DOI:
10.1039/D1RA05820G
(Paper)
RSC Adv., 2022,
12, 8719-8730
Pomegranate seed polyphenol-based nanosheets as an efficient inhibitor of amyloid fibril assembly and cytotoxicity of HEWL†
Received
31st July 2021
, Accepted 9th March 2022
First published on 21st March 2022
Abstract
Poor water solubility and low bioavailability are considered as two main factors restricting therapeutic applications of natural polyphenols in relation to various disorders including amyloid-related diseases. Among various strategies developed to overcome these limitations, nanonization has attracted considerable attention. Herein, we compared the potency of bulk and nano forms of the polyphenolic fraction of pomegranate seed (PFPS) for modulating Hen Egg White Lysozyme (HEWL) amyloid fibril formation. Prepared PFPS nanosheets using direct oxidative pyrolysis were characterized by employing a range of spectroscopic and microscopic techniques. We found that the nano form can inhibit the assembly process and disintegrate preformed fibrils of HEWL much more effective than the bulk form of PFPS. Moreover, MTT-based cell viability and hemolysis assays showed the capacity of both bulk and nano forms of PFPS in attenuating HEWL amyloid fibril-induced toxicity, where the nano form was more effective. On the basis of thioflavin T results, a delay in the initiation of amyloid fibril assembly of HEWL appears to be the mechanism of action of PFPS nanosheets. We suggest that the improved efficiency of PFPS nanosheets in modulating the HEWL fibrillation process may be attributed to their increased surface area in accord with the surface-assistance model. Our results may present polyphenol-based nanosheets as a powerful approach for drug design against amyloid-related diseases.
Introduction
Among approaches developed for amyloid fibrillation inhibition, use of naturally-occurring molecules, especially certain dietary polyphenols, has attracted a great attention.1 However, there are a number of challenges that limit development of natural polyphenols as therapeutic agents relating to amyloid-related diseases. These include poor water solubility and stability, low absorption upon oral administration, and rapid metabolism under physiological conditions. Moreover, because of their hydrophobic nature, most of polyphenols are not expected to cross the blood–brain barrier (BBB), where they may demonstrate their neuroprotective effects.2,3 In recent years, nanonization, the process of size reduction of particles to nanoscale, especially with a sheet-like structure,4,5 has become one of the interesting research topics. In the case of polyphenols, nanonization has been considered as a promising approach to increase water solubility and bioavailability, and improve the pharmacological effects of these compounds.6–8 Interestingly, many recent reports show that nanoformulation of various natural polyphenols significantly improves their affinity for amyloidogenic species as well as their capacity to bypass tissue filtration and cross the BBB, where they effectively influence amyloid fibril formation and even target amyloid plaques.9–13 Pomegranate (Punica granatum L.) is an ancient edible fruit with a very diverse chemical composition including flavonoids, polyphenols, minerals, fatty acids, tannins, anthocyanins, and complex carbohydrates.14 It has been reported that extracts from different parts of pomegranate exhibit therapeutic properties and target a wide range of diseases including various types of cancer, cardiovascular diseases,15 diabetes,16 AIDS, obesity, and neurodegenerative disorders.17 These activities are mostly considered to be related to ellagitannins (including punicalagins), and various classes of polyphenolic compounds such as flavonoids, anthocyanidins, anthocyanins, flavonols, and flavones.17 Although several mechanisms are suggested to be related to the therapeutic benefits of pomegranate, it's antioxidant, anticarcinogenic, and anti-inflammatory properties have attracted the most attention.17 Regarding to the neurodegenerative diseases, some investigators have found that pomegranate extracts/constituents could be used as a novel protectant.18–26 However, poor water solubility, low bioavailability, and inability to cross BBB are the main challenges restricting their therapeutic applications, leading to attempts to solve these problems.27,28 Recently, we reported that polyphenolic fraction of propolis, in the nanosheet form, exhibits an improved capacity for modulating the assembly process as well as clearance of preformed fibrils of bovine insulin, and concluded that increased water solubility and surface area upon nanosheet formation might be involved.29 This finding suggests that nanoformulation of polyphenolic compounds, naturally occurring in fruits and plants, could be a promising approach for drug design relating to the neurological disorders. To further confirm this proposition, self-polymerization of the polyphenolic fraction of pomegranate seed (PFPS) was performed through direct pyrolysis. The bulk and nano forms of PFPS (PFPS nanosheets) were structurally and morphologically characterized. This was followed by employing a range of amyloid-specific techniques to compare the efficacy of bulk and nano forms of PFPS in modulating hen egg white lysozyme (HEWL) amyloid fibrillation process. HEWL is one of the most extensively studied proteins which assemble into amyloid fibrils under in vitro conditions.30 Moreover, structural homology with human lysozyme, which it's amyloid deposits are related to systemic amyloidosis,31 has made HEWL an attractive model for in vitro fibrillation studies. Obtained results clearly demonstrated that PFPS nanosheets, compared to their respective bulk form, were much more effective in inhibiting HEWL amyloid fibrillation and clearance of preformed fibrils, indicating improved anti-amyloidogenic properties of PFPS in the nano form.
Materials and methods
Materials
HEWL, Thioflavin T (ThT), Congo red (CR), Nile red (NR), 3-(4,5-dimethyl tiazol-2-yl)-2,5 diphenyl tetrazolium bromide (MTT), and dialysis tube were purchased from Sigma (St. Louis, MO, USA). SH-SY5Y cells were a gift from Dr Karima (Shahid Beheshti University of Medical Sciences, Tehran, Iran). The cell culture medium (DMEM-F12), fetal bovine serum (FBS), and penicillin–streptomycin antibiotics were purchased from Gibco BRL (Life Technology, Paisley, Scotland). n-Hexane was purchased from CARLO ERBA, France. All other chemicals were obtained from Merck (Darmstadt, Germany) and were reagent grade.
Extraction of polyphenolic fraction of pomegranate seed (PFPS)
Pomegranates were collected from the Mazandaran, Iran. Fifty grams of the dried pomegranate seeds were grinded finely, and then it's fatty acid content was extracted with Soxhlet using n-hexane for 24 h. To extract PFPS, the resultant powder was dried and then dispersed in 100 mL of deionized water (DW), stirred for 24 h, and filtered on a Whatman filter paper to eliminate non-soluble materials. Finally, the suspension was evaporated in an oven at 60 °C, and the resultant brown powder (Fig. S1A†) was grinded finely and stored in a dark place for subsequent experiments.
Preparation of PFPS nanosheets
PFPS nanosheets were prepared using direct oxidative pyrolysis, as previously reported with slight modifications.29 Briefly, one gram of the PFPS powder was added into a 10 mL beaker and heated to 200 °C for 2.5 min. A color change from brown to dark brown indicates successful pyrolysis and nanosheet formation. Then, the sample was quickly neutralized by adding 200 mL of 250 mM NaOH and stirred at room temperature (RT) for 8 h. The obtained suspension was filtered on a Whatman filter paper and dialyzed (using cut-off 12 kDa dialysis tube) against DW until neutral pH was achieved. The resultant dialyzed solution, containing the purified PFPS nanosheets, was dried in an oven at 60 °C. Finally, the obtained dark brown powder (Fig. S1B†) was grinded finely and stored in a dark place for subsequent experiments.
Characterization of bulk and nano forms of PFPS
Nanonization progress was confirmed by UV-Vis, FT-IR, and X-ray photoelectron spectroscopies. The UV-Vis spectrum of bulk and nano forms of PFPS (20 μg mL−1) was recorded using a spectrophotometer (UV-2550, Shimadzu). For FT-IR spectroscopy, samples were dried in a vacuum oven at 60 °C. The related spectrums were obtained from 400 to 4000 cm−1 using a Bruker spectrometer (EQUINOX 55 Bruker, Germany) by preparing their KBr pellets. X-ray photoelectron spectroscopy (XPS) analysis was performed using a K-alpha X-ray photoelectron spectrometer. The fluorescence emission spectra of bulk and nano forms of PFPS were measured in aqueous solution (10 μg mL−1) by a fluorescence spectrophotometer (Cary Eclipse VARIAN, Mulgrave, Australia) with a slit width of 5 nm for both excitation and emission. The thermal stability of bulk and nano forms of PFPS was determined by a thermal gravimetric analyzer (NETZSCH STA 409 PC/PG). The samples were dried in a vacuum oven at 60 °C and thermal gravimetric analysis (TGA) was performed as described.29 The surface zeta potential of PFPS nanosheets (20 μg mL−1) in a range of pH (from 2 to 12) was recorded using a zeta potential analyzer (Brookhaven Instrument, Holtsville, NY 11742-1896, USA). Finally, the microscopic features of PFPS nanosheets were analyzed using electron and atomic force microscopies. Briefly, aliquots of an aqueous suspension of PFPS nanosheets (∼20 μg mL−1) were placed on a carbon-coated copper grid and dried at RT. The samples were analyzed using a Philips CM300 transmission electron microscope (TEM). An aliquot of aqueous suspension was dropped on a clean glass slide and dried in the oven at 60 °C, coated with a gold layer and imaged by the field emission scanning electron microscopy (FE-SEM, Hitachi S4160 operating at 20 kV). Finally, for atomic force microscopy (AFM), aliquots of PFPS nanosheets solution (diluted with DW to a final concentration of 15 μg mL−1) were placed on a freshly cleaved mica and dried at RT. Images were acquired as reported previously.29
Sample preparation
Protein sample was dissolved in 50 mM glycine buffer (pH 2.2) in a final concentration of 7.5 mg mL−1 (∼500 μM). The stock solutions of bulk and nano forms of PFPS were prepared at 5 and 100 mg mL−1, respectively, using DW as solvent, and were stored at −20 °C until use (Fig. S1C†). The molar ratios of bulk/nano forms of PFPS to the HEWL used in this study were 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 150, 1
150, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50, and 1
50, and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 33.
33.
