Open Access Article
Open Access ArticleSpatiotemporal monitoring of intracellular metabolic dynamics by resonance Raman microscopy with isotope labeling†
Yusuke Yonamine *a,
Kotaro Hiramatsu
*a,
Kotaro Hiramatsu bcd,
Takuro Ideguchi
bcd,
Takuro Ideguchi cde,
Takuro Ito
cde,
Takuro Ito f,
Tomomi Fujiwarag,
Yoshiko Miura
f,
Tomomi Fujiwarag,
Yoshiko Miura g,
Keisuke Goda
g,
Keisuke Goda bfhi and
Yu Hoshino
bfhi and
Yu Hoshino *g
*g
aResearch Institute for Electronic Science, Hokkaido University, Kita 21, Nishi 10, Kita-ku, Sapporo 001-0021, Japan. E-mail: yonamine@poly.es.hokudai.ac.jp
bDepartment of Chemistry, The University of Tokyo, 7-3-1 Hongo, Bunkyo-ku, Tokyo 113-0033, Japan
cResearch Centre for Spectrochemistry, The University of Tokyo, 7-3-1 Hongo, Bunkyo-ku, Tokyo 113-0033, Japan
dPRESTO, Japan Science and Technology Agency, 7-3-1 Hongo, Bunkyo-ku, Tokyo 113-0033, Japan
eInstitute for Photon Science and Technology, The University of Tokyo, 7-3-1 Hongo, Bunkyo-ku, Tokyo 113-0033, Japan
fJapan Science and Technology Agency, 4-1-8 Honcho, Kawaguchi-shi, Saitama 332-0012, Japan
gDepartment of Chemical Engineering, Kyushu University, 744 Motooka, Fukuoka 819-0395, Japan. E-mail: yhoshino@chem-eng.kyushu-u.ac.jp
hInstitute of Technological Sciences, Wuhan University, Hubei 430072, China
iDepartment of Bioengineering, University of California, Los Angeles, California 90095, USA
First published on 28th April 2020
Abstract
Cellular metabolites are valuable in a diverse range of applications. For example, the unicellular green alga Haematococcus lacustris produces as a secondary metabolite the carotenoid pigment astaxanthin (AXT), which is widely used in nutraceutical, cosmetic, and food industries due to its strong antioxidant activity. In order to enhance the productivity of H. lacustris, spatial and temporal understanding of its metabolic dynamics is essential. Here we show spatiotemporal monitoring of AXT production in H. lacustris cells by resonance Raman microscopy combined with stable isotope labeling. Specifically, we incorporated carbon dioxide (13CO2) labeled with a stable isotope (13C) into H. lacustris cells through carbon fixation and traced its conversion to 13C-AXT using our resonance Raman microscope. We incubated H. lacustris cells under various conditions by switching, pulsing, and replacing 13CO2 and 12CO2. By measurement of these cells we determined the fixation time of 13C-carbon, visualized the intracellular localization of 13C- and 12C-AXTs, and revealed the dynamic consumption–production equilibrium of the accumulated AXT. This work is a valuable step in the development of effective screening criteria for high AXT-producing H. lacustris cells.
Introduction
The observation and analysis of intracellular dynamics is essential to the study of cell biology.1–6 For example, organelles undergo dynamic morphological changes during autophagy1 or cell death2 by apoptosis or necrosis. Microtubules3 and nucleolus4 form a wide range of spatial configurations in cell-cycle transitions including cell division, differentiation, and morphogenesis. Mitochondria mount metabolic defenses against intracellular pathogens in response to infection.5,6 Beyond basic science, cellular metabolites produced by animal cells and microbes are widely used in the pharmaceutical, nutraceutical, cosmetic, and food industries,7–10 and increasing metabolite production efficiency by cell engineering remains a major goal. For instance, antibodies produced by Chinese Hamster Ovary (CHO) cells are used as therapeutic agents,7 and secondary metabolites produced by microalgae cells are used as biofuels8,9 and nutritional supplements.10 Metabolic fluxes in a living cell are highly regulated and varied in response to environmental changes, bioactive substances, and transitions of the cell cycle. Accordingly, the spatiotemporal observation of intracellular metabolic dynamics, including production, accumulation, and transportation, is foundational to the understanding of cellular metabolism. It is also effective for developing screening criteria for applications such as those above as well as for regenerative medicine and cell therapy.11–13Fluorescence microscopy, with the help of fluorescent probes,14,15 is widely used to identify the intracellular localization and distribution of metabolites with subcellular resolution. However, such fluorescent labeling inevitably comes with several fundamental drawbacks when observing intracellular metabolic dynamics. First, the hydrophobicity of small-molecule fluorescent probes16 and the bulkiness of fluorescent protein labels17,18 often disturb natural biological functions and lead to cellular toxicity. Second, intracellular fluorescent staining with fluorescent dyes suffers from non-specific binding and misdistribution, leading to potential errors in quantitative live imaging.19,20 Third, sequential labelling such as pulse-chase analysis using protein tags21 is not applicable to the study of small metabolites. For these reasons, fluorescence microscopy is not always suitable for monitoring intracellular metabolic dynamics.
