Open Access Article
Open Access ArticleAmphiphilic cationic cyclodextrin nanovesicles: a versatile cue for guiding cell adhesion†
Francesco
Valle‡
 *ab,
Silvia
Tortorella‡
*ab,
Silvia
Tortorella‡
 c,
Angela
Scala
c,
Angela
Scala
 d,
Annalaura
Cordaro
de,
Marianna
Barbalinardo
a,
Fabio
Biscarini
d,
Annalaura
Cordaro
de,
Marianna
Barbalinardo
a,
Fabio
Biscarini
 fg and
Antonino
Mazzaglia
fg and
Antonino
Mazzaglia
 *e
*e
aConsiglio Nazionale delle Ricerche, Istituto per lo Studio dei Materiali Nanostrutturati (CNR-ISMN), Via P. Gobetti 101, 40129, Bologna, Italy. E-mail: francesco.valle@cnr.it
bConsorzio Interuniversitario per lo Sviluppo dei Sistemi a Grande Interfase (CSGI), Firenze, Italy
cDipartimento di Chimica Industriale “Toso Montanari”, Università di Bologna “Alma Mater Studiorum”, Via Zamboni 33, 40126, Bologna, Italy
dDipartimento di Scienze Chimiche, Biologiche, Farmaceutiche ed Ambientali, Università degli Studi di Messina, Viale F. Stagno D'Alcontres, 31, 98166, Messina, Italy
eConsiglio Nazionale delle Ricerche, Istituto per lo Studio dei Materiali Nanostrutturati (CNR-ISMN) c/o Dipartimento di Scienze Chimiche, Biologiche, Farmaceutiche ed Ambientali, Università degli Studi di Messina, Viale F. Stagno D'Alcontres, 31, 98166, Messina, Italy. E-mail: antonino.mazzaglia@cnr.it
fUniversità di Modena e Reggio Emilia, Dipartimento di Scienze della Vita, Via Campi 103, 41125 Modena, Italy
gIstituto Italiano di Tecnologia, Center for Translational Neurophysiology, Via Fossato di Mortara 17-19, 4412, Ferrara, Italy
First published on 12th November 2020
Abstract
It is well known that amphiphilic cationic β-cyclodextrins (amβCDs) form nanovesicles able to release their cargo in aqueous solution upon applying different stimuli. In addition they can be selectively positioned onto substrates by unconventional soft lithography. This makes them a powerful tool for designing environments where different cues can be externally supplied to the cells helping to achieve good control of their fate. Lithographically controlled wetting (LCW) of amβCD nanovesicles loaded with fluorescein isothiocyanate (FITC), amβCD/FITC, has been used here to fabricate geometrically functionalized surfaces, thus achieving multiscale control of the cell environment. The amβCD functionalization was strongly influenced by the surface energy of the underlying substrates that, according to their hydrophobicity, orient the amβCD in a different way, thus “offering” different portions to the cells. The structure of the pattern was characterized both over large scales exploiting the FITC fluorescence and at the nanoscale by atomic force microscopy. Cell guidance and aCD/FITC cell internalization were demonstrated in human neuroblastoma SHSY5Y cells.
Introduction
Cell behavior is influenced by the 3D environment dynamically sensed by the cell.1 This includes topographical–chemical–mechanical stimuli, electromagnetic fields, gradients of chemo-attractants and haptotaxis.2–5Multiscale control of the cell environment is crucial to many aspects of cell research and medical applications. Observed changes in the gene expression profile of primary cells in vitro, compared to that of cells in organs for example, raised serious concerns in the pharmaceutical industry, because drug-screening technology is largely based on cell culture data. Regenerative medicine requires the control of stem cell commitment at the level of individual cells and colonies6 and the correct tissue regeneration can be either hindered or promoted by means of an accurate positioning of the cells and by the local supply of molecules (differentiation agents, adhesion molecules).7 Long-term applications including diagnostics, loco-regional therapy of neurological diseases, and sorting and localization of cancer cells before metastasis occurs cannot be achieved with traditional culture plates or scaffolds that do not mimic the multiscale cell environment. The mimicking of the natural environment of the cells in an organ should achieve control of cell–cell contacts that are one of the key factors in cell behavior.8 In addition, endocrine signaling should also be taken into account and included in the biomimicking system.9,10 Such an approach requires spatial and dimensional control of the structures making up the environment along with temporal control of the cues at the surface and in the solution. Investigation of selected cues has highlighted the importance of topographical, chemical, and more recently mechanical/viscoelastic interactions of the cell and the surface.11–13
In this framework cell patterning has received in the recent past some attention both to define technological pathways for building complex environments and to study the cell behaviour under the chosen conditions. The first attempts were made by patterning adhesion promoting or antifouling molecules to obtain a good cell positioning;14,15 then the fabrication strategies tackled higher cellular processes such as differentiation by patterning topographical–chemical–mechanical stimuli.16–18
To date, molecular recognition has been fruitfully exploited for surface functionalization; in particular, the host–guest interactions between covalent or non-covalent immobilized β-cyclodextrin (βCD) derivatives and suitable groups or biochemical moieties (i.e. carbohydrate, biotin, etc.) have been reported.19,20 In this context, polymeric adhesive membranes functionalized with ferrocene and cyclodextrins (CDs) were investigated,21 and more recently, surfaces patterned with CD-based polymers have been proposed as supramolecular glues, exploiting the host–guest inclusion between CD and azobenzene polymer brushes22 or photoresponsive arylazopyrazoles.23 Liposomes and amphiphilic βCD vesicles were also selectively positioned on surfaces by microcontact printing using a coiled-coil binding motif.24 In this direction, some of us reported the electrical release of dopamine and levodopa mediated by amphiphilic cationic βCD (amβCD) immobilized on polycrystalline gold,25 while the controlled and tunable entrapment and release of cargo by polymer-shelled vesicles prepared from amphiphilic βCD by using gigahertz acoustics was proposed for addressing site-specific therapeutic activity.26
