Open Access Article
Open Access ArticleSize and density adjustment of nanostructures in nanochannels for screening performance improvement†
Dagui Wang‡
a,
Hongli Cheng‡b,
Cheng Chea,
Xiaoqing Wua,
Yuezhan Fengc,
Pengcheng Gao *a and
Fan Xia
*a and
Fan Xia *a
*a
aEngineering Research Center of Nano-Geomaterials of Ministry of Education, Faculty of Materials Science and Chemistry, China University of Geosciences, Wuhan 430074, P. R. China
bState Key Laboratory of Material Processing and Die & Mould Technology, School of Material Sciences and Engineering, Huazhong University of Science and Technology (HUST), Wuhan, 430074, P. R. China
cKey Laboratory of Advanced Materials Processing & Mold (Ministry of Education), National Engineering Research Center for Advanced Polymer Processing Technology, Zhengzhou University, Zhengzhou 450002, China
First published on 11th January 2021
Abstract
Biomimetic solid-state nanochannel/nanopore with flexible geometric structures, mechanical robustness and multifunctional surfaces have attracted extensive attention in separation, catalysis, drug delivery and other fields. Nanostructures have been introduced in nanoconfines to compress substances passthrough for high-efficient screening. However, precise controls of the nanostructure's growth in nanoconfines is rare. Herein, we developed a method to control size and number density of nanoparticles in nanochannels by adjusting polydopamine reducing conditions, achieving (1) particle size increasing, density increasing; (2) particle size increasing, density decreasing; (3) particle size increasing, density invariant; (4) particle size invariant, density increasing. The nanoparticles compressed the space of functional molecules decorated on them. Increasing size and density of nanoparticle enhanced the steric hinderance of functional molecules decorated on them and improved the wetting and chirality screening through nanochannels.
Biological pores are widespread in the membrane structure of organisms, which play a vital role in substances transport, energy transmission, signal exchange and other life processes.1–6
It is well-known that the high-efficient screening properties of biological pores stem from the numerous protein synaptophysin with fine structures and the precise functions on the surface of biological pores.7–10 Inspired by biological pores, solid-state nanochannels (SSNs) have been developed rapidly and prosperously, which show the superior flexibility in terms of geometry, robustness, surface chemical diversity.11–14 Functional elements (FEs) have been further introduced to improve the sensibility and selectivity of SSNs due to their strong effects on steric hinderance, electrostatic field and wettability in the confined channels at nanometric,15,16 which are widely applied in sensing,17–19 catalysis,20 drug delivery,21 and energy conversion.22–24 Simultaneously, the geometry structure of SSNs affects the performance of FEs. Kang et al. indicated that an improvement in the selectivity of K+ and Na+ by tuning pore diameter of graphite in combination with applying voltage.25 Zhao et al. designed a Cu2+detection by using an asymmetric polyglutamic acid-modified nanochannel with the smaller pore diameter approximate to hydrate Cu2+ radius.26 More recently, researches focused on the performance improvement of SSNs by introducing nanostructures for loading FEs and further compressing the space in between FEs and the inner wall of SSNs.27–30 For instance, Omar et al. developed a method to grow Au nanoparticles (AuNPs) reduced by polydopamine (PDA) in the nanopores which improved the rectification of a nanofluidic diode in a pH-dependent manner.31 Tang et al. reported a novel strategy to modify AuNPs with magnetic Fe3O4 before decorating aptamer on SSNs for sensing carcinoembryonic antigen.32 We further used the PDA-reduced AuNPs to decrease the pore diameter down to 10 nm locally. The compressed space enhances the interaction between DNA probes on AuNPs and DNA targets pass-by, that the single-base mismatch recognition of linear DNA with high specificity.33–35 It can be expected that the regulation of nanostructures in SSNs, such as size, geometry, surface chemistry and number density, will lead to more efficient regulation on substances transport by FEs. However, till now, the related studies have been rare.
