Open Access Article
Open Access ArticleEnzyme cascade-amplified immunoassay based on the nanobody–alkaline phosphatase fusion and MnO2 nanosheets for the detection of ochratoxin A in coffee†
Zeling Zhanga,
Benchao Sua,
Huan Xua,
Zhenyun Heb,
Yuling Zhouc,
Qi Chena,
Zhichang Suna,
Hongmei Cao a and
Xing Liu
a and
Xing Liu *a
*a
aSchool of Food Science and Engineering, Hainan University, 58 Renmin Avenue, Haikou 570228, China. E-mail: zhangzeling@hainanu.edu.cn; benchao312@hainanu.edu.cn; xuhuan.hnu@foxmail.com; qichen@hainanu.edu.cn; sunzhichang11@163.com; hmcao@hainanu.edu.cn
bHainan College of Economics and Business, Haikou 571129, China. E-mail: zhenyun89@foxmail.com
cHainan Institute for Food Control, Haikou 570314, China. E-mail: zhouyuling0607@163.com
First published on 21st June 2021
Abstract
Ochratoxin A (OTA) is a common food contaminant with multiple toxicities and thus rapid and accurate detection of OTA is indispensable to minimize the threat of OTA to public health. Herein a novel enzyme cascade-amplified immunoassay (ECAIA) based on the mutated nanobody–alkaline phosphatase fusion (mNb–AP) and MnO2 nanosheets was established for detecting OTA in coffee. The detection principle is that the dual functional mNb–AP could specifically recognize OTA and dephosphorylate the ascorbic acid-2-phosphate (AAP) into ascorbic acid (AA), and the MnO2 nanosheets mimicking the oxidase could be reduced by AA into Mn2+ and catalyze the 3,3′,5,5′-tetramethyl benzidine into blue oxidized product for quantification. Using the optimal conditions, the ECAIA could be finished within 132.5 min and shows a limit of detection of 3.38 ng mL−1 (IC10) with an IC50 of 7.65 ng mL−1 and a linear range (IC20–IC80) of 4.55–12.85 ng mL−1. The ECAIA is highly selective for OTA. Good recovery rates (84.3–113%) with a relative standard deviation of 1.3–3% were obtained and confirmed by high performance liquid chromatography with a fluorescence detector. The developed ECAIA was demonstrated to be a useful tool for the detection of OTA in coffee which provides a reference for the analysis of other toxic small molecules.
1. Introduction
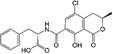
Ochratoxin A (OTA) is a mycotoxin and principally produced through the secondary metabolism of Aspergillus and Penicillium strains.1,2 As a typical storage mycotoxin, improper storage conditions will lead to the production of OTA, which is widely found in cereal, coffee, and meat. The teratogenicity, carcinogenicity, mutagenicity, and genotoxicity of OTA have been confirmed.3 In 1993, OTA was listed as a possible human carcinogen (group 2B) by the International Agency for Research on Cancer (IARC).4 To reduce the risk of a series of health problems caused by accidental consumption of food contaminated by OTA, it is urgent and necessary to establish rapid and accurate screening systems for OTA.So far, there have been many mature methods for the detection of OTA, such as gas chromatography, high performance liquid chromatography, and liquid chromatography-tandem mass spectrometry.5–9 Although these methods have high sensitivity and good specificity, the shortcomings of tedious and time-consuming sample pre-treatment and the requirements on the professional operators and sophisticated instruments restrict their widespread use. As an alternative, enzyme-linked immunosorbent assay (ELISA) has many advantages, such as ease operation, speediness, low cost, and high efficiency in practical application.10 Since the sensitivity of the traditional ELISA is relatively low, various nanomaterials such as luminescent nanomaterials and nanozymes are employed to enhance the method sensitivity.11,12 As a novel artificial enzyme, nanozyme is very attractive due to its advantages of ease of preparation, high stability, ease of surface modification, and low cost. Nanozymes could mimic natural enzymes (such as oxidase, peroxidase and catalase) and be applied for biosensors and immunoassays.12 The manganese dioxide (MnO2) nanosheets is a kind of two-dimensional nanosheet-based nanozyme with the merits of good water solubility, simple preparation, high biocompatibility, and stability.13 The MnO2 nanosheets mimic horse radish peroxide for catalyzing the 3,3′,5,5′-tetramethyl benzidine (TMB) and its oxidation activity could be tuned using the reducing agent ascorbic acid (AA).14,15 This property could be used for sensing alkaline phosphatase (AP) since the AP can catalyze the dephosphorylation of ascorbic acid-2-phosphate (AAP) into AA.16 Inspired by this idea, it is very likely to develop immunoassays using MnO2 nanosheets for detecting OTA because the AP is an enzyme commonly used for labeling the antibody.
In regard of the conjugation of AP to antibody, the commonly adopted chemical coupling methods are inevitable to bring the problems of randomness of coupling ratio, declined activity of antibody and enzyme.17 The genetically engineered antibody technology provides ideas for solving the above problems. Nanobody (Nb), also known single-domain antibody, is a highly concerned genetically engineered antibody.18 The Nb mainly includes the variable domain of the heavy chain antibody (VHH) in Camelidae and the variable domain of new antigen receptor (VNAR) in sharks.19,20 Due to the striking merits of high soluble expression, ease of genetic manipulation, and good thermostability, various Nbs and Nb-based fusions have been generated for application in food, environment, and biomedical analysis, of which the Nb–AP fusion retaining good activities of antibody and enzyme is particularly prominent in immunoassays.21–32
To expand the application of Nb–AP fusion in immunoassays, we constructed an Nb–AP fusion specific for OTA and developed an enzyme cascade-amplified immunoassay based on Nb–AP fusion and MnO2 nanosheets for colorimetric detection of OTA in coffee as seen in Scheme 1. The construction of the colorimetric sensing system for Nb–AP and optimization of the experimental conditions as well as evaluation of the enzyme cascade-amplified immunoassay were introduced in detail. As far as we know, there are few studies integrating the Nb–AP fusion and MnO2 nanosheets in an immunoassay for enzyme cascade-amplified detection of OTA and other toxic small molecules in food.
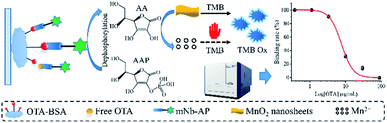 | ||
| Scheme 1 Schematic illustration of the enzyme cascade-amplified immunoassay based on Nb–AP fusion and MnO2 nanosheets for colorimetric detection of OTA in coffee. | ||
2. Experimental
2.1 Materials and reagents
AAP was obtained from Psaitong (Beijing, China). AA was obtained from Sinopharm (Beijing, China). Colormixed protein marker and nickel-nitrilotriacetic acid (Ni-NTA) Sepharose were obtained from Solarbio (Beijing, China). OTA, ochratoxin B (OTB), ochratoxin C (OTC), zearalenone (ZEN), aflatoxin B1 (AFB1), fumonisin B1 (FB1), and deoxynivalenol (DON) standards were procured from Pribolab (Qingdao, China). 3,3′,5,5′-Tetramethyl benzidine (TMB), p-nitrophenylphosphate (pNPP), ovalbumin (OVA), bovine serum albumin (BSA), and 96-well microplates were obtained from Sangon Biotech (Shanghai, China). Tetramethylammonium hydroxide (TMA·OH) and manganese chloride tetrahydrate (MnCl2·4H2O) were purchased from Aladdin (Shanghai, China). Polyvinylidene fluoride membrane was obtained from Millipore (Bearrica, MA). OTA–BSA conjugates were prepared previously in the laboratory. The recombinant pET25b-Nb-G53Q&S102D vector containing the mutated Nb gene for OTA and the recombinant pecan45-Nb28-AP vector were prepared in previous works.24,332.2 Construction of the recombinant expression vector for mutated nanobody–alkaline phosphatase fusion
The recombinant expression vector encoding the gene fragment of mutated nanobody–alkaline phosphatase fusion (mNb–AP) was constructed using the primers listed in Table S1† as follows. Firstly, the mNb gene fragment was amplified from the pET25b-Nb-G53Q&S102D vector by PCR using the primer pair of NA-NF and NA-NR containing Hind III restriction enzyme site and linker sequence. The PCR was performed using 30 cycles of denaturation (98 °C, 10 s), annealing (58 °C, 30 s) and extension (72 °C, 1 min). Afterwards, the AP gene fragment was amplified from the pecan45-Nb28-AP vector by PCR using the primer pair of NA-AF and NA-AR containing linker sequence and Not I restriction enzyme site. The PCR conditions were the same as that for mNb. Then, the purified mNb gene and AP gene were assembled through splicing by overlapped extension-polymerase chain reaction (SOE-PCR) using the primer pair of NA-NF and NA-AR. The SOE-PCR was performed by 7 cycles of denaturation (98 °C, 10 s) and annealing (68 °C, 2 min), and then 30 cycles of denaturation (98 °C, 10 s), annealing (58 °C, 15 s) and annealing (72 °C, 2 min).After purification, the amplified target genes were digested with Hind III and Not I and ligated into the similarly digested pET25b(+) vector to generate the pET25b/mNb–AP plasmid. The ligation products were then transformed into the chemically competent cells of E. coli DH5α and the transformation products were spread on the plate of solid LB medium with 50 μg mL−1 ampicillin for incubation (37 °C, 12 h). Single colonies grown on the plate were stochastically selected for colony PCR analysis and sequencing confirmation.
2.3 Expression and characterization of the mutated nanobody–alkaline phosphatase fusion
The confirmed expression vector of mNb–AP was extracted and transformed into the E. coli BL21(DE3) strain for auto-expression as previously reported.27 After expression, the bacteria were collected from the medium and broke by ultrasonication for releasing the soluble proteins secreted into the periplasm of bacteria cells. The mNb–AP was purified from the soluble proteins using an Ni-NTA column for further characterization. SDS-PAGE and western blot were used to evaluate the expression and purity of the mNb–AP in accordance to our previous work.24To measure the antibody activity and enzymatic activity of the mNb–AP, two indirect ELISAs were performed similarly except for the different signal amplification methods. Briefly, the 96-well microplate was incubated (37 °C, 2 h) with the coating antigen OTA–BSA conjugate (0.5 μg mL−1 in TBS, 100 μL per well). After washing three times of TBS containing 0.05% Tween-20 (TBST), the plate was blocked with 300 μL per well of 3% (m/v) skimmed milk powder (SMP)/TBS solution (37 °C, 1 h) and washed with three times of TBST. Then, 100 μL per well of the mNb–AP solutions with serial concentrations (0.07–4.6 μg mL−1 in TBS) was added for incubation (37 °C, 30 min) and washed again. For antibody activity test, 100 μL per well of the HRP/mouse anti-his tag mAb conjugate (0.33 μg mL−1 in TBS) was injected for incubation (37 °C, 30 min). The rinsed plate was then incubated (37 °C, 5 min) with 100 μL per well of the TMB substrate for color reaction. The detection signal (OD450) was generated by the addition of 2 M H2SO4 solution (50 μL per well) and measured on a microplate reader. While for enzymatic activity analysis, the plate was then directly incubated (37 °C, 10 min) with 100 μL per well of the pNPP substrate (1 mg mL−1 in 1 M diethanolamine solution containing 0.5 mM MgCl2, pH 9.8). The reaction was promptly terminated by 3 M NaOH solution (50 μL per well) and the absorbance at 405 nm (OD405) was recorded by the microplate reader.
2.4 Procedures of enzyme cascade-amplified immunoassay for OTA
The MnO2 nanosheets were synthesized and characterized as described in the ESI.† Then the enzyme cascade-amplified immunoassay based on mNb–AP and MnO2 nanosheets (ECAIA) for detecting OTA was performed as follows. First, the 96-well microplate was coated with OTA–BSA (2 μg mL−1 in TBS, 100 μL per well) and incubated at 37 °C for 2 h. After washing three times with TBST, the plate was incubated (37 °C, 1 h) with 3% (m/v) SMP/TBS solution. After washing again, the mixture of 50 μL per well of OTA standard solution and 50 μL per well of mNb–AP (1 μg mL−1) were incubated in the plate (37 °C, 30 min). Afterwards, the plate was rinsed and incubated (37 °C, 90 min) with 30 μL per well of 100 μM AAP, followed by incubation (37 °C, 150 s) with the mixture of 150 μL NaAC-HAC (40 mM, pH 3.8) and 10 μL 1.1 mM MnO2 nanosheets. After the final incubation (37 °C, 10 min) with 50 μL per well of 1 mM TMB chromogenic solution, the absorbance at 650 nm was measured on a multimode reader. For quantitative analysis, the competitive inhibition standard curve of OTA was constructed by plotting the binding rate (%) versus the logarithmic concentration of OTA. The binding rate (%) was calculated as 100 × (A − A′)/(A0 − A′), where A is the absorbance in presence of OTA and mNb–AP, A′ is the absorbance of blank (absent of OTA and mNb–AP), and A0 is the absorbance in presence of mNb–AP and absence of OTA.2.5 Selectivity of the ECAIA for OTA
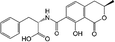
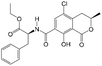
To evaluate the selectivity of the developed ECAIA for OTA, the cross-reactivity (CR) of the mNb–AP with OTA structural analogs (OTB and OTC) and some common cereal mycotoxins (AFB1, DON, FB1, ZEN) was determined by the ECAIA. The CR (%) was calculated as CR (%) = 100 × (IC50 of OTA)/(IC50 of other mycotoxins).2.6 Spiked sample analysis and validation
The spike-and-recovery study was performed to evaluate the accuracy and precision of the ECAIA in analyzing real coffee specimen. One gram of milled bake coffee bean with no OTA contamination tested by LC-MS/MS was weight into the 5 mL centrifuge tube and spiked with three levels of OTA standard (400, 700, and 1100 μg kg−1). Then 1 mL of 50% (v/v) methanol/0.5× TBS solution was added for each 15 min of vortex mixing and ultrasonic extraction. Afterwards, the supernatant was separated from the mixture by frozen centrifugation (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g, 15 min), and passed through a 0.22 μm filter. The filtrate was directly used for ECAIA analysis after dilution. To confirm the effectiveness of the developed method, the spiked coffee samples were simultaneously determined by the high performance liquid chromatography with fluorescence detector (HPLC-FLD). The pretreatment of coffee samples for HPLC-FLD is similar to that for ECAIA except that the obtained filtrate was further dried under nitrogen and resolved in pure methanol before analysis.
000g, 15 min), and passed through a 0.22 μm filter. The filtrate was directly used for ECAIA analysis after dilution. To confirm the effectiveness of the developed method, the spiked coffee samples were simultaneously determined by the high performance liquid chromatography with fluorescence detector (HPLC-FLD). The pretreatment of coffee samples for HPLC-FLD is similar to that for ECAIA except that the obtained filtrate was further dried under nitrogen and resolved in pure methanol before analysis.
3. Results and discussion
3.1 Construction, expression, and characterization of mNb–AP
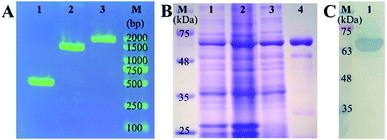
For the construction of expression vector of mNb–AP, the amplified fragments of mNb–linker (approximately 400 bp, lane 1), linker–AP (approximately 1000 bp, lane 2), and mNb–linker–AP (approximately 1400 bp, lane 3) were confirmed by DNA agarose gel electrophoresis as seen in Fig. 1A. After confirmation by colony PCR and DNA sequencing, the recombinant plasmid was transformed into the strain of E. coli BL21(DE3) for auto-induced expression. As shown in Fig. 1B, the target protein of approximately 66 kDa appeared in the total bacteria protein before induction (lane 1), which can be ascribed to the background expression caused by the trace lactose in the culture medium. After auto-induced expression, the content of mNb–AP in the total bacteria protein was increased (lane 2). The soluble mNb–AP in supernatant of the bacteria lysate (lane 3) was purified and the resultant target protein was obtained with a high purity (lane 4). As shown in Fig. 1C, the purified mNb–AP was further analyzed by western blot, and an only clear target protein band was detected (lane 1), indicating that there has no degradation of mNb–AP and no endogenous alkaline phosphatase contamination. Furthermore, the antibody activity and enzymatic activity of the mNb–AP were also determined by an indirect ELISA. As seen in Fig. S1,† both signal valules for mNb (OD450) and AP (OD405) rose rapidly with the increasing concentration of mNb–AP, which demonstrated the well-retained antibody activity and enzymatic activity of the fusion.3.2 Construction and optimization of the colorimetric sensing system for mNb–AP
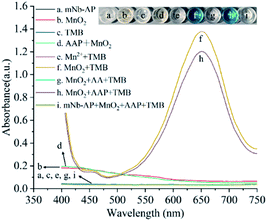
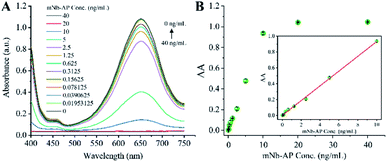
The colorimetric sensing system of mNb–AP is the key for the colorimetric method for detecting AFP based on mNb–AP and MnO2 nanosheets. The colorimetric sensing system was designed to contain MnO2 nanosheets, AAP, and TMB, and different experiments were performed to verify its feasibility. As shown in Fig. 2, both the mNb–AP (curve a) and the individual component of the sensing system, MnO2 nanosheets (curve b) and TMB (curve c), have no significant characteristic absorption peak with the wavelength in the range of 400–750 nm. When the MnO2 nanosheets was mixed with the TMB solution, a distinct characteristic absorption peak appeared at the wavelength of 650 nm. Accordingly, the solution color was changed from colorless and transparent (well c in inset) to blue (well f in inset). This result indicated the oxidizability of MnO2 nanosheets for TMB. However, the MnO2 nanosheets can't oxidize the TMB at the presence of 150 μM AA (curve g) since the MnO2 nanosheets were reduced and disintegrated into the non-oxidizing Mn2+ by AA as seen in Fig. S3† and curve e. The colorimetric sensing system has a high sensitivity for AA (LOD = 0.0723 μM), which is critical for highly sensitive detection of mNb–AP (Fig. S4†). There has no significant variation in the absorption band and color of MnO2 nanosheets with or without the addition of AAP (curve b, d and well b, d). However, the AAP could cause a slight decline of the characteristic absorption peak at 650 nm for the mixture of TMB and MnO2 nanosheets (curve f, h). This result confirmed that the AAP is weakly reductive to MnO2 nanosheets. Furthermore, after incubation with mNb–AP, the characteristic absorption peak at 650 nm for the mixture of TMB and MnO2 nanosheets disappeared completely with the change of the mixture color from blue to colorless and transparent (curve h, i and well h, i). This phenomenon was attributed to the dephosphorylation of mNb–AP and indicated that the colorimetric sensing system was available for detecting mNb–AP.After optimization of a series of experimental conditions (Fig. S5†), the colorimetric sensing system achieved the optimal performance for detecting mNb–AP based on 1.1 mM MnO2 nanosheets, 150 mM AAP, 150 s incubation of MnO2 nanosheets, and 90 min incubation of AAP. Fig. 3A shows a predicted and consecutive drop of the characteristic absorption peak at 650 nm with the increasing concentrations of mNb–AP (0–40 ng mL−1). As seen in Fig. 3B, a good correlation (R2 = 0.99868) was acquired between the A (A = A0 − A) and the concentration of mNb–AP (0.01953–10 ng mL−1), where A0 and A denote the absorbance at 650 nm in the absence and presence of mNb–AP, respectively. The linear regression equation was y = 0.00117 + 0.09301x, and the LOD (defined as the average response of 20 blank samples plus 3-fold standard deviations) was determined to be 0.0113 ng mL−1 of mNb–AP. Thus, the above results indicated the availability of the constructed colorimetric sensing system in highly sensitive detection of mNb–AP.
3.3 Optimization of the ECAIA for OTA
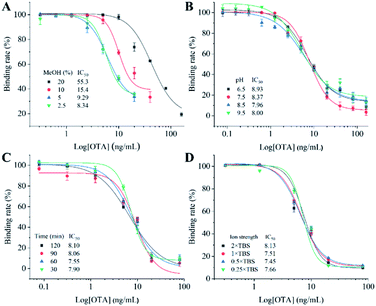
To acquire the optimal detection performance of the ECAIA, the optimum concentration of mNb–AP and coating antigen (OTA–BSA) was determined as 1 μg mL−1 and 2 μg mL−1 by the checkerboard titration (Table S2†) and indirect competitive ELISA (Fig. S6†), respectively. After that, various experimental conditions including methanol concentration, pH, competitive reaction time, and ion strength were further optimized using the half maximal inhibition concentration (IC50) as the evaluation criterion. The methanol is a frequently used solvent for OTA. As seen in Fig. 4A, the IC50 rose dramatically (8.34–55.3 ng mL−1) with the increasing methanol concentration (2.5–20%), indicating that the methanol can significantly influence the interaction of OTA–BSA and mNb–AP. Thus, 2.5% methanol was selected for the lowest IC50. Then, assay buffers with four different pH were used for ECAIA. Fig. 4B shows that the IC50 gradually declined with the pH in range of 6.5–8.5 and slightly increased when the pH further increased. Therefore, the optimal pH value was determined as 8.5. Subsequently, the competitive reaction time for ECAIA was optimized. As seen Fig. 4C, there was no significant effect on the performance of ECAIA under diverse reaction time. To speed the detection, 30 min of competitive reaction time was selected. Afterwards, effects of four TBS solutions (0.25×, 0.5×, 1×, and 2× TBS) with various ion strength on the ECAIA were evaluated. As shown in Fig. 4D, the IC50 slightly varied in range of 7.45–7.66 ng mL−1 when the ion strength didn't exceed that of 1× TBS. The 0.5× TBS was finally selected because of the lowest IC50. Thus, the optimal experimental conditions were determined as follows: 2.5% methanol, pH 8.5, 30 min of competitive reaction time, and 0.5× TBS.3.4 Analytical performance of the ECAIA for OTA
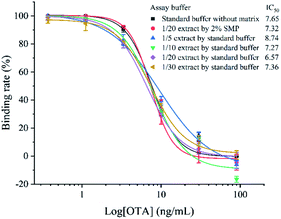
Using the optimized working conditions, the ECAIA was performed and shows a limit of detection of 3.38 ng mL−1 (IC10) with an IC50 of 7.65 ng mL−1 and a linear range (IC20–IC80) of 4.55–12.85 ng mL−1 as observed in Fig. 5. Compared with other reported methods based on nanomaterials for detecting OTA, the developed ECAIA shows a certain advantage in detection sensitivity or speed, although it exhibits a relatively narrow linear detection range.To evaluate the selectivity of the developed ECAIA for OTA, CRs of the mNb–AP with six substitutes of OTA were determined by the ECAIA. As seen in Table 1, the ECAIA has a low CR with the structural analogs of OTA (5.6% of OTB and 17.1% of OTC), and ignorable CRs (<1%) were obtained with other four common cereal mycotoxins (FB1, AFB1, ZEN and DON). These results demonstrated the high selectivity of the ECAIA for OTA.
3.5 Spiked sample analysis and validation
To minimize the matrix effects that are often encountered in food specimen analysis by immunoassay, we tried the dilution method to eliminate the coffee matrix interference on the ECAIA. As shown in Fig. 5, although at least 10-fold dilution of the coffee extract can facilitate the reduction of matrix effect with the IC50 (7.27 ng mL−1) drawing near that (7.65 ng mL−1) from the standard buffer without matrix, the standard competitive inhibition curves of the diluted extracts still can't overlap well with that of the standard buffer. In regard of this problem, we evaluated the effects of several shielding reagents including BSA, OVA, fish gelatin, and SMP on reducing matrix interference as described in theESI† (results seen in Tables S4 and S5†). After 20-fold dilution of the coffee extract by 2% SMP, its standard competitive inhibition overlapped well with that from the standard buffer as seen in Fig. 5, indicating that the matrix effect from coffee was minimized through 20-fold dilution using 2% SMP.The accuracy and precision of the established ECAIA for OTA was then evaluated by the recovery experiments using OTA-free coffee samples. As shown in Table 2, repetitive experiments indicated that the intra-assay recovery rate ranged from 84.3% to 113% with the relative standard deviation (RSD) in the range of 1.3–2.1%, and the inter-assay recovery rate ranged between 95.9% and 105% with the RSD of 2.1–3%. The HPLC-FLD was conducted to validate the results of the ECAIA. It exhibited a range of 94.9–105% for within-run recovery rate and 0.8–2.3% for RSD and 95–98.5% for between-run recovery rate with RSD of 1.5–6.1%. These results demonstrated that the ECAIA is applicable for accurate and precise detection of OTA in real coffee samples.
| Spiked OTA (μg kg−1) | ECAIA | HPLC-FLD | |||
|---|---|---|---|---|---|
| Recovery ± SD (%) | RSD (%) | Recovery ± SD (%) | RSD (%) | ||
| a Each assay was conducted three times on the same day.b Assays were conducted on three different days. | |||||
| Intra-assay (n = 3)a | 400 | 99.5 ± 1.4 | 1.4 | 105 ± 2.4 | 2.3 |
| 700 | 113 ± 2.4 | 2.1 | 94.9 ± 1.3 | 1.4 | |
| 1100 | 84.3 ± 1.1 | 1.3 | 97.2 ± 0.8 | 0.8 | |
| Intra-assay (n = 3)b | 400 | 105 ± 2.7 | 2.6 | 96.6 ± 5.9 | 6.1 |
| 700 | 101 ± 2.1 | 2.1 | 98.5 ± 2.3 | 2.3 | |
| 1100 | 95.9 ± 2.9 | 3.0 | 95.0 ± 1.4 | 1.5 | |
4. Conclusions
In this study, a mutated nanobody–alkaline phosphatase fusion (mNb–AP) was constructed and applied to develop an enzyme cascade-amplified immunoassay (ECAIA) using MnO2 nanosheets as the oxidase mimic for sensitive and colorimetric detection of OTA. The dual functional mNb–AP is the core element of the ECAIA, which retains good antigen-binding capacity and enzymatic activity. The constructed and optimized highly sensitive sensing system of mNb–AP, which is comprised of AAP, TMB, and MnO2 nanosheets, finally contributed to the ECAIA for OTA. The established ECAIA was demonstrated to be sensitive and selective detection of OTA in coffee samples with good accuracy and precision. Overall, this work further verified the immense potential of nanobody in analysis of toxic small molecules in food and broaden the use of MnO2 nanosheets for sensing detection of hazardous materials.Author contributions
Zeling Zhang: methodology, investigation, Writing – original draft. Benchao Su: data curation. Huan Xu: validation. Zhenyun He: supervision. Yuling Zhou: supervision. Qi Chen: funding acquisition. Zhichang Sun: funding acquisition. Hongmei Cao: resources. Xing Liu: conceptualization, resources, writing – review & editing, supervision, project administration, funding acquisition.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This research was funded by the Key Research and Development Project of Hainan Province (grant number ZDYF2020157), the Natural Science Foundation of Hainan Province (grant number 320RC509 and 2019RC119), and the National Natural Science Foundation of China (grant numbers 31760493 and 31901800). The APC was funded by the Key Research and Development Project of Hainan Province (grant number ZDYF2020157).References
- K. J. Van Der Merwe, P. S. Steyn, L. Fourie, D. B. Scott and J. J. Theron, Nature, 1965, 205, 1112–1113 CrossRef CAS PubMed.
- S. Amézqueta, S. Schorr-Galindo, M. Murillo-Arbizu, E. González-Peñas, A. López de Cerain and J. P. Guiraud, Food Control, 2012, 26, 259–268 CrossRef.
- F. Malir, V. Ostry, A. Pfohl-Leszkowicz, J. Malir and J. Toman, Toxins, 2016, 8, 191 CrossRef.
- International Agency for Research on Cancer (IARC), 1993.
- X. Zhang, M. Li, Z. Cheng, L. Ma, L. Zhao and J. Li, Food Chem., 2019, 297, 124850 CrossRef CAS PubMed.
- E. Moez, D. Noel, S. Brice, G. Benjamin, A. Pascaline and M. Didier, Food Chem., 2020, 310, 125851 CrossRef CAS PubMed.
- A.-K. Rausch, R. Brockmeyer and T. Schwerdtle, Food Chem., 2021, 338, 127801 CrossRef CAS.
- E. Vasconcelos Soares Maciel, K. Mejía-Carmona and F. M. Lanças, Molecules, 2020, 25, 2756 CrossRef PubMed.
- X. Li, W. Ma, Z. Ma, Q. Zhang and H. Li, Crit. Rev. Food Sci. Nutr., 2021, 1–32, DOI:10.1080/10408398.2021.1885340.
- J. Singh and A. Mehta, Food Sci. Nutr., 2020, 8, 2183–2204 CrossRef CAS.
- X. Fu, L. Chen and J. Choo, Anal. Chem., 2017, 89, 124–137 CrossRef CAS PubMed.
- Q. Wang, H. Wei, Z. Zhang, E. Wang and S. Dong, TrAC, Trends Anal. Chem., 2018, 105, 218–224 CrossRef CAS.
- M. Wu, P. Hou, L. Dong, L. Cai, Z. Chen, M. Zhao and J. Li, Int. J. Nanomed., 2019, 14, 4781–4800 CrossRef CAS PubMed.
- J. Wu, Q. Yang, Q. Li, H. Li and F. Li, Anal. Chem., 2021, 93, 4084–4091 CrossRef CAS PubMed.
- F. Tian, R. Fu, J. Zhou, Y. Cui, Y. Zhang, B. Jiao and Y. He, Sens. Actuators, B, 2020, 321, 128604 CrossRef CAS.
- Q. Yang, X. Wang, H. Peng, M. Arabi, J. Li, H. Xiong, J. Choo and L. Chen, Sens. Actuators, B, 2020, 302, 127176 CrossRef CAS.
- V. C. W. Tsang, R. M. Greene and J. B. Pilcher, J. Immunoassay, 1995, 16, 395–418 CrossRef CAS PubMed.
- S. Muyldermans, Annu. Rev. Biochem., 2013, 82, 775–797 CrossRef CAS PubMed.
- C. Hamers-Casterman, T. Atarhouch, S. Muyldermans, G. Robinson, C. Hammers, E. B. Songa, N. Bendahman and R. Hammers, Nature, 1993, 363, 446–448 CrossRef CAS.
- A. S. Greenberg, D. Avila, M. Hughes, A. Hughes, E. C. McKinney and M. F. Flajnik, Nature, 1995, 374, 168–173 CrossRef CAS PubMed.
- Y. Wang, P. Li, Z. Majkova, C. R. Bever, H. J. Kim, Q. Zhang, J. E. Dechant, S. J. Gee and B. D. Hammock, Anal. Chem., 2013, 85, 8298–8303 CrossRef CAS PubMed.
- X. Liu, Y. Xu, Y.-h. Xiong, Z. Tu, Y.-p. Li, Z.-y. He, Y.-l. Qiu, J.-h. Fu, S. J. Gee and B. D. Hammock, Anal. Chem., 2014, 86, 7471–7477 CrossRef CAS PubMed.
- J. Wang, C. R. S. Bever, Z. Majkova, J. E. Dechant, J. Yang, S. J. Gee, T. Xu and B. D. Hammock, Anal. Chem., 2014, 86, 8296–8302 CrossRef CAS PubMed.
- X. Liu, Y. Xu, D.-b. Wan, Y.-h. Xiong, Z.-y. He, X.-x. Wang, S. J. Gee, D. Ryu and B. D. Hammock, Anal. Chem., 2015, 87, 1387–1394 CrossRef CAS PubMed.
- J. Wang, Z. Majkova, C. R. S. Bever, J. Yang, S. J. Gee, J. Li, T. Xu and B. D. Hammock, Anal. Chem., 2015, 87, 4741–4748 CrossRef CAS PubMed.
- Z. Sun, J. Lv, X. Liu, Z. Tang, X. Wang, Y. Xu and B. D. Hammock, Anal. Chem., 2018, 90, 10628–10634 CrossRef CAS PubMed.
- Z. Tang, X. Wang, J. Lv, X. Hu and X. Liu, Food Control, 2018, 92, 430–436 CrossRef CAS.
- W. Ren, Z. Li, Y. Xu, D. Wan, B. Barnych, Y. Li, Z. Tu, Q. He, J. Fu and B. D. Hammock, J. Agric. Food Chem., 2019, 67, 5221–5229 CrossRef CAS PubMed.
- Z. Sun, X. Wang, Z. Tang, Q. Chen and X. Liu, Ecotoxicol. Environ. Saf., 2019, 171, 382–388 CrossRef CAS PubMed.
- K. Wang, N. Vasylieva, D. Wan, D. A. Eads, J. Yang, T. Tretten, B. Barnych, J. Li, Q. X. Li, S. J. Gee, B. D. Hammock and T. Xu, Anal. Chem., 2019, 91, 1532–1540 CrossRef CAS PubMed.
- Y. He, Y. Ren, B. Guo, Y. Yang, Y. Ji, D. Zhang, J. Wang, Y. Wang and H. Wang, Food Chem., 2020, 310, 125942 CrossRef CAS PubMed.
- B. Su, Y. Wang, H. Pei, Z. Sun, H. Cao, C. Zhang, Q. Chen and X. Liu, Anal. Methods, 2020, 12, 4742–4748 RSC.
- X. Wang, Q. Chen, Z. Sun, Y. Wang, B. Su, C. Zhang, H. Cao and X. Liu, Int. J. Biol. Macromol., 2020, 151, 312–321 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Table S1: Primers for the construction of recombinant expression vector for mNb–AP, Fig. S1: Analysis of antibody activity and enzymatic activity of the mNb–AP by an indirect ELISA, Fig. S2: Morphology and stability analysis of the MnO2 nanosheets, Fig. S3: UV-vis absorption spectra of MnO2 nanosheets, AA, and MnO2 nanosheets + AA, Fig. S4: Absorption spectra of the MnO2 nanosheets/TMB solution of the colorimetric sensing system induced by various concentrations of AA and the relationship between the difference of absorbance at 650 nm and the AA concentration, Fig. S5: Optimization of the colorimetric sensing system of mNb–AP, Table S2: Optimization of the concentrations of OTA–BSA and mNb–AP by the checkerboard titration, Fig. S6: Optimization of the coating antigen concentration by the indirect competitive ELISA, Table S3: Analytical performance comparison of the ECAIA with other reported methods based on nanomaterials for detecting OTA, Table S4: Evaluation of the effects of different shielding reagents on reducing matrix interference from coffee, Table S5: Optimization of the concentration of SMP. See DOI: 10.1039/d1ra03615g |
| This journal is © The Royal Society of Chemistry 2021 |