Open Access Article
Open Access ArticleKey aurophilic motif for robust quantum-tunneling-based characterization of a nucleoside analogue marker†
Takafumi
Furuhata
 a,
Yuki
Komoto
a,
Yuki
Komoto
 b,
Takahito
Ohshiro
b,
Masateru
Taniguchi
b,
Takahito
Ohshiro
b,
Masateru
Taniguchi
 *b,
Ryosuke
Ueki
*b,
Ryosuke
Ueki
 a and
Shinsuke
Sando
a and
Shinsuke
Sando
 *ac
*ac
aDepartment of Chemistry and Biotechnology, Graduate School of Engineering, The University of Tokyo, 7-3-1 Hongo, Bunkyo-ku, Tokyo 113-8656, Japan. E-mail: ssando@chembio.t.u-tokyo.ac.jp
bThe Institute of Scientific and Industrial Research, Osaka University, 8-1 Mihogaoka, Ibaraki, Osaka 567-0047, Japan. E-mail: taniguti@sanken.osaka-u.ac.jp
cDepartment of Bioengineering, Graduate School of Engineering, The University of Tokyo, 7-3-1 Hongo, Bunkyo-ku, Tokyo 113-8656, Japan
First published on 7th September 2020
Abstract
A quantum sequencer offers a scalable electrical platform for single-molecule analysis of genomic events. A thymidine (dT) analog exhibiting uniquely high single-molecule conductance is a key element in capturing DNA synthesis dynamics by serving as a decodable marker for enzymatic labeling of nascent strands. However, the current design strategies of dT analogs that focus on their molecular orbital energy levels require bulky chemical modifications to extend the π-conjugation, which hinders polymerase recognition. We report herein a polymerase-compatible dT analog that is highly identifiable in quantum sequencing. An ethynyl group is introduced as a small gold-binding motif to differentiate the nucleobase–gold electronic coupling, which has been an overlooked factor in modifying nucleobase conductance. The resulting C5-ethynyl-2′-deoxyuridine exhibits characteristic signal profiles that allowed its correct identification at a 93% rate while maintaining polymerase compatibility. This study would expand the applicability of quantum sequencing by demonstrating a robust nucleoside marker with high identifiability.
Introduction
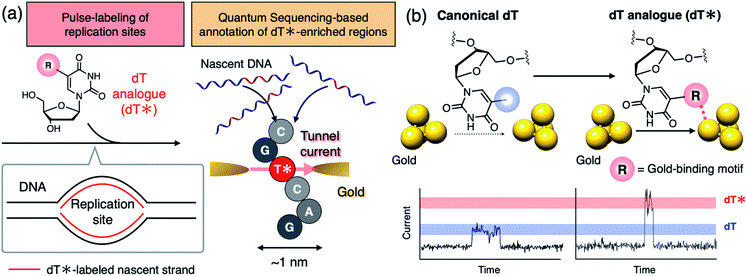
Single-molecule sequencing has attracted increasing interest as a powerful methodology for analyzing the heterogeneity of nucleic acid biosynthesis resulting from DNA replication and transcription.1–3 Thymidine (dT) and uridine (U) analogs that exhibit characteristic signal profiles in a single-molecule sequencer play an important role in capturing the dynamic aspects of these events by serving as bio-compatible markers to pulse-label elongation sites of nascent DNA or RNA (Fig. 1a, left). Those nucleoside analogs are applied to cells at a specific time-point of cellular activities, such as cell division and stimulus–response, to mark nascent DNA or RNA strands synthesized at that time. The following single-molecule decoding of the DNA and RNA not only identifies the sequences of the individual labeled strands including those of minor populations but also affords the length of DNA and the amount of RNA synthesized during the pulse-labeling period.1–3 Thus, dT or U analogs decodable in a single-molecule sequencer are keys to reveal the dynamics of DNA and RNA synthesis for individual sequences. Recently, Müller et al. adopted C5-bromo-2′-deoxyuridine (BrdU) as a dT analog to analyze the strand-specific movements of DNA replication forks using a biological nanopore single-molecule sequencer to unveil the heterogeneous landscape of the DNA replication dynamics in budding yeast.1 Single-molecule analysis of nucleic acid biosynthesis has now been extended to RNA demonstrating the sequence-specific dynamics of its synthesis and degradation.3 However, existing single-molecule sequencers do not have sufficient throughput for comprehensive analysis of DNA and RNA in higher eukaryotes.4 More scalable and durable single-molecule methodology is needed to achieve further high-throughput analysis.The quantum sequencer has emerged as a solid-state framework for electrical single-molecule sequencing.5–7 The method directly characterizes single nucleotides,8,9 even noncanonical ones,9–12 based on the tunnel current flowing through a single nucleotide trapped between a pair of nanogap electrodes. The potential compatibility of quantum sequencing with massive integration on hardware would make it a scalable platform for single-molecule analysis of DNA and RNA biosynthesis when combined with appropriate dT or U analogs which exhibit tunnel current profiles that can be distinguished from those of the canonical nucleosides (dA: 2′-deoxyadenosine, dC: 2′-deoxycytidine, dG: 2′-deoxyguanosine, and dT) (Fig. 1).
A previous study has reported the design of a highly conductive 2′-deoxyuridine (dU) derivative by modulating the energy gap between its HOMO and the Fermi level of gold to allow statistical identification of the nucleoside based on tunnel current intensity.13 However, the HOMO-based design is accompanied by introduction of a rigid and bulky aromatic ring to expand the conjugated π-system of the nucleobase, which generally impairs polymerase compatibility.14 Thus, no robust strategy exists for developing a conductive dT analog with minimal chemical modification that retains its polymerase compatibility as a genetic alphabet “T”.
Herein, we report a polymerase-compatible dT analog that can be accurately identified in quantum sequencing. To significantly increase the conductance of the dT analog with minimal modification, we introduce gold-binding motifs at the C5 position of dU to differentiate the nucleobase–gold electronic coupling that contributes to charge transport at the metal–molecule interface (Fig. 1b). Current measurements of the dT analogs reveal that introduction of the ethynyl group, which involves multiple orbital interactions including electron donation and back-donation upon binding to gold, is particularly effective in increasing nucleoside conductance. The distinct signal profiles of C5-ethynyl dU (EtdU) allowed its discrimination from the canonical nucleosides with more than 90% correct assignment rates, while the ethynyl group at the C5 position of dU is small enough to retain polymerase compatibility. Our results demonstrate the potential of EtdU as a robust nucleoside marker for the quantum sequencing-based analysis of DNA synthesis.
Results
Design of a series of dU derivatives modified with gold-binding motifs
There are two molecular design requirements from physicochemical and biochemical points of view to develop a dT analog that is efficiently decodable in quantum sequencing for DNA synthesis analysis (Fig. 1). From a physicochemical point of view, the dT analog should exhibit tunnel current outputs that are sufficiently distinctive relative to those of canonical nucleosides (dA, dC, dG, and dT) to render it highly identifiable. From a biochemical point of view, the dT analog should be easily incorporated into DNA without disrupting the DNA replication process.A previous study has addressed the physicochemical requirement by designing a conductive dU derivative with strong current intensity by reducing the energy gap between its HOMO and the Fermi level of gold nanogap electrodes.13 However, the HOMO-based approach requires modification of the nucleobase with an aromatic ring to expand its π-conjugation. Introduction of the rigid and bulky substituent generally impedes polymerase recognition of the nucleoside as a genetic alphabet “T”,14 and thus does not satisfy the biochemical requirement.
Electronic coupling between the nucleobase and nanogap electrodes is the other important factor contributing to charge transport in single-molecule conduction.15,16 In this study, we focus on this property as a promising factor to differentiate nucleoside conductance with minimal chemical modification. Electronic coupling is characterized by the orbital overlap between the molecule and the electrode. The conductivity of aromatic rings trapped between metal gap electrodes is dramatically influenced by gold-binding exocyclic substituents.17,18 This suggests that even structurally small functional groups can dramatically alter nucleoside conductance by changing the nucleobase–gold electronic coupling while maintaining polymerase compatibility of the dT analog. We hypothesize that the introduction of an exocyclic gold-binding group that is absent in canonical dT will increase nucleobase conductance by altering the gold–nucleobase electronic coupling (Fig. 1b).
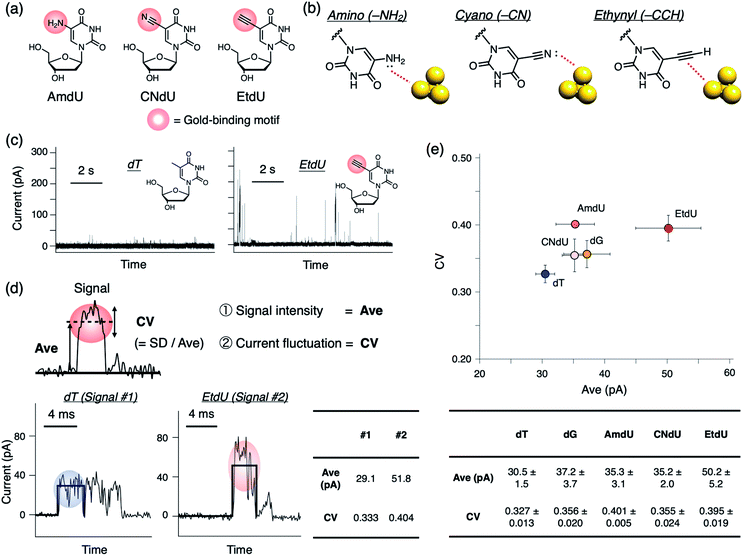
As candidates for a highly conductive dT analog, we designed a series of dU derivatives modified with different gold-binding motifs at the C5 position where small structural modifications are often tolerated in polymerase recognition.19,20Fig. 2a shows the chemical structures of the analogs evaluated in this study: C5-amino dU (AmdU), C5-cyano dU (CNdU), and C5-ethynyl dU (EtdU). Selection of the functional groups was based on their ability to bind reversibly to gold, but with characteristic modes of interaction (Fig. 2b). The amino group can strongly interact with gold by the lone pair on the nitrogen atom that is conjugated with the aromatic ring.21 The cyano group also has a nitrogen lone pair as a potential gold-binding moiety,22 although it is not conjugated with the aromatic ring. The ethynyl group has structure similar to that of the cyano group, but its mode of interaction is different. Although it lacks a lone pair, the carbon–carbon triple bond interacts strongly with gold via multiple interactions that include π-electron donation to gold and back-donation from gold to the π* orbital of the ethynyl group.23 By comparing dT analogs with these modifications, the gold-binding motifs effective in differentiating the nucleoside analog conductance from those of the canonical nucleosides would be extracted.
Evaluation of the tunnel current profiles of the dU derivatives
To evaluate the conductance of the dT analog and canonical nucleosides, we performed current measurements at 0.1 V bias voltage using a 0.6 nm gold gap prepared by the mechanically controlled break junction (MCBJ) method as previously reported.24 When a nucleoside sample was applied, positive current pulses were observed as electrical signals (Fig. 2c and S1†). We adopted the average value of an individual current pulse (Ave) as a measure of detected conductance (Fig. 2d). Boxplots of Ave values constructed from all detected signals showed that the Ave profiles of the dT analogs had large distributions which were overlapped with each other especially in the low-value region (Fig. S1†). There were distinct differences in the frequency of large Ave values among the nucleosides. Therefore, we assume that the profiles in the large-value region represent the characteristic conductance of each nucleoside as previously reported.12 For simplicity, the 75th percentile (p75), which is a statistical parameter in the large-value region, was adopted as the representative indicator of Ave. Comparison of averaged p75 values obtained from more than four independent measurement sets shows that the modified dT analogs have greater Ave values than canonical dT (Fig. 2e). EtdU exhibits the particularly high conductance, even when compared with dG, which is the most conductive canonical nucleoside12,25 (Fig. 2e and S2†).We also found that each nucleoside exhibits a characteristic profile in the coefficient of variation (CV) of the current values recorded during an individual current pulse (Fig. 2d and e). The CV is a dimensionless quantity calculated by normalizing the standard deviation (SD) by the average value (Ave). Therefore, the CV value represents the scale of current fluctuation in a signal pulse relative to the signal amplitude. Though the boxplots of CV values constructed from all detected signals were overlapped between the nucleosides due to the wide distribution (Fig. S1†), statistical comparison of p75 shows that the CV values of the dT analogs tend to be greater than that of canonical dT (Fig. 2e). EtdU and AmdU also exhibit greater CV values than those of canonical dC, dA, and dG (Fig. 2e and S2†). These observations indicate that the current fluctuation represented by the CV value can be used as a feature to characterize nucleosides in addition to the signal intensity represented by Ave. EtdU exhibits particularly large Ave and CV profiles (Fig. 2e and S2†), which indicates its ability to be distinguished from the canonical nucleosides via two-dimensional (2D) statistical processing of Ave and CV.
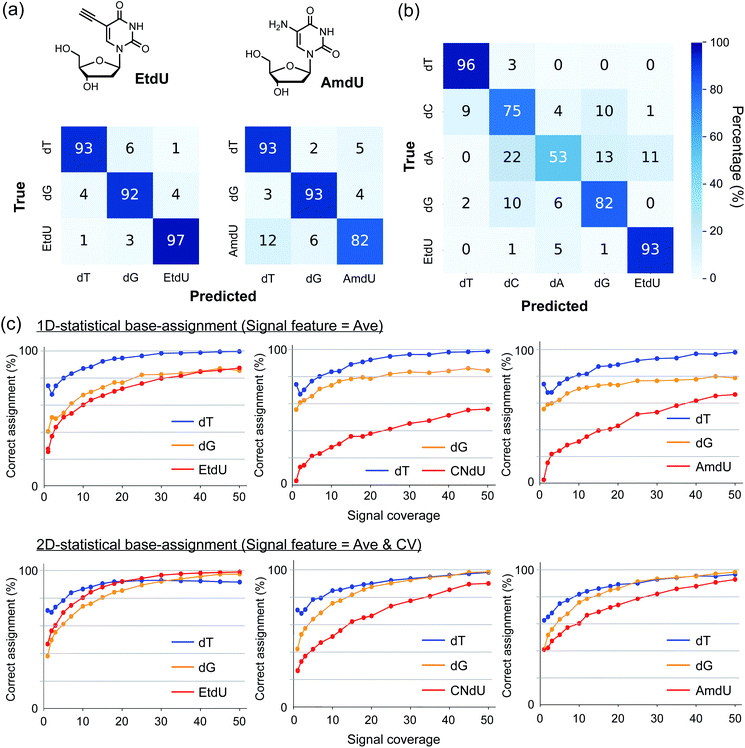
Two-dimensional statistical base-assignment of EtdU using Ave and CV values
To evaluate the ability of EtdU to be identified in quantum sequencing, we undertook a base-assignment program based on 2D statistical processing of the signals in terms of Ave and CV (Fig. S3; see ESI† for procedural details). The results show that EtdU was identified with a greater than 95% correct assignment rate with thirty-signal coverage in the classification of dT, dG, and EtdU, while the corresponding assignment rate was 82% for AmdU (Fig. 3a). In particular, the correct assignment of EtdU reached 93% with forty-signal coverage even when classifying five nucleosides containing all the canonical nucleosides (Fig. 3b). Thus, EtdU is accurately distinguished from the canonical nucleosides due to its characteristic Ave and CV profiles. Strikingly, the correct assignment of EtdU by 2D processing of Ave and CV exceeded 90% with much less signal coverage than the 1D statistical processing of Ave (Fig. 3c). Although the high identifiability of EtdU is attributed primarily to its high Ave profile, as shown by the 1D statistical processing of Ave (Fig. 3c), this result indicates that both Ave and CV changed upon introduction of the ethynyl group concertedly contribute to the accurate identification of EtdU.dT analogs that can be detected by single-molecule techniques have been used as nucleoside markers to enzymatically label nascent DNA strands for strand-specific annotation of DNA replication patterns.1,2,26,27 Envisioning the potential use of EtdU as a nucleoside marker for quantum sequencing-based analysis of DNA synthesis, we further evaluated the capability of EtdU for enzymatic incorporation into DNA as a genetic alphabet “T”. EtdU has been used to label nascent DNA strands in cellular contexts for microscopic imaging.28–30 We performed primer extension with EtdU triphosphate using a polymerase with proofreading activity to evaluate the enzymatic incorporation rate of EtdU. The production of full-length (41 bp) DNA was confirmed in a similar manner between the cases adopting dT and EtdU as a genetic alphabet “T” (Fig. S4†). We also carried out PCR amplification of the human TNFα gene (702 bp) to confirm the applicability of EtdU triphosphate as a substrate for the replication of longer DNA sequences and established that PCR products using dT triphosphate and EtdU triphosphate were amplified in a similar way (Fig. S4†). These results indicate that EtdU is enzymatically incorporated into DNA with high efficiency.
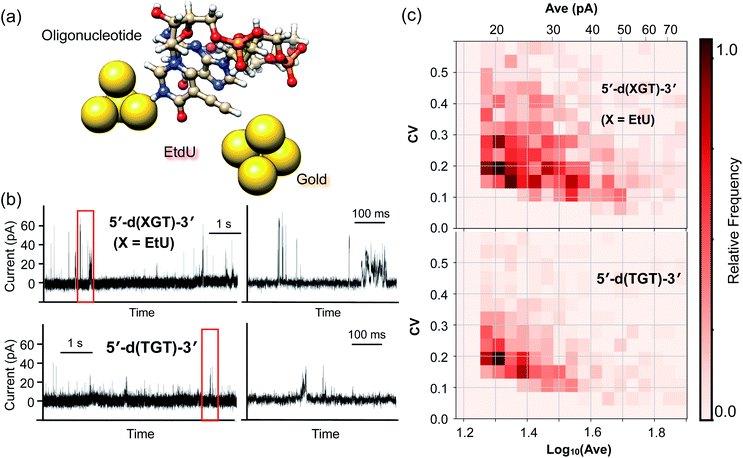
Tunnel current profiles of the EtdU-incorporated oligonucleotide sequence
The high identifiability of EtdU (Fig. 3) and its efficient enzymatic incorporation into DNA (Fig. S4†) suggest the potential utility of EtdU as a DNA replication marker. To further validate the applicability of EtdU towards quantum sequencing-based DNA replication analysis, we investigated its detectability in the DNA strand by evaluating the signal profile of an oligonucleotide containing EtdU (Fig. 4a). Here, there are two levels of criteria to be confirmed; (1) EtdU is discriminable from the unmodified dT, which is essential to judge whether a decoded sequence of interest is a nascent strand whose dT is replaced with EtdU, and (2) EtdU exhibits higher conductivity compared with dG that is the most conductive among the canonical nucleosides (Fig. S2†)12,25 in order to verify the statistical discriminability of EtdU in the oligonucleotide from all the canonical nucleosides including dA and dC. To test whether EtdU exhibits characteristic signal profiles compared with dT and dG, we designed a 3-mer oligonucleotide containing dT, dG and EtdU as a model sequence (5′-d(XGT)-3′, X = EdU) and conducted the current measurement (Fig. 4a and b).For statistical analysis of signals, we extracted the current plateaus that correspond to each of the single trapping events of the nucleobases (Fig. S5†). At the present stage, it is not necessarily easy to conclusively assign a nucleoside to each single current plateau due to the overlap of the Ave and CV distributions between the nucleosides (Fig. S2†). However, the statistical comparison of the signal features shows that the count of current plateaus with high intensity (Ave) or large fluctuation (CV) increased for the 5′-d(XGT)-3′ sequence compared with the oligonucleotide lacking EtdU (5′-d(TGT)-3′) (Fig. 4c and S5†). More specifically, current plateaus with an Ave value greater than 40 pA or a CV value greater than 0.3 were characteristic of 5′-d(XGT)-3′ and were attributed to the presence of EtdU in the oligonucleotide. When compared with “mono”-EtdU (Fig. S1†), the 5′-d(XGT)-3′ sequence provided signals with both large Ave and large CV values less frequently, supposedly because certain molecular orientations of EtdU that produce such signals were inhibited in the oligonucleotide due to increased steric hindrance near the gap electrode. However, this result indicates that EtdU exhibits characteristic signals with statistically greater signal intensities or larger current fluctuations compared with the canonical nucleosides in the oligonucleotide.
Discussion
We find the dT analog that can be accurately identified in the quantum sequencing towards single-molecule analysis of DNA synthesis (Fig. 3). To this aim, we introduced gold-binding motifs at the C5 position of dU to differentiate gold–nucleobase electronic coupling which has been an overlooked factor in modifying nucleobase conductance.12,13 This strategy has led to the increase of the nucleobase conductance with minimal modifications. In particular, it has been demonstrated that the ethynyl group is a key structural motif in dramatically differentiating the signal profiles of the dT analog from those of canonical nucleosides while maintaining polymerase compatibility. C5-ethynyl dU (EtdU) was correctly identified at a 93% rate with forty-signal coverage in the classification of five nucleosides including all canonical nucleosides in addition to EtdU (Fig. 3b). The high identifiability of EtdU and its efficient enzymatic incorporation into DNA (Fig. S4†)28–30 collectively indicate that EtdU will be a powerful marker of DNA replication sites decodable in quantum sequencing-based analysis.Considering that EtdU would be applied to the living cells in the practical use, it is also important whether EtdU can be used without significant cytotoxicity. Though EtdU exhibits cytotoxicity and causes cell cycle arrest upon long-term treatment over one cell cycle,31,32 the effect on the cell viability is negligible in the short-term treatment.31 Since the time to pulse-label DNA replication sites should be much shorter than a single cell cycle to site-specifically mark DNA strands elongated at a specific time point, EtdU may be applicable without significant perturbation of cellular functions. Actually, EtdU has been widely applied to living cells as a chemical marker for microscopic imaging of DNA replication.28–30 Müller et al. have pioneered the comprehensive profiling of DNA replication dynamics by capturing genome replication of the budding yeast using a single-molecule sequencer based on the protein nanopore.1 The quantum sequencer may offer a more scalable and durable solid-state platform towards single-molecule profiling of the DNA replication dynamics in higher eukaryotes in combination with EtdU.
Though the combination of EtdU and the quantum sequencer is promising as discussed above, it is also the fact that there are rooms for the quantum sequencers to be improved at the current stage. The large overlap of the signal features between nucleobases makes it difficult to accurately identify dA and dC (Fig. 3b) and to reliably assign a nucleobase to each current plateau of oligonucleotide signals. This issue can be attributed to the random orientation of nucleobases passing near the nanogap electrodes. For the practical application, it is necessary to integrate a methodology to control the orientation and the unidirectional flow of DNA near the nano-gap with the quantum sequencer. Regarding this point, promising nanotechnologies have been proposed to control the dynamics of DNA in a nano-space so far.33–35
In this study, the ethynyl group is the most effective among the substituents tested in increasing nucleoside conductance (Fig. 2e and S1†). This observation is consistent with the fact that p-diethynylbenzene exhibits greater conductance than the p-diaminobenzene and p-dicyanobenzene evaluated as model aromatic compounds (Fig. S6†). The unmistakable increase in molecular conductance upon ethynyl modification in the quantum sequencer can be attributed to the unique mode of alkyne coordination to metal electrodes, as previously studied for acetylene.36 The interaction of acetylene with metal electrodes involves multiple molecular orbitals including π and π* orbitals of the carbon–carbon triple bond engaged in electron-donating and back-donating interactions. In that case, the tunneling current is mediated by the multiple orbitals that interact with the metal electrodes, which concertedly contribute to charge transport through the metal–molecule interface. The combined involvement of multiple orbitals in charge transport through the ethynyl–gold interface might impart high conductivity to ethynyl-modified molecules (EtdU and p-diethynylbenzene).
In addition to current intensity, we also find that the scale of current fluctuation represented by the CV is characteristic of each dT analog (Fig. 2e). This observation indicates that the scale of current fluctuation is also an important feature that facilitates identification of the molecules. For example, EtdU was discriminated from the canonical nucleosides with greater accuracy and efficiency via 2D characterization using Ave and CV values compared to 1D characterization using only Ave values (Fig. 3c and S7†). In addition, though the conductance of AmdU was less different from that of dT and dG than that of EtdU was (Fig. 2e and S2†), the correct assignment of AmdU exceeded 90% in classification of dT, dG, and AmdU via 2D-signal characterization with fifty-signal coverage probably owing to its characteristic CV profile (Fig. 3c). Therefore, rational control of current fluctuations should lead to further improvements in developing dT analogs that are decodable by quantum sequencing. From a chemical perspective, this would be the next challenge, which might be tackled by the design and evaluation of a series of dT analogs that have different exocyclic substituents with varied fluctuations of the orbital interactions with gold and orbital anisotropies to elucidate their correlations with the current fluctuation.
Conclusions
We have developed a polymerase-compatible dT analog with high identifiability in quantum sequencing via 2D characterization of signal profiles in terms of current intensities and fluctuations. The dT analog, C5-ethynyl dU (EtdU), was correctly identified at a 93% rate in the classification of five nucleosides including all canonical ones based on its characteristic signal profile. The ease of identifying EtdU in quantum sequencing is attributed to the increase in conductance brought about by ethynyl group introduced to differentiate its nucleobase–gold electronic coupling from that of the canonical nucleosides. The structurally small ethynyl group also retains the polymerase compatibility of EtdU as a genetic alphabet “T”. The fact that the nucleobase conductance was tuned by a small aurophilic motif offers chemical insight to the design of nucleoside analogs according to users' demands in DNA replication analysis and other applications including RNA synthesis and chemical modification analysis, when combined with available labeling methodologies of nascent RNA strands3,30,37,38 and modified nucleobases.39–41 Therefore, this study represents an important step towards expanding the applicability of quantum sequencing to single-molecule analysis of genomic events.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We thank the Takeda Super Clean Room at the University of Tokyo for the use of SAMCO FA-1. We also are grateful for the One-stop Sharing Facility Center for Future Drug Discoveries at the University of Tokyo for the use of micrOTOF II. This work was supported by a grant by KAKENHI (No. 19K22244) to S. S. from the Japan Society for the Promotion of Science (JSPS), a grant by KAKENHI (No. 17J10215), a Grant-in-Aid for JSPS Fellows to T. F. from JSPS, and a grant by KAKENHI (No. 26220603) to M. T. from JSPS. The MCBJ instrument was developed with a CREST grant from Intelligent Measurement Analysis (No. JPMJCR1666).References
- C. A. Müller, M. A. Boemo, P. Spingardi, B. M. Kessler, S. Kriaucionis, J. T. Simpson and C. A. Nieduszynski, Nat. Methods, 2019, 16, 429–436 CrossRef.
- M. Hennion, J.-M. Arbona, C. Cruaud, F. Proux, B. Le Tallec, E. Novikova, S. Engelen, A. Lemainque, B. Audit and O. Hyrien, bioRxiv, 2018, 426858 Search PubMed.
- K. C. Maier, S. Gressel, P. Cramer and B. Schwalb, bioRxiv, 2019, 601856 Search PubMed.
- A. Ameur, W. P. Kloosterman and M. S. Hestand, Trends Biotechnol., 2019, 37, 72–85 CrossRef.
- M. Di Ventra and M. Taniguchi, Nat. Nanotechnol., 2016, 11, 117–126 CrossRef.
- M. Zwolak and M. Di Ventra, Nano Lett., 2005, 5, 421–424 CrossRef.
- J. Lagerqvist, M. Zwolak and M. Di Ventra, Nano Lett., 2006, 6, 779–782 CrossRef.
- M. Tsutsui, M. Taniguchi, K. Yokota and T. Kawai, Nat. Nanotechnol., 2010, 5, 286–290 CrossRef.
- S. Huang, J. He, S. Chang, P. Zhang, F. Liang, S. Li, M. Tuchband, A. Fuhrmann, R. Ros and S. Lindsay, Nat. Nanotechnol., 2010, 5, 868–873 CrossRef.
- M. Tsutsui, K. Matsubara, T. Ohshiro, M. Furuhashi, M. Taniguchi and T. Kawai, J. Am. Chem. Soc., 2011, 133, 9124–9128 CrossRef CAS.
- J. Im, S. Sen, S. Lindsay and P. Zhang, ACS Nano, 2018, 12, 7067–7075 CrossRef CAS.
- T. Furuhata, T. Ohshiro, G. Akimoto, R. Ueki, M. Taniguchi and S. Sando, ACS Nano, 2019, 13, 5028–5035 CrossRef CAS.
- T. Furuhata, T. Ohshiro, Y. Izuhara, T. Suzuki, R. Ueki, M. Taniguchi and S. Sando, ChemBioChem, 2020, 21, 335–339 CrossRef CAS.
- S. Obeid, A. Baccaro, W. Welte, K. Diederichs and A. Marx, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 21327–21331 CrossRef CAS.
- F. Chen and N. J. Tao, Acc. Chem. Res., 2009, 42, 429–438 CrossRef CAS.
- T. A. Su, M. Neupane, M. L. Steigerwald, L. Venkataraman and C. Nuckolls, Nat. Rev. Mater., 2016, 1, 16002 CrossRef CAS.
- F. Chen, X. Li, J. Hihath, Z. Huang and N. Tao, J. Am. Chem. Soc., 2006, 128, 15874–15881 CrossRef CAS.
- Z. Li, M. Smeu, M. A. Ratner and E. Borguet, J. Phys. Chem. C, 2013, 117, 14890–14898 CrossRef CAS.
- M. Kuwahara, J. I. Nagashima, M. Hasegawa, T. Tamura, R. Kitagata, K. Hanawa, S. I. Hososhima, T. Kasamatsu, H. Ozaki and H. Sawai, Nucleic Acids Res., 2006, 34, 5383–5394 CrossRef CAS.
- B. L. Cavanagh, T. Walker, A. Norazit and A. C. B. Meedeniya, Molecules, 2011, 16, 7980–7993 CrossRef CAS.
- L. Venkataraman, J. E. Klare, I. W. Tam, C. Nuckolls, M. S. Hybertsen and M. L. Steigerwald, Nano Lett., 2006, 6, 458–462 CrossRef CAS.
- A. Mishchenko, L. A. Zotti, D. Vonlanthen, M. Bürkle, F. Pauly, J. C. Cuevas, M. Mayor and T. Wandlowski, J. Am. Chem. Soc., 2011, 133, 184–187 CrossRef CAS.
- F. Ferraro, J. F. Pérez-Torres and C. Z. Hadad, J. Phys. Chem. C, 2015, 119, 7755–7764 CrossRef CAS.
- M. Tsutsui, M. Taniguchi and T. Kawai, Appl. Phys. Lett., 2008, 93, 163115 CrossRef.
- T. Ohshiro, K. Matsubara, M. Tsutsui, M. Furuhashi, M. Taniguchi and T. Kawai, Sci. Rep., 2012, 2, 501 CrossRef.
- F. De Carli, N. Menezes, W. Berrabah, V. Barbe, A. Genovesio and O. Hyrien, Small Methods, 2018, 2, 1800146 CrossRef.
- C. J. Merrick, D. Jackson and J. F. X. Diffley, J. Biol. Chem., 2004, 279, 20067–20075 CrossRef CAS.
- A. Salic and T. J. Mitchison, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 2415–2420 CrossRef CAS.
- H. Yamakoshi, K. Dodo, M. Okada, J. Ando, A. Palonpon, K. Fujita, S. Kawata and M. Sodeoka, J. Am. Chem. Soc., 2011, 133, 6102–6105 CrossRef CAS.
- J. T. Ngo, S. R. Adams, T. J. Deerinck, D. Boassa, F. Rodriguez-Rivera, S. F. Palida, C. R. Bertozzi, M. H. Ellisman and R. Y. Tsien, Nat. Chem. Biol., 2016, 12, 459–465 CrossRef CAS.
- S. Diermeier-Daucher, S. T. Clarke, D. Hill, A. Vollmann-Zwerenz, J. A. Bradford and G. Brockhoff, Cytometry, Part A, 2009, 75, 535–546 CrossRef.
- H. Hua and S. E. Kearsey, Nucleic Acids Res., 2011, 39, e60 CrossRef CAS.
- U. F. Keyser, J. R. Soc., Interface, 2011, 8, 1369–1378 CrossRef CAS.
- B. Luan, G. Stolovitzky and G. Martyna, Nanoscale, 2012, 4, 1068–1077 RSC.
- K. Yokota, M. Tsutsui and M. Taniguchi, RSC Adv., 2014, 4, 15886–15899 RSC.
- T. Nakazumi, S. Kaneko, R. Matsushita and M. Kiguchi, J. Phys. Chem. C, 2012, 116, 18250–18255 CrossRef CAS.
- S. Nainar, B. J. Cuthbert, N. M. Lim, W. E. England, K. Ke, K. Sophal, R. Quechol, D. L. Mobley, C. W. Goulding and R. C. Spitale, Nat. Methods, 2020, 17, 311–318 CrossRef CAS.
- M. Rabani, J. Z. Levin, L. Fan, X. Adiconis, R. Raychowdhury, M. Garber, A. Gnirke, C. Nusbaum, N. Hacohen, N. Friedman, I. Amit and A. Regev, Nat. Biotechnol., 2011, 29, 436–442 CrossRef CAS.
- E.-A. Raiber, R. Hardisty, P. van Delft and S. Balasubramanian, Nat. Rev. Chem., 2017, 1, 0069 CrossRef CAS.
- M. Berney and J. F. McGouran, Nat. Rev. Chem., 2018, 2, 332–348 CrossRef CAS.
- X. Shu, J. Cao, M. Cheng, S. Xiang, M. Gao, T. Li, X. Ying, F. Wang, Y. Yue, Z. Lu, Q. Dai, X. Cui, L. Ma, Y. Wang, C. He, X. Feng and J. Liu, Nat. Chem. Biol., 2020, 16, 887–895 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available: Detailed procedures including experimental settings of the electrical device, current measurement, analytical procedures, enzymatic incorporation of nucleotides and chemical synthesis. See DOI: 10.1039/d0sc03946b |
| This journal is © The Royal Society of Chemistry 2020 |