Herein, we reported a method to control size and number density of PDA-reduced AuNPs in anodic aluminum oxide (AAO) SSNs by adjusting PDA reducing conditions. We obtained the SSNs modified with higher number density of AuNPs with bigger size by increasing PDA concentration in precursor and HCl concentration for the pre-etching of AAO surface. In the SSNs, the interactions between FEs and foreign substances on the AuNPs was further enhanced through the increase of AuNPs size and number density, which discriminated the transport of hydrophobic and hydrophilic molecules using hydrophobic FEs. Meanwhile, the transport of bull serum albumin (BSA) in a D-Cys and L-CyS modified SSNs was distinguished.
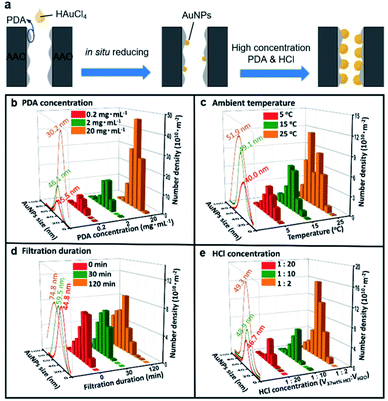
We investigated the reducing condition of PDA on the size and number density of AuNPs in SSNs, including (1) PDA concentration, (2) ambient temperature, (3) hydrochloric acid concentration and (4) filtration duration (Fig. 1a, Table S1, ESI†). The size of AuNPs varying monotonically or synchronically, are precisely regulated by adjusting the reaction conditions within the AAO channels with the pore diameter of 85 ± 15 nm (Fig. S1–S6, ESI†). Specifically, on the one hand, we can synchronically control the particle size and the number density of AuNPs on the inner wall of AAO membrane. Specifically, by increasing the PDA concentration, the size of AuNPs increases slightly, then decreases, while the number density of AuNPs increases as shown in Fig. 1b. Or, by raising the ambient temperature, the particle size and the number density of the AuNPs increases (Fig. 1c). On the other hand, we also can decorate the AAO membrane with the particle size or number density monotonically. By prolonging the filtration duration, the particles increase, while the number density keeps no change (Fig. 1d). The PDA in nanochannels serve as reductant and reduce HAuCl4 into AuNPs. By prolong the filtration duration, the size of AuNPs changes from 44.8 nm to 74.8 nm. The possible reason is that the long-time filtration will offer more enough PDA in nanochannel and realize sufficient reduction reaction of AuNPs. Thus, the large AuNPs emerge in nanochannels. Furthermore, by increasing the concentration of HCl, the number density of AuNPs increases, while the size of AuNPs has nearly no change as shown in Fig. 1e. In the present work, the variation of the AuNPs' size and number density would be realized by coordinating adjustment these four experimental conditions, from 30 nm to 80 nm and 109 to 1012 m−2, respectively. According to the results, we found the appropriate preparation conditions for larger and more AuNPs that were 2 mg mL−1 PDA concentration, VHCl![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) VH2O = 1
VH2O = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 and 25 °C ambient temperature (this method is used below).
2 and 25 °C ambient temperature (this method is used below).
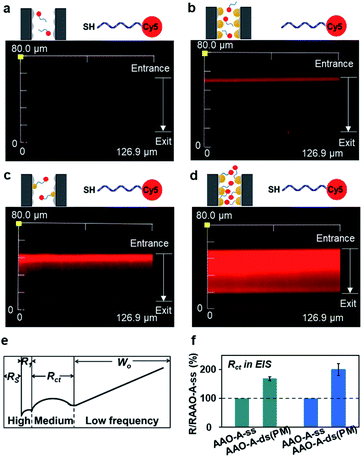
We next investigated the impact from the AuNPs on the distribution of FEs using laser confocal scanning microscopy (LCSM). The DNA was used as FEs due to their programmable structures and modification by numerous functional groups. The SSNs were firstly immersed in the buffer containing 1 μM Cy5-modified DNA for overnight (Sequence 1 in Table S2, ESI†). Then, the as-prepared SSNs were rinsed thoroughly. As shown in Fig. 2a, in a SSN without any AuNPs, nearly all SSNs were dark. When using Cy5-modified DNA without the thiol (–SH), shown in the fluorescence 2a, exists only at the outer surface of SSNs.