HEWL amyloid fibril formation and characterization
For amyloid fibrillation induction, aliquots of HEWL, containing 20 μM thioflavin T (ThT) and various concentrations (0, 50, 150, and 225 μg mL−1) of bulk or nano forms of PFPS, were transferred into a non-binding black surface with clear bottom polystyrene 96-well plate (Corning Incorporated, 2 Alfred Road, Kennebunk ME 04043 USA) with a final volume of 200 μL of assay solution in each well. The plate was sealed with crystal clear sealing tape (Axygen, Corning Incorporated USA) and loaded into a Synergy Hybrid Multi-Mode microplate reader (BioTek Instruments, Winooski, VT 05404-0998, USA) followed by incubation at 57 °C without agitation. The fluorescence signal was measured at 10 h intervals, with excitation at 440 nm and emission at 485 nm for 8 days. In all experiments, ThT fluorescence measurement was done in triplicate and the mean of the three measurements was determined. The acquired data from ThT fluorescence measurements were fitted to the sigmoid curve and kinetics parameters were determined according to the equation described by Nielsen et al.32 For Nile red (NR) fluorescence measurements, aliquots of protein solutions were removed at the end of incubation time and diluted to a final concentration of 5 μM, using H2O containing 50 μM NR. Samples were excited at 530 nm and emission spectra were recorded from 550–800 nm. For Congo red (CR) binding assay, aliquots of incubated HEWL (final concentration of 5 μM), were added to 950 μL of CR solution (20 μM). After 30 min of incubation at RT, absorbance spectra were recorded between 400 and 600 nm. The amount of bound CR was quantified according to the equation described by Klunk et al.33 Far-UV CD spectra were recorded in the range of 190–260 nm using an AVIV 215 spectropolarimeter (Aviv Associates, Lakewood, NJ, USA), in a final concentration of 30 μM in glycine buffer (50 mM, pH 2.2). Finally, 10 μL of the incubated samples (diluted to a final concentration of 25 μM) were mixed with 10 μL of ThT solution (100 μM) and incubated for 10 min in a dark place at RT. The mixture was added on a clean glass slide and air-dried. The images were captured on a fluorescence microscope (Zeiss, Germany) at 20× magnification.
SDS-PAGE analysis
Sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) was performed without boiling treatment. Aliquots (20 μL) corresponding to HEWL incubated under amyloidogenic conditions for 8 days, either alone or in the presence of various concentrations of bulk or nano forms of PFPS, were mixed with 10 μL of the sample buffer and applied onto the gel (15%). Electrophoresis was performed at constant voltage at 100 V. Gel was stained with 0.025% (w/v) Coomassie brilliant blue followed by multiple destaining.
Remodeling activity of bulk and nano forms of PFPS
Matured fibrils of HEWL were prepared by incubation of the protein solutions under amyloidogenic conditions for sufficient time, as described before. Then, aliquots (200 μL) of preformed amyloid fibrils were incubated alone or with various concentrations of bulk or nano forms of PFPS (150, 300, and 600 μg mL−1) at 37 °C for 96 h. The disaggregating potency of bulk and nano forms of PFPS was investigated by ThT assay and fluorescence microscopy imaging as described above.
In vitro cell viability assay
Human neuroblastoma SH-SY5Y cells, were cultured in DMEM-F12 medium as reported previously.34 For cytotoxicity experiments, cells were treated with increasing amounts (0–2000 μg mL−1) of bulk or nano forms of PFPS, or HEWL amyloid fibrils (0–50 μM) and incubated for 24 h. To evaluate the protective effects of bulk/nano forms of PFPS against toxicity induced by HEWL amyloid fibrils, the aliquots of incubated samples, aged without or with various concentrations of bulk/nano forms of PFPS under amyloidogenic conditions, were added to the cells (in a final concentration of 20 μM) and left for 24 h. Cells treated with 50 mM glycine buffer (pH 2.2) were used as control. Cell viability was assessed using the conventional MTT reduction assay. Results were expressed as percentage of MTT reduction relative to the control cells, assuming that the absorbance of control cells was 100%.
In vitro hemolysis assay
The hemolytic activity of bulk/nano forms of PFPS (0–400 μg mL−1), or HEWL amyloid fibrils (0–50 μM) was evaluated as described previously.29 To evaluate the protective effects of bulk/nano forms of PFPS against permeabilization induced by HEWL amyloid fibrils, the cell suspensions (1% hematocrit) were incubated alone or with various concentrations of incubated samples for 3 h at 37 °C while being gently stirred. The samples were then centrifuged at 1000 g for 10 min, followed by measuring the absorbance of supernatants at 540 nm. The percentage of hemolysis was calculated as a fraction of the maximum effect (1% (v/v) Triton X-100): (measured signal-blank signal)/(maximum signal-blank signal). For microscopy imaging, 10 μL of resultant pellets (diluted 3 fold with phosphate buffered saline) were added on a clean glass slide and air-dried. The images were captured on an optical microscope (Nikon, Japan) at 20× magnification.
Statistical analysis
All assays were performed two or three times with triplicate repeats. Data are expressed as percentage of values in untreated control cells, and each value represents the mean ± SD (n = 3). The significant differences between the means of the treated and untreated groups were calculated by unpaired Student's t-test and p-values less than 0.01 were considered significant. *p < 0.01, **p < 0.001 were significantly different from control cells/erythrocytes. #p < 0.01 was significantly different from those exposed only to HEWL amyloid fibrils.
Results and discussion
Extraction of PFPS, synthesis and characterization of PFPS nanosheets
The high fatty acid content of pomegranate seed was first removed by Soxhlet using n-hexane as solvent. It's polyphenolic content was then extracted by DW as described above. Indeed, this simple method provides an efficient way to extract the relative hydrophilic polyphenols of pomegranate seed, with the aim to prepare final hydrophilic water-soluble nanosheets. Combustion, pyrolysis, oxidative coupling, carbonization and hydrothermal carbonization are well-known methods to obtain value-added products from naturally-occurring polyphenols. However, due to existence of various polyphenols, in different content and chemical structure, investigating the mechanism of reactions occurring in these processes have many challenges.35 The PFPS nanosheets were fabricated through the pyrolysis process in the presence of atmospheric oxygen, leading to the fusion of polyphenolic species by reacting oxygen-containing functional groups like hydroxyl and carboxyl groups and forming the related ether and ester bridges. It worth to note that, decarboxylation of compounds and decomposition of ester-containing polyphenols during the process would diminish the possibility of polyphenols self-condensation.36 But, that's not the whole story, the phenoxy radicals formed in polyphenols structure show a resonance in which the radical character is localized on the ortho- and para-carbons as well as the phenolic oxygen, which can lead to self-condensation of polyphenols via the C–O and C–C bonds.37 The charge densities calculations have revealed that 41.3% and 39.1% of the spin densities are located on the phenolic and para-carbons, respectively,38 indicating that the polyphenols are mostly self-condensed through these two sites. The UV-Vis absorption spectrum of bulk form of PFPS showed a maximum absorption peak at 283 nm (corresponding to the typical π–π* transition of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C) with a shoulder at about 350 nm (corresponding to the n–π* transition of C
C) with a shoulder at about 350 nm (corresponding to the n–π* transition of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O bond) (Fig. 1A), indicating the presence of flavonoids/polyphenols.39,40 Upon undergoing nanonization, however, we observed a significant enhancement in the absorption spectrum with a blue shift (Fig. 1A). Despite confinement of the chromophore regions (i.e., short ranged ordered sp2 domains) by atoms with sp3 hybridization, it seems that the number of these regions is increased, likely due to the fusion of polyphenolic compounds during the nanonization process, leading to increased UV-Vis absorption of PFPS nanosheets (Fig. 1A). One notable characteristic of the bulk and nano forms of PFPS is their excitation wavelength-dependent emission. While the bulk form of PFPS shows some fluorescence properties (Fig. S2†), we observed a considerable emission for PFPS nanosheets (Fig. 1B). This may be attributed to the increased number of chromophore regions as described above, leading to a significant fluorescence with various emission patterns (Fig. 1B and S3†), a sign of non-uniformity in the size of chromophore regions in the PFPS nanosheets.41 As shown in Fig. 1B, the maximum emission of PFPS nanosheets is around 460 nm when excited at 350 nm. The emission peak shifts to shorter wavelengths (up to excitation at 320 nm) and longer wavelengths (up to excitation at 450 nm). The shift is more obvious when the fluorescence intensity is normalized at different excitation wavelengths (Fig. S3†). This excitation wavelength-dependent emission property of nano form is due to the inhomogeneity of the particle sizes and plenteous emissive traps of surface states formed by the hydroxy, carboxy and other oxygen-based functional groups.42,43 We believe that the prepared nanostructures have non-directional nature due to C–C cross linking phenolic moieties of polyphenols. Consequently, the configuration of interconnected carbon network would be tilted or rotated leading to isotopically bundled three-dimensional architectures. This would generate a non-graphitized phase of hard carbon with abundant structural defects after pyrolysis. Therefore, these structural defects along with those derived from partially carbonization can lead to short-range-ordered sp2-carbon domains of hard carbons. Moreover, due to the complicated mixture of polyphenols, it seems that there are various ordered sp2-carbon domains of different sizes which can result in excitation wavelength-dependent emission. We believe that the fluorescence property of PFPS nanosheets may provide these structures with the potential to use as imaging probes for protein aggregates detection. The FT-IR spectroscopy was employed to determine the functional groups in bulk and nano forms of PFPS. For the bulk form, the characteristic peaks are 3000–3200 (aromatic C–H), 2850–2925 (aliphatic C–H), 1475, 1531, and 1600 (aromatic C
