 CrossRef CAS PubMed.
- J. Ides, D. Thomae, L. Wyffels, C. Vangestel, J. Messagie, J. Joossens, F. Lardon, P. Van der Veken, K. Augustyns, S. Stroobants and S. Staelens, Nucl. Med. Biol., 2014, 41, 477–487 CrossRef CAS PubMed.
- Y. C. Su, T. C. Cheng, Y. L. Leu, S. R. Roffler, J. Y. Wang, C. H. Chuang, C. H. Kao, K. C. Chen, H. E. Wang and T. L. Cheng, Mol. Cancer Ther., 2014, 13, 2852–2863 CrossRef CAS PubMed.
- C. Vangestel, D. Thomae, J. Van Soom, J. Ides, L. Wyffels, P. Pauwels, S. Stroobants, P. Van der Veken, V. Magdolen, J. Joossens, K. Augustyns and S. Staelens, Contrast Media Mol. Imaging, 2016, 11, 448–458 CrossRef CAS PubMed.
- N. Fukumitsu, S. H. Yeh, L. G. Flores Ii, U. Mukhopadhyay, D. Young, K. Ogawa, H. J. Jeong, W. Tong and J. G. Gelovani, Contrast Media Mol. Imaging, 2018, 3, e3612027 Search PubMed.
- A. Mizutani, M. Kobayashi, K. I. Fujita, K. Takahashi, T. Hokama, H. Takasu, K. Nishi, R. Nishii, N. Shikano, K. Fukuchi and K. Kawai, Nucl. Med. Commun., 2018, 39, 825–833 CrossRef CAS PubMed.
- T. M. Bateman, J. Nucl. Cardiol., 2012, 19, 3–11 CrossRef PubMed.
- M. Ali, B. Pulli and J. W. Chen, Curr. Cardiovasc. Imaging Rep., 2014, 7, 9258 CrossRef PubMed.
- B. P. Rempel, E. W. Price and C. P. Phenix, Mol. Imaging, 2017, 16, 1–30 CrossRef CAS PubMed.
- S. Unnikrishnan and A. L. Klibanov, AJR, Am. J. Roentgenol., 2012, 199, 292–299 CrossRef PubMed.
- F. Kiessling, S. Fokong, P. Koczera, W. Lederle and T. Lammers, J. Nucl. Med., 2012, 53, 345–348 CrossRef CAS PubMed.
- J. L. Su, B. Wang, K. E. Wilson, C. L. Bayer, Y. S. Chen, S. Kim, K. A. Homan and S. Y. Emelianov, Expert Opin. Med. Diagn., 2010, 4, 497–510 CrossRef PubMed.
- S. Wang, J. Lin, T. Wang, X. Chen and P. Huang, Theranostics, 2016, 6, 2394–2413 CrossRef CAS PubMed.
- Q. Miao and K. Pu, Bioconjugate Chem., 2016, 27, 2808–2823 CrossRef CAS PubMed.
- D. Wu, L. Huang, M. S. Jiang and H. Jiang, Int. J. Mol. Sci., 2014, 15, 23616–23639 CrossRef.
- L. V. Wang and S. Hu, Science, 2012, 335, 1458–1462 CrossRef CAS PubMed.
- C. Liu, S. Li, Y. Gu, H. Xiong, W. T. Wong and L. Sun, Mol. Imaging Biol., 2018, 20, 919–929 CrossRef CAS PubMed.
- H. Qin, Y. Zhao, J. Zhang, X. Pan, S. Yang and D. Xing, Biol. Med., 2016, 12, 1765–1774 CAS.
- L. L. Li, Q. Zeng, W. J. Liu, X. F. Hu, Y. Li, J. Pan, D. Wan and H. Wang, ACS Appl. Mater. Interfaces, 2016, 8, 17936–17943 CrossRef CAS PubMed.
- D. Zhang, G. B. Qi, Y. X. Zhao, S. L. Qiao, C. Yang and H. Wang, Adv. Mater., 2015, 27, 6125–6130 CrossRef CAS PubMed.
- K. Yang, L. Zhu, L. Nie, X. Sun, L. Cheng, C. Wu, G. Niu, X. Chen and Z. Liu, Theranostics, 2014, 4, 134–141 CrossRef CAS PubMed.
- J. Levi, S. R. Kothapalli, S. Bohndiek, J. K. Yoon, A. Dragulescu-Andrasi, C. Nielsen, A. Tisma, S. Bodapati, G. Gowrishankar, X. Yan, C. Chan, D. Starcevic and S. S. Gambhir, Clin. Cancer Res., 2013, 19, 1494–1502 CrossRef CAS PubMed.
- A. Dragulescu-Andrasi, S. R. Kothapalli, G. A. Tikhomirov, J. Rao and S. S. Gambhir, J. Am. Chem. Soc., 2013, 135, 11015–11022 CrossRef CAS PubMed.
- C. Liu, Y. Yang, Z. Qiu, Y. Huang and L. Sun, Ultrasonics Symposium IEEE, 2015 Search PubMed.
- W. Ding, Z. Ji and D. Xing, Appl. Phys. Lett., 2017, 110, e183701 CrossRef.