The amphiphilic βCD vesicles may in addition form various nanocomplexes in aqueous dispersions either with chemotherapeutic27 or photosensitizer drugs28–30 or a proper combination of both.31 Hence, amphiphilic βCD nanovesicles are able to act at two levels: locally changing the chemical–physical properties of the surface32 and delivering the drug hosted both in their cavities and within the bilayer to the cells.31 By tailoring the different moieties of βCD (the hydrophilic portions, the hydrophobic ones and the terminal groups that can be exposed to the solution), the orientation assumed by amphiphilic βCD nanovesicles interacting with surfaces can be suitably exploited to tune the chemical–physical properties of the surface. In addition, being dispersible in water they can be used as biocompatible building blocks for soft lithography.33
These techniques in the last decade turned out to be a useful toolbox for generating patterns able to guide the adhesion and migration of cells;34,35 different mechanical properties or topographical features have been locally patterned,36–38 as well as chemical cues such as adhesion molecules,39 small peptides or whole proteins.11,40,41
Beyond the proof-of-concepts, several important questions related to the complexity of the cell environment remain still at an infancy level: what is the effect of combined cues? Is there a synergy or a compensation between different cues? How can cell behavior on a surface be reprogrammed by a dynamic change of the environment (e.g. adhesion or detachment upon switching interfacial energy)? Are there correlations among different cues? How does the cell fate emerge from combined cues? Other important aspects concern the adhesion and the migration driven by the environmental stimuli. In this work we developed a platform easily tunable as a proof of concept with the aim of addressing some of these questions. We patterned surfaces with amβCD nanovesicles, exploiting the ability of these nanostructures to entrap dyes and release them into the biological environment.42–44 Specifically, we selected amβCD able to form nanovesicles in water,45 with high charge affinity for glass coverslips, and we loaded fluorescein isothiocyanate (FITC) as a drug model. The unmistakable green fluorescence makes FITC a useful dye in many bioanalytical processes, both for characterizing the pattern and for detecting and monitoring the cell internalization of the nanocomplexes. To the best of our knowledge, this approach can be preparatory to directly investigate cell behavior upon their spatial organization on surfaces patterned with nanodrugs by means of fluorescence microscopy.
Materials and methods
Materials
Heptakis[2-ω-amino-oligo(ethyleneglycol)-6-deoxy-6-hexadecylthio]-β-cyclodextrin heptahydrochloride salt (amβCD, SC16NH2, corresponding to the exact masses with 7–14 units of ethylene oxide (EO) and 7 units of ethylene amino; exact mass: [M11EO] = 3545.20; mol. wt = 3551.2; found: 3546.25, and nEO = 1–2 in Fig. 1; MALDI is reported in Fig. S1†) was synthesized and characterized by NMR (Varian 500 MHz) in CDCl3 and MALDI-TOF according to the general procedures.45 Fluorescein 5-isothiocyanate (FITC, MW = 389.38), MEM Non-Essential Amino Acids (NEAA), Dulbecco's phosphate buffered saline (DPBS), and Dulbecco's modified Eagle's medium/Nutrient Mixture F-12 Ham (DMEM/F-12), L-glutamine, penicillin and streptomycin were purchased from Sigma-Aldrich. Fetal bovine serum (FBS) was purchased from Gibco. The nanovescicle dispersions used for spectroscopic characterization were prepared in ultrapure microfiltered water (Galenica Senese). Dichloromethane (DCM) of spectrophotometric grade was purchased from Romil. All other reagents and solvents were of the highest available commercial grade.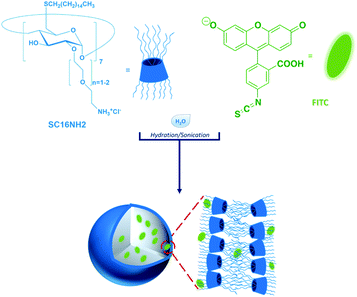 | ||
| Fig. 1 Sketch of the nanosystem used in this work. Vesicles are formed by amβCD structured in a bilayer and incorporating the FITC dye. | ||
Preparation and spectroscopic characterization of aqueous dispersions of amβCD/FITC nanovesicles
Nanovesicles of amβCD and amβCD/FITC were prepared with a 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 molar ratio of [amβCD]
1 molar ratio of [amβCD]![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) [FITC] ([amβCD] = 120 μM, [FITC] = 6 μM) according to a previously reported procedure.45 Briefly, SC16NH2 (1.1 mg) was dissolved in DCM and evaporated overnight to form a thin film which was hydrated with an FITC solution in ultrapure water ([FITC] = 6 μM) at 50 °C followed by 5 h of sonication in an ultrasonic bath. amβCD/FITC was purified from free dye by gel filtration on a 30 × 2 cm Sephadex G-25 column using H2O as an eluent. Entrapment efficiency (EE%) was estimated from the difference of fluorescence emission spectra before and after column purification, using a calibration curve by fluorescence emission of FITC ([FITC] = 1–10 μM). Spectroscopic characterization was carried out by dispersing amβCD, FITC and amβCD/FITC in ultrapure water and in physiological aqueous solution (NaCl, 0.9% w/w), respectively. The pH of amβCD/FITC, measured with a Metrohm 744 pH meter, in ultrapure water and physiological aqueous solution (NaCl, 0.9% w/w) was 5.2 and 6.30 (±0.01), respectively.
[FITC] ([amβCD] = 120 μM, [FITC] = 6 μM) according to a previously reported procedure.45 Briefly, SC16NH2 (1.1 mg) was dissolved in DCM and evaporated overnight to form a thin film which was hydrated with an FITC solution in ultrapure water ([FITC] = 6 μM) at 50 °C followed by 5 h of sonication in an ultrasonic bath. amβCD/FITC was purified from free dye by gel filtration on a 30 × 2 cm Sephadex G-25 column using H2O as an eluent. Entrapment efficiency (EE%) was estimated from the difference of fluorescence emission spectra before and after column purification, using a calibration curve by fluorescence emission of FITC ([FITC] = 1–10 μM). Spectroscopic characterization was carried out by dispersing amβCD, FITC and amβCD/FITC in ultrapure water and in physiological aqueous solution (NaCl, 0.9% w/w), respectively. The pH of amβCD/FITC, measured with a Metrohm 744 pH meter, in ultrapure water and physiological aqueous solution (NaCl, 0.9% w/w) was 5.2 and 6.30 (±0.01), respectively.
Steady-state fluorescence and excitation measurements were performed on a Jasco model FP-750 spectrofluorimeter by using a 1 cm path length quartz cell at room temperature (r.t. ≅ 25 °C). Spectra were not corrected by extinction. Dynamic Light Scattering (DLS) was carried out using a Zetasizer Nano ZS (Malvern Instrument, Malvern, U.K.) utilizing a Non-Invasive Back-Scattering (NIBS) technique. The measurements were performed at a 173° angle with respect to the incident beam at 25 ± 1 °C for each dispersion. The deconvolution of the measured correlation curve to an intensity size distribution was achieved by using a non-negative least-squares algorithm. The ζ-potential values were determined using a Zetasizer Nano ZS from Malvern Instruments equipped with a He–Ne laser at power P = 4.0 mW and λ = 633 nm.
Atomic force microscopy imaging was performed in the SPM@ISMN facility using a Multimode VIII microscope equipped with a Nanoscope V controller (Bruker, Santa Barbara, CA). Images were collected with a microscope operated in PeakForce Tapping mode using an SNL10 cantilever with a nominal spring constant of 0.24 N m−1. Images were analyzed with Gwyddion46 open source software used in this work for background subtraction and profile analysis.
Patterning of nanovesicles
For patterning experiments, amβCD was stored at 4 °C as a thin film in glass vials. amβCD film was reconstituted in physiological solution (NaCl 0.9% w/w) by sonication at 50 °C for 5 h (final concentration 120 μM). amβCD/FITC was prepared by hydration of amβCD organic film with FITC solution (NaCl 0.9% w/w) as described above.amβCD and amβCD/FITC nanovesicles were patterned on the surface of glass coverslips (Corning, Sigma-Aldrich) by Lithographically Controlled Wetting (LCW) as previously described.34 Electron microscope grids (Pelco, Ted Pella) were used as patterning stamps due to the large variability both in the motifs and in the size of the features available. Coverslips (diameter d = 0.8 cm) were cleaned by 1 h dipping in 37% HCl, sonication for 15 min in acetone, 15 min in isopropanol, and 15 min in ethanol and drying in a stream of nitrogen; sterilization was performed both by UV lamp treatment overnight and in an autoclave. A portion of the substrates was then additionally exposed for 1 minute to an O2 plasma at 60 W. The patterns were then coated with 10 nm gold and then imaged by Scanning Electron Microscopy using an FEG-SEM Hitachi S4000.
Cell culture and morphology
Human neuroblastoma SHSY5Y cells (ECACC 94030304; Sigma Aldrich, St. Louis, MO, USA) were cultured in Dulbecco's modified Eagle's medium/Nutrient Mixture F-12 Ham (DMEM F-12) with 2 mM L-glutamine, 100 IU mL−1 penicillin, 100 μg mL−1 streptomycin and supplemented 15% fetal bovine serum (FBS). Cells were maintained under standard conditions at 37 °C in a humidified atmosphere (95% humidity, 5% CO2) and split regularly (rate 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10) by trypsinization. SHSY5Y cells were seeded (7500 cells per cm2) and cultured on glass samples in 24-well plates, for 48 hours. For the actin and nucleus staining, cells were fixed with 4% paraformaldehyde in Dulbecco's phosphate-buffered saline DPBS 1× and washed with DPBS 1×. They were then permeabilized with 0.01% Triton-X 100. The cells were labelled with TRITC-conjugated phalloidin to map the local orientation of actin filaments followed by rinsing with DPBS 1×. Nuclear counterstaining was performed by incubation with DAPI for 3 min, followed by rinsing with DPBS 1×. Samples were examined using a Nikon Eclipse 80i microscope equipped for fluorescence analysis.
10) by trypsinization. SHSY5Y cells were seeded (7500 cells per cm2) and cultured on glass samples in 24-well plates, for 48 hours. For the actin and nucleus staining, cells were fixed with 4% paraformaldehyde in Dulbecco's phosphate-buffered saline DPBS 1× and washed with DPBS 1×. They were then permeabilized with 0.01% Triton-X 100. The cells were labelled with TRITC-conjugated phalloidin to map the local orientation of actin filaments followed by rinsing with DPBS 1×. Nuclear counterstaining was performed by incubation with DAPI for 3 min, followed by rinsing with DPBS 1×. Samples were examined using a Nikon Eclipse 80i microscope equipped for fluorescence analysis.
Statistical analysis
All data represented the mean ± standard deviation (SD) of at least three independent culture experiments; a blank control was always included in every multi-well plate. Statistical significance was determined using a one-way analysis of variance with Tukey's test for multiple comparisons using Origin 8 software (OriginLab Corporation). Differences were considered significant when p < 0.005 and p < 0.001. Analysis of cell adhesion was performed using ImageJ software (NIH); AFM image elaboration was performed using open source software Gwyddion (Department of Nanometrology, Czech Metrology Institute).Results and discussion
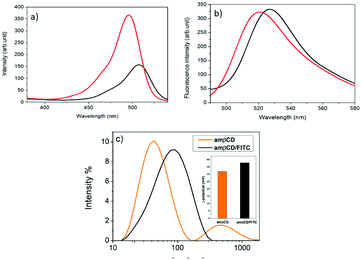
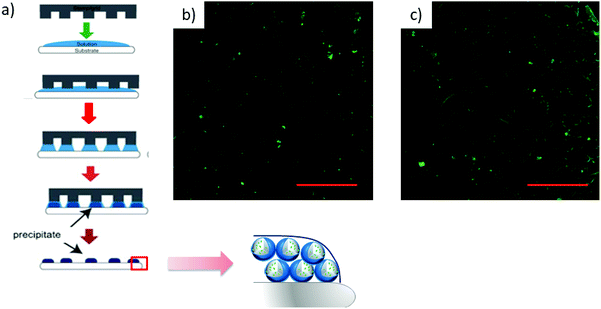
amβCD/FITC nanovesicles (Fig. 1) were prepared by a solvent-evaporation method, hydrating an organic film of amβCD carrier with an aqueous solution of the guest (FITC), followed by sonication and purification by gel permeation chromatography. The coincident fluorescence emission spectra of amβCD/FITC before and after purification (data not shown) suggested that FITC EE% was ≅ 100%. Initially amβCD/FITC nanocomplexes were characterized in an aqueous dispersion using steady-state fluorescence spectra (Fig. 2). The excitation spectrum of free FITC showed a band centered at 496 nm, which was shifted to a longer wavelength (λ = 507 nm) upon interaction with amβCD nanovesicles. The decrease of intensity and the significant red-shift clearly indicated the formation of amβCD/FITC complexes. Emission spectra of the free dye exhibited a band centered at 521 nm which was shifted to 527 nm in amβCD/FITC confirming the FITC entrapment within amβCD nanovesicles. However, the fluorescence emission intensity was virtually not changed, suggesting that FITC is plausibly complexed in amβCD in a not self-aggregated form according to other previous reports on dyes entrapped in other cationic amphiphilic βCD47 or nanoparticles.48 Moreover, fluorescence emission spectra recorded in the presence of Triton X (Fig. S2†) indicated that FITC was probably entrapped more in the lumen of the amβCD nanovesicles than in the layer. Indeed, upon addition of Triton X to the amβCD/FITC aqueous dispersion, the FITC fluorescence emission increased, without affecting the wavelength of the emission maximum. This could be tentatively ascribed to the leaking of FITC from the vesicle aqueous core and its dispersion in aqueous solution as free molecules, following the disintegration of nanovesicles mediated by Triton X.42 DLS (Fig. 2 and Table S1†) revealed the presence of small and large amβCD aggregates (∼60 and 600 nm sized) with a positive ζ-potential (32 ± 3 mV). A similar value (ζ = 38 ± 3 mV) was confirmed in amβCD/FITC which has a size of ∼100 nm, pointing to a moderate increase of the size of the smaller aggregates while the larger sized population nearly totally disappeared (see Table S1†). All these results suggested a certain rearrangement of amβCD nanovesicles upon FITC entrapment, even if their external charge surface seemed to be almost unaffected.amβCD nanovesicles were patterned using lithographically controlled wetting (LCW)49 (Fig. 3) based on the controlled formation of the menisci of a solution pinned between the surface to be functionalized and the protrusions of the stamp. amβCD vesicles were deposited where the menisci formed, thus reproducing the positive pattern made by the protrusions of the stamp on the surface (Fig. 3a). Specifically, using both metallic linear and hexagonal copper grids, a glass surface can be functionalized with the same pattern over a macroscopic scale (Fig. 3b and c).