On the other hand, Raman microscopy together with stable isotope (SI) labeling22–37 provides significant advantages in monitoring intracellular metabolite dynamics. First, since SI probes can be incorporated and metabolized in cells similarly to the original substrates, their use does not lead to cellular toxicity, interference with biological functions, or non-specific binding. Second, the metabolic process from incorporation to production can be tracked by exposing cells to an SI-labeled substrate and detecting SI-labeled products. Third, by analysis of the peaks of Raman spectra, Raman mapping quantifies the ratio of SI-labeled products to unlabeled products at spatial positions of interest. Though Raman microscopy with SI-labeled substrates has revealed the cellular anabolic processes of biopolymers including proteins,24–31 DNAs,26–28 and lipid droplets,32–35 the spatiotemporal dynamics of secondary metabolites has not been monitored to date despite the usefulness of these metabolites in a wide range of applications.38–41
In this paper, we show spatiotemporal monitoring of intracellular metabolic dynamics, for screening the production of secondary metabolites, using resonance Raman microscopy with an SI-labeled substrate exposed to various conditions (Fig. 1). Specifically, we investigated the biosynthesis process of a commercially valuable carotenoid, astaxanthin (AXT), produced by the unicellular photosynthetic alga Haematococcus lacustris,42 a species known to accumulate AXT under induced stress, usually accompanied by a drastic morphological transformation from green motile cells to red non-motile cyst cells.43 While fluorescence from AXT is weak,44 resonance Raman microscopy can provide an enhanced Raman signal from AXT45–48 and can be used to selectively identify AXT in single H. lacustris cells from their primary cellular constituents including proteins, nucleotides, and lipids. We incorporated carbon dioxide (13CO2) labeled by an SI (13C) into H. lacustris cells through carbon fixation and traced its conversion to 13C-AXT by resonance Raman microscopy with the SI. We incubated H. lacustris cells under various conditions by switching, pulsing, and replacing 13CO2 and 12CO2. By measurement of these cells we determined the fixation time of 13C-carbon, visualized the intracellular localization of 13C- and 12C-AXTs, and revealed the dynamic consumption–production equilibrium of the accumulated AXT. We believe this work represents a valuable step in the development of effective screening criteria for high AXT-producing H. lacustris cells.
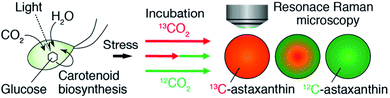 | ||
| Fig. 1 Schematic of our approach. We monitored the production of AXT in a single H. lacustris cell by resonance Raman microscopy with 13CO2 and 12CO2 incubation. | ||
Results and discussion
Resonance Raman imaging of H. lacustris cells accumulating 13C-AXT
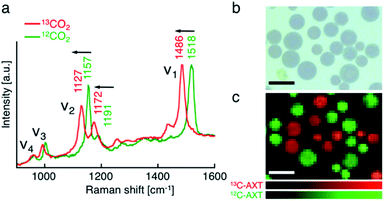
First, we performed resonance Raman imaging of H. lacustris cells incubated with 13CO2 long enough for 13CO2 to convert to 13C-AXT. We induced intracellular production of AXT for 5 days under nitrogen deficiency and strong light irradiation and measured the resonance Raman spectra of the cells. We obtained typical resonance Raman spectra of carotenoids including ν1–ν4 peaks from the resultant red cyst cells (Fig. 2a, red).43 The ν1–ν4 peaks are assigned to vibrational modes as follows: ν1, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C stretching; ν2, C–C stretching/C–H in-plane bending; ν3, CH3 rocking; ν4, C–H out-of-plane wagging.45,49 Spectra consist mainly of peaks corresponding to AXT, as it is the major ingredient (>90%) of carotenoid production by H. lacustris cells.50 The AXT spectra of cells incubated with 13CO2 are shifted to lower wavenumbers compared to those of cells incubated with 12CO2 (Fig. 2a). This red shift is attributed to the substitution of 12C by the heavier isotope 13C, resulting in a lower frequency of molecular vibration. In particular, large shifts were observed in ν1 and ν2 peaks, which are assigned to vibrational modes of carbon–carbon bonds. The largest shift (32 cm−1) was observed in the ν1 peak, which shifted from 1518 cm−1 (12C: green) to 1486 cm−1 (13C: red). These results indicate that 13CO2 was incorporated into H. lacustris cells through carbon fixation and that 13C-carbon was converted to AXT through the non-mevalonate pathway and subsequent carotenogenesis.51
C stretching; ν2, C–C stretching/C–H in-plane bending; ν3, CH3 rocking; ν4, C–H out-of-plane wagging.45,49 Spectra consist mainly of peaks corresponding to AXT, as it is the major ingredient (>90%) of carotenoid production by H. lacustris cells.50 The AXT spectra of cells incubated with 13CO2 are shifted to lower wavenumbers compared to those of cells incubated with 12CO2 (Fig. 2a). This red shift is attributed to the substitution of 12C by the heavier isotope 13C, resulting in a lower frequency of molecular vibration. In particular, large shifts were observed in ν1 and ν2 peaks, which are assigned to vibrational modes of carbon–carbon bonds. The largest shift (32 cm−1) was observed in the ν1 peak, which shifted from 1518 cm−1 (12C: green) to 1486 cm−1 (13C: red). These results indicate that 13CO2 was incorporated into H. lacustris cells through carbon fixation and that 13C-carbon was converted to AXT through the non-mevalonate pathway and subsequent carotenogenesis.51
We then demonstrated discrimination of H. lacustris cells accumulating 13C- or 12C-AXT by resonance Raman imaging based on the spectral shift. We mixed in equal number H. lacustris cells that accumulated 13C- and those that accumulated 12C-AXT, and then analyzed the cell mixture by Raman mapping. Raman images were generated by classical least squares modeling of the 13C- and 12C-AXT spectra obtained during the long-term induction. At each image pixel position, a linear combination of factors was calculated to estimate the measured spectrum. 13C- and 12C-AXT in the images were color-coded as red and green, respectively. As a result, the two types of cells were clearly discriminated (Fig. 2c). This result shows the advantage of Raman imaging over bright field microscopy (Fig. 2b) for detecting targets labeled with stable isotopes.