- J. Weber, P. C. Beard and S. E. Bohndiek, Nat. Methods, 2016, 13, 639–650 CrossRef CAS PubMed.
- D. Razansky, N. C Deliolanis, C. Vinegoni and V. Ntziachristos, Curr. Pharm. Biotechnol., 2012, 13, 504–522 CAS.
- D. E. Lee, H. Koo, I. C. Sun, J. H. Ryu, K. Kim and I. C. Kwon, Chem. Soc. Rev., 2012, 41, 2656–2672 RSC.
- Y. Sun, X. Zeng, Y. Xiao, C. Liu, H. Zhu, H. Zhou, Z. Chen, F. Xu, J. Wang, M. Zhu, J. Wu, M. Tian, H. Zhang, Z. Deng, Z. Cheng and X. Hong, Chem. Sci., 2018, 9, 2092–2097 RSC.
- H. Lee, W. J. Akers, P. P. Cheney, W. B. Edwards, K. Liang, J. P. Culver and S. Achilefu, J. Biomed. Opt., 2009, 14, e040507 Search PubMed.
- L. Josephson, M. F. Kircher, U. Mahmood, Y. Tang and R. Weissleder, Bioconjugate Chem., 2002, 13, 554–560 CrossRef CAS PubMed.
- X. Xia, M. Yang, L. K. Oetjen, Y. Zhang, Q. Li, J. Chen and Y. Xia, Nanoscale, 2011, 3, 950–953 RSC.
- M. Razavian, S. Tavakoli, J. Zhang, L. Nie, L. W. Dobrucki, A. J. Sinusas, A. Michael, R. Simon and M. M. Sadeghi, J. Nucl. Med., 2011, 52, 1795–1802 CrossRef CAS PubMed.
- M. Razavian, J. Zhang, L. Nie, S. Tavakoli, N. Razavian, L. W. Dobrucki, A. J. Sinusas, D. S. Edwards, M. Azure and M. M. Sadeghi, J. Nucl. Med., 2010, 51, 1107–1115 CrossRef CAS PubMed.
- Z. Brady, M. L. Taylor, M. Haynes, M. Whitaker, A. Mullen, L. Clews, M. Partridge, R. J. Hicks and J. V. Trapp, Australas. Phys. Eng. Sci. Med., 2008, 31, 90–109 CrossRef CAS PubMed.
- Y. Xu, M. Tian, H. Zhang, Y. Xiao, X. Hong and Y. Sun, Chin. Chem. Lett., 2018, 29, 1093–1097 CrossRef CAS.
- F. Ding, S. Chen, W. Zhang, Y. Tu and Y. Sun, Bioorg. Med. Chem., 2017, 25, 5179–5184 CrossRef CAS PubMed.
- A. Louie, Chem. Rev., 2010, 110, 3146–3195 CrossRef CAS PubMed.
- L. Tu, Y. Xu, Q. Ouyang, X. Li and Y. Sun, Chin. Chem. Lett., 2019 DOI:10.1016/j.cclet.2019.05.022.
- F. Ding, Y. Fan, Y. Sun and F. Zhang, Adv. Healthcare Mater., 2019, e1900260 CrossRef PubMed.
- K. Kalantar-Zadeh, C. K. Yao, K. J. Berean, N. Ha, J. Z. Ou, S. A. Ward, N. Pillai, J. Hill, J. J. Cottrell, F. R. Dunshea, C. McSweeney, J. G. Muir and P. R. Gibson, Gastroenterology, 2016, 150, 37–39 CrossRef PubMed.
- M. Mimee, P. Nadeau, A. Hayward, S. Carim, S. Flanagan, L. Jerger, J. Collins, S. McDonnell, R. Swartwout, R. J. Citorik, V. Bulovic, R. Langer, G. Traverso, A. P. Chandrakasan and T. K. Lu, Science, 2018, 360, 915–918 CrossRef CAS PubMed.
- K. Kalantar-Zadeh, K. J. Berean, N. Ha, A. F. Chrimes, K. Xu, D. Grando, J. Z. Ou, N. Pillai, J. L. Campbell, R. Brkljača, K. M. Taylor, R. E. Burgell, C. K. Yao, S. A. Ward, C. S. McSweeney, J. G. Muir and P. R. Gibson, Nat. Electron., 2018, 1, 79–87 CrossRef.
- J. Z. Ou, J. J. Cottrell, N. Ha, N. Pillai, C. K. Yao, K. J. Berean, S. A. Ward, D. Grando, J. G. Muir, C. J. Harrison, U. Wijesiriwardana, F. R. Dunshea, P. R. Gibson and K. Kalantar-Zadeh, Sci. Rep., 2016, 6, e33387 CrossRef PubMed.
- P. J. van der Schaar, J. F. Dijksman, H. Broekhuizen-de Gast, J. Shimizu, N. van Lelyveld, H. Zou, V. Iordanov, C. Wanke and P. D. Siersema, Gastrointest. Endosc., 2013, 78, 520–528 CrossRef PubMed.
- C. Steiger, A. Abramson, P. Nadeau, A. P. Chandrakasan, R. Langer and G. Traverso, Nat. Rev. Mater., 2019, 4, 83–98 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2019 |