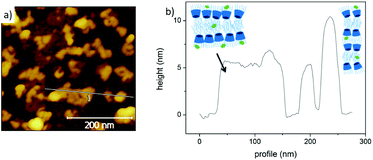
Atomic force microscopy (AFM) (Fig. 4) was used to characterize at high resolution the amβCD/FITC nanovesicles patterned on the surfaces (see Fig. 3). Fig. 4 shows the shape assumed by the nanovesicles adsorbed on the glass during LCW; the image and the section analysis confirm that under these conditions they mostly do not preserve their spherical shape50 but they open and likely release the liquid entrapped in their lumen, thus forming either a bilayer or double bilayer. Scanning Electron Microscopy (SEM) was also used to provide information about the fabricated amβCD/FITC nanovesicle pattern, visible at a large scale in Fig. S3a† and more in detail in Fig. S3b.†51
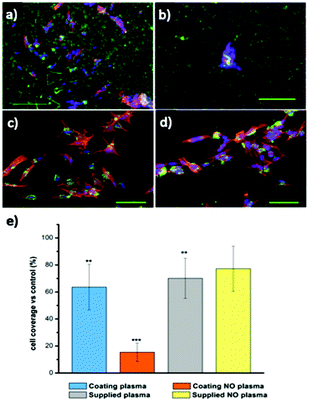
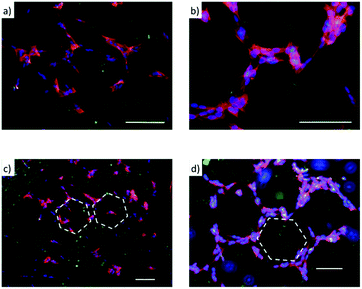
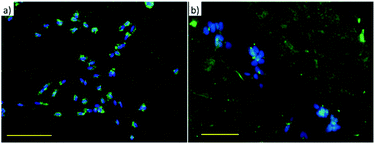
The amβCDs used here to entrap FITC are cationic amphiphilic building blocks; thus the functionalization of the glass substrates by amβCD/FITC nanovesicles is locally influenced by surface chemistry of the glass substrate that drives their orientation. To test whether this effect might affect cell adhesion and growth, glass coverslips were functionalised, without any lateral confinement, by drop casting the amβCD/FITC aqueous dispersion onto their surfaces. Then, amβCD/FITC was adsorbed both on a very hydrophilic grass surface obtained upon O2 plasma treatment (Fig. S4a†) and on a more hydrophobic one consisting of the glass simply cleaned with ethanol/acetone (Fig. S4b†). A rather homogeneous coating was thus obtained with different amβCD/FITC portions exposed to the solvent. The amβCD/FITC coating yields a good cell adhesion and proliferation (Fig. 5a) when it was formed on the very hydrophilic glass coverslip; conversely the cells did not adhere and proliferate when the coating was done on the bare glass (Fig. 5b). To exclude the possibility of ascribing this difference to the direct interaction of the cells with the underlying surface of glass treated in different ways, cells were seeded directly onto the hydrophilic and hydrophobic coverslips (without any amβCD/FITC coating) and no differences were visible as shown in Fig. 5c and d. It can thus be hypothesized that the amβCD/FITC when adsorbed on a more hydrophobic surface exposes cues able to interfere with the formation of the focal adhesion points. This is probably due to the orientation assumed that exposes the oligoethylene-glycol (OEG) portions to the solvent. OEG is in fact known to be an effective antifouling agent preventing the adsorption of biomolecules and the adhesion of cells.52 In addition, the samples were also supplied with an amβCD/FITC aqueous dispersion, revealing the uptake of amβCD/FITC by the adhered cells. This indicates that the effect shown in Fig. 5a and b was due to the way the glass shaped the amβCD/FITC layer. The reported results are quantitatively summed up in Fig. 5e where it can be seen that only the amβCD/FITC coated hydrophobic glass inhibits cell adhesion. Combining the patterning of the nanocarriers with these different surfaces, one can envision fabricating surfaces able to confine or guide the adhesion and migration of cells. This can be achieved by designing surfaces where regions allowing the cell adhesion (as in Fig. 5a, c and d) are flanked by antifouling ones (as in Fig. 5b). Soft lithography (as sketched in Fig. 3) was thus used for patterning amβCD/FITC, with the aim to locally guide cell adhesion and proliferation. Fig. 6 shows how SH-SY5Y cells respond to these surfaces; where the pattern was fabricated on plasma treated glass (Fig. 6a), the cells grew throughout the surface, and no discrimination between the presence or absence of the amβCD/FITC on the surfaces was evident due to the similar cell adhesion propensity shown in Fig. 5e.
On the other hand, in Fig. 6b the presence of two types of areas with markedly different cell adhesion propensities leads SH-SY5Y to be confined to the portions of the surface where the amβCD/FITC is not present, due to the tendency of the cells to move away from the regions where the adhesion is depleted by the surface chemistry. In this case the presence of neighboring areas where the amβCD/FITC is absent or present under the rather anti-fouling conditions (amβCD adsorbed on raw glass) led to a strong confinement of the cells. This is in our view a valuable result because it adds to the presently explored cell patterning experiments the possibility to achieve the guidance with molecular assemblies able to act also as a delivery system. This can be appreciated in Fig. 7 that describes the colocalization of the vesicles and the cells. As reported in Fig. 4, it seems that vesicles held a double bilayer structure and just leaked the internal fluid when patterned. It is likely that these vesicles upon rehydration in the cell culture media swell again and can reach the cells. A similar effect seems more pronounced when amβCD/FITC is supplied from the aqueous dispersion to the culture media (Fig. 7a). On the other hand, the patterned amβCD/FITC is only mildly internalized by the cells (Fig. 7b); this effect is probably due to the release in the solution from the surface of the nanovesicles that maintained their structure within the pattern in the proximity of the cells.
The delivery from the pattern can be seen as an additional cue that can be given to the cell besides the topographic one (control of the adhesion); cells are thus effectively guided and positioned in a precise location where they could receive a chemical/biological stimulus for further uptake and transport of the nanocarrier, indicating a net increase of the complexity in the artificial environment provided to the cells.
Conclusions
The possibility to fabricate smart surfaces able to guide cell adhesion and migration combining amphiphilic cationic βCD nanocarriers and soft lithography techniques was investigated. Concerning the advantage brought by our approach with respect to the questions listed above, we would like to underline that here we fabricated a platform as a proof-of-concept demonstrating the combination of two different cues: the surface treatment, leading to an external control of the cell positioning and the patterning with drug delivery systems based on amphiphilic CD which can act within the cellular machinery. Lithographically controlled wetting allowed us to pattern amβCD/FITC nanovesicles within defined portions of a surface and to orient them according to the surface features. Also, the cell internalization of the drug delivered by nanocarriers present on the surface was shown. In particular a more hydrophilic surface (obtained by O2 plasma treatment) combined with the amβCD pattern permitted the adhesion of SH-SY5Y cells while a more hydrophobic surface (obtained without O2 plasma treatment) inhibited the cell adhesion where the surface is rich in amβCD. From this perspective, the spatial distribution of amβCD nanovesicles on the treated surface could be exploited to confine or to guide cells to different areas. As amβCD nanovesicles can be tailored with further cues (i.e. drugs, cell targeting groups, and stimuli-responsive functionalities), according to the surface arrangement of amphiphilic cationic βCD, spatial control of the involved cues in cellular pathways could be achieved. Altogether, we believe that this strategy could be useful for building microenvironments where a variety of stimuli can be supplied also dynamically to the cells as cues exposed by the patterned surfaces and/or molecules released and internalized by the cells according to their position on the surface.Author contributions
A. M. suggested this work. F. V., F. B. and A. M. conceived the experiments. F. V. and S. T. followed the patterning experiments and performed the imaging. S. T. and M. B. performed the cell culture experiments. S. T. performed the fabrication experiments. A. M., A. S. and A. C. performed the synthesis and the characterization of the nanocarriers. F. V. and A. M. prepared the original draft and F. B. reviewed it.Conflicts of interest
The authors declare no conflicts of interest.Acknowledgements
We are greatful to Dr Luisa Sturiale (IPCB-CNR, Catania) who recorded the Maldi spectrum. The authors thank the SPM@ISMN facility for the support in image collection and analysis. This work was supported by EU projects ESF EUROCORES (10-EuroBioSAS-FP-009), Intelligent Cell Surfaces (ICS), Italian flagship NANOMAX project N-CHEM and PON and PON03PE_00216_1 Drug Delivery. FV was partly supported by the Horizon 2020 Framework Programme under the grant FETOPEN-801367 evFOUNDRY.Notes and references
- H. Honda, M. Tanemura and T. Nagai, J. Theor. Biol., 2004, 226, 439–453 CrossRef.
- D. Falconnet, G. Csucs, H. M. Grandin and M. Textor, Biomaterials, 2006, 27, 3044–3063 CrossRef CAS.
- F. Valle, M. Sandal and B. Samorì, Phys. Life Rev., 2007, 4, 157–188 CrossRef.
- G. Cellot, E. Cilia, S. Cipollone, V. Rancic, A. Sucapane, S. Giordani, L. Gambazzi, H. Markram, M. Grandolfo, D. Scaini, F. Gelain, L. Casalis, M. Prato, M. Giugliano and L. Ballerini, Nat. Nanotechnol., 2009, 4, 126–133 CrossRef CAS.
- I. Tonazzini, E. Bystrenova, B. Chelli, P. Greco, P. Stoliar, A. Calò, A. Lazar, F. Borgatti, P. D'Angelo, C. Martini and F. Biscarini, Biophys. J., 2010, 98, 2804–2812 CrossRef CAS.
- N. S. Hwang, S. Varghese, H. J. Lee, Z. Zhang, Z. Ye, J. Bae, L. Cheng and J. Elisseeff, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 20641 CrossRef CAS.
