3D hydrogel mimics of the tumor microenvironment: the interplay among hyaluronic acid, stem cells and cancer cells†
Sara
Amorim
 *ab,
Diana
Soares da Costa
*ab,
Diana
Soares da Costa
 ab,
Iva
Pashkuleva
ab,
Iva
Pashkuleva
 ab,
Celso A.
Reis
cdef,
Rui L.
Reis
ab,
Celso A.
Reis
cdef,
Rui L.
Reis
 ab and
Ricardo A.
Pires
ab and
Ricardo A.
Pires
 *ab
*ab
a3B's Research Group, I3Bs – Research Institute on Biomaterials, Biodegradables and Biomimetics, University of Minho, Headquarters of the European Institute of Excellence on Tissue Engineering and Regenerative Medicine, AvePark, Parque de Ciência e Tecnologia, Zona Industrial da Gandra, 4805-017 Barco, Portugal. E-mail: sara.amorim@i3bs.uminho.pt; rpires@i3bs.uminho.pt
bICVS/3B's - PT Government Associate Laboratory, Braga/Guimarães, Portugal
ci3S, University of Porto, Portugal
dIPATIMUP, Porto, Portugal
eDepartment of Pathology and Oncology, Faculty of Medicine, Porto University, Portugal
fInstitute of Biomedical Sciences Abel Salazar, University of Porto, Portugal
First published on 28th October 2020
Abstract
The present work reports on a 3D model of the tumor microenvironment that contains hyaluronic acid (HA) and alginate, and demonstrates the utility of this model to study the effect of HA size on the crosstalk between cancer cells and mesenchymal stem cells (MSCs). The system incorporates a core that contains HA of specific size (i.e. 6.4, 741 or 1500 kDa) with encapsulated epithelial MKN45 cancer cells and a shell with MSCs that mimic the presence of stem cells next to the tumor site. It was found that short HA (i.e. 6.4 kDa) promotes the invasion of cancer cells from the core to the shell, whereas longer HA (i.e. 741 and 1500 kDa) recruits the MSCs into the core, i.e. the tumor site, where a reduction of the formation of cancer cell aggregates was observed. In summary, the developed 3D model recapitulates some key tumor features related to the effect of HA size on both cancer cell invasiveness and MSC behavior at the tumor site.
Introduction
The interactions of cancer cells (CCs) with the components of the surrounding microenvironment, such as proteins, glycoproteins, proteoglycans, signaling molecules and different cell types, create a complex tumor microenvironment (TME).1–3 Changes in the TME impact cancer progression: unbalanced synthesis and degradation of its components lead to altered stiffness, elasticity, permeability,4 and biochemical composition.5,6 One of the TME components that plays a critical role in cancer invasiveness is hyaluronic acid (HA).7,8 The abnormal synthesis of HA by the synthases and the deregulated digestion by hyaluronidases lead to the accumulation of HA with different molecular weights (Mws) in the basement membrane of cancer cells.7 The accumulated HA can trigger various signaling pathways (e.g. via interaction with HA-specific transmembrane receptor CD44) that affect cell proliferation, migration or even latency and apoptosis.6 The cell response is dependent on the Mw of HA:9,10 low Mw HA (<100 kDa) is pro-inflammatory and promotes CC invasion and metastasis, while high Mw (>1000 kDa) HA induces CC latency.7,11 The TME is also governed by the crosstalk between CCs and healthy cells from the surrounding microenvironment (e.g. mesenchymal stem cells – MSCs, macrophages and fibroblasts, among others).12–14 MSCs are linked to tumor progression but their role is controversial. Some authors show that MSCs induce tumor growth by disrupting cell-to-cell contact and promoting CC invasion.15 Other works suggest that MSCs inhibit the invasiveness of CCs by reducing their drug resistance.16 In fact, the MSCs’ activity depends on their density. It has been reported that a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 MSC
1 MSC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CC ratio induces upregulation of metastatic genes; whereas a lower ratio (e.g. 1
CC ratio induces upregulation of metastatic genes; whereas a lower ratio (e.g. 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2) results in tumor remission.17–19 The presence of MSCs can also influence the CC aggregation: the MSCs added to pre-formed aggregates caused disorganization of the structures, while the MSCs present since the beginning of the formation of CC spheroids enhanced spatial organization and homogeneity.20
2) results in tumor remission.17–19 The presence of MSCs can also influence the CC aggregation: the MSCs added to pre-formed aggregates caused disorganization of the structures, while the MSCs present since the beginning of the formation of CC spheroids enhanced spatial organization and homogeneity.20
The complexity of the TME as well as the mismatch between different animal models and the human cellular environment impose the development of 3D models that mimic the biochemical and mechanical features of the TME as a valuable tool to study cancer progression and/or the efficiency of different treatments.21,22 Herein, we developed a 3D co-culture system that recapitulates some important features of the TME: the core is the cancer site with encapsulated CCs exposed to HA of specific Mws that are known to elicit different CC responses, e.g. migration or latency, and the surrounding shell contains healthy MSCs that can crosstalk with the CCs present at the cancer site (i.e. core of the hydrogel). We demonstrate that this system can replicate the key features of cancer progression and invasiveness.
Materials and methods
Materials
Sodium hyaluronates (HA) with Mws of 6.4, 741 and 1500 kDa were acquired from Lifecore (USA) and sodium alginate (Alg, UP VLVG, >75 kDa) from Pronova (Norway). The CD44 monoclonal antibody (Ascites, AM06286SU-N) was purchased from ACRIS. The FITC-labelled CD90 (Thy-1) monoclonal antibody (eBio5E10 (5E10)) was obtained from eBioscience™. The other antibodies, rabbit monoclonal to Vimentin [EPR3776], rabbit monoclonal to E-cadherin [EP700Y] and rabbit monoclonal to alpha smooth muscle actin [E184], were obtained from Abcam. The secondary antibodies IRDye® 800CW Goat-anti-Rabbit and IRDye® 800CW Goat-anti-Mouse were obtained from LI-COR Biosciences.Methods
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio of CM
1 ratio of CM![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) fresh αMEM).
fresh αMEM).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 350 in 1% w/v BSA/TBS-CaCl2), followed by rabbit anti-mouse Alexa Fluor-594 (1
350 in 1% w/v BSA/TBS-CaCl2), followed by rabbit anti-mouse Alexa Fluor-594 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 in 1% w/v BSA/TBS-CaCl2) and CD90-FITC (1
500 in 1% w/v BSA/TBS-CaCl2) and CD90-FITC (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 250 in 1% w/v BSA/TBS-CaCl2), respectively. Nuclei were counter-stained with DAPI (1 mg mL−1 in 1% BSA in TBS-CaCl2). After 1 h of incubation with the antibodies, the gels were washed with TBS-CaCl2 and observed under a confocal laser scanning microscope (TCS SP8, Leica, Germany).
250 in 1% w/v BSA/TBS-CaCl2), respectively. Nuclei were counter-stained with DAPI (1 mg mL−1 in 1% BSA in TBS-CaCl2). After 1 h of incubation with the antibodies, the gels were washed with TBS-CaCl2 and observed under a confocal laser scanning microscope (TCS SP8, Leica, Germany).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g, 16 min, 4 °C), and the supernatant was further analysed by western blotting (WB). Each protein, under Laemmli buffer, was denatured at 37 °C for 1 h 30 min and at 95 °C for 5 min prior to use. The lysates, containing 20 μg of protein, were dissolved using 4–12% Bis-Tris Protein Gels (Invitrogen NuPAGE, Thermo Fisher Scientific, USA) and transferred to nitrocellulose membranes (Thermo Fischer Scientific, USA). The membranes were incubated in 4% (m/v) BSA in Tris-buffered saline-Tween (TBS-T, Cell Signalling Technology) and probed with α-smooth muscle actin (1
000g, 16 min, 4 °C), and the supernatant was further analysed by western blotting (WB). Each protein, under Laemmli buffer, was denatured at 37 °C for 1 h 30 min and at 95 °C for 5 min prior to use. The lysates, containing 20 μg of protein, were dissolved using 4–12% Bis-Tris Protein Gels (Invitrogen NuPAGE, Thermo Fisher Scientific, USA) and transferred to nitrocellulose membranes (Thermo Fischer Scientific, USA). The membranes were incubated in 4% (m/v) BSA in Tris-buffered saline-Tween (TBS-T, Cell Signalling Technology) and probed with α-smooth muscle actin (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5000), Vimentin (1
5000), Vimentin (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000) and E-cadherin (1
1000) and E-cadherin (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5000) antibodies. After 3 cycles of 5 min washing with TBS-T, the membranes were incubated with IRDye®800CW anti-Rabbit or anti-Mouse (1
5000) antibodies. After 3 cycles of 5 min washing with TBS-T, the membranes were incubated with IRDye®800CW anti-Rabbit or anti-Mouse (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10
10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000) for 1 h and imaged on an Odyssey Infrared Imaging System (LI-COR Biosciences, USA).
000) for 1 h and imaged on an Odyssey Infrared Imaging System (LI-COR Biosciences, USA).
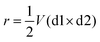 | (1) |
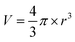 | (2) |
The quantification of fluorescence from the confocal images and densitometric analysis of the WB membranes were performed using ImageJ software (Version 2.0.0-rc-34/1.50a).
Results and discussion
Development of a core–shell 3D hydrogel
Mimicking the TME is challenging due to its complexity.13 Cell spheroids and 3D scaffolds are the current in vitro TME models used in cancer studies. Whereas spheroids are based on clustering of one (e.g. CCs) or more cell types (e.g. cancer-associated fibroblasts (CAFs) and MSCs), they lack the dynamic crosstalk between CCs and the surrounding environment;26 the 3D hydrogel models mimic the features of the extracellular matrix (ECM) composition by using specific components of the ECM, with controlled porosity and mechanical properties.27Here, we developed a core–shell 3D cancer model that resembles the complexity of the TME, combining (1) the presence of HA of different Mws in the ECM of the cancer tissue; (2) its influence on the behaviour of gastric CCs (i.e. MKN45) at the cancer site and of healthy bone marrow (bm) MSCs in the neighbouring regions; and (3) the interplay among HA, CCs and healthy bmMSCs (Fig. 1A).
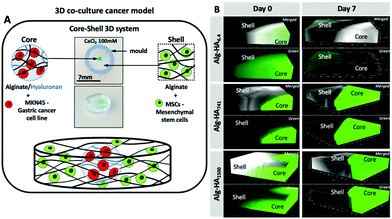 | ||
| Fig. 1 (A) Graphical illustration of the developed 3D core–shell system used for the co-culture of cancer MKN45 cells and bone marrow mesenchymal stem cells (bmMSCs). Hydrogel images are shown in Fig. S1.† (B) Confocal microscopy images showing the diffusion of the HA (green) from the core to the shell (bottom images, where green corresponds to HA-FITC incorporated in the Alg core; top images merged channels from the transmitted and fluorescence images of the Alg/HA-FITC). | ||
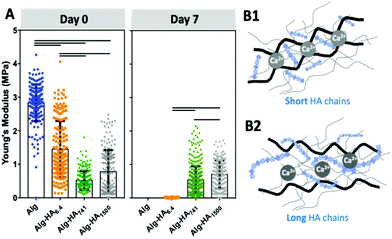
The proposed system is composed of a core hydrogel's microparticle that contains HA of different Mws, blended with the biocompatible Alg to generate the 3D polymer network cross-linked under mild conditions, i.e. with Ca2+.28 We performed an initial assessment of the stability of this system and its ability to maintain the HA in the core of the 3D hydrogel. This was executed using labelled HA (i.e. HA-FITC) making possible to confirm its incorporation into the core of the construct (Fig. 1B) by fluorescence microscopy. As expected, short HA (i.e. 6.4 kDa) is more diffusive: fluorescence can be seen beyond the core of the system even at very short immersion times. This diffusion can explain the similarity between the mechanical properties and stability of Alg and Alg-HA6.4 cores (Fig. 2A). However, in general, we observe a reduction of stiffness when HA is combined with Alg, with a higher impact as the HA's Mw increases. HA contributes differently to the stability of the hydrogels, with a clear distinction between shorter HA chains (i.e. HA6.4) and longer HA chains (i.e. HA741 and HA1500). Our results are consistent with the ability of the longer chains of HA to partially block the ionic crosslinking of Alg, leading to the formation of hydrogels of lower stiffness (Fig. 2B2). In contrast, the shorter chains of HA6.4 are not able to significantly block the activity of the Ca2+ ions, presenting a limited impact on the Alg cross-linking (Fig. 2B1).29,30 Another important point to consider is the very high viscosity of HA1500 that strengthens the Alg matrix, partially reverting the loss of stiffness observed for the core composed of Alg-HA741.
After 7 days of immersion (5% CO2 incubator at 37 °C in cell culture medium, but in the absence of cells), the Alg and Alg-HA6.4 spheres lose their integrity due to the release of Ca2+ (i.e. the Alg crosslinking agent) from the hydrogel network, resulting in the disruption of the Alg matrix.31
Longer HA chains (i.e. 741 and 1500 kDa) remained within the core even after 7 days of immersion (Fig. 1B) due to the interpenetrating network formed by Alg and the lower mobility and diffusion of the HA of high Mw compared with those of the low Mw polymer. Overall, the generated Alg-HA741 and Alg-HA1500 networks had lower stiffness values as compared to Alg (Fig. 2A); however they were more stable after 7 days of immersion. In fact, their mechanical properties did not change significantly over time, while the Alg and Alg-HA6.4 disassembled during the same time period.
The incorporation of MKN45 cells influences the mechanical properties and stability of the hydrogels’ core
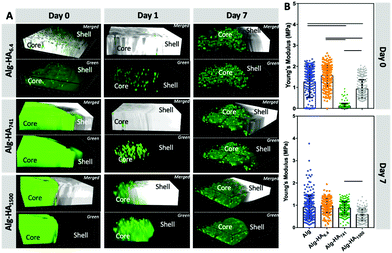
The stiffness of 3D matrices affects the cellular adhesion and proliferation.32–35 In turn, cells also modulate the stiffness of the surrounding environment through the secretion of different proteins: the higher deposition of the ECM components in the TME usually leads to a stiffening of the cancer site.36Indeed, we observed that the MKN45 cells encapsulated in the construct's core affected its mechanical properties (Fig. 3). The MKN45 cells formed aggregates with increased cell viability in the presence of HA (Fig. 4A – core). This result can be explained with the recognition of HA by the MKN45 cells.37 Indeed, confocal microscopy showed a scattered fluorescence signal indicating HA reorganization in the presence of cells (Fig. 3A). At day 0, this change is visible only for the short and diffusive HA6.4, i.e. immediately after cell encapsulation. However, after 1 day of culture, similar redistribution is also visible for the longer HA741 and HA1500. In these cases (especially for HA1500), a blurred fluorescence co-exists with the scattered signal suggesting the role of HA in two processes: interpenetrating network formation (blurred fluorescence) and cell–HA interactions.
Cellular encapsulation reduced the stiffness of the hydrogels (Fig. 2Avs.Fig. 3B, at day 0) but the dependence on the HA size was similar to the one obtained for the hydrogels without cells. The main difference was observed after 7 days of culture: the MKN45 cells significantly improved the stability of the Alg and Alg-HA6.4 hydrogels. These results are consistent with the ability of the encapsulated cells to proliferate and produce ECM under these experimental conditions leading to improved stability of the 3D hydrogel structure over time.38
The use of Alg to generate the 3D polymer network (through crosslinking with Ca2+) is compatible with cell culture conditions.37 In addition, Alg also lacks adhesive epitopes and, thus, can serve as a bioinert background that allows the evaluation of the bioactivity of the added HA. Such an approach is advantageous because HA can be immobilized in the 3D construct without any modification. The core containing HA was loaded with CCs (mimicking the cancer site), which were embedded in a hydrogel disc, to which healthy bmMSCs were encapsulated (mimicking the surrounding environment). Moreover, the proposed core–shell model, where CCs are confined in the hydrogel's core, mimic the hypoxic tumor physiological conditions and reduced flow of nutrients.38 This system was used to evaluate the impact of HA's Mw on cancer invasiveness and the influence of the surrounding healthy cells.
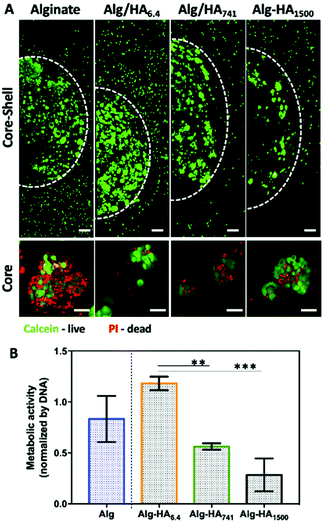
The viability of the cancer (i.e. MKN45) and healthy (i.e. bmMSCs) cells, cultured on the developed 3D system, was evaluated after 10 days of cell culture. Our Live/Dead staining results show high viability of both cell types in the core and shell of the 3D system (Fig. 4A, S2† – split channels). The formation of MKN45 cell clusters is visible in the core of the hydrogel, with increased cell death in the Alg-only sphere. The addition of HA into the core improves cellular viability, which could be related to the internalization of HA by the MKN45 cells, as suggested by the fluorescence images obtained using HA-FITC (Fig. 3A, days 1 and 7).37 Moreover, the MKN45 cells seeded on HA of 6.4 kDa present higher metabolic activity when compared to the hydrogels presenting HA of higher Mw, showing the ability of the HA Mw to modulate the proliferation and viability of cells (Fig. 4B).
Cancer cell invasion and protein expression in the presence of mesenchymal stem cells
The influence of MSCs on CC behavior is mediated through the release of pro-inflammatory cytokines and/or growth factors (e.g. TGF-beta1, VEGF and IL-6).39 However, direct MSC-CC contact can also occur and lead to the disruption of CC agglomerates mainly through the cleavage of the cell–cell junctions, which are maintained by E-cadherin.15,40 The presence of MSCs in the TME also increases the sensitivity of CCs to anti-cancer drugs.41 The MSC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CC ratio is also critical for any outcome, including changes in CC invasiveness and tumor progression.18,26
CC ratio is also critical for any outcome, including changes in CC invasiveness and tumor progression.18,26
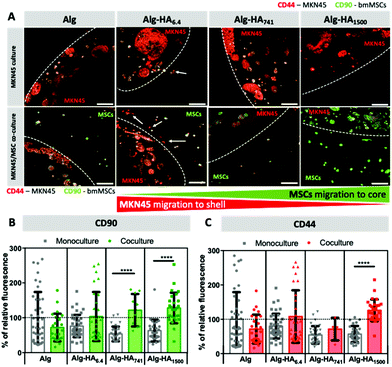
To mimic the complexity of the TME, we encapsulated MSCs in the shell of the construct, which was designed to be in direct contact with the core (representing the cancer site) containing the encapsulated CCs. To track the movement of each cell type, we tagged cells for the mesenchymal stem cell marker, CD90,42 and the transmembrane HA receptor, CD44, in all studied cells.43 Both cell types express CD90 and CD44 (Fig. S3†), but the relative expression is very different and can be used to distinguish MKN45 cells from bmMSCs. Our results show that MKN45 cells (encapsulated in the Alg-HA6.4 core) escape into the Alg shell. When the core contained longer HA chains (i.e. 741 and 1500 kDa), no MKN45 invasion was observed but bmMSCs migrated from the shell into the core of the construct (Fig. 5A and S4†).
In the TME, the secretion of pro-inflammatory cytokine Interleukin-6 (IL-6) is linked to tumorigenesis, cancer cell proliferation and metastasis.44 Herein, we observed a significant increase of the IL-6 protein level in the medium from the co-cultures of the core–shell hydrogels containing Alg-HA6.4 (Fig. 6). This is in good agreement with the migration pattern of MKN45 cells, which showed an invasive character in the presence of HA of low Mw. The absence of IL-6 expression on MKN45 monoculture (Fig. S7†), as previously observed by others,45 shows that bmMSCs in the tumour vicinity trigger the aggressive character of the gastric CCs, which may be correlated with poor prognosis of cancer and resistance to chemotherapy.45
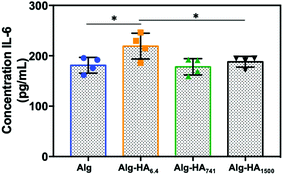 | ||
| Fig. 6 IL-6 secretion measured by ELISA from the cells seeded in the hydrogels under co-culture conditions. Statistical differences are marked * for p < 0.05. | ||
The invasion of MKN45 cells from the Alg-HA6.4 core into the shell was also observed in the absence of bmMSCs (Fig. 5A; monoculture, white arrows) and this can be associated with (1) fast solubilization of the core structure, through the release of Ca2+ cations (as previously shown in Fig. 1 and 2), which leads to the reorganization of the core matrix increasing the invasion of MKN45 cells; (2) the HA gradient generated upon the diffusion of HA into the shell (Fig. 1B) acting as directional cues for CC migration; and (3) the internalization of HA6.4 by CCs, as suggested by the fluorescence images of HA-FITC on the CC sites (core, Fig. 3A) which could act as a signaling mechanism to induce CCs’ invasion.38,46 bmMSCs cultured alone did not migrate from the shell to the HA-rich core (Fig. S4†); however, in the presence of MKN45 cells, it appears that the stem cells colonized Alg-HA1500 (Fig. 5A). The observed result (high relative intensity of green in the core, Fig. 5A and B) can be also due to increased CD90 expression by MKN45 in the presence of bmMSCs in the vicinity of the CCs. The same variation of CD90 expression was observed for the MKN45 cell cultures supplemented with medium from bmMSC expansion (Fig. S6†). This result showed that MSC-secretome alone is able to modulate MKN45 behavior but the direct MSC–MKN45 contact amplifies this response.
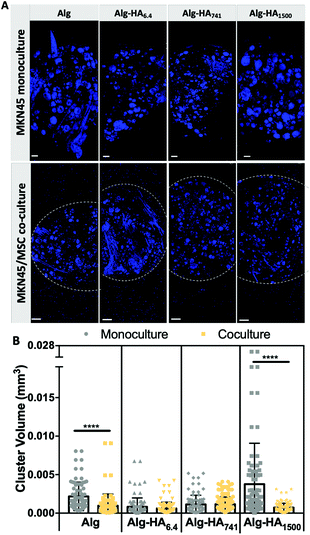
Disruption of MKN45 cell–cell contacts in the presence of MSCs
The ability of bmMSCs to modulate the cell-to-cell contact in tumors was assessed from the capacity of MKN45 cells to form aggregates when cultured alone or together with bmMSCs (Fig. 7). We observed significantly smaller MKN45 aggregates in Alg and Alg-HA1500 in the presence of bmMSCs (Fig. 7B). We believe that the co-culture on an Alg-only system induces a faster degradation of the hydrogel, compromising cellular adhesion and proliferation. On the other hand, bmMSCs cultured in the shell of the Alg-HA1500 rich-core system promote the formation of smaller CC aggregates and therefore inhibit tumor growth. In fact, it has been previously reported that the same tumor inhibition occurred when a small, but relevant, number of MSCs (a CC![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) MSCs ratio of 2
MSCs ratio of 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) were injected into the tumor site.47
1) were injected into the tumor site.47
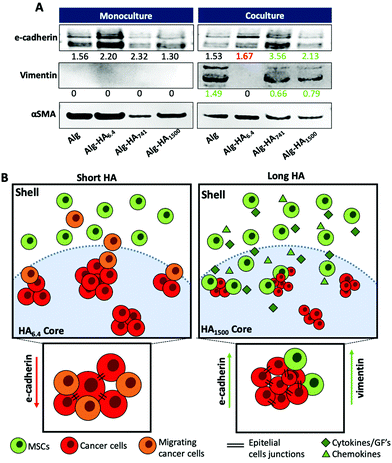
The mesenchymal/epithelial markers expressed by the MKN45 cells encapsulated in the core of the construct was evaluated by western blotting (WB, Fig. 8). Under co-culture conditions, the cells in the core enhance the expression of vimentin, a mesenchymal marker42 for the Alg, Alg-HA741 and Alg-HA1500 hydrogels’ cores. It is noteworthy that vimentin was not expressed by MKN45 cells cultured alone or supplemented with bmMSC-conditioned media (Fig. S8†). This observation is consistent with the idea that either vimentin is derived from the epithelial to mesenchymal transition of the MKN45 cells or the bmMSCs attracted to the core are the source of this vimentin expression. Overall, our results are consistent with the latter possibility as bmMSCs are observed in the core of the hydrogel formulated with HA of higher Mw (Fig. 5A). Moreover, the presence of HA of high Mw has been reported to induce CCs to produce chemotactic factors, such as cytokines and growth factors, which act as chemoattractants for MSCs, further supporting our results that show the recruitment of bmMSCs to the core of the 3D system.48–50
For the same samples (i.e. Alg, Alg-HA741 and Alg-HA1500) we also observed overexpression of E-cadherin by the cells in the core of the 3D system showing an increment of the cell–cell contacts. This result indicates the promotion of the epithelial transition of MKN45 cells when they are in contact with bmMSCs. The absence of vimentin expression and a downregulation of E-cadherin (responsible for the epithelial cell–cell junctions) in the MKN45 cells encapsulated in the Alg-HA6.4 core suggest that the CCs are not in direct contact with MSCs in the core of the hydrogel, but instead CC invasion into the shell is induced (loss of E-cadherin expression), as previously reported.51
Conclusions
We developed a 3D core–shell model that is able to recapitulate a series of mechanical, biochemical and biological features of a gastric TME. Using co-culturing conditions (MKN45 cells at the tumour site and bmMSCs in its periphery) we were able to mimic the influence of healthy bmMSCs on the CCs’ behavior. We further demonstrate that the HA's Mw at the cancer site is able to modulate CCs’ behavior. Low Mw HA, e.g. HA6.4, induces a migratory phenotype in MKN45 cells, which also internalize HA immediately after cell seeding. In contrast, HA of high Mw, e.g. HA1500, does not promote an invasive behavior on MKN45 CCs; instead, it attracts bmMSCs to the cancer site, reducing the growth of CC clusters. Overall, with the developed core–shell 3D model, we were able to mimic the TME and assess the influence of the biochemical features of specific components of the cancer ECM, e.g. HA and its Mw, as well as the presence of bmMSCs, on CCs’ behaviour.Additional statement
All experiments were performed in accordance with the Guidelines of the Code of Ethical Conduct of the University of Minho and Experiments were approved by the Ethics Committee of Hospital da Prelada (Ref. DC 05/2015). Informed consent was obtained from the human participants in this study.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We acknowledge the financial support from the European Commission and the Horizon 2020 – WIDESPREAD programme, under the grant agreement number #668983-FORECAST. SA acknowledges the Portuguese Foundation for Science and Technology for her PhD grant (SFRH/BD/112075/2015).References
- L. L. Hui and Y. Chen, Cancer Lett., 2015, 368, 7–13 CrossRef CAS.
- M. Wang, J. Zhao, L. Zhang, F. Wei, Y. Lian, Y. Wu, Z. Gong, S. Zhang, J. Zhou, K. Cao, X. Li, W. Xiong, G. Li, Z. Zeng and C. Guo, J. Cancer, 2017, 8, 761–773 CrossRef CAS.
- S. S. Pinho and C. A. Reis, Nat. Rev. Cancer, 2015, 15, 540–555 CrossRef CAS.
- A. V. Taubenberger, L. J. Bray, B. Haller, A. Shaposhnykov, M. Binner, U. Freudenberg, J. Guck and C. Werner, Acta Biomater., 2016, 36, 73–85 CrossRef CAS.
- F. A. Venning, L. Wullkopf and J. T. Erler, Front. Oncol., 2015, 5, 224 Search PubMed.
- S. Mereiter, A. M. Martins, C. Gomes, M. Balmana, J. A. Macedo, K. Polom, F. Roviello, A. Magalhaes and C. A. Reis, FEBS Lett., 2019, 593, 1675–1689 CrossRef CAS.
- T. Chanmee, P. Ontong and N. Itano, Cancer Lett., 2016, 375, 20–30 CrossRef CAS.
- L. P. Setälä, M. I. Tammi, R. H. Tammi, M. J. Eskelinen, P. K. Lipponen, U. M. Ågren, J. Parkkinen, E. M. Alhava and V. M. Kosma, Br. J. Cancer, 1999, 79, 1133–1138 CrossRef.
- C. Walker, E. Mojares and A. Del Río Hernández, Int. J. Mol. Sci., 2018, 19(10), 3028 CrossRef.
- Z. K. Price, N. A. Lokman and C. Ricciardelli, Cancers, 2018, 10(12), 482 CrossRef CAS.
- C. Yang, M. Cao, H. Liu, Y. He, J. Xu, Y. Du, Y. Liu, W. Wang, L. Cui, J. Hu and F. Gao, J. Biol. Chem., 2012, 287, 43094–43107 CrossRef CAS.
- F. Chen, X. Zhuang, L. Lin, P. Yu, Y. Wang, Y. Shi, G. Hu and Y. Sun, BMC Med., 2015, 13, 45 CrossRef.
- H. Sawayama, T. Ishimoto and H. Baba, J. Cancer Metastasis Treat., 2018, 4, 10 CrossRef.
- S. Mereiter, M. Balmana, D. Campos, J. Gomes and C. A. Reis, Cancer Cell, 2019, 36, 6–16 CrossRef CAS.
- A. Dittmer, K. Hohlfeld, J. Lützkendorf, L. P. Müller and J. Dittmer, Cell. Mol. Life Sci., 2009, 66, 3053–3065 CrossRef CAS.
- A. Y. Khakoo, S. Pati, S. A. Anderson, W. Reid, M. F. Elshal, I. I. Rovira, A. T. Nguyen, D. Malide, C. A. Combs, G. Hall, J. Zhang, M. Raffeld, T. B. Rogers, W. Stetler-Stevenson, J. A. Frank, M. Reitz and T. Finkel, J. Exp. Med., 2006, 203, 1235–1247 CrossRef CAS.
- W. Zhu, M. Wang, Y. Fu, N. J. Castro, S. W. Fu and L. G. Zhang, Acta Biomater., 2015, 14, 164–174 CrossRef CAS.
- B. L. Yen and M.-L. Yen, J. Cancer Mol., 2008, 4(1), 5–9 CAS.
- F. Djouad, C. Bony, F. Apparailly, P. Louis-Plence, C. Jorgensen and D. Noel, Transplantation, 2006, 82, 1060–1066 CrossRef.
- A. H. Klopp, L. Lacerda, A. Gupta, B. G. Debeb, T. Solley, L. Li, E. Spaeth, W. Xu, X. Zhang, M. T. Lewis, J. M. Reuben, S. Krishnamurthy, M. Ferrari, R. Gaspar, T. A. Buchholz, M. Cristofanilli, F. Marini, M. Andreeff and W. A. Woodward, PLoS One, 2010, 5, e12180 CrossRef.
- I. W. Mak, N. Evaniew and M. Ghert, Am. J. Transl. Res., 2014, 6, 114–118 Search PubMed.
- S. Melissaridou, E. Wiechec, M. Magan, M. V. Jain, M. K. Chung, L. Farnebo and K. Roberg, Cancer Cell Int., 2019, 19, 16 CrossRef.
- D. S. Ferreira, A. P. Marques, R. L. Reis and H. S. Azevedo, Biomater. Sci., 2013, 1, 952–964 RSC.
- D. S. da Costa, R. A. Pires, A. M. Frias, R. L. Reis and I. Pashkuleva, J. Mater. Chem., 2012, 22, 7172–7178 RSC.
- S. Amorim, A. Martins, N. M. Neves, R. L. Reis and R. A. Pires, J. Mater. Chem. B, 2014, 2, 6939–6946 RSC.
- L. P. Ferreira, V. M. Gaspar and J. F. Mano, Biomaterials, 2018, 185, 155–173 CrossRef CAS.
- G. Rijal and W. Li, Sci. Adv., 2017, 3, e1700764 CrossRef.
- F. Hached, C. Vinatier, P. G. Pinta, P. Hulin, C. Le Visage, P. Weiss, J. Guicheux, A. Billon-Chabaud and G. Grimandi, Stem Cells Int., 2017, 2017, 9303598 Search PubMed.
- M. J. Costa, A. M. Marques, L. M. Pastrana, J. A. Teixeira, S. M. Sillankorva and M. A. Cerqueira, Food Hydrocolloids, 2018, 81, 442–448 CrossRef CAS.
- I. Braccini and S. Perez, Biomacromolecules, 2001, 2, 1089–1096 CrossRef CAS.
- G. Chan and D. J. Mooney, Acta Biomater., 2013, 9, 9281–9291 CrossRef CAS.
- H. N. Kim and N. Choi, BioChip J., 2019, 13, 8–19 CrossRef CAS.
- M. Cavo, M. Fato, L. Peñuela, F. Beltrame, R. Raiteri and S. Scaglione, Sci. Rep., 2016, 6, 35367 CrossRef CAS.
- J. E. Kim, D. S. Reynolds, M. H. Zaman and M. Mak, Integr. Biol., 2018, 10, 232–241 CrossRef CAS.
- C. Liu, Y. Liu, X. X. Xu, H. Wu, H. G. Xie, L. Chen, T. Lu, L. Yang, X. Guo, G. W. Sun, W. Wang, X. J. Ma and X. He, Exp. Cell Res., 2015, 330, 123–134 CrossRef CAS.
- H. Yu, J. K. Mouw and V. M. Weaver, Trends Cell Biol., 2011, 21, 47–56 CrossRef.
- A. Canibano-Hernandez, L. Saenz Del Burgo, A. Espona-Noguera, G. Orive, R. M. Hernandez, J. Ciriza and J. L. Pedraz, Mol. Pharm., 2017, 14, 2390–2399 CrossRef CAS.
- M. J. Kratochvil, A. J. Seymour, T. L. Li, S. P. Paşca, C. J. Kuo and S. C. Heilshorn, Nat. Rev. Mater., 2019, 4, 606–622 CrossRef CAS.
- S. M. Ridge, F. J. Sullivan and S. A. Glynn, Mol. Cancer, 2017, 16, 31 CrossRef.
- L. P. Ferreira, V. M. Gaspar, R. Henrique, C. Jeronimo and J. F. Mano, Biotechnol. J., 2017, 12, 12 Search PubMed.
- A. Dittmer, A. Fuchs, I. Oerlecke, B. Leyh, S. Kaiser, J. W. Martens, J. Lutzkendorf, L. Muller and J. Dittmer, Int. J. Oncol., 2011, 39, 689–696 CAS.
- R. Secunda, R. Vennila, A. M. Mohanashankar, M. Rajasundari, S. Jeswanth and R. Surendran, Cytotechnology, 2015, 67, 793–807 CrossRef CAS.
- S. Amorim, D. S. da Costa, D. Freitas, C. A. Reis, R. L. Reis, I. Pashkuleva and R. A. Pires, Sci. Rep., 2018, 8, 16058 CrossRef.
- D. T. Fisher, M. M. Appenheimer and S. S. Evans, Semin. Immunol., 2014, 26, 38–47 CrossRef CAS.
- I. H. Ham, H. J. Oh, H. Jin, C. A. Bae, S. M. Jeon, K. S. Choi, S. Y. Son, S. U. Han, R. A. Brekken, D. Lee and H. Hur, Mol. Cancer, 2019, 18, 68 CrossRef.
- R. Stern, A. A. Asari and K. N. Sugahara, Eur. J. Cell Biol., 2006, 85, 699–715 CrossRef CAS.
- F. Djouad, C. Bony, F. Apparailly, P. Louis-Plence, C. Jorgensen and D. Noël, Transplantation, 2006, 82, 1060–1066 CrossRef.
- H. Y. Lee and I. S. Hong, Cancer Sci., 2017, 108, 1939–1946 CrossRef CAS.
- E. Spaeth, A. Klopp, J. Dembinski, M. Andreeff and F. Marini, Gene Ther., 2008, 15, 730–738 CrossRef CAS.
- Y. Rattigan, J.-M. Hsu, P. J. Mishra, J. Glod and D. Banerjee, Exp. Cell Res., 2010, 316, 3417–3424 CrossRef CAS.
- Y. F. Zhao, S. P. Qiao, S. L. Shi, L. F. Yao, X. L. Hou, C. F. Li, F. H. Lin, K. Guo, A. Acharya, X. B. Chen, Y. Nie and W. M. Tian, ACS Appl. Mater. Interfaces, 2017, 9, 9327–9338 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0bm00843e |
| This journal is © The Royal Society of Chemistry 2021 |