O bond) (Fig. 1A), indicating the presence of flavonoids/polyphenols.39,40 Upon undergoing nanonization, however, we observed a significant enhancement in the absorption spectrum with a blue shift (Fig. 1A). Despite confinement of the chromophore regions (i.e., short ranged ordered sp2 domains) by atoms with sp3 hybridization, it seems that the number of these regions is increased, likely due to the fusion of polyphenolic compounds during the nanonization process, leading to increased UV-Vis absorption of PFPS nanosheets (Fig. 1A). One notable characteristic of the bulk and nano forms of PFPS is their excitation wavelength-dependent emission. While the bulk form of PFPS shows some fluorescence properties (Fig. S2†), we observed a considerable emission for PFPS nanosheets (Fig. 1B). This may be attributed to the increased number of chromophore regions as described above, leading to a significant fluorescence with various emission patterns (Fig. 1B and S3†), a sign of non-uniformity in the size of chromophore regions in the PFPS nanosheets.41 As shown in Fig. 1B, the maximum emission of PFPS nanosheets is around 460 nm when excited at 350 nm. The emission peak shifts to shorter wavelengths (up to excitation at 320 nm) and longer wavelengths (up to excitation at 450 nm). The shift is more obvious when the fluorescence intensity is normalized at different excitation wavelengths (Fig. S3†). This excitation wavelength-dependent emission property of nano form is due to the inhomogeneity of the particle sizes and plenteous emissive traps of surface states formed by the hydroxy, carboxy and other oxygen-based functional groups.42,43 We believe that the prepared nanostructures have non-directional nature due to C–C cross linking phenolic moieties of polyphenols. Consequently, the configuration of interconnected carbon network would be tilted or rotated leading to isotopically bundled three-dimensional architectures. This would generate a non-graphitized phase of hard carbon with abundant structural defects after pyrolysis. Therefore, these structural defects along with those derived from partially carbonization can lead to short-range-ordered sp2-carbon domains of hard carbons. Moreover, due to the complicated mixture of polyphenols, it seems that there are various ordered sp2-carbon domains of different sizes which can result in excitation wavelength-dependent emission. We believe that the fluorescence property of PFPS nanosheets may provide these structures with the potential to use as imaging probes for protein aggregates detection. The FT-IR spectroscopy was employed to determine the functional groups in bulk and nano forms of PFPS. For the bulk form, the characteristic peaks are 3000–3200 (aromatic C–H), 2850–2925 (aliphatic C–H), 1475, 1531, and 1600 (aromatic C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1730 (C
C), 1730 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O of ester and/or aldehyde groups), 1664 (ketone group), 1027, 1070, and 1232 cm−1 (C–O) (Fig. S4†). The assigned peak sequences at 690–900 cm−1 are for “out of plane”’ (oop) bending, indicating aromatic ring substitutes.29 Upon undergoing nanonization, the spectrum pattern was changed due to the fusion of polyphenolic molecules and the production of a new structure. As shown in Fig. S4,† broad peaks at 1554–1760 (aromatic C
O of ester and/or aldehyde groups), 1664 (ketone group), 1027, 1070, and 1232 cm−1 (C–O) (Fig. S4†). The assigned peak sequences at 690–900 cm−1 are for “out of plane”’ (oop) bending, indicating aromatic ring substitutes.29 Upon undergoing nanonization, the spectrum pattern was changed due to the fusion of polyphenolic molecules and the production of a new structure. As shown in Fig. S4,† broad peaks at 1554–1760 (aromatic C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C and C
C and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1000–1200 (C–O), and 3600 cm−1 (O–H) indicate the fundamental structural changes in the polyphenolic structures leading to the formation of various oxygen-based functional groups in the course of nanonization process.29 These structural changes were further confirmed by XPS analysis. The C 1s spectrum of bulk form was deconvoluted into four peaks including 284.5 eV (aromatic and aliphatic C
O), 1000–1200 (C–O), and 3600 cm−1 (O–H) indicate the fundamental structural changes in the polyphenolic structures leading to the formation of various oxygen-based functional groups in the course of nanonization process.29 These structural changes were further confirmed by XPS analysis. The C 1s spectrum of bulk form was deconvoluted into four peaks including 284.5 eV (aromatic and aliphatic C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 286.07 eV (C–O), 287.51 eV (C
C), 286.07 eV (C–O), 287.51 eV (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), and 288.71 eV (O–C
O), and 288.71 eV (O–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) (Fig. 1C). Upon nanonization, however, the C 1s spectrum of PFPS nanosheets was deconvoluted into three peaks including 284.5 eV (aromatic and aliphatic C
O) (Fig. 1C). Upon nanonization, however, the C 1s spectrum of PFPS nanosheets was deconvoluted into three peaks including 284.5 eV (aromatic and aliphatic C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 285.93 eV (C–O), and 287.72 eV (C
C), 285.93 eV (C–O), and 287.72 eV (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) (Fig. 1D).29,44–46 Comparison of the XPS spectrums and extracted data revealed a decrement in the intensity of C–O and C
O) (Fig. 1D).29,44–46 Comparison of the XPS spectrums and extracted data revealed a decrement in the intensity of C–O and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O peaks, and disappearance of the O–C
O peaks, and disappearance of the O–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O peak as well, upon nanonization (Fig. 1C, D and Table 1). Thus, the high abundance of C
O peak as well, upon nanonization (Fig. 1C, D and Table 1). Thus, the high abundance of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C, compared to C–O and C
C, compared to C–O and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, groups may provide another reason for the notable fluorescence properties of PFPS nanosheets (Fig. 1B and S3†). These observations suggest that PFPS molecules can be fused and combined by eliminating oxygen-containing small molecules such as water and alcohol. Moreover, it seems that the carboxylic groups in PFPS nanosheets undergo decarboxylation and produce CO2 during the pyrolysis process. As depicted in Fig. S5,† the surface zeta potential of PFPS nanosheets follows a pH-dependent decrease from +1.14 to −22.99 and then increased to −14.77 mV, indicating an isoelectric point around 1.7. In almost the whole pH range, the negative surface charge may be related to deprotonation of various functional groups existing in the structure of PFPS nanosheets.29 The increased of zeta potential by changing pH from 7.3 to 11.5 is likely related to the ion screening effect.47,48 The TEM and AFM images revealed around and sheet morphology with a diameter ranging from 400 to 600 nm and an average thickness of 2–3 nm (Fig. 2). The formation of stacked nanosheets may suggest the involvement of both lateral and vertical hydrogen bonds, mediated by the presence of oxygen-based functional groups. The FE-SEM images were also confirmed their potential to aggregate while drying (Fig. S6†). The thermal stability of the bulk and nano forms of PFPS was studied by TGA. Four main weight losses at temperatures below 100 °C (elimination of physically adsorbed water), between 130–250 °C and 250–367 °C (likely due to removal of flavonoids/polyphenols), and above 500 °C (probably due to the decomposition of resistant aromatic structures of biogenic salts such as carbonates29,49), was observed for the bulk form of PFPS (Fig. 3). In the case of nano form, we observed three main weight losses (Fig. 3). The first one below 100 °C relating to the elimination of physically adsorbed water. The second one ranging from 206–480 °C, is more likely due to the removal of functional groups, existing on the surface of PFPS nanosheets, and physically adsorbed organic polyphenols, accounting for 29.10 wt% or, in other words 34.03 wt% of the total weight of
PFPS nanosheets (Fig. 3). The third weight loss observed above 660 °C may attribute to the decomposition of main skeleton of PFPS nanosheets. TGA results suggest increased thermal stability of PFPS nanosheets, especially at high temperatures.
O, groups may provide another reason for the notable fluorescence properties of PFPS nanosheets (Fig. 1B and S3†). These observations suggest that PFPS molecules can be fused and combined by eliminating oxygen-containing small molecules such as water and alcohol. Moreover, it seems that the carboxylic groups in PFPS nanosheets undergo decarboxylation and produce CO2 during the pyrolysis process. As depicted in Fig. S5,† the surface zeta potential of PFPS nanosheets follows a pH-dependent decrease from +1.14 to −22.99 and then increased to −14.77 mV, indicating an isoelectric point around 1.7. In almost the whole pH range, the negative surface charge may be related to deprotonation of various functional groups existing in the structure of PFPS nanosheets.29 The increased of zeta potential by changing pH from 7.3 to 11.5 is likely related to the ion screening effect.47,48 The TEM and AFM images revealed around and sheet morphology with a diameter ranging from 400 to 600 nm and an average thickness of 2–3 nm (Fig. 2). The formation of stacked nanosheets may suggest the involvement of both lateral and vertical hydrogen bonds, mediated by the presence of oxygen-based functional groups. The FE-SEM images were also confirmed their potential to aggregate while drying (Fig. S6†). The thermal stability of the bulk and nano forms of PFPS was studied by TGA. Four main weight losses at temperatures below 100 °C (elimination of physically adsorbed water), between 130–250 °C and 250–367 °C (likely due to removal of flavonoids/polyphenols), and above 500 °C (probably due to the decomposition of resistant aromatic structures of biogenic salts such as carbonates29,49), was observed for the bulk form of PFPS (Fig. 3). In the case of nano form, we observed three main weight losses (Fig. 3). The first one below 100 °C relating to the elimination of physically adsorbed water. The second one ranging from 206–480 °C, is more likely due to the removal of functional groups, existing on the surface of PFPS nanosheets, and physically adsorbed organic polyphenols, accounting for 29.10 wt% or, in other words 34.03 wt% of the total weight of
PFPS nanosheets (Fig. 3). The third weight loss observed above 660 °C may attribute to the decomposition of main skeleton of PFPS nanosheets. TGA results suggest increased thermal stability of PFPS nanosheets, especially at high temperatures.