In contract, when using Cy5-modified DNA with thiol, the most of SSNs were lighted by Cy5 fluorescence, which indicated the immobilization of FEs at the AuNPs through specific Au–S interactions. In the SSN with higher number density and larger AuNPs, red domain covers nearly the whole region of the SSNs. It demonstrated that the thiol-modified FEs increased with the number density of AuNPs in SSNs.
The interactions between FEs and foreign substances taken place on the AuNPs were investigated taking the hybridization between thiol-modified DNA and their perfectly matched DNA chains as foreign substances as an example. Electrochemical Impedance Spectroscopy (EIS) is applied to characterize the variation of steric hindrance, in virtue of the higher sensitivity to the DNA grafting and hybridization than the I–V characterization.33 A typical two-electrode system was carried out and the equivalent circuit to simulate the impedance spectra was listed (Fig. S7, ESI†). Here, we set the charge-transfer resistances (Rct) of AAO–AuPNs (AAO–A) immobilized with single-stranded DNA (AAO–A–ss) as 100%. The real values are shown in Fig. S7 (ESI†). Interestingly, after the hybridization between ss-DNA and their perfectly matched DNA chains (AAO–A–ds (PM)), the increment of Rct is 101% for the AAO membrane with higher density of bigger AuNPs, which is greater than the increment of Rct as 69% for the AAO membrane with lower density of smaller AuNPs. Obviously, the larger size and higher density of AuNPs enhanced the interactions between FEs and foreign substances taken place on the AuNPs in SSNs.
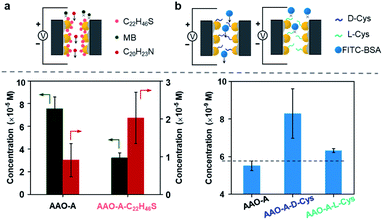
SSNs are an important platform for substances screening, on account of that their narrow spaces enhance the interaction with the foreign substances. We investigated the screening property of the SSNs with larger size and higher number density AuNPs. The AuNPs were modified with hydrophobic alkane. We investigated the concentration of hydrophilic methylene blue (MB) and hydrophobic amitriptyline (C22H46S). Schematic of our permeation experiment is shown in Fig. S8.† We applied the +2 V drive voltage to the permeate and the feed compartment, and the directionality drive lasted 40 min. We tested the ultraviolet (UV) absorption value of MB and C20H23N in permeate compartment, using the curve of standard concentration to calculate the concentrations (Fig. S9 and S10, ESI†). As can be seen in Fig. 3a, MB diffuses through AAO–A faster than AAO–A–C22H46S, and C20H23N in reverse. It illustrated that the transmission of hydrophilic molecules in AAO was inhibited, and the transmission of hydrophobic molecules was promoted after the secondary modification of hydrophobic molecules, which improved the screening properties.
To verify the screening property, we did another experiment that fluorescein isothiocyanate-bovine serum albumin (FITC-BSA) diffused through AuNP-decorated AAO membrane with secondary modification of L/D-cysteine (L/D-Cys).36 We also tested the UV absorption value of FITC-BSA in permeate compartment, using the curve of standard concentration to calculate the concentrations (Fig. S11, ESI†). FITC-BSA diffuses through AAO (≈6.5 nM) faster than AAO–A (≈5.5 nM) in Fig. 3. The increasing steric hindrance of AAO membranes after AuNPs modification blocked FITC-BSA diffusing directionally. However, FITC-BSA diffuses through AAO–A–L-Cys (≈8.0 nM) and AAO–A–D-Cys (≈6.4 nM) faster than AAO–A. Interestingly, we also found that FITC-BSA diffusion showed the selection of chirality in AAO–A–L/D-Cys.
In conclusion, we reported a strategy for controlling size and number density of AuNPs decorated on the inner walls of nanochannels. The appropriate preparation conditions are 2 mg mL−1 PDA concentration, VHCl![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) VH2O = 1
VH2O = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 and 25 °C ambient temperature, which can get higher number density AuNPs with larger size. It can impact on the distribution of FEs and the interaction between FEs and foreign substances taken place on the AuNPs. By introducing larger and more AuNPs into nanochannels, it can increase the steric hindrance of channels, in order to increase the probability of the interactions between FEs and the foreign substances. It achieves the purpose of screening performance improvement. Through further modification of hydrophobic and chiral molecule on the AuNPs, the SSNs show the selection of wetting and chirality in SSNs, respectively. Such a strategy offers a platform for high-efficient interactions in a nanoconfine.