- M. Barbalinardo, M. Di Giosia, I. Polishchuk, G. Magnabosco, S. Fermani, F. Biscarini, M. Calvaresi, F. Zerbetto, G. Pellegrini, G. Falini, B. Pokroy and F. Valle, J. Mater. Chem. B, 2019, 7, 5808–5813 RSC.
- T. Reya, S. J. Morrison, M. F. Clarke and I. L. Weissman, Nature, 2001, 414, 105–111 CrossRef CAS.
- A. Apelqvist, H. Li, L. Sommer, P. Beatus, D. J. Anderson, T. Honjo, M. Hrabe de Angelis, U. Lendahl and H. Edlund, Nature, 1999, 400, 877–881 CrossRef CAS.
- C. Stoecklin, Z. Yue, W. W. Chen, R. de Mets, E. Fong, V. Studer and V. Viasnoff, Adv. Biosyst., 2018, 2, 1700237 CrossRef.
- B. Chelli, M. Barbalinardo, F. Valle, P. Greco, E. Bystrenova, M. Bianchi and F. Biscarini, Interface Focus, 2014, 4, 20130041 CrossRef.
- D. E. Discher, P. Janmey and Y. L. Wang, Science, 2005, 310, 1139–1143 CrossRef CAS.
- A. Tampieri, M. Sandri, E. Landi, D. Pressato, S. Francioli, R. Quarto and I. Martin, Biomaterials, 2008, 29, 3539–3546 CrossRef CAS.
- L. Lauer, C. Klein and A. Offenhäusser, Biomaterials, 2001, 22, 1925–1932 CrossRef CAS.
- N. Sgarbi, D. Pisignano, F. Di Benedetto, G. Gigli, R. Cingolani and R. Rinaldi, Biomaterials, 2004, 25, 1349–1353 CrossRef CAS.
- Y. Liu, L. Zhang, J. Wei, S. Yan, J. Yu and X. Li, J. Mater. Chem. B, 2014, 2, 3029–3040 RSC.
- D. Mohammed, G. Pardon, M. Versaevel, C. Bruyère, L. Alaimo, M. Luciano, E. Vercruysse, B. L. Pruitt and S. Gabriele, Cell. Mol. Bioeng., 2020, 13, 87–98 CrossRef CAS.
- M. Ventre, F. Valle, M. Bianchi, F. Biscarini and P. A. Netti, Langmuir, 2012, 28, 714–721 CrossRef CAS.
- A. González-Campo, S.-H. Hsu, L. Puig, J. Huskens, D. N. Reinhoudt and A. H. Velders, J. Am. Chem. Soc., 2010, 132, 11434–11436 CrossRef.
- S. Lamping, C. Buten and B. J. Ravoo, Acc. Chem. Res., 2019, 52, 1336–1346 CAS.
- J. Guo, C. Yuan, M. Guo, L. Wang and F. Yan, Chem. Sci., 2014, 5, 3261–3266 RSC.
- O. Roling, L. Stricker, J. Voskuhl, S. Lamping and B. J. Ravoo, Chem. Commun., 2016, 52, 1964–1966 RSC.
- S. Lamping, L. Stricker and B. J. Ravoo, Polym. Chem., 2019, 10, 683–690 RSC.
- J. Voskuhl, C. Wendeln, F. Versluis, E.-C. Fritz, O. Roling, H. Zope, C. Schulz, S. Rinnen, H. F. Arlinghaus, B. J. Ravoo and A. Kros, Angew. Chem., Int. Ed. Engl., 2012, 51, 12616–12620 CrossRef CAS.
- G. Foschi, F. Leonardi, A. Scala, F. Biscarini, A. Kovtun, A. Liscio, A. Mazzaglia and S. Casalini, Nanoscale, 2015, 7, 20025–20032 RSC.
- Y. Lu, W. C. de Vries, N. J. Overeem, X. Duan, H. Zhang, H. Zhang, W. Pang, B. J. Ravoo and J. Huskens, Angew. Chem., Int. Ed. Engl., 2019, 58, 159–163 CrossRef CAS.
- F. Quaglia, L. Ostacolo, A. Mazzaglia, V. Villari, D. Zaccaria and M. T. Sciortino, Biomaterials, 2009, 30, 374–382 CrossRef CAS.
- C. Conte, A. Scala, G. Siracusano, N. Leone, S. Patanè, F. Ungaro, A. Miro, M. T. Sciortino, F. Quaglia and A. Mazzaglia, RSC Adv., 2014, 4, 43903–43911 RSC.
- A. Galstyan, U. Kauscher, D. Block, B. J. Ravoo and C. A. Strassert, ACS Appl. Mater. Interfaces, 2016, 8, 12631–12637 CrossRef CAS.
- J. Voskuhl, U. Kauscher, M. Gruener, H. Frisch, B. Wibbeling, C. A. Strassert and B. J. Ravoo, Soft Matter, 2013, 9, 2453–2457 RSC.
- C. Conte, A. Scala, G. Siracusano, G. Sortino, R. Pennisi, A. Piperno, A. Miro, F. Ungaro, M. T. Sciortino, F. Quaglia and A. Mazzaglia, Colloids Surf., B, 2016, 146, 590–597 CrossRef CAS.
- H. R. Zope, F. Versluis, A. Ordas, J. Voskuhl, H. P. Spaink and A. Kros, Angew. Chem., Int. Ed. Engl., 2013, 52, 14247–14251 CrossRef CAS.
- M. Cavallini, A. Calò, P. Stoliar, J. C. Kengne, S. Martins, F. C. Matacotta, F. Quist, G. Gbabode, N. Dumont, Y. H. Geerts and F. Biscarini, Adv. Mater., 2009, 21, 4688–4691 CrossRef CAS.
- F. Valle, B. Chelli, M. Bianchi, P. Greco, E. Bystrenova, I. Tonazzini and F. Biscarini, Adv. Eng. Mater., 2010, 12, B185–B191 CrossRef.
- H. M. Osorio, S. Martín, M. C. López, S. Marqués-González, S. J. Higgins, R. J. Nichols, P. J. Low and P. Cea, Beilstein J. Nanotechnol., 2015, 6, 1145–1157 CrossRef CAS.
- D. E. Discher, D. J. Mooney and P. W. Zandstra, Science, 2009, 324, 1673 CrossRef CAS.
- F. Biscarini, M. Bianchi, B. Chelli, F. Valle, C. Dionigi, E. Bystrenova and P. Greco, Adv. Eng. Mater., 2012, 14, B208–B215 CrossRef.
- K. A. Mosiewicz, L. Kolb, A. J. van der Vlies and M. P. Lutolf, Biomater. Sci., 2014, 2, 1640–1651 RSC.
- M. Barbalinardo, D. Gentili, F. Lazzarotto, F. Valle, M. Brucale, M. Melucci, L. Favaretto, M. Zambianchi, A. I. Borrachero-Conejo, E. Saracino, V. Benfenati, D. Natalini, P. Greco, M. G. Di Carlo, G. Foschi and M. Cavallini, Small Methods, 2018, 2, 1700377 CrossRef.
- D. A. Heller, V. Garga, K. J. Kelleher, T.-C. Lee, S. Mahbubani, L. A. Sigworth, T. R. Lee and M. A. Rea, Biomaterials, 2005, 26, 883–889 CrossRef CAS.
- E. Ostuni, R. Kane, C. S. Chen, D. E. Ingber and G. M. Whitesides, Langmuir, 2000, 16, 7811–7819 CrossRef CAS.
- B. J. Ravoo and R. Darcy, Angew. Chem., Int. Ed. Engl., 2000, 39, 4324–4326 CrossRef CAS.
- S. K. Nalluri and B. J. Ravoo, Angew. Chem., Int. Ed. Engl., 2010, 49, 5371–5374 CrossRef CAS.
- A. Mazzaglia, A. Valerio, V. Villari, A. Rencurosi, L. Lay, S. Spadaro, L. Monsù Scolaro and N. Micali, New J. Chem., 2006, 30, 1662–1668 RSC.
- R. Donohue, A. Mazzaglia, B. J. Ravoo and R. Darcy, Chem. Commun., 2002, 2864–2865, 10.1039/b207238f.
- D. Nečas and P. Klapetek, Cent. Eur. J. Phys., 2012, 10, 181–188 Search PubMed.
- A. Mazzaglia, N. Angelini, D. Lombardo, N. Micali, S. Patané, V. Villari and L. M. Scolaro, J. Phys. Chem. B, 2005, 109, 7258–7265 CrossRef CAS.
- A. Imhof, M. Megens, J. J. Engelberts, D. T. N. de Lang, R. Sprik and W. L. Vos, J. Phys. Chem. B, 1999, 103, 1408–1415 CrossRef CAS.
- M. Cavallini, D. Gentili, P. Greco, F. Valle and F. Biscarini, Nat. Protoc., 2012, 7, 1668–1676 CrossRef CAS.
- A. Ridolfi, M. Brucale, C. Montis, L. Caselli, L. Paolini, A. Borup, A. T. Boysen, F. Loria, M. J. C. van Herwijnen, M. Kleinjan, P. Nejsum, N. Zarovni, M. H. M. Wauben, D. Berti, P. Bergese and F. Valle, bioRxiv, 2019, 854539, DOI:10.1101/854539.
- C. Montis, S. Busatto, F. Valle, A. Zendrini, A. Salvatore, Y. Gerelli, D. Berti and P. Bergese, Adv. Biosyst., 2018, 2, 1700200 CrossRef.
- M. Castronovo, S. Radovic, C. Grunwald, L. Casalis, M. Morgante and G. Scoles, Nano Lett., 2008, 8, 4140–4145 CrossRef CAS.
Footnotes |
| † Electronic supplementary information (ESI) available: Fig. S1: MALDI-TOF analysis of cationic aCD (SC16NH2); Fig. S2: fluorescence emission spectra of FITC and FITC/aCD; Fig. S3: SEM images of the patterned aCD/FITC; Fig. S4: contact angle of the glass coverslips prior to and after exposure for one minute to O2 plasma. See DOI: 10.1039/d0na00623h |
| ‡ ST and FV contributed equally. |
| This journal is © The Royal Society of Chemistry 2020 |