Resonance Raman spectroscopy of H. lacustris cells incubated with a mixture of 13CO2 and 12CO2
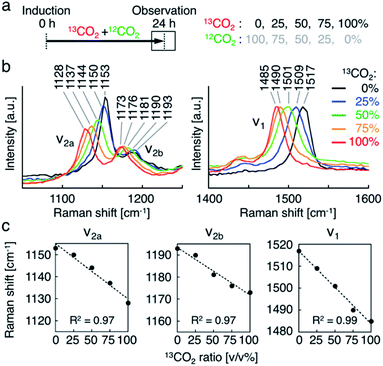
Second, we investigated the resonance Raman spectral shift of AXT in H. lacustris cells incubated with 13CO2 and 12CO2 mixed in various ratios. We induced production of AXT in the cells under various 13CO2 ratios (13CO2: 0, 25, 50, 75, and 100 vol%) for 24 h (Fig. 3a). The Raman peaks of AXT (ν1 and ν2) gradually shifted toward lower wavenumber positions as the 13CO2 ratio increased (Fig. 3b). When plotting the ratio of 13CO2 versus the wavenumber of each peak (ν1, ν2a, and ν2b), all showed a linear relationship (Fig. 3c). The slope (m) of the ν1 peak, attributed to C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C stretching, has a larger absolute value (ν1: m = −0.33) than that of the ν2 peaks, which include a contribution from C–C stretching (ν2a: m = −0.25, ν2b: m = −0.22). These results are congruent with observed resonance Raman spectral shifts of carotenoids (mainly β-carotene) in cyanobacterial cells incubated with a 13C source.22 H. lacustris cells fix 13CO2 and 12CO2 gases with equal probability, since stable isotopes have the same biochemical properties.52 Consequently, the carbon ratio of 13C and 12C in an AXT molecule produced in a cell is presumably the same as the ratio of 13CO2 and 12CO2 in the culturing atmosphere. On the other hand, the ratio of the 13C carbon–carbon bond wavenumber (σ13C) to the 12C carbon–carbon bond wavenumber (σ12C) corresponds to the square root of the ratio of the reduced masses, given that the wavenumber is defined as: σ = √k/2πc√μ, where k is the spring constant of the corresponding chemical bond and μ is the reduced mass.30 In fact, all of the ratios of resonance Raman peaks (σ13C-100%/σ12C-100%) are constant (ν1, 0.979; ν2a, 0.983; ν2b, 0.978; Fig. 3b and c) and approximately consistent with the theoretical value (σ13C/σ12C = 0.960). These results show that the resonance Raman spectra of AXT in H. lacustris cells incubated with a mixture of 13CO2 and 12CO2 shift to lower wavenumbers in a linear manner corresponding to the 13CO2 ratio in the culturing atmosphere.
C stretching, has a larger absolute value (ν1: m = −0.33) than that of the ν2 peaks, which include a contribution from C–C stretching (ν2a: m = −0.25, ν2b: m = −0.22). These results are congruent with observed resonance Raman spectral shifts of carotenoids (mainly β-carotene) in cyanobacterial cells incubated with a 13C source.22 H. lacustris cells fix 13CO2 and 12CO2 gases with equal probability, since stable isotopes have the same biochemical properties.52 Consequently, the carbon ratio of 13C and 12C in an AXT molecule produced in a cell is presumably the same as the ratio of 13CO2 and 12CO2 in the culturing atmosphere. On the other hand, the ratio of the 13C carbon–carbon bond wavenumber (σ13C) to the 12C carbon–carbon bond wavenumber (σ12C) corresponds to the square root of the ratio of the reduced masses, given that the wavenumber is defined as: σ = √k/2πc√μ, where k is the spring constant of the corresponding chemical bond and μ is the reduced mass.30 In fact, all of the ratios of resonance Raman peaks (σ13C-100%/σ12C-100%) are constant (ν1, 0.979; ν2a, 0.983; ν2b, 0.978; Fig. 3b and c) and approximately consistent with the theoretical value (σ13C/σ12C = 0.960). These results show that the resonance Raman spectra of AXT in H. lacustris cells incubated with a mixture of 13CO2 and 12CO2 shift to lower wavenumbers in a linear manner corresponding to the 13CO2 ratio in the culturing atmosphere.
Resonance Raman imaging of H. lacustris cells incubated with atmosphere switching from 13CO2 to 12CO2
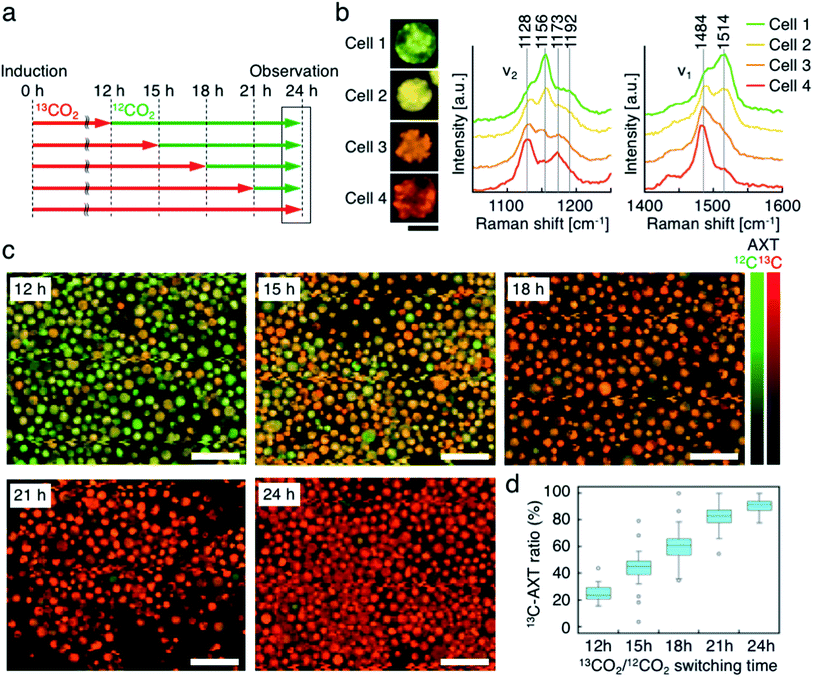
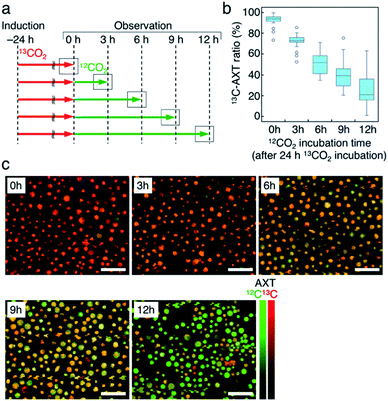
Third, we switched the incubation atmosphere from 13CO2 to 12CO2 during AXT induction, and then investigated the ratios of 13C- and 12C-AXT in H. lacustris cells by resonance Raman imaging. We varied the time to switch from 13CO2 to 12CO2 during the 24 h incubation period to investigate cells incubated with 13CO2 for 12 h, 15 h, 18 h, 21 h, and 24 h (Fig. 4a). In most cases, resonance Raman spectra of the H. lacustris cells were a superposition of the 13C-AXT spectrum (peaks: ν2, 1128 cm−1 and 1173 cm−1; ν1, 1484 cm−1; in red) and the 12C-AXT spectrum (peaks: ν2, 1156 cm−1 and 1192 cm−1; ν1, 1514 cm−1; in green) with varying intensities of the two spectra (representative four spectra are shown in Fig. 4b). This is in contrast to the gradual red shift observed in spectra of cells incubated with a mixture of 13CO2 and 12CO2 (Fig. 3). The results of this atmosphere switching experiment indicate that cells contain both 100% 13C-AXT and 100% 12C-AXT in various proportions, with no or a negligible amount of AXT molecules composed of both 13C- and 12C-carbons. This can be attributed to the rapid conversion rate from carbon source to AXT molecule, due to the small molecular size of AXT. In other words, after switching from 13CO2 to 12CO2, the 13C-carbon source was consumed rapidly for AXT production, and then the carbon source quickly switched to 12C-carbon, resulting in productions of 100% 13C-AXT and 100% 12C-AXT. In resonance Raman images generated by the classical least squares modeling of the 13C- and 12C-AXT spectra, H. lacustris cells are represented as a gradation of red (13C-AXT) and green (12C-AXT) in an RGB color model (e.g., Cell 1, yellow-green; Cell 2, yellow; Cell 3, orange; Cell 4, red-orange; Fig. 4b), since they include both 100% 13C-AXT and 100% 12C-AXT. On the other hand, AXT inside of a cell was distributed non-uniformly, suggesting that the AXT-accumulation process was still in a transition period to maturation after AXT-induction at 24 h (Fig. 4b). Notably, localization of 13C-AXT (red) and 12C-AXT (green) was observed inside some cells (Fig. S1†), revealing the progression of carbon fixation and conversion to AXT.We then observed the H. lacustris cells with low-magnification resonance Raman images and investigated the comprehensive changes of 13C-AXT in the cells (Fig. 4c). As the incubation time with 13CO2 increased, green color (12C-AXT) decreased and red color (13C-AXT) increased, as observed in resonance Raman images of cells incubated with 13CO2 for 12 h, 15 h, 18 h, and 21 h (Fig. 4c). This gradual change in color indicates that H. lacustris cells have CO2 fixation activity during the 12 h to 21 h period and then complete the conversion to AXT by the end of the incubation (24 h). On the other hand, comparing the images at 21 h and 24 h, no significant color change is observed, showing only red color (13C-AXT) without green color (12C-AXT). This result indicates that the conversion process from 12CO2 fixation to 12C-AXT production was incomplete during this period (21–24 h). In order to quantitatively support this, we measured resonance Raman spectra of 30 cells (at the center of the cells) randomly selected from individual images and calculated the 13C-AXT ratios by spectral separation using a multivariate curve resolution–alternating least squares (MCR-ALS) algorithm (Fig. 4d). From these spectra, we confirmed that 13C-AXT accumulated until 21 h and became saturated afterwards. We then investigated the activity of CO2 fixation and AXT production after 24 h. After 24 h of 13CO2, the incubation atmosphere was switched from 13CO2 to 12CO2 at 6 hour intervals, and the total AXT-induction time was 48 h (Fig. S2a†). In these cases, accumulation rate of 13C-AXT decreased (Fig. S2b and c†), which indicates that the conversion activity from 24 h to 48 h was lower than that of the 12 h to 24 h period. As demonstrated above, the active conversion period from CO2 fixation to AXT production can be probed by switching from 13CO2 to 12CO2 at various times and comparing the difference of 13C- or 12C-AXT accumulation by resonance Raman imaging.