2 and 25 °C ambient temperature, which can get higher number density AuNPs with larger size. It can impact on the distribution of FEs and the interaction between FEs and foreign substances taken place on the AuNPs. By introducing larger and more AuNPs into nanochannels, it can increase the steric hindrance of channels, in order to increase the probability of the interactions between FEs and the foreign substances. It achieves the purpose of screening performance improvement. Through further modification of hydrophobic and chiral molecule on the AuNPs, the SSNs show the selection of wetting and chirality in SSNs, respectively. Such a strategy offers a platform for high-efficient interactions in a nanoconfine.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work is supported by the National Natural Science Foundation of China (21525523, 21974126, 21874121, 51803194). This research is supported by the Zhejiang Provincial Natural Science Foundation of China under grant no. LY19B030001. The project is supported by the Open-end Funds from Engineering Research Center of Nano-Geomaterials of Ministry of Education (NGM2019KF013) and the Fundamental Research Funds for National Universities, China University of Geosciences (Wuhan). This research work was supported by the Open Funds of the State Key Laboratory of Electroanalytical Chemistry (SKLEAC202003) and the National Key Research and Development Program of China (2018YFE0206900).Notes and references
- E. Perozo, D. M. Cortes, P. Sompornpisut, A. Kloda and B. Martinac, Nature, 2002, 418, 942 CrossRef CAS.
- B. Guo, Y. Sheng, K. Zhou, Q. Liu, L. Liu and H.-C. Wu, Angew. Chem., Int. Ed., 2018, 57, 3602 CrossRef CAS.
- C. Zhou, L. Mei, Y.-S. Su, L.-H. Yeh, X. Zhang and S. Qian, Sens. Actuators, B, 2016, 229, 305–314 CrossRef CAS.
- R. S. Wei, V. Gatterdam, R. Wieneke, R. Tampé and U. Rant, Nat. Nanotechnol., 2012, 7, 257 CrossRef CAS.
- H. Zhang, Y. Tian and L. Jiang, Chem. Commun., 2013, 49, 10048 RSC.
- J. Zhang, Y. Yuan, T. Ye, S. Tan, N. Liu, J. Qian, S. Huang and F. Xia, Chem. Commun., 2020, 56, 10855 RSC.
- F. Alber, S. Dokudovskaya, L. M. Veenhoff, W. Zhang, J. Kipper, D. Devos, A. Suprapto, O. Karni-Schmidt, R. Williams, B. T. Chait, A. Sali and M. P. Rout, Nature, 2007, 450, 695 CrossRef CAS.
- Y.-L. Ying and Y.-T. Long, J. Am. Chem. Soc., 2019, 141, 15720 CrossRef CAS.
- X. Lin, Q. Yang, L. H. Ding and B. Su, ACS Nano, 2015, 9, 11266 CrossRef CAS.
- C. Zhu, Y. Teng, G. Xie, P. Li, Y. Qian, B. Niu, P. Liu, W. Chen, X. Kong, L. Jiang and L. Wen, Chem. Commun., 2020, 56, 8123 RSC.
- Z. Zhang, L. Wen and L. Jiang, Chem. Soc. Rev., 2018, 47, 322 RSC.
- Z. Zhu, X. Duan, Q. Li, R. Wu, Y. Wang and B. Li, J. Am. Chem. Soc., 2020, 142, 4481 CrossRef CAS.
- Y. –L. Ying, Y. –J. Li, J. Mei, R. Gao, Y. –X. Hu, Y. –T. Long and H. Tian, Nat. Commun., 2018, 9, 3657 CrossRef.
- M. Lepoitevin, T. Ma, M. Bechelany, J. –M. Janot and S. Balme, Adv. Colloid Interface Sci., 2017, 250, 195 CrossRef CAS.
- Z. Chen, T. Sun and G. Qing, J. Mater. Chem. B, 2019, 7, 3710 RSC.
- F. Yan, L. Yao, K. Chen, Q. Yang and B. Su, J. Mater. Chem. A, 2019, 7, 2385 RSC.
- L. Lin, J. Yan and J. Li, Anal. Chem., 2014, 86, 10546 CrossRef CAS.
- Y. Jiang, Y. Feng, J. Su, J. Nie, L. Cao, L. Mao, L. Jiang and W. Guo, J. Am. Chem. Soc., 2017, 139, 18739 CrossRef CAS.
- Z. Sun, F. Zhang, X. Zhang, D. Tian, L. Jiang and H. Li, Chem. Commun., 2015, 51, 4823 RSC.
- J. Ma, K. Li, Z. Li, Y. Qiu, W. Si, Y. Ge, J. Sha, L. Liu, X. Xie, H. Yi, Z. Ni, D. Li and Y. Chen, J. Am. Chem. Soc., 2019, 141, 4264 CrossRef CAS.
- Y. Sun, F. Zhang, Z. Sun, M. Song, D. Tian and H. Li, Chem.–Eur. J., 2016, 22, 4355 CrossRef CAS.
- J. Yang, X. Hu, X. Kong, P. Jia, D. Ji, D. Quan, L. Wang, Q. Wen, D. Lu, J. Wu, L. Jiang and W. Guo, Nat. Commun., 2019, 10, 1171 CrossRef.
- W. Xin, Z. Zhang, X. Huang, Y. Hu, T. Zhou, C. Zhu, X. –Y. Kong, L. Jiang and L. Wen, Nat. Commun., 2019, 10, 3876 CrossRef.
- S. F. Buchsbaum, G. Nguyen, S. Howorka and Z. S. Siwy, J. Am. Chem. Soc., 2014, 136, 9902 CrossRef CAS.
- Y. Kang, Z. Zhang, H. Shi, J. Zhang, L. Liang, Q. Wang, H. Agrenb and Y. Tub, Nanoscale, 2014, 6, 10666 RSC.
- X. Zhao, S. Wang, M. R. Younis, X. Xia and C. Wang, Anal. Chem., 2018, 90, 896 CrossRef CAS.
- L. Shi, C. Mu, T. Gao, W. Chai, A. Sheng, T. Chen, J. Yang, X. Zhu and G. Li, J. Am. Chem. Soc., 2019, 141, 8239 CAS.
- Q. Liu, K. Xiao, L. Wen, H. Lu, Y. Liu, X. Kong, G. Xie, Z. Zhang, Z. Bo and L. Jiang, J. Am. Chem. Soc., 2015, 137, 11976 CrossRef CAS.
- J. Zhang, N. Liu, B. Wei, X. Ou, X. Xu, X. Lou and F. Xia, Chem. Commun., 2015, 51, 10146 RSC.
- X. He, Y. Liu, K. Zhang, F. Wu, P. Yu and L. Mao, Angew. Chem., Int. Ed., 2018, 130, 4680 CrossRef.
- P. Gonzalo, T. Jimena, K. Wolfgang, T. Christina, T. María and A. Omar, J. Am. Chem. Soc., 2015, 137, 6011 CrossRef.
- H. Tang, H. Wang, C. Yang, D. Zhao, Y. Qian and Y. Li, Anal. Chem., 2020, 92, 3042 CrossRef CAS.
- P. Gao, L. Hu, N. Liu, Z. Yang, X. Lou, T. Zhai, H. Li and F. Xia, Adv. Mater., 2016, 28, 460 CrossRef CAS.
- T. Yan, F. Yang, S. Qi, X. Fan, S. Liu, N. Ma, Q. Luo, Z. Dong and J. Liu, Chem. Commun., 2019, 55, 2509 RSC.
- F. Zhu, G. Yang, M. K. Dhinakaran, R. Wang, M. Song and H. Li, Chem. Commun., 2019, 55, 12833 RSC.
- F. Zhang, Y. Sun, D. Tian and H. Li, Angew. Chem., Int. Ed., 2017, 56, 7186 CrossRef CAS.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d0ra10097h |
| ‡ Dagui Wang and Hongli Cheng equally contributed to this work. |
| This journal is © The Royal Society of Chemistry 2021 |