 Open Access Article
Open Access ArticleSelective strategies for antibacterial regulation of nanomaterials
Jinliang Ma *ab,
Kexin Lia and
Shaobin Gu
*ab,
Kexin Lia and
Shaobin Gu *a
*a
aCollege of Food and Bioengineering, Henan University of Science and Technology, Luoyang, Henan 471023, China. E-mail: majin1987610@126.com; shaobingu@haust.edu.cn
bInstitute of Molecular Medicine, Renji Hospital, School of Medicine, Shanghai Jiao Tong University, Shanghai 200127, China
First published on 9th February 2022
Abstract
Recalcitrant bacterial infection, as a worldwide challenge, causes large problems for human health and is attracting great attention. The excessive antibiotic-dependent treatment of infections is prone to induce antibiotic resistance. A variety of unique nanomaterials provide an excellent toolkit for killing bacteria and preventing drug resistance. It is of great importance to summarize the design rules of nanomaterials for inhibiting the growth of pathogenic bacteria. We completed a review involving the strategies for regulating antibacterial nanomaterials. First, we discuss the antibacterial manipulation of nanomaterials, including the interaction between the nanomaterial and the bacteria, the damage of the bacterial structure, and the inactivation of biomolecules. Next, we identify six main factors for controlling the antibacterial activity of nanomaterials, including their element composition, size dimensions, surface charge, surface topography, shape selection and modification density. Every factor possesses a preferable standard for maximizing antibacterial activity, providing universal rules for antibacterial regulation of nanomaterials. We hope this comprehensive review will help researchers to precisely design and synthesize nanomaterials, developing intelligent antibacterial agents to address bacterial infections.
1. Introduction
Infectious diseases arising from pathogenic bacteria pose serious threats to public health, leading to increased morbidity and mortality.1,2 A variety of antibiotics have been widely utilized to control the growth of bacteria by inhibiting cell wall synthesis and the functions of essential proteins, DNA and RNA since the first discovery of penicillin in 1928.3,4 However, with excessive use of the available antibiotics, the treated bacteria have started to adapt to adverse living conditions through gene mutation and transfer, resulting in antibiotic resistance, which is a worldwide challenge.5,6 This escalating challenge has contributed to an urgent need to develop new antibacterial agents to alleviate the “superbugs” emerging from antibiotic resistance. In addition to designing and synthesizing new antibiotics to address the increasing challenge,7 researchers are devoting more efforts to look for more alternative strategies in a variety of fields,8,9 such as the improved antibiotic sensitization of bacteria based on biological regulation,10,11 the innovative development of antimicrobial peptides and protein toxins,12,13 and the personalized design of antibacterial nanomaterials.14,15Especially, due to their unique physiochemical properties, antibacterial nanomaterials have provided a robust toolkit for combating drug-resistant pathogenic bacteria and avoiding the emergence of antibiotic resistance, these materials include metal-based substances,16–19 polymeric nanostructures,20–22 and carbon-based nanomaterials.23 Their antibacterial activity arises from damage of the bacterial membrane and inactivation of biomolecules to impede the normal cellular function of the bacteria.24 For example, in metal nanoparticles (NPs), most of the studied silver NPs (AgNPs) possess excellent antibacterial activity against a series of pathogenic bacteria, contributing to their potential application in medical devices and consumer products.25,26 Their antimicrobial activity is exerted due to the silver cations released from the AgNPs.15,27,28 During the antibacterial reaction, the contact surface area and charge on the NPs are two main factors for regulating the binding of the NPs with the cell membrane, destroying the membrane integrity.29–31 In addition, AgNP-induced formation of reactive oxygen species (ROS) leads to the death of the bacteria.32 The stability of metal NPs is important to realize their antibacterial properties, where a too unstable or too stable structure may not be preferable.33–35 Polymeric nanostructures obtained via self-assembly, such as phosphonium-functionalized polymer micelles and povidone-iodine-based polