Mass spectrometric detection of enantioselectivity in three-component complexation, copper(II)-chiral tetradentate ligand-free amino acid in solution†
Takashi
Nakakoji
a,
Hirofumi
Sato
b,
Daisuke
Ono
b,
Hiroyuki
Miyake
*a,
Satoshi
Shinoda
 a,
Hiroshi
Tsukube
a,
Hideya
Kawasaki
a,
Hiroshi
Tsukube
a,
Hideya
Kawasaki
 c,
Ryuichi
Arakawa
c and
Motohiro
Shizuma
c,
Ryuichi
Arakawa
c and
Motohiro
Shizuma
 *b
*b
aDepartment of Chemistry, Graduate School of Science, Osaka City University, 3-3-138 Sugimoto, Sumiyoshi-ku, Osaka 558-8585, Japan. E-mail: miyake@sci.osaka-cu.ac.jp
bOsaka Research Institute of Industrial Science and Technology, 1-6-50 Morinomiya, Joto-ku, Osaka 536-8553, Japan. E-mail: shizuma@omtri.or.jp
cFaculty of Chemistry, Materials and Bioengineering, Kansai University, 3-3-35 Yamatecho, Suita, Osaka 564-8680, Japan
First published on 11th November 2019
Abstract
By using electrospray ionization mass spectrometry coupled with the use of deuterium-labelled quasienantiomers, enantioselective coordination of a free amino acid as the second ligand of a copper(II) complex with a novel chiral tetradentate ligand was evaluated quantitatively in a short measurement time.
Metal complexes with chiral amino acid derivatives as the stereogenic centres have been developed to exhibit various promising functions. Asymmetric catalytic ability,1–4 stimulus-responsive helicity control,5–10 stereoselective encapsulations in organic zeolite11 and metal organic frameworks (MOFs),12 circular polarized luminescence induction13,14 and collagen synthesis in a cell were typical examples.15 Chiral molecular recognition of the free amino acid is one of the most important functions,16 because it is applicable to understanding biological metabolism/molecular recognition systems, and developing sophisticated industrial systems. In this paper, we report for the first time detecting enantioselective coordination of a free amino acid as the second ligand of a metal complex with a simple chiral tetradentate ligand5 by using electrospray ionization mass spectrometry (ESI-MS) coupled with the use of deuterium-labelled quasienantiomers.17–20 In general, a chiral discrimination ability of a chiral host toward chiral guest in solution is evaluated by a spectroscopic titration method using NMR, UV-vis and CD measurements. However, these methods basically require high concentration and are time consuming. Compared to these titration methods, the ESI-MS method can determine chiral discrimination ability of the complex toward free amino acids more rapidly at smaller scale.
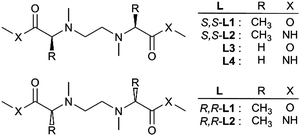
As the suitable chiral tetradentate ligand of a chiral metal complex, simple S,S-chiral ligands (S,S-L1 and S,S-L2) and the corresponding R,R-enantiomers (R,R-L1 and R,R-L2) were designed (Chart 1) and synthesized using the conventional method.5 Achiral ligands (L3 and L4) were also synthesized as the reference compounds.
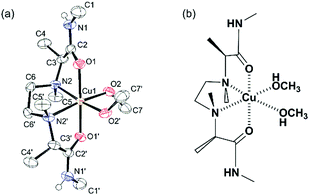
This type of ligand coordinates with a metal cation in the tetradentate fashion, and the resulting metal complex can provide two binding sites suitable for chiral recognition of a bidentate amino acid as the second ligand. In fact, the X-ray crystal analysis of the copper(II) complex with S,S-L2 showed that tetradentate S,S-L2 coordinated with the copper cation in the helical cis-α Λ form and two further methanol molecules occupied two remaining binding sites of the copper cation (Fig. 1). These methanol molecules could be replaced by a bidentate amino acid to form a three-component complex. In order to evaluate enantioselective complexation with chiral amino acid, a three-component complex ion composed of a metal cation, ligand (L) and amino acid is required to be detected with high sensitivity in ESI-MS. Firstly, metal cations providing 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 complex ions with the achiral ligand (L3) in a mass spectrum were systematically examined using the following metal salts: CrCl3, MnCl2, FeCl3, CoCl2, NiCl2, CuCl2, NiCl2 and LaCl3. In the case of MnCl2, CoCl2, NiCl2, CuCl2 and ZnCl2, 1
1 complex ions with the achiral ligand (L3) in a mass spectrum were systematically examined using the following metal salts: CrCl3, MnCl2, FeCl3, CoCl2, NiCl2, CuCl2, NiCl2 and LaCl3. In the case of MnCl2, CoCl2, NiCl2, CuCl2 and ZnCl2, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 complex ions of [M(L3) + Cl]+ were observed (Fig. S7, ESI†). In particular, CuCl2 gave a highly intense ESI mass spectrum without contaminating peaks and unassignable signals. Therefore, the copper cation was selected as the suitable metal ion. Second, different anions were screened using the copper salts CuCl2, Cu(CH3COO)2, Cu(NO3)2, Cu(acac)2 and Cu(OTf)2 in the presence of L3 and S-Ala. As a result, it was found that CuCl2 provided desirable ions, [CuII(L3)(S-Ala–H)]+ as the base peak with high intensity and with almost no contaminating ions including cluster peaks corresponding to [CuII(L3)(S-Ala–H)2] complex in the mass spectrum (Fig. S8, ESI†).
1 complex ions of [M(L3) + Cl]+ were observed (Fig. S7, ESI†). In particular, CuCl2 gave a highly intense ESI mass spectrum without contaminating peaks and unassignable signals. Therefore, the copper cation was selected as the suitable metal ion. Second, different anions were screened using the copper salts CuCl2, Cu(CH3COO)2, Cu(NO3)2, Cu(acac)2 and Cu(OTf)2 in the presence of L3 and S-Ala. As a result, it was found that CuCl2 provided desirable ions, [CuII(L3)(S-Ala–H)]+ as the base peak with high intensity and with almost no contaminating ions including cluster peaks corresponding to [CuII(L3)(S-Ala–H)2] complex in the mass spectrum (Fig. S8, ESI†).
We chose the MS/enantiomer-labelled (MS/EL) method18 to discriminate chiral compounds rapidly with high sensitivity. In this method, an equimolar mixture of deuterium-labelled S- and unlabelled R-enantiomers of chiral amino acid (AA) ([2H]n-S-AA and R-AA, n = number of deuterium atoms) were used to evaluate the chiral discrimination ability of a chiral CuII-(L) complex toward chiral amino acid on the basis of the relative peak intensity of diastereomeric three-component complex ions, [CuII(L) + ([2H]n-S-AA–H)]+ and [CuII(L) + (R-AA–H)]+, observed in mass spectra.
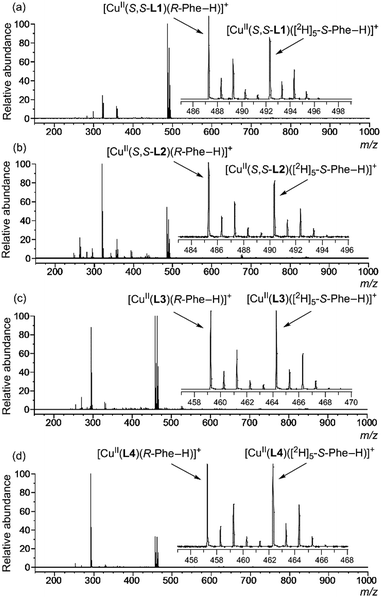
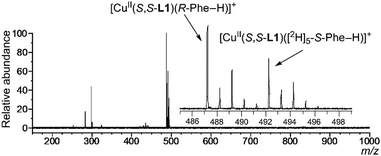
Sample solutions for the MS/EL method were prepared by mixing a methanol solution of CuCl2 and ligand (S,S-L1, S,S-L2, L3 or L4) and an aqueous solution of quasiracemic mixture of amino acid (an equimolar mixture of deuterium-labelled S-AA and unlabelled R-AA) and K2CO3. Fig. 2 shows the typical mass spectra in which phenylalanine (Phe) was used as the quasiracemic mixture of amino acid. In the case of achiral ligands L3 and L4, the relative peak intensity values of two diastereomeric complex ion peaks (I[CuII(L)(R-Phe–H)]+/I[CuII(L)([2H]5-S-Phe–H)]+ = IR/IS value) were unity due to no selectivity (Fig. 2(c) and (d)). In the case of chiral ligands S,S-L1 and S,S-L2, the IR/IS values were larger than 1.00 (Fig. 2(a) and (b)), indicating that R-Phe preferentially coordinates with the CuII-S,S-L1 and CuII-S,S-L2 complexes compared to S-Phe. Inversely, S-Phe preferentially coordinates with CuII-R,R-L2, which is the enantiomer of CuII-S,S-L2 (Fig. S9, ESI†). The multiplication of each IR/IS value (1.55 × 0.63 = 0.98) from [CuII(S,S-L2)(Phe–H)]+ and [CuII(R,R-L2)(Phe–H)]+ under the same sample concentration conditions and spectral measurement conditions was unity. It shows that differentiation of the peak intensity related to the diastereomeric complex ions is caused not by random complexation but by highly controlled stereo-specific complexation. Thus, the chiral discrimination ability of the complex ion toward amino acid corresponds to the IR/IS values.
As the IR/IS value reflects the concentration ratio of the diastereomeric three-component complex ions [CuII(S,S-L1)(R-AA–H)]+ and [CuII(S,S-L1)([2H]n-S-AA–H)]+, the value changes depending on the concentration ratio of the quasiracemic mixture of amino acid to the CuII-S,S-L1 complex (Fig. S11, ESI†). Amount of amino acid to be added was optimized to evaluate chiral discrimination ability (IR/IS value). As a result, CuCl2/L1/R-AA/[2H]n-S-AA = 1.2/1.0/2.0/2.0 (mole ratio) system ([L1]0 = 7.14 × 10−5 M) was found to be the most suitable condition. Under this condition, the highest IR/IS value for each amino acid was obtained which was stable during the MS measurement. When the mole ratio of the quasiracemic amino acid was more than 1.0 equivalents, the IR/IS value became slightly larger and the signal-to-noise ratio became smaller due to formation of several interfering chemical species such as [CuII(AA–H)2] and S,S-L1 (Fig. S10, ESI†).
The formation of [CuII(AA–H)2] was also confirmed by other spectroscopic analyses. As illustrated in Fig. S3 and S4 (ESI†), UV-vis spectral changes were induced by reacting chiral ligand (S,S-L) with CuCl2 in methanol, indicating the generation of the [CuII(S,S-L)]2+ complex. Adding a solution of equimolar S-Phe into the resulting [CuII(S,S-L1)]2+ solution in methanol clarified that the [CuII(S,S-L1)(S-Phe–H)]+ complex was produced (Fig. S5, ESI†). Further adding S-Phe changed UV-vis and CD spectra. Both titration studies, UV-vis and MS measurements (Fig. S10g, ESI†), suggest that the chiral tetradentate ligand S,S-L1 was dissociated from the [CuII(S,S-L1)(S-Phe–H)]+ complex and [CuII(S-Phe–H)2] was generated simultaneously. Its UV-vis and CD spectra also gave close agreement with that of in situ generated [CuII(S-Phe–H)2] (Fig. S6, ESI†).21
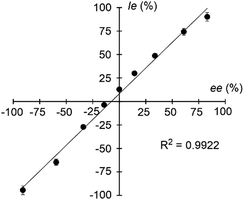
Signals of the complex ions ([CuII(S,S-L)(R-AA–H)]+ and [CuII(S,S-L)([2H]n-S-AA–H)]+) were clearly detected in all cases of the nine given amino acids, Ala, Leu, Val, Met, Orn, Lys, Phe, Hyp and Trp (Fig. 3 and Fig. S12, ESI†).22 The IR/IS values are summarized in Table 1. The IR/IS values were larger than 1.00 for most of the given amino acids, indicating R-selectivity. However, CuII-S,S-L1-Leu, CuII-S,S-L2-Ala, CuII-S,S-L2-Leu and CuII-S,S-L2-Orn gave 0.95–1.05, meaning no selectivity. Especially, the IR/IS values for Val, Phe, Hyp and Trp were larger than those for the other amino acids. These amino acids have bulky substituents at the α-position, suggesting that the main factor of chiral discrimination toward amino acids is steric repulsion between the chiral ligand and the substituents of amino acids. IR/IS values changed depending on enantiomer excess (ee) of added Phe with linear correlation (Fig. 4). Thus, the MS/EL method is suitable for evaluation of the concentration ratio, [CuII(S,S-L2)(R-Phe–H)]+/[CuII(S,S-L2)([2H]5-S-Phe–H)]+, in solution, precisely.
| Complex | AA | I R /I S | Complex | AA | I R /I S |
|---|---|---|---|---|---|
| a All errors measuring the same sample were within 5%. Intensity of overlapped isotopic peak of the lower m/z signal on the higher one was corrected based on the natural abundance of isotope. | |||||
| [Cu(S,S-L1)]2+ | Ala | 1.09 | [Cu(S,S-L2)]2+ | Ala | 1.01 |
| Leu | 1.01 | Leu | 0.97 | ||
| Val | 1.69 | Val | 1.56 | ||
| Met | 1.20 | Met | 1.07 | ||
| Orn | 1.21 | Orn | 1.02 | ||
| Lys | 1.22 | Lys | 1.16 | ||
| Phe | 1.68 | Phe | 1.62 | ||
| Hyp | 3.11 | Hyp | 1.41 | ||
| Trp | 1.86 | Trp | 1.31 | ||
As mentioned above, IR/IS values obtained under the optimized Cu–ligand–amino acid concentration conditions reflects the concentration ratio of the diastereomeric three-component complex ions [CuII(S,S-L1)(R-AA–H)]+ and [CuII(S,S-L1)([2H]n-S-AA–H)]+, meaning IR/IS ≈ KR/KS.18,23 This concludes that free energy difference between diastereomers ΔΔG(R/S) is also affected by IR/IS value. Then, we studied structures of the diastereomeric complex ions, [CuII(S,S-L2)(R-Phe–H)]+ and [CuII(S,S-L2)(S-Phe–H)]+, in methanol by DFT calculations in which four conformers for the [CuII(S,S-L2)(R-Phe–H)]+ complex and three conformers for [CuII(S,S-L2)(S-Phe–H)]+ were obtained (see each Boltzmann population in Table S1, ESI†). Total population percentage of [CuII(S,S-L2)(R-Phe–H)]+ and [CuII(S,S-L2)(S-Phe–H)]+ in solution was calculated as 62.7% and 37.3%, respectively (Table S1, ESI†); the complex ion [CuII(S,S-L2)(R-Phe–H)]+ was slightly more stable than [CuII(S,S-L2)(S-Phe–H)]+. Fig. 5 shows the most stable structure for each complex ion. This calculated population ratio of the diastereomeric complex ions (62.7/37.3 = 1.68) is in good agreement with the IR/IS value (1.62) observed in the MS/EL method.
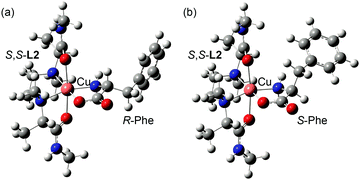 | ||
| Fig. 5 Molecular structures of the most stable conformer of (a) [CuII(S,S-L2)(R-Phe–H)]+ and (b) [CuII(S,S-L2)(S-Phe–H)]+ optimized by DFT calculation at B3LYP/LANL2DZ. | ||
As described above, we have succeeded in quantitative evaluation of the enantioselective coordination behaviours of free amino acids with chiral organometallics by mass spectrometry. Several researches have been conducted on the optical resolution of amino acids with metal complex since Gillard et al. reported in 1968.24–27 Although mass spectroscopic techniques have been widely used in determining the enantiomeric purity of chiral compounds,28–30 ESI-MS method is the most convenient and sensitive for detecting metal complex ion, which exist in the equilibrium between diastereomeric metal complexes in solution. Therefore, establishment of a new system for evaluating the enantiomeric purity of amino acids more easily and quickly based on the research results described here will enable microanalysis of free amino acid in the human body, which will be widely used in the medical field.
This work was partially supported by JSPS KAKENHI Grant Numbers JP18K0595 to M. S. and JP19K05505 to H. M.
Conflicts of interest
There are no conflicts to declare.Notes and references
- A. Sato, M. Yoshida and S. Hara, Chem. Commun., 2008, 6242–6244 RSC.
- Y. Kawato, N. Takahashi, N. Kumagai and M. Shibasaki, Org. Lett., 2010, 12, 1484–1487 CrossRef CAS PubMed.
- M. Yoshida, M. Narita and S. Hara, J. Org. Chem., 2011, 76, 8513–8517 CrossRef CAS.
- M. M. Sheeba, M. M. Tamizh, L. J. Farrugia and R. Karvembu, J. Organomet. Chem., 2017, 831, 45–49 CrossRef CAS.
- H. Miyake, K. Yoshida, H. Sugimoto and H. Tsukube, J. Am. Chem. Soc., 2004, 126, 6524–6525 CrossRef CAS.
- H. Miyake, H. Sugimoto, H. Tamaki and H. Tsukube, Chem. Commun., 2005, 4291–4293 RSC.
- H. Miyake, H. Kamon, I. Miyahara, H. Sugimoto and H. Tsukube, J. Am. Chem. Soc., 2008, 130, 792–793 CrossRef CAS PubMed.
- S. Zahn and J. W. Canary, Science, 2000, 288, 1404–1407 CrossRef CAS.
- H. Miyake, H. Hikita, M. Itazaki, H. Nakazawa, H. Sugimoto and H. Tsukube, Chem. – Eur. J., 2008, 14, 5393–5396 CrossRef CAS.
- J. Gregoliński, M. Hikita, T. Sakamoto, H. Sugimoto, H. Tsukube and H. Miyake, Inorg. Chem., 2016, 55, 633–643 CrossRef.
- R. Nomura, H. Mitsukuri, A. Ohtaka and O. Shimomura, Trans. Mater. Res. Soc. Jpn., 2008, 33, 399–402 CAS.
- S. Wang, M. Wahiduzzaman, L. Davis, A. Tissot, W. Shepard, J. Marrot, C. Martineau-Corcos, D. Hamdane, G. Maurin, S. Devautour-Vinot and C. Serre, Nat. Commun., 2018, 9, 1–8 CrossRef.
- P. Atkinson, Y. Bretonniere, D. Parker and G. Muller, Helv. Chim. Acta, 2005, 88, 391–405 CrossRef CAS.
- B. T. Nguyen, A. J. Ingram and G. Muller, Chirality, 2016, 28, 325–331 CrossRef CAS.
- H. Tsukube, Y. Noda, Y. Kataoka, H. Miyake, S. Shinoda, A. Kojima-Yuasa, Y. Nishida and I. Matsui-Yuasa, Dalton Trans., 2008, 4038–4043 RSC.
- J. Wang, H.-B. Liu, Z. Tong and C.-S. Ha, Coord. Chem. Rev., 2015, 303, 139–184 CrossRef CAS.
- A. Horeau and A. Nouaille, Tetrahedron Lett., 1990, 31, 2707–2710 CrossRef CAS.
- M. Sawada, Y. Takai, H. Yamada, S. Hirayama, T. Kaneda, T. Tanaka, K. Kamada, T. Mizooku, S. Takeuchi, K. Ueno, K. Hirose, Y. Tobe and K. Naemura, J. Am. Chem. Soc., 1995, 117, 7726–7736 CrossRef CAS.
- M. Sawada, Mass Spectrom. Rev., 1997, 16, 73–90 CrossRef CAS.
- M. Shizuma, Chiral Recognition in Mass Spectrometry, Focusing on FAB Mass Spectrometry, in Chiral Recognition in the Gas Phase, ed. A. Zehnacker, CRC Press, New York, 2010, ch. 5, pp. 61–86 Search PubMed.
- It was unable to estimate the stability constants of the complexation of [Cu(S,S-L)(CH3OH)2]2+ with S-Phe in solution, since the curve fitting of the titration plot could not be performed precisely due to the formation of the [CuII(S-Phe–H)2] complex. Therefore, enantioselectivity could not be evaluated by UV-vis and CD measurements.
- Structures of deuterium-labelled S-amino acids are shown in Table S2 (ESI†).
- K R and KS are defined as described in page S9 of ESI†.
- R. D. Gillard, P. R. Mitchell and H. L. Roberts, Nature, 1968, 217, 949–950 CrossRef CAS.
- S. Tashiro, Y. Ogura, S. Tsuboyama, K. Tsuboyama and M. Shionoya, Inorg. Chem., 2011, 50, 4–6 CrossRef CAS.
- J. Chin, S. S. Lee, K. J. Lee, S. Park and D. H. Kim, Nature, 1999, 401, 254–257 CrossRef CAS.
- A. Fujihara, N. Maeda and S. Hayakawa, Chirality, 2015, 27, 349–352 CrossRef CAS.
- W. A. Tao, D. Zhang, D. Wang, F. Thomas, P. D. Cooks and R. G. Cooks, Anal. Chem., 1999, 71, 4427–4429 CrossRef CAS.
- D. Schröder and H. Schwarz, Int. J. Mass Spectrom., 2004, 231, 139–146 CrossRef.
- Y. Takai, K. Iguchi, H. Yamada, M. Shizuma, R. Arakawa and M. Sawada, J. Mass Spectrom., 2006, 41, 266–268 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available: Experimental procedures including general methods and materials, synthesis of ligands, UV and CD spectra of complexes in solution, X-ray crystal structure analysis, DFT calculation, and mass spectra. CCDC 1912468. For ESI and crystallographic data in CIF or other electronic format see DOI: 10.1039/c9cc07231d |
| This journal is © The Royal Society of Chemistry 2020 |