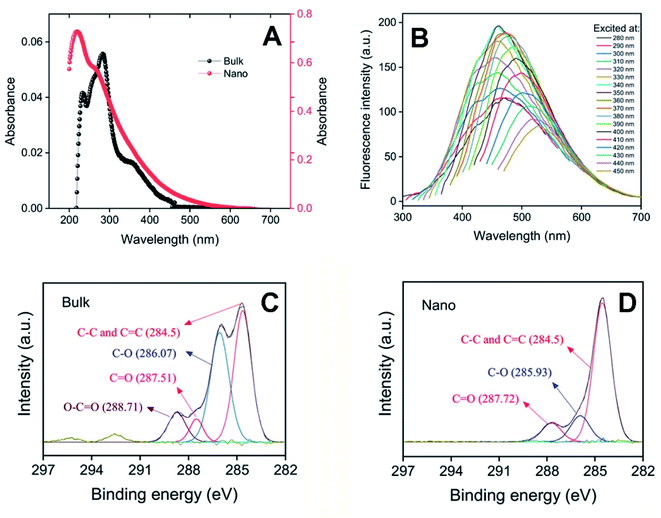 |
| | Fig. 1 Spectroscopic characterization of bulk and nano forms of PFPS. (A) UV-Vis absorption spectra of bulk (left axis) and nano (right axis) forms of PFPS (20 μg mL−1 aqueous solutions). (B) Fluorescence emission spectra of PFPS nanosheets, excited at different wavelengths. (C and D) C 1s spectra of bulk and nano forms of PFPS, respectively. | |
Table 1 Obtained data from XPS analysis of C 1s in bulk and nano forms of PFPS
| Bond type |
Bulk |
Nano |
| Peak energy bond, eV |
Height ratio |
Area ratio |
FWHM, eV |
Peak energy bond, eV |
Height ratio |
Area ratio |
FWHM, eV |
C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C, C–C C, C–C |
284.64 |
1 |
1 |
1.32 |
284.55 |
1 |
1 |
1.38 |
| C–O |
286.07 |
0.83 |
0.89 |
1.41 |
285.93 |
0.19 |
0.2 |
1.45 |
| C=O |
287.51 |
0.18 |
0.13 |
1.01 |
287.72 |
0.14 |
0.15 |
1.47 |
O–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O O |
288.71 |
0.23 |
0.23 |
1.32 |
— |
— |
— |
— |
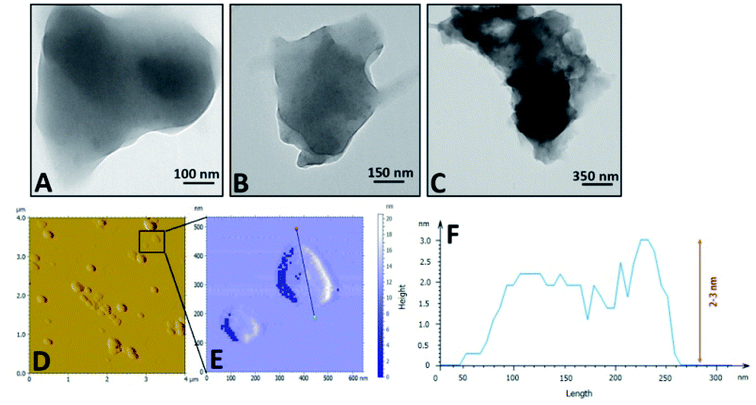 |
| | Fig. 2 Microscopic characterization of PFPS nanosheets. (A–C) TEM images; and (D and E) AFM imagesand (F) related line profile of PFPS nanosheets, respectively. | |
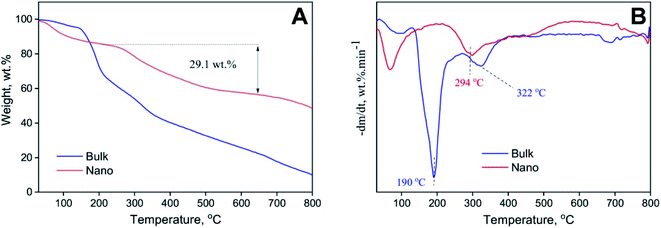 |
| | Fig. 3 Thermal stability of bulk and nano forms of PFPS. (A) TGA; and (B) DTG thermograms of bulk and nano forms of PFPS. | |
Effect of bulk and nano forms of PFPS on fibrillation process of HEWL
ThT fluorescence assay was employed to monitor the effect of bulk and nano forms of PFPS on the amyloid fibril formation of HEWL. From data depicted in Fig. 4A and B, it can clearly conclude that PFPS, in either bulk and nano forms, can modulate HEWL fibrillation process dose-dependently. However, at a same concentration, nano form of PFPS was found much more effective. As shown in Fig. 4A, in the presence of various concentrations of bulk form a dose-dependent decrease in ThT fluorescence intensity, with some increase in the nucleation phase duration (Table 2), was observed. In samples treated with nano form of PFPS, however, decrease in ThT fluorescence intensity was accompanied with a more significant elongation of the nucleation phase, as in the presence of 225 μg mL−1 PFPS nanosheets the fibrillation process of HEWL was completely inhibited without any detectible lag time (Fig. 4B and Table 2). These results suggest that PFPS, in the nanosheet form, exhibits enhanced capacity to delay and inhibit amyloid fibril assembly of HEWL. This improved efficiency of PFPS nanosheets was further confirmed by CR binding assay (Fig. S7†). The higher anti-amyloidogenic properties of PFPS nanosheets, compared to its respective bulk form, were further confirmed by fluorescence microscopy and SDS-PAGE analysis. The morphology of structures produced alone or in the presence of increasing concentrations of bulk/nano forms of PFPS were examined by fluorescence microscopy. As shown in Fig. 4C, incubation of protein samples for 8 days under amyloidogenic condition is sufficient to induce the formation of mature amyloid fibrils. The presence of bulk form of PFPS caused a dose-dependent inhibition of amyloid fibrils formation (Fig. 4C). In samples containing 225 μg mL−1 bulk form of PFPS, the formation of such fibrillar structures was prominently prevented, with the appearance of short amyloid fibrils (Fig. 4C). In samples treated with PFPS nanosheets, however, the extent of inhibition was more effective, as in samples containing 225 μg mL−1 PFPS nanosheets, no detectible structure was seen (Fig. 4C). The SDS-PAGE profile of incubated samples is depicted in Fig. 5. For the protein samples incubated alone a band trapped in the well of stacking gel, corresponding to high molecular weight amyloid fibrils, was seen. The presence of either bulk or nano forms of PFPS, however, caused a dose-dependent transition from large aggregates into low molecular weight intermediates and monomeric species; where nanosheets were more effective (Fig. 5). This observation further confirms the higher efficiency of nano form of PFPS to delay the initiation of HEWL fibrillation process, in accord with ThT fluorescence results (Fig. 4A and B). On the basis of obtained results, we suggest that in the presence of high concentrations of PFPS nanosheets, formation of mature fibrils is strongly inhibited. These results are in accord with our recent report,29 suggesting that nano form of polyphenolic compounds, in comparison with their respective bulk form, may exhibit improved capacity to modulate the assembly process of various amyloidogenic peptides and proteins. The size-dependent modulating effect of PFPS on HEWL fibrillation process is in accord with the surface-assistance model, reported by other studies.50–52 While the bulk form of PFPS was somewhat water-soluble (up to 5 mg mL−1), upon nanosheet formation, we were able to prepare a 100 mg mL−1 aqueous solution of PFPS. Clearly, this improved water solubility would increase capacity of PFPS nanosheets for hydrogen bonding/electrostatic interactions with surrounding molecules including HEWL monomers. This can lead to more efficient interaction with HEWL monomers of nano, compared to bulk, form of PFPS. Additionally, we suggest that increased surface area as well as chemical modifications introduced upon nanonization may also be involved. On the other hand, due to increased surface area, PFPS nanosheets exhibit a significantly high protein absorption capacity. Accordingly, nano form of PFPS may deplete a large fraction of monomers form the solution leading to decreased solution concentration and aggregation rate of the protein.50
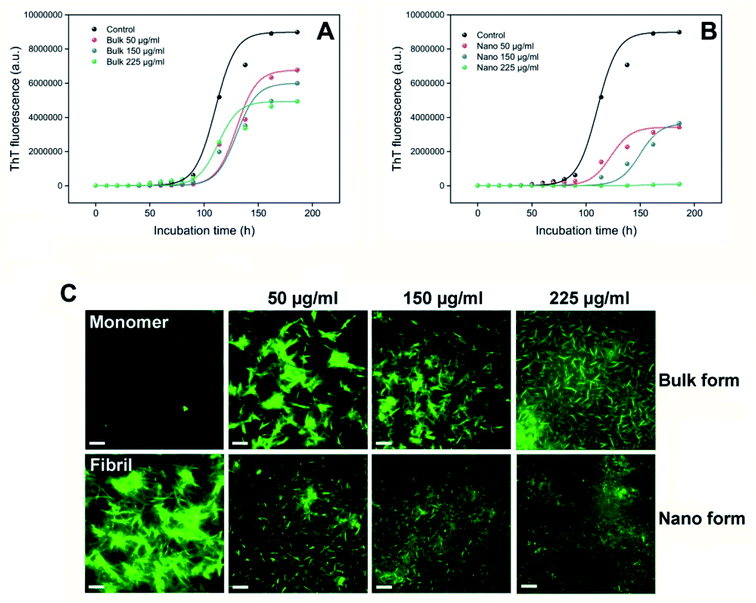 |
| | Fig. 4 Effect of bulk and nano forms of PFPS on amyloid fibrillation of HEWL. (A and B) Kinetics of HEWL amyloid fibril formation monitored by increasing fluorescence intensity of ThT at 485 nm. (C) Fluorescence microscopy images of HEWL samples incubated without or with various concentrations of the bulk or nano forms of PFPS for 8 days. The scale bars represent 500 nm. | |
Table 2 Effect of bulk and nano forms of PFPS on the kinetics parameters of HEWL amyloid fibrillation
| |
Lag time (h) |
t50 (h) |
Amplitude × 10−3 |
| The lag time could not be determined in the presence of 225 μg mL−1 nano form of PFPS since aggregation was not observed. |
| Control |
79.44 ± 1.81 |
110.88 ± 1.34 |
8990 ± 272.88 |
| Bulk 50 μg mL−1 |
101.89 ± 3.29 |
130.07 ± 3.91 |
6770 ± 77.88 |
| Bulk 150 μg mL−1 |
102.92 ± 1.50 |
128.21 ± 3.31 |
6000 ± 92.73 |
| Bulk 225 μg mL−1 |
90.12 ± 2.10 |
113.55 ± 1.19 |
4940 ± 71.18 |
| Nano 50 μg mL−1 |
102.11 ± 1.63 |
122.61 ± 1.15 |
3430 ± 43.20 |
| Nano 150 μg mL−1 |
128.51 ± 3.00 |
148.61 ± 2.03 |
3660 ± 77.88 |
| Nano 225 μg mL−1 |
—a |
165.1 ± 2.88 |
95.94 ± 1.49 |
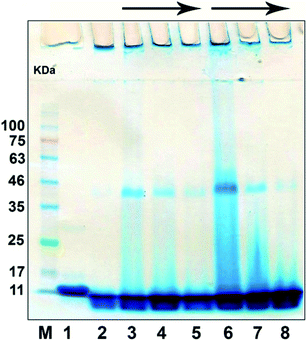 |
| | Fig. 5 SDS-PAGE analysis of HEWL samples incubated alone or with increasing concentrations of the bulk or nano forms of PFPS for 8 days. The lanes represent native HEWL (lane 1), samples incubated alone (lane 2) or with various concentrations of bulk (lanes 3–5) or nano (lanes 6–8) forms of PFPS. The arrows indicate the increasing concentration of the bulk/nano forms of PFPS corresponding to 50, 150, or 225 μg mL−1. Left lane represents protein marker. | |
Effect of bulk and nano forms of PFPS on structural/conformational changes of HEWL
In continuous of the study, the structural/conformational changes of HEWL in the absence and presence of bulk and nano forms of PFPS were examined using far-UV CD and NR fluorescence experiments. As depicted in Fig. 6, incubation of HEWL alone under amyloidogenic conditions caused significant conformational changes characterized by a gradual transition from a predominant α-helix to a β-sheet structure and a considerable enhancement in NR fluorescence signal with a pronounced blue shift of the spectrum from 663 nm to 632 nm. However, the presence of either bulk or nano forms of PFPS prevented such structural/conformational changes; where PFPS nanosheets showed more efficiency (Fig. 6). It worth to note that, although secondary structural transitions caused by amyloidogenic conditions were not completely inhibited in the presence of neither bulk nor nano forms of PFPS (Fig. 6A and B), we observed an effective suppression of the exposure of hydrophobic regions, along with a considerable red shift, in the samples incubated with 225 μg mL−1 PFPS nanosheets (Fig. 6C and D). This observation indicates that the presence of PFPS nanosheets has resulted in prevention of solvent-exposure of hydrophobic regions.
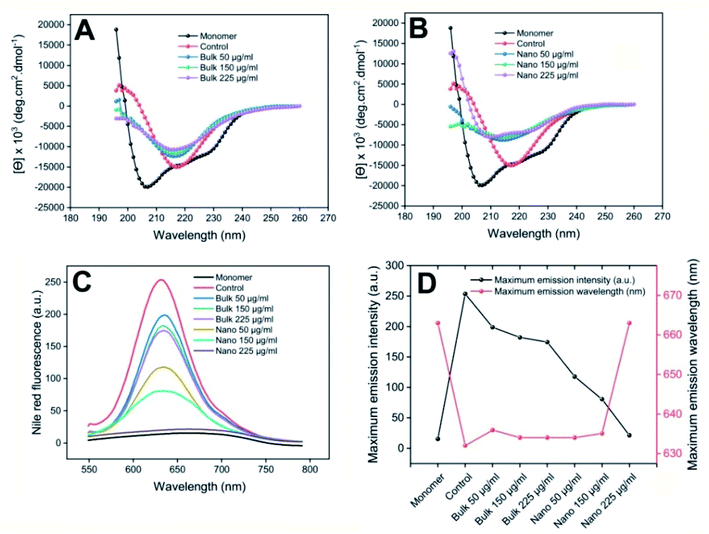 |
| | Fig. 6 Effect of bulk and nano forms of PFPS on structural/conformational changes of HEWL. (A and B) Far-UV CD spectra of HEWL samples incubated with the bulk or nano forms of PFPS, respectively. (C) Changes in surface hydrophobicity of HEWL samples incubated with the bulk or nano forms of PFPS, respectively, as measured by NR fluorescence assay. (D) Left-hand and right-hand y axes show maximum emission intensity and maximum emission wavelength of incubated samples, respectively. Further descriptions are provided in the text. | |
Remodeling activity of bulk and nano forms of PFPS
The potency of bulk and nano forms of PFPS to disaggregate preformed fibrils of HEWL was investigated by ThT assay and fluorescence microscopy imaging. As illustrated in Fig. 7A, the both bulk and nano forms of PFPS were able to disaggregate preformed fibrils of HEWL in a dose-dependent manner. Interestingly, decrease in fluorescence intensity of ThT was very fast, reaching a maximum decrement during the first few hours of incubation (Fig. 7A and B). However, with the same concentration, nano form of PFPS can more efficiently disaggregate the amyloid fibrils (Fig. 7A and B). For instance, the performance of 150 μg mL−1 nano form is better than 600 μg mL−1 bulk form of PFPS (Fig. 7A and B). As shown in Fig. 7D, further incubation of HEWL fibrils at 37 °C led to generation of very long fibrils assembled in large bundles, indicating the continuation of amyloid fibril assembly. While disintegration of the HEWL fibrils mediated by either bulk or nano forms of PFPS was considerable, nanosheets were more effective (Fig. 7E–H). As illustrated in Fig. 7H, in the presence of 600 μg mL−1 PFPS nanosheets we almost observed complete disintegration of amyloid fibrils without any detectable fibrillar structure. Through bearing a larger surface area, we speculate that via broad interaction and binding to fibrillar aggregates, PFPS nanosheets may disintegrate fibrillar structures more effective that the bulk form of PFPS, further clarifying surface-assistance model.50–52 Moreover, modification of surface functional groups upon nanonization, evidenced by various spectroscopic measurements, may also involve in facilitating absorption of fibrillar aggregates at the surface of PFPS nanosheets. However, further studies are needed to validate this proposition.
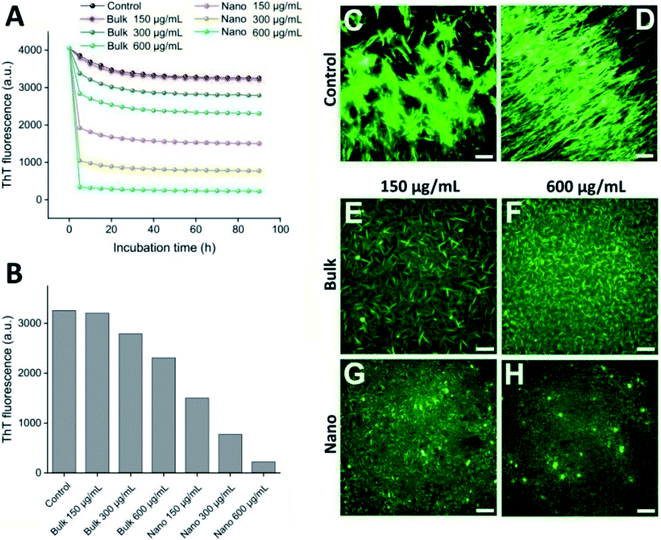 |
| | Fig. 7 Remodeling of preformed fibrils of HEWL by bulk or nano forms of PFPS. (A) Remodeling kinetics of preformed fibrils of HEWL demonstrated by decreasing fluorescence intensity of ThT at 485 nm. (B) An enlarged view of the changes in the fluorescence emission intensity of incubated samples after 1.5 h incubation. (C–H) Fluorescence microscopy images of preformed fibrils of HEWL before (C) or after incubation at 37 °C either alone (D) or with 150 (E) or 600 (F) μg mL−1 bulk, or 150 (G) or 600 (H) μg mL−1 nano forms of PFPS for 96 h. The scale bars represent 500 nm. | |
Effect of bulk and nano forms of PFPS on cytotoxicity and hemolytic activity of HEWL amyloid fibrils
The protective effect of PFPS nanosheets, in comparison with the respective bulk form, on toxicity of HEWL aggregates was evaluated using MTT-based cell viability and hemolysis assays. The non-toxic doses of bulk/nano forms of PFPS were determined based on the viability of SH-SY5Y human neuroblastoma cells. We found slight, if any, toxicity in relation to the bulk form of PFPS when applied at very high concentrations; whereas PFPS nanosheets were totally nontoxic (Fig. S8†). Results clearly show that HEWL fibrils, dose-dependently, reduce the cell viability (Fig. S9†). While a dose-dependent decrease in HEWL fibril-induced cytotoxicity was observed in the samples treated with either bulk or nano forms of PFPS, those containing PFPS nanosheets showed more significant viability (∼73%, compared to 37%, increase in the cell viability of samples containing 225 μg mL−1 nano or bulk forms of PFP, respectively) (Fig. 8), indicating that substantial inhibition of HEWL fibrillation process correlates with reduced cellular toxicity. These protective effects were further corroborated by hemolysis assay. First, the effect of bulk/nano forms of PFPS on the integrity of erythrocytes membrane was investigated. We observed some permeabilization of erythrocytes membrane relating to PFPS nanosheets, when applied at high concentrations (Fig. S10†), indicating higher membrane binding capacity of nano PFPS correlates with their increased surface area. Similar to cell toxicity, a dose-dependent release of hemoglobin was observed upon treatment with HEWL amyloid fibrils of erythrocytes (Fig. S11†). This was accompanied with substantial aggregation at high concentration (Fig. S11,† inset), in accord with a previous report.53 In the samples containing either bulk or nano forms of PFPS, however, a dose-dependent decrease in HEWL fibril-induced hemolysis was observed; where those containing nano form of PFPS were more protective (∼72%, compared to 63%, decrease in the hemolysis rate of samples containing 225 μg mL−1 nano or bulk forms of PFP, respectively) (Fig. 9A). Interestingly, neither bulk nor nano forms of PFPS were effective in preventing erythrocytes aggregation induced by HEWL aggregates (Fig. 9D–I), suggesting that membrane aggregation does not necessarily lead to permeabilization. Altogether, obtained data indicates that PFPS nanosheets, in comparison with the respective bulk form, can more effectively mitigate fibrillation and disaggregate preformed fibrils of HEWL and related toxicity. This is proposed to occur by interfering with the initial steps of fibrillation process and constraining the protein in a native-like state with significantly buried hydrophobic regions.
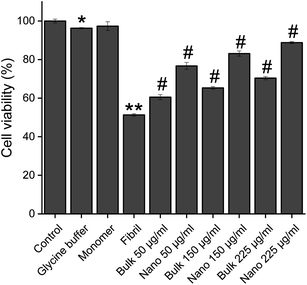 |
| | Fig. 8 Effect of the bulk and nano forms of PFPS on cytotoxicity induced by HEWL aggregates. Further descriptions are provided in the text. *p < 0.01, **p < 0.001, significantly different from control cells. #p < 0.001, significantly different from cells exposed only to HEWL amyloid fibrils. | |
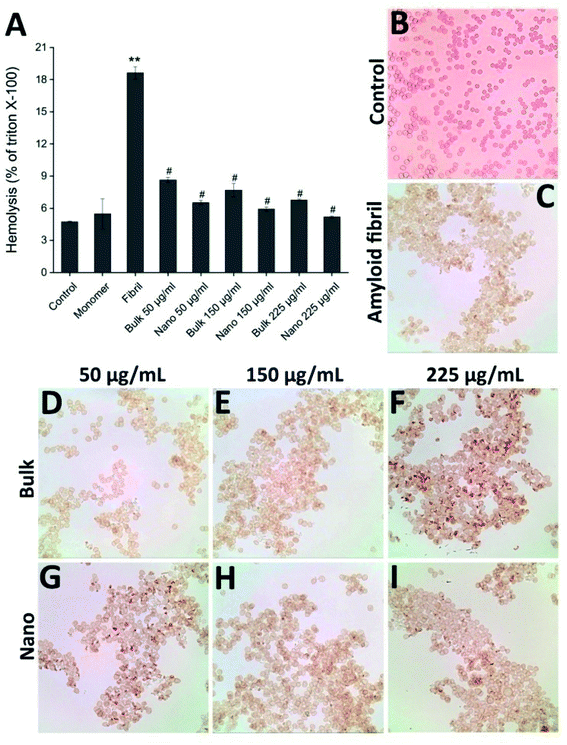 |
| | Fig. 9 Effect of the bulk and nano forms of PFPS on hemolytic activity of HEWL aggregates. (A) The percentage of hemolysis as a fraction of Triton X-100. (B–I) Optical microscopy images of samples incubated with HEWL aggregates aged either alone or with different concentrations of bulk or nano forms of PFPS. Further descriptions are provided in the text. **p < 0.001, significantly different from control erythrocytes. #p < 0.001, significantly different from erythrocytes exposed only to HEWL amyloid fibrils. | |
Conclusion
In the present study, we demonstrated that nano form of PFPS is much more effective than the respective bulk form of PFPS in modulating fibrillation process, as well as clearance of preformed fibrils of HEWL. The obtained results suggest that delay in initiation of amyloid fibril assembly, likely due to increased water solubility and surface area upon undergoing nanonization process, is the mechanism by which PFPS nanosheets modulate amyloid fibrillation of HEWL. While there are many reports describing the effects of polyphenol-based nanoparticles on assembly process of various peptides and proteins,9,11,54,55 we believe that our work has characteristics that distinguish it from the others. First, to avoid the profound challenges introduced by foreign additives including metals and polymers (commonly used as core materials for preparation of polyphenolic nanoparticles),56–59 we employed direct pyrolysis to self-polymerize PFPS. Second, the PFPS nanosheets are water-soluble nano-sized particles (without limitations such as poor water solubility and low bioavailability that greatly restrict therapeutic application of natural polyphenols), and compared to the bulk form, show considerable capacity to prevent assembly process of HEWL, as at PFPS nanosheets:HEWL molar ratios of as low as 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 33 the fibril formation process was completely inhibited. Beside these, our preliminary experiments showed the potential of these nanosheets to use as imaging probe for detection and targeting intracellular protein aggregates. While further investigations are needed to validate these results, we suggest that polyphenol-based nanosheets have the potency to use as a straightforward strategy to design potent inhibitor relating to amyloid-related diseases.
33 the fibril formation process was completely inhibited. Beside these, our preliminary experiments showed the potential of these nanosheets to use as imaging probe for detection and targeting intracellular protein aggregates. While further investigations are needed to validate these results, we suggest that polyphenol-based nanosheets have the potency to use as a straightforward strategy to design potent inhibitor relating to amyloid-related diseases.
Author contributions
A.-A. M. supervised the project, N. N. supervised preparation and characterization of the bulk and nano forms of PFPS; A.-A. M. and N. N. designed the experiments and analyzed obtained data; V. H. and A.-A. M. conducted experiments; A.-A. M., N. N., and A. M. wrote the manuscript; all authors read the manuscript and commented on it.
Conflicts of interest
The authors declare that they have no conflict of interest.
Acknowledgements
This work was supported by grants from the Research Council of the Institute for Advanced Studies in Basic Sciences (IASBS), Zanjan, Iran. The authors thank the departments of chemistry and physics of the IASBS for their valuable instrumental support.
References
- Y. Porat, A. Abramowitz and E. Gazit, Chem. Biol. Drug Des., 2006, 67, 27–37 CrossRef CAS PubMed.
- J. Chen, H. Lin and M. Hu, J. Pharmacol. Exp. Ther., 2003, 304, 1228–1235 CrossRef CAS PubMed.
- M. Singh, M. Arseneault, T. Sanderson, V. Murthy and C. Ramassamy, J. Agric. Food Chem., 2008, 56, 4855–4873 CrossRef CAS PubMed.
- C. Tan, X. Cao, X. J. Wu, Q. He, J. Yang and X. Zhang, et al., Chem. Rev., 2017, 117, 6225–6331 CrossRef CAS PubMed.
- C. Tan and H. Zhang, Chem. Soc. Rev., 2015, 44, 2713–2731 RSC.
- F. L. O. Da Silva, M. B. D. F. Marques, K. C. Kato and G. Carneiro, Expet Opin. Drug Discov., 2020, 15, 853–864 CrossRef CAS PubMed.
- H. Chen, C. Khemtong, X. Yang, X. Chang and J. Gao, Drug Discov. Today, 2011, 16, 354–360 CrossRef CAS PubMed.
- K. Riehemann, S. W. Schneider, T. A. Luger, B. Godin, M. Ferrari and H. Fuchs, Nanomedicine—challenge and perspectives, Angew. Chem., Int. Ed. Engl., 2009, 48, 872–897 CrossRef CAS PubMed.
- K. Debnath, S. Shekhar, V. Kumar, N. R. Jana and N. R. Jana, ACS Appl. Mater. Interfaces, 2016, 8, 20309–20318 CrossRef CAS PubMed.
- Y. Zhao, J. Cai, Z. Liu, Y. Li, C. Zheng and Y. Zheng, et al., Nano Lett., 2019, 19, 674–683 CrossRef CAS PubMed.
- H. Mohammad-Beigi, D. Morshedi, S. A. Shojaosadati, J. N. Pedersen, A. T. Marvian and F. Aliakbari, et al., RSC Adv., 2016, 6, 85312–85323 RSC.
- B. Yang, Y. Dong, F. Wang and Y. Zhang, Molecules, 2020, 25, 4613 CrossRef CAS PubMed.
- S. Z. Moradi, S. Momtaz, Z. Bayrami, M. H. Farzaei and M. Abdollahi, Front. Bioeng. Biotechnol., 2020, 8, 238 CrossRef PubMed.
- S. Sreekumar, H. Sithul, P. Muraleedharan, J. M. Azeez and S. Sreeharshan, BioMed Res. Int., 2014, 2014, 686921 CrossRef PubMed.
- D. Wang, C. Özen, I. M. Abu-Reidah, S. Chigurupati, J. K. Patra and J. O. Horbanczuk, et al., Front. Pharmacol., 2018, 9, 544 CrossRef PubMed.
- Y. Khajebishak, L. Payahoo, M. Alivand and B. Alipour, J. Cell. Physiol., 2019, 234, 2112–2120 CrossRef CAS PubMed.
- J. S. Jurenka, Altern. Med. Rev., 2008, 13, 128–144 Search PubMed.
- S. J. Choi, J. H. Lee, H. J. Heo, H. Y. Cho, H. K. Kim and C. J. Kim, et al., J. Med. Food, 2011, 14, 695–701 CrossRef CAS PubMed.
- S. Subash, N. Braidy, M. M. Essa, A. B. Zayana, V. Ragini and S. Al-Adawi, et al., Nutrition, 2015, 31, 223–229 CrossRef CAS PubMed.
- L. Rojanathammanee, K. L. Puig and C. K. Combs, J. Nutr., 2013, 143, 597–605 CrossRef CAS PubMed.
- R. E. Hartman, A. Shah, A. M. Fagan, K. E. Schwetye, M. Parsadanian and R. N. Schulman, et al., Neurobiol. Dis., 2006, 24, 506–515 CrossRef CAS PubMed.
- N. Braidy, M. M. Essa, A. Poljak, S. Selvaraju, S. Al-Adawi and T. Manivasagm, et al., Oncotarget, 2016, 7, 64589 CrossRef PubMed.
- M. M. Essa, S. Subash, M. Akbar, S. Al-Adawi and G. J. Guillemin, PLoS One, 2015, 10, e0120964 CrossRef PubMed.
- M. Kujawska, M. Jourdes, M. Kurpik, M. Szulc, M. Szaefer and P. Chmielarz, et al., Int. J. Mol. Sci., 2019, 21, 202 CrossRef PubMed.
- V. Tapias, J. R. Cannon and J. T. Greenamyre, Neurobiol. Aging, 2014, 35, 1162–1176 CrossRef PubMed.
- M. C. Morzelle, J. M. Salgado, M. Telles, D. Mourelle, P. Bachiega and H. S. Buck, et al., PLoS One, 2016, 11, e0166123 CrossRef PubMed.
- M. Mizrahi, Y. Friedman-Levi, L. Larush, K. Frid, O. Binyamin and D. Dori, et al., Nanomed.: Nanotechnol. Biol. Med., 2014, 10, 1353–1363 CrossRef CAS PubMed.
- M. S. Almuhayawi, W. S. Ramadan, S. Harakeh, S. K. Al Jaouni, D. J. Bharali and S. A. Mousa, et al., Saudi J. Biol. Sci., 2020, 27, 1710–1716 CrossRef CAS PubMed.
- M. Ramezani, M. D. Hesami, Y. Rafiei, E. R. Ghareghozloo, A. A. Meratan and N. Nikfarjam, ACS Appl. Bio Mater., 2021, 4, 3547–3560 CrossRef CAS PubMed.
- M. R. Krebs, D. K. Wilkins, E. W. Chung, M. C. Pitkeathly, A. K. Chamberlain and J. Zurdo, et al., J. Mol. Biol., 2000, 300, 541–549 CrossRef CAS PubMed.
- M. B. Pepys, P. N. Hawkins, D. R. Booth, D. M. Vigushin, G. A. Tennent and A. K. Soutar, et al., Nature, 1993, 362, 553–557 CrossRef CAS PubMed.
- L. Nielsen, R. Khurana, A. Coats, S. Frokjaer, J. Brange and S. Vyas, et al., Biochemistry, 2001, 40, 6036–6046 CrossRef CAS PubMed.
- W. E. Klunk, J. W. Pettegrew and D. J. Abraham, J. Histochem. Cytochem., 1989, 37, 1273–1281 CrossRef CAS PubMed.
- B. Katebi, M. Mahdavimehr, A. A. Meratan, A. Ghasemi and M. Nemat-Gorgani, Arch. Biochem. Biophys., 2018, 659, 22–32 CrossRef CAS PubMed.
- J. S. Kim, Bioresour. Technol., 2015, 178, 90–98 CrossRef CAS PubMed.
- G. H. Chio, M. G. Nam, S. J. Kwak, S. H. Kim, H. Chang and C. S. Shin, et al., Energy Environ. Sci., 2021, 14, 3203–3215 RSC.
- M. Altarawneh, B. Z. Dlugogorski, E. M. Kennedy and J. C. Mackie, Prog. Energy Combust. Sci., 2009, 35, 245–274 CrossRef CAS.
- R. Janoschek and W. M. F. Fabian, J. Mol. Struct., 2003, 661–662, 635–645 CrossRef CAS.
- K. R. Markham and T. J. Mabry, in The flavonoids, Springer, Boston, MA, 1975, pp. 45–77 Search PubMed.
- X. Tang, H. Yu, B. Bui, L. Wang, C. Xing and S. Wang, et al., Bioact. Mater., 2020, 6, 1541–1554 CrossRef PubMed.
- Y. P. Sun, B. Zhou, Y. Lin, W. Wang, K. S. Fernando and P. Pathak, et al., J. Am. Chem. Soc., 2006, 128, 7756–7757 CrossRef CAS PubMed.
- H. Ding, S. B. Yu, J. S. Wei and H. M. Xiong, ACS Nano, 2016, 10, 484–491 CrossRef CAS PubMed.
- P. Zhu, K. Tan, Q. Chen, J. Xiong and L. Gao, Chem. Mater., 2019, 31, 4732–4742 CrossRef CAS.
- C. Petit, M. Seredych and T. J. Bandosz, J. Mater. Chem., 2009, 19, 9176–9185 RSC.
- C. Botas, P. Álvarez, C. Blanco, R. Santamaría, M. Granda and P. Ares, et al., Carbon, 2012, 50, 275–282 CrossRef CAS.
- H. He, J. Klinowski, M. Forster and A. Lerf, Chem. Phys. Lett., 1998, 287, 53–56 CrossRef CAS.
- F. Liu, M. H. Jang, H. D. Ha, J. H. Kim, Y. H. Cho and T. S. Seo, Adv. Mater., 2014, 25, 3657–3662 CrossRef PubMed.
- S. Hassanzadeh, K. H. Adolfsson and M. Hakkarainen, RSC Adv., 2015, 5, 57425–57432 RSC.
- T. Carballo, M. V. Gil, X. Gómez, F. González-Andrés and A. Morán, Biodegradation, 2008, 19, 815–830 CrossRef PubMed.
- J. Wang, Y. Cao, Q. Li, L. Liu and M. Dong, Chem.–Eur. J., 2015, 21, 9632–9637 CrossRef CAS PubMed.
- T. Cedervall, I. Lynch, S. Lindman, T. Berggård, E. Thulin and H. Nilsson, et al., Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 2050–2055 CrossRef CAS PubMed.
- S. Linse, C. Cabaleiro-Lago, W. F. Xue, I. Lynch, S. Lindman and E. Thulin, et al., Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 8691–8696 CrossRef CAS PubMed.
- B. Huang, J. He, J. Ren, X. Y. Yan and C. M. Zeng, Biochemistry, 2009, 48, 5794–5800 CrossRef CAS PubMed.
- S. Palmal, A. R. Maity, B. K. Singh, S. Basu, N. R. Jana and N. R. Jana, Chem. - Eur. J., 2014, 20, 6184–6191 CrossRef CAS PubMed.
- A. Basu, S. Kundu, A. Das, C. Basu, S. Bhayye and S. Das, et al., Mater. Adv., 2020, 1, 1142–1150 RSC.
- N. Swilam and K. A. Nematallah, Sci. Rep., 2020, 10, 1–11 CrossRef PubMed.
- M. C. Moulton, L. K. Braydich-Stolle, M. N. Nadagouda, S. Kunzelman, S. M. Hussain and R. S. Varma, Nanoscale, 2010, 2, 763–770 RSC.
- J. Liang, F. Li, Y. Fang, W. Yang, X. An and L. Zhao, et al., Colloids Surf., B, 2011, 82, 297–301 CrossRef CAS PubMed.
- R. Indiarto, L. P. A. Indriana, R. Andoyo, E. Subroto and B. Nurhadi, Eur. Food Res. Technol., 2022, 248, 1–24 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1ra05820g |
|
| This journal is © The Royal Society of Chemistry 2022 |
Click here to see how this site uses Cookies. View our privacy policy here.  Open Access Article
Open Access Article *a,
Vahid Hassania,
Atiyeh Mahdavia and
Nasser Nikfarjam
*a,
Vahid Hassania,
Atiyeh Mahdavia and
Nasser Nikfarjam b
b
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 150, 1
150, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50, and 1
50, and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 33.
33.
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C) with a shoulder at about 350 nm (corresponding to the n–π* transition of C
C) with a shoulder at about 350 nm (corresponding to the n–π* transition of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O bond) (Fig. 1A), indicating the presence of flavonoids/polyphenols.39,40 Upon undergoing nanonization, however, we observed a significant enhancement in the absorption spectrum with a blue shift (Fig. 1A). Despite confinement of the chromophore regions (i.e., short ranged ordered sp2 domains) by atoms with sp3 hybridization, it seems that the number of these regions is increased, likely due to the fusion of polyphenolic compounds during the nanonization process, leading to increased UV-Vis absorption of PFPS nanosheets (Fig. 1A). One notable characteristic of the bulk and nano forms of PFPS is their excitation wavelength-dependent emission. While the bulk form of PFPS shows some fluorescence properties (Fig. S2†), we observed a considerable emission for PFPS nanosheets (Fig. 1B). This may be attributed to the increased number of chromophore regions as described above, leading to a significant fluorescence with various emission patterns (Fig. 1B and S3†), a sign of non-uniformity in the size of chromophore regions in the PFPS nanosheets.41 As shown in Fig. 1B, the maximum emission of PFPS nanosheets is around 460 nm when excited at 350 nm. The emission peak shifts to shorter wavelengths (up to excitation at 320 nm) and longer wavelengths (up to excitation at 450 nm). The shift is more obvious when the fluorescence intensity is normalized at different excitation wavelengths (Fig. S3†). This excitation wavelength-dependent emission property of nano form is due to the inhomogeneity of the particle sizes and plenteous emissive traps of surface states formed by the hydroxy, carboxy and other oxygen-based functional groups.42,43 We believe that the prepared nanostructures have non-directional nature due to C–C cross linking phenolic moieties of polyphenols. Consequently, the configuration of interconnected carbon network would be tilted or rotated leading to isotopically bundled three-dimensional architectures. This would generate a non-graphitized phase of hard carbon with abundant structural defects after pyrolysis. Therefore, these structural defects along with those derived from partially carbonization can lead to short-range-ordered sp2-carbon domains of hard carbons. Moreover, due to the complicated mixture of polyphenols, it seems that there are various ordered sp2-carbon domains of different sizes which can result in excitation wavelength-dependent emission. We believe that the fluorescence property of PFPS nanosheets may provide these structures with the potential to use as imaging probes for protein aggregates detection. The FT-IR spectroscopy was employed to determine the functional groups in bulk and nano forms of PFPS. For the bulk form, the characteristic peaks are 3000–3200 (aromatic C–H), 2850–2925 (aliphatic C–H), 1475, 1531, and 1600 (aromatic C
O bond) (Fig. 1A), indicating the presence of flavonoids/polyphenols.39,40 Upon undergoing nanonization, however, we observed a significant enhancement in the absorption spectrum with a blue shift (Fig. 1A). Despite confinement of the chromophore regions (i.e., short ranged ordered sp2 domains) by atoms with sp3 hybridization, it seems that the number of these regions is increased, likely due to the fusion of polyphenolic compounds during the nanonization process, leading to increased UV-Vis absorption of PFPS nanosheets (Fig. 1A). One notable characteristic of the bulk and nano forms of PFPS is their excitation wavelength-dependent emission. While the bulk form of PFPS shows some fluorescence properties (Fig. S2†), we observed a considerable emission for PFPS nanosheets (Fig. 1B). This may be attributed to the increased number of chromophore regions as described above, leading to a significant fluorescence with various emission patterns (Fig. 1B and S3†), a sign of non-uniformity in the size of chromophore regions in the PFPS nanosheets.41 As shown in Fig. 1B, the maximum emission of PFPS nanosheets is around 460 nm when excited at 350 nm. The emission peak shifts to shorter wavelengths (up to excitation at 320 nm) and longer wavelengths (up to excitation at 450 nm). The shift is more obvious when the fluorescence intensity is normalized at different excitation wavelengths (Fig. S3†). This excitation wavelength-dependent emission property of nano form is due to the inhomogeneity of the particle sizes and plenteous emissive traps of surface states formed by the hydroxy, carboxy and other oxygen-based functional groups.42,43 We believe that the prepared nanostructures have non-directional nature due to C–C cross linking phenolic moieties of polyphenols. Consequently, the configuration of interconnected carbon network would be tilted or rotated leading to isotopically bundled three-dimensional architectures. This would generate a non-graphitized phase of hard carbon with abundant structural defects after pyrolysis. Therefore, these structural defects along with those derived from partially carbonization can lead to short-range-ordered sp2-carbon domains of hard carbons. Moreover, due to the complicated mixture of polyphenols, it seems that there are various ordered sp2-carbon domains of different sizes which can result in excitation wavelength-dependent emission. We believe that the fluorescence property of PFPS nanosheets may provide these structures with the potential to use as imaging probes for protein aggregates detection. The FT-IR spectroscopy was employed to determine the functional groups in bulk and nano forms of PFPS. For the bulk form, the characteristic peaks are 3000–3200 (aromatic C–H), 2850–2925 (aliphatic C–H), 1475, 1531, and 1600 (aromatic C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1730 (C