Resonance Raman imaging of 13C-AXT in H. lacustris cells labeled by pulse incubation with 13CO2
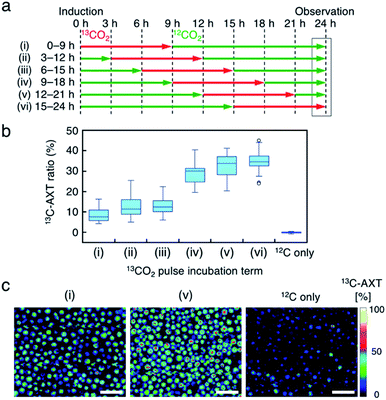
Fourth, we labeled AXT in H. lacustris cells by pulse incubation, where the duration of incubation with 13CO2 was shorter, and detected 13C-AXT by resonance Raman imaging to specify the carbon fixation time. In the aforementioned incubations with the switching of 13CO2 to 12CO2, whether 13CO2 was fixed at earlier or later times could not be determined, since the 13C-AXT indicated by the red color accumulated during the 13CO2 incubation. In this experiment, we incubated the cells with 13CO2 for a constant “pulse” of time (9 h), with the remaining incubation under 12CO2 (15 h), for a total AXT induction time of 24 h. The 13CO2 pulse incubations were shifted by 3 h in the timeline (Fig. 5a) to probe the carbon fixation time. We calculated the 13C-AXT ratios of 30 cells randomly selected for each 13CO2 pulse timing (Fig. 5b). The plot shows that 13CO2 fixation and the resultant 13C-AXT production are significantly higher in the late 13C-induction phases ((iv) 9–18 h, (v) 12–21 h, (vi) 15–24 h), while both are lower in the early 13C-induction phases ((i) 0–9 h, (ii) 3–12 h, (iii) 6–15 h). Notably, there is a large gap between conditions (iii) and (iv), indicating that CO2 is actively fixed and converted into AXT after 15 h. Among the late pulse incubations, conditions (v) and (vi) show higher 13C-AXT ratios than condition (iv) (t-test; p < 0.05), whereas no significant difference was observed (t-test; p = 0.20) between (v) and (vi). These results indicate that the activity of carbon-fixation and conversion is high during the 15 to 21 h period of incubation.We then compared the 13C-AXT ratios of H. lacustris cells in the same resonance Raman image frame. We represent only 13C-AXT in pseudo color (Fig. 5c) to highlight 13C-carbon fixed during the pulse incubation, rather than representing both 13C-AXT (red) and 12C-AXT (green) in the RGB color model (Fig. S3a†). Cells that actively fixed 13CO2 and converted it to 13C-AXT are clearly detected in the image of condition (v) 12–21 h, while such cells were scarcely observed in the image of condition (i) 0–9 h or the image of the control condition, which was incubation under only 12CO2. These results demonstrate that this method of pulse incubation can be useful for screening active H. lacustris cells which show a high efficiency of carbon-fixation and AXT-conversion.
Resonance Raman imaging of H. lacustris cells after replacing 13C-AXT by 12C-AXT by 12CO2 incubation
Lastly, we accumulated 13C-AXT in H. lacustris cells and then replaced it with 12C-AXT by incubating the cells under 12CO2. We induced 13C-AXT production in H. lacustris cells for 24 h by incubation under 13CO2 and then replaced the atmosphere with 12CO2 for different durations (3 h increments) before observation (Fig. 6a). Calculation of the 13C-AXT ratios of 30 cells randomly selected for each condition showed a large dispersion of 13C-AXT ratios, which reflects the difference in 12C-AXT production activity among the cells (Fig. 6b). Interestingly, we observed that 13C-AXT in some cells was completely replaced with 12C-AXT after 12 h (Fig. 6b and c), suggesting that AXT is in dynamic equilibrium between consumption and production. This information on carbon flow cannot be obtained by merely detecting non-labeled AXT and thus demonstrates the advantage of combining resonance Raman imaging with 13C-labeling. Furthermore, we observed heterogeneity of 13C-AXT (red) and 12C-AXT (green) in a single cell after incubation with 12CO2, but the AXTs became homogenized as the time of incubation with 12CO2 was extended (Fig. 7). Specifically, 12C-AXT infiltrated from the edge of the cell towards its center (Fig. 7a). In fact, the resonance Raman spectra around the cell periphery confirmed that the ratio of 12C-AXT was high at the edge and low at the center (Fig. 7b). This observation can be attributed to the transportation of newly produced 12C-AXT precursors to lipid droplets around the center.53,54 In an intermediate cell during encystment, chloroplasts, which produce an AXT precursor, are located around the cell periphery, while oil droplets, which serve as a “sink” for AXT, are located around the nucleus at the center.53,54 The newly produced 100% 12C-AXT precursors are delivered to the oil droplets from the periphery, converted to 12C-AXT by an enzyme inside, and mixed with pre-existing 100% 13C-AXT. Consequently, a gradient of 12C-AXT and 13C-AXT forms, causing heterogeneity in a single cell. These results demonstrate that the distribution of AXTs produced at different times can be visualized in a single cell by resonance Raman imaging combined with 13C- and 12C-labeling.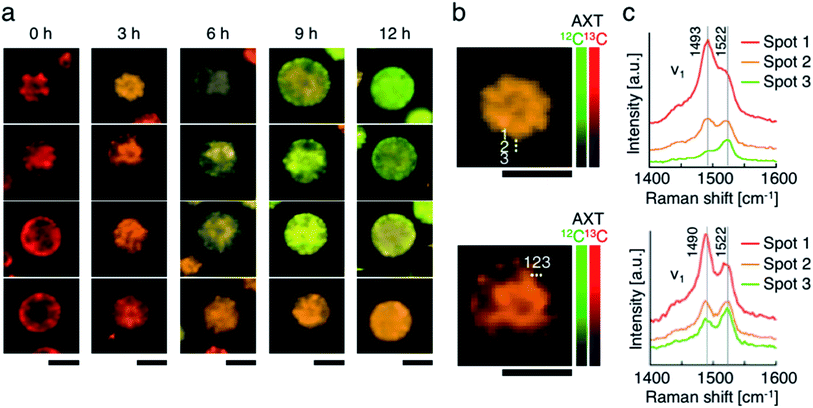 | ||
| Fig. 7 Resonance Raman imaging of the heterogeneity of 13C-AXT and 12C-AXT in a single H. lacustris cell. (a) Resonance Raman images of H. lacustris cells incubated with 12CO2 after incubation with 13CO2. The incubation conditions used here are identical to those in Fig. 6(b and c). Magnified resonance Raman images (b) and spectra (c) of H. lacustris cells incubated with 12CO2 for 3 h after their accumulation of 13C-AXT. The spectra correspond to the spots in the images of (b). 13C- and 12C-AXT are color-coded in red (13C) and green (12C), respectively. Scale bars: 20 μm. Color bars: intensities of Raman spectra of 13C- (red) and 12C-AXT (green). | ||