C), 1730 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O of ester and/or aldehyde groups), 1664 (ketone group), 1027, 1070, and 1232 cm−1 (C–O) (Fig. S4†). The assigned peak sequences at 690–900 cm−1 are for “out of plane”’ (oop) bending, indicating aromatic ring substitutes.29 Upon undergoing nanonization, the spectrum pattern was changed due to the fusion of polyphenolic molecules and the production of a new structure. As shown in Fig. S4,† broad peaks at 1554–1760 (aromatic C
O of ester and/or aldehyde groups), 1664 (ketone group), 1027, 1070, and 1232 cm−1 (C–O) (Fig. S4†). The assigned peak sequences at 690–900 cm−1 are for “out of plane”’ (oop) bending, indicating aromatic ring substitutes.29 Upon undergoing nanonization, the spectrum pattern was changed due to the fusion of polyphenolic molecules and the production of a new structure. As shown in Fig. S4,† broad peaks at 1554–1760 (aromatic C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C and C
C and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1000–1200 (C–O), and 3600 cm−1 (O–H) indicate the fundamental structural changes in the polyphenolic structures leading to the formation of various oxygen-based functional groups in the course of nanonization process.29 These structural changes were further confirmed by XPS analysis. The C 1s spectrum of bulk form was deconvoluted into four peaks including 284.5 eV (aromatic and aliphatic C
O), 1000–1200 (C–O), and 3600 cm−1 (O–H) indicate the fundamental structural changes in the polyphenolic structures leading to the formation of various oxygen-based functional groups in the course of nanonization process.29 These structural changes were further confirmed by XPS analysis. The C 1s spectrum of bulk form was deconvoluted into four peaks including 284.5 eV (aromatic and aliphatic C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 286.07 eV (C–O), 287.51 eV (C
C), 286.07 eV (C–O), 287.51 eV (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), and 288.71 eV (O–C
O), and 288.71 eV (O–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) (Fig. 1C). Upon nanonization, however, the C 1s spectrum of PFPS nanosheets was deconvoluted into three peaks including 284.5 eV (aromatic and aliphatic C
O) (Fig. 1C). Upon nanonization, however, the C 1s spectrum of PFPS nanosheets was deconvoluted into three peaks including 284.5 eV (aromatic and aliphatic C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 285.93 eV (C–O), and 287.72 eV (C
C), 285.93 eV (C–O), and 287.72 eV (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) (Fig. 1D).29,44–46 Comparison of the XPS spectrums and extracted data revealed a decrement in the intensity of C–O and C
O) (Fig. 1D).29,44–46 Comparison of the XPS spectrums and extracted data revealed a decrement in the intensity of C–O and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O peaks, and disappearance of the O–C
O peaks, and disappearance of the O–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O peak as well, upon nanonization (Fig. 1C, D and Table 1). Thus, the high abundance of C
O peak as well, upon nanonization (Fig. 1C, D and Table 1). Thus, the high abundance of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C, compared to C–O and C
C, compared to C–O and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, groups may provide another reason for the notable fluorescence properties of PFPS nanosheets (Fig. 1B and S3†). These observations suggest that PFPS molecules can be fused and combined by eliminating oxygen-containing small molecules such as water and alcohol. Moreover, it seems that the carboxylic groups in PFPS nanosheets undergo decarboxylation and produce CO2 during the pyrolysis process. As depicted in Fig. S5,† the surface zeta potential of PFPS nanosheets follows a pH-dependent decrease from +1.14 to −22.99 and then increased to −14.77 mV, indicating an isoelectric point around 1.7. In almost the whole pH range, the negative surface charge may be related to deprotonation of various functional groups existing in the structure of PFPS nanosheets.29 The increased of zeta potential by changing pH from 7.3 to 11.5 is likely related to the ion screening effect.47,48 The TEM and AFM images revealed around and sheet morphology with a diameter ranging from 400 to 600 nm and an average thickness of 2–3 nm (Fig. 2). The formation of stacked nanosheets may suggest the involvement of both lateral and vertical hydrogen bonds, mediated by the presence of oxygen-based functional groups. The FE-SEM images were also confirmed their potential to aggregate while drying (Fig. S6†). The thermal stability of the bulk and nano forms of PFPS was studied by TGA. Four main weight losses at temperatures below 100 °C (elimination of physically adsorbed water), between 130–250 °C and 250–367 °C (likely due to removal of flavonoids/polyphenols), and above 500 °C (probably due to the decomposition of resistant aromatic structures of biogenic salts such as carbonates29,49), was observed for the bulk form of PFPS (Fig. 3). In the case of nano form, we observed three main weight losses (Fig. 3). The first one below 100 °C relating to the elimination of physically adsorbed water. The second one ranging from 206–480 °C, is more likely due to the removal of functional groups, existing on the surface of PFPS nanosheets, and physically adsorbed organic polyphenols, accounting for 29.10 wt% or, in other words 34.03 wt% of the total weight of
PFPS nanosheets (Fig. 3). The third weight loss observed above 660 °C may attribute to the decomposition of main skeleton of PFPS nanosheets. TGA results suggest increased thermal stability of PFPS nanosheets, especially at high temperatures.
O, groups may provide another reason for the notable fluorescence properties of PFPS nanosheets (Fig. 1B and S3†). These observations suggest that PFPS molecules can be fused and combined by eliminating oxygen-containing small molecules such as water and alcohol. Moreover, it seems that the carboxylic groups in PFPS nanosheets undergo decarboxylation and produce CO2 during the pyrolysis process. As depicted in Fig. S5,† the surface zeta potential of PFPS nanosheets follows a pH-dependent decrease from +1.14 to −22.99 and then increased to −14.77 mV, indicating an isoelectric point around 1.7. In almost the whole pH range, the negative surface charge may be related to deprotonation of various functional groups existing in the structure of PFPS nanosheets.29 The increased of zeta potential by changing pH from 7.3 to 11.5 is likely related to the ion screening effect.47,48 The TEM and AFM images revealed around and sheet morphology with a diameter ranging from 400 to 600 nm and an average thickness of 2–3 nm (Fig. 2). The formation of stacked nanosheets may suggest the involvement of both lateral and vertical hydrogen bonds, mediated by the presence of oxygen-based functional groups. The FE-SEM images were also confirmed their potential to aggregate while drying (Fig. S6†). The thermal stability of the bulk and nano forms of PFPS was studied by TGA. Four main weight losses at temperatures below 100 °C (elimination of physically adsorbed water), between 130–250 °C and 250–367 °C (likely due to removal of flavonoids/polyphenols), and above 500 °C (probably due to the decomposition of resistant aromatic structures of biogenic salts such as carbonates29,49), was observed for the bulk form of PFPS (Fig. 3). In the case of nano form, we observed three main weight losses (Fig. 3). The first one below 100 °C relating to the elimination of physically adsorbed water. The second one ranging from 206–480 °C, is more likely due to the removal of functional groups, existing on the surface of PFPS nanosheets, and physically adsorbed organic polyphenols, accounting for 29.10 wt% or, in other words 34.03 wt% of the total weight of
PFPS nanosheets (Fig. 3). The third weight loss observed above 660 °C may attribute to the decomposition of main skeleton of PFPS nanosheets. TGA results suggest increased thermal stability of PFPS nanosheets, especially at high temperatures.
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C, C–C
C, C–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O
O

![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 33 the fibril formation process was completely inhibited. Beside these, our preliminary experiments showed the potential of these nanosheets to use as imaging probe for detection and targeting intracellular protein aggregates. While further investigations are needed to validate these results, we suggest that polyphenol-based nanosheets have the potency to use as a straightforward strategy to design potent inhibitor relating to amyloid-related diseases.
33 the fibril formation process was completely inhibited. Beside these, our preliminary experiments showed the potential of these nanosheets to use as imaging probe for detection and targeting intracellular protein aggregates. While further investigations are needed to validate these results, we suggest that polyphenol-based nanosheets have the potency to use as a straightforward strategy to design potent inhibitor relating to amyloid-related diseases.