Conclusions
In this paper, we demonstrated spatiotemporal monitoring of AXT production in H. lacustris cells, incubated under varying 13CO2 and 12CO2 atmospheric conditions, by resonance Raman imaging. First, we induced AXT production in H. lacustris by incubating the cells with 13CO2 for enough time to produce 13C-AXT, the resonance Raman spectrum of which shows red shifts compared to that of 12C-AXT. This result shows that 13CO2 was incorporated into H. lacustris cells through carbon fixation and that 13C-carbon was converted to AXT through cellular metabolism. Based on the spectral difference between 13C- and 12C-AXT, we clearly discriminated H. lacustris cells accumulating 13C- or 12C-AXT by resonance Raman imaging. Second, we investigated the resonance Raman spectral shift of AXT in H. lacustris cells when incubated with a mixture of 13CO2 and 12CO2 gases in various ratios. We observed that the resonance Raman spectra shifted to positions at lower wavenumbers in a linear manner corresponding to the 13CO2 ratio of the culturing atmosphere. Third, we switched the incubation atmosphere from 13CO2 to 12CO2 at various times and then investigated the ratios of 13C- and 12C-AXT in the cells by resonance Raman imaging. The results indicated that the cells contained both 100% 13C-AXT and 100% 12C-AXT in various proportions. By comparing via resonance Raman imaging the differences in 13C- and 12C-AXT accumulation, the active conversion period from CO2 fixation to AXT production could be probed. Fourth, we labeled AXT with 13C by incubating cells under 13CO2 for short pulses of time, to specify the active time of carbon fixation and conversion. Lastly, we accumulated 13C-AXT in H. lacustris cells in advance and subsequently replaced it with 12C-AXT by incubating the cells under 12CO2 for different durations. We observed a high variation between cells in the rate at which 13C-AXT decreased, reflecting individual differences in 12C-AXT production activity among cells. In some cells, 13C-AXT was completely replaced with 12C-AXT, suggesting that AXT is in dynamic equilibrium between consumption and production. Furthermore, localizations of 13C-AXT and 12C-AXT in a single cell were observed by magnified resonance Raman imaging.Our method provides spatiotemporal information for single cells, including the conversion process from substrate to metabolite, the distribution process of the metabolite, and the turnover of the metabolite in dynamic equilibrium. By virtue of non-cytotoxic labelling and non-invasive imaging, long-term monitoring of metabolic dynamics in living cells can be achieved. Furthermore, by integrating our method with technologies for single cell isolation,55,56 the intracellular metabolism of an identified cell can be monitored. When combined with state-of-the-art Raman spectroscopy techniques that have higher spatiotemporal resolution, such as stimulated Raman scattering (SRS)57–60 or coherent anti-Stokes Raman scattering (CARS),61–63 the process of metabolite distribution can be monitored in a single cell with further detail. Leveraging such information for biotechnological production of target metabolites could provide significant value in terms of productivity. Furthermore, using Raman-activated cell sorting (RACS)64–66 will enable screening for cells with high metabolite productivity from a genetically diverse cell population.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported primarily by the ImPACT Program of the CSTI (Cabinet Office, Government of Japan) and partly by the JSPS Core-to-Core Program and White Rock Foundation. The authors thank Prof. Tomohisa Hasunuma at Kobe University for discussion.References
- N. Mizushima, T. Yoshimori and B. Levine, Cell, 2010, 140, 313–326 CrossRef CAS PubMed.
- O. Kepp, L. Galluzzi, M. Lipinski, J. Yuan and G. Kroemer, Nat. Rev. Drug Discovery, 2011, 10, 221–237 CrossRef CAS PubMed.
- J. Zenker, M. D. White, R. M. Templin, R. G. Parton, O. Thorn-Seshold, S. Bissiere and N. Plachta, Science, 2017, 357, 925–928 CrossRef CAS PubMed.
- A. Sakaue-Sawano, H. Kurokawa, T. Morimura, A. Hanyu, H. Hama, H. Osawa, S. Kashiwagi, K. Fukami, T. Miyata, H. Miyoshi, T. Imamura, M. Ogawa, H. Masai and A. Miyawaki, Cell, 2008, 132, 487–498 CrossRef CAS PubMed.
- L. Pernas, C. Bean, J. C. Boothroyd and L. Scorrano, Cell Metab., 2018, 27, 886–897.e4 CrossRef CAS PubMed.
- T. Yoshizumi, T. Ichinohe, O. Sasaki, H. Otera, S. Kawabata, K. Mihara and T. Koshiba, Nat. Commun., 2014, 5, 4713 CrossRef CAS PubMed.
- J. Y. Kim, Y. G. Kim and G. M. Lee, Appl. Microbiol. Biotechnol., 2012, 93, 917–930 CrossRef CAS PubMed.
- M. S. Cooper, W. R. Hardin, T. W. Petersen and R. A. Cattolico, J. Biosci. Bioeng., 2010, 109, 198–201 CrossRef CAS PubMed.
- K. Yamada, H. Suzuki, T. Takeuchi, Y. Kazama, S. Mitra, T. Abe, K. Goda, K. Suzuki and O. Iwata, Sci. Rep., 2016, 6, 26327 CrossRef CAS PubMed.
- M. A. Borowitzka, J. Appl. Phycol., 2013, 25, 743–756 CrossRef CAS.
- J. Nielsen and J. D. Keasling, Cell, 2016, 164, 1185–1197 CrossRef CAS PubMed.
- J. C. Caicedo, S. Cooper, F. Heigwer, S. Warchal, P. Qiu, C. Molnar, A. S. Vasilevich, J. D. Barry, H. S. Bansal, O. Kraus, M. Wawer, L. Paavolainen, M. D. Herrmann, M. Rohban, J. Hung, H. Hennig, J. Concannon, I. Smith, P. A. Clemons, S. Singh, P. Rees, P. Horvath, R. G. Linington and A. E. Carpenter, Nat. Methods, 2017, 14, 849–863 CrossRef CAS PubMed.
- K. D. Piatkevich, E. E. Jung, C. Straub, C. Linghu, D. Park, H. J. Suk, D. R. Hochbaum, D. Goodwin, E. Pnevmatikakis, N. Pak, T. Kawashima, C. T. Yang, J. L. Rhoades, O. Shemesh, S. Asano, Y. G. Yoon, L. Freifeld, J. L. Saulnier, C. Riegler, F. Engert, T. Hughes, M. Drobizhev, B. Szabo, M. B. Ahrens, S. W. Flavell, B. L. Sabatini and E. S. Boyden, Nat. Chem. Biol., 2018, 14, 352–360 CrossRef CAS PubMed.
- J. W. Lichtman and J. A. Conchello, Nat. Methods, 2005, 2, 910–919 CrossRef CAS PubMed.
- Z. Liu, L. D. Lavis and E. Betzig, Mol. Cell, 2015, 58, 644–659 CrossRef CAS PubMed.
- L. Wang, M. S. Frei, A. Salim and K. Johnsson, J. Am. Chem. Soc., 2019, 141, 2770–2781 CrossRef CAS PubMed.
- H.-S. Liu, M.-S. Jan, C.-K. Chou, P.-H. Chen and N.-J. Ke, Biochem. Biophys. Res. Commun., 1999, 260, 712–717 CrossRef CAS PubMed.
- M. R. Depaoli, H. Bischof, E. Eroglu, S. Burgstaller, J. Ramadani-Muja, T. Rauter, M. Schinagl, M. Waldeck-Weiermair, J. C. Hay, W. F. Graier and R. Malli, Pharmacol. Ther., 2019, 202, 98–119 CrossRef CAS PubMed.
- M. C. Wang, W. Min, C. W. Freudiger, G. Ruvkun and X. S. Xie, Nat. Methods, 2011, 8, 135–138 CrossRef CAS PubMed.
- S. Yamashiro, D. Taniguchi, S. Tanaka, T. Kiuchi, D. Vavylonis and N. Watanabe, Biophys. J., 2019, 116, 142–150 CrossRef CAS PubMed.
- A. Gautier, A. Juillerat, C. Heinis, I. R. Correa Jr, M. Kindermann, F. Beaufils and K. Johnsson, Chem. Biol., 2008, 15, 128–136 CrossRef CAS PubMed.
- M. Li, D. P Canniffe, P. J. Jackson, P. A. Davison, S. FitzGerald, M. J. Dickman, J. G. Burgess, C. N. Hunter and W. E. Huang, ISME J., 2012, 6, 875–885 CrossRef CAS PubMed.
- D. McIlvenna, W. E. Huang, P. Davison, A. Glidle, J. Coopera and H. Yin, Lab Chip, 2016, 16, 1420–1429 RSC.
- H. N. Noothalapati Venkata and S. Shigeto, Chem. Biol., 2012, 19, 1373–1380 CrossRef CAS PubMed.
- Y. Shen, F. Xu, L. Wei, F. Hu and W. Min, Angew. Chem., Int. Ed., 2014, 53, 5596–5599 CrossRef CAS PubMed.
- Y. Wang, Y. Ji, E. S. Wharfe, R. S. Meadows, P. March, R. Goodacre, J. Xu and W. E. Huang, Anal. Chem., 2013, 85, 10697–10701 CrossRef CAS PubMed.
- M. Li, W. E. Huang, C. M. Gibson, P. W. Fowler and A. Jousset, Anal. Chem., 2013, 85, 1642–1649 CrossRef CAS PubMed.
- P. Kubryk, J. S. Kölschbach, S. Marozava, T. Lueders, R. U. Meckenstock, R. Niessner and N. P. Ivleva, Anal. Chem., 2015, 87, 6622–6630 CrossRef CAS PubMed.
- H.-J. van Manen, A. Lenferink and C. Otto, Anal. Chem., 2008, 80, 9576–9582 CrossRef CAS PubMed.
- L. Wei, Y. Yu, Y. Shen, M. C. Wang and W. Min, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 11226–11231 CrossRef CAS PubMed.
- L. Wei, Y. Shen, F. Xu, F. Hu, J. K. Harrington, K. L. Targoff and W. Min, ACS Chem. Biol., 2015, 10, 901–908 CrossRef CAS PubMed.
- H.-J. van Manen, Y. M. Kraan, D. Roos and C. Otto, Proc. Natl. Acad. Sci. U. S. A., 2005, 102, 10159–10164 CrossRef CAS PubMed.
- D. Zhang, M. N. Slipchenko and J.-X. Cheng, J. Phys. Chem. Lett., 2011, 2, 1248–1253 CrossRef CAS PubMed.
- J. Li and J.-X. Cheng, Sci. Rep., 2014, 4, 6807 CrossRef CAS PubMed.
- F. Hu, L. Wei, C. Zheng, Y. Shen and W. Min, Analyst, 2014, 139, 2312–2317 RSC.
- D. Berry, E. Mader, T. K. Lee, D. Woebken, Y. Wang, D. Zhu, M. Palatinszky, A. Schintlmeister, M. C. Schmid, B. T. Hanson, N. Shterzer, I. Mizrahi, I. Rauch, T. Decker, T. Bocklitz, J. Popp, C. M. Gibson, P. W. Fowler, W. E. Huang and M. Wagner, Proc. Natl. Acad. Sci. U. S. A., 2014, 112, 194–203 CrossRef PubMed.
- Y. Yonamine, Y. Suzuki, T. Ito, Y. Miura, K. Goda, Y. Ozeki and Y. Hoshino, ChemBioChem, 2017, 18, 2063–2068 CrossRef CAS PubMed.
- M. A. Borowitzka, J. Appl. Phycol., 1995, 7, 3–15 CrossRef CAS.
- M. Donia and M. T. Hamann, Lancet Infect. Dis., 2003, 3, 338–348 CrossRef CAS PubMed.
- B. Singh, T. K. Bhat and B. Singh, J. Agric. Food Chem., 2003, 51, 5579–5597 CrossRef CAS PubMed.
- A. L. Demain, Appl. Microbiol. Biotechnol., 1999, 52, 455–463 CrossRef CAS PubMed.
- M. Guerin, M. E. Huntley and M. Olaizola, Trends Biotechnol., 2003, 21, 210–216 CrossRef CAS PubMed.
- S. Boussiba, Physiol. Plant., 2000, 108, 111–117 CrossRef CAS.
- K. Ukibe, T. Katsuragi, Y. Tani and H. Takagi, FEMS Microbiol. Lett., 2008, 286, 241–248 CrossRef CAS PubMed.
- B. Robert, Photosynth. Res., 2009, 101, 147–155 CrossRef CAS PubMed.
- A. Kaczor, K. Turnau and M. Baranska, Analyst, 2011, 136, 1109–1112 RSC.
- A. M. Collins, H. D. Jones, D. Han, Q. Hu, T. E. Beechem and J. A. Timlin, PLoS One, 2011, 6, e24302 CrossRef CAS PubMed.
- S. R. Fagerer, T. Schmid, A. J. Ibanez, M. Pabst, R. Steinhoff, K. Jefimovs, P. L. Urban and R. Zenobi, Analyst, 2013, 138, 6732–6736 RSC.
- S. Sato and M. Tasumi, J. Raman Spectrosc., 1983, 14, 310–321 CrossRef.
- L. Recht, N. Topfer, A. Batushansky, N. Sikron, Y. Gibon, A. Fait, Z. Nikoloski, S. Boussiba and A. Zarka, J. Biol. Chem., 2014, 289, 30387–30403 CrossRef CAS PubMed.
- K. Grunewald, J. Hirschberg and C. Hagen, J. Biol. Chem., 2001, 276, 6023–6029 CrossRef CAS PubMed.
- B. Koletzko, T. Sauerwald and H. Demmelmair, Eur. J. Pediatr., 1997, 156, S12–S17 CrossRef CAS PubMed.
- U. Pick, A. Zarka, S. Boussiba and L. Davidi, Planta, 2019, 249, 31–47 CrossRef CAS PubMed.
- M. Wayama, S. Ota, H. Matsuura, N. Nango, A. Hirata and S. Kawano, PLoS One, 2013, 8, e53618 CrossRef CAS PubMed.
- A. E. Vasdekis and G. Stephanopoulos, Metab. Eng., 2015, 27, 115–135 CrossRef CAS PubMed.
- N. Ota, Y. Yonamine, T. Asai, Y. Yalikun, T. Ito, Y. Ozeki, Y. Hoshino and Y. Tanaka, Anal. Chem., 2019, 91, 9631–9639 CrossRef CAS PubMed.
- B. G. Saar, C. W. Freudiger, J. Reichman, C. M. Stanley, G. R. Holtom and X. S. Xie, Science, 2010, 330, 1368–1370 CrossRef CAS PubMed.
- Y. Ozeki, W. Umemura, Y. Otsuka, S. Satoh, H. Hashimoto, K. Sumimura, N. Nishizawa, K. Fukui and K. Itoh, Nat. Photonics, 2012, 6, 845–851 CrossRef CAS.
- Y. Wakisaka, Y. Suzuki, O. Iwata, A. Nakashima, T. Ito, M. Hirose, R. Domon, M. Sugawara, N. Tsumura, H. Watarai, T. Shimobaba, K. Suzuki, K. Goda and Y. Ozeki, Nat. Microbiol., 2016, 1, 16124 CrossRef CAS PubMed.
- Y. Suzuki, K. Kobayashi, Y. Wakisaka, D. Deng, S. Tanaka, C.-J. Huang, C. Lei, C.-W. Sun, H. Liu, Y. Fujiwaki, S. Lee, A. Isozaki, Y. Kasai, T. Hayakawa, S. Sakuma, F. Arai, K. Koizumi, H. Tezuka, M. Inaba, K. Hiraki, T. Ito, M. Hase, S. Matsusaka, K. Shiba, K. Suga, M. Nishikawa, M. Jona, Y. Yatomi, Y. Yalikun, Y. Tanaka, T. Sugimura, N. Nitta, K. Goda and Y. Ozeki, Proc. Natl. Acad. Sci. U. S. A., 2019, 116, 15842–15848 CrossRef CAS PubMed.
- C. L. Evans, E. O. Potma, M. Pouris'haag, D. Cote, C. P. Lin and X. S. Xie, Proc. Natl. Acad. Sci. U. S. A., 2005, 102, 16807–16812 CrossRef CAS PubMed.
- K. Hashimoto, M. Takahashi, T. Ideguchi and K. Goda, Sci. Rep., 2016, 6, 21036 CrossRef CAS PubMed.
- M. Tamamitsu, Y. Sakaki, T. Nakamura, G. K. Podagatlapalli, T. Ideguchi and K. Goda, Vib. Spectrosc., 2016, 91, 163–169 CrossRef.
- D. A. Watson, L. O. Brown, D. F. Gaskill, M. Naivar, S. W. Graves, S. K. Doorn and J. P. Nolan, Cytometry, Part A, 2008, 73, 119–128 CrossRef PubMed.
- Q. Zhang, P. Zhang, H. Gou, C. Mou, W. E. Huang, M. Yang, J. Xu and B. Ma, Analyst, 2015, 140, 6163–6174 RSC.
- K. Hiramatsu, T. Ideguchi, Y. Yonamine, S.-W. Lee, Y. Luo, K. Hashimoto, T. Ito, M. Hase, J.-W. Park, Y. Kasai, S. Sakuma, T. Hayakawa, F. Arai, Y. Hoshino and K. Goda, Sci. Adv., 2019, 5, eaau0241 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Experimental section and supporting figures. See DOI: 10.1039/d0ra02803g |
| This journal is © The Royal Society of Chemistry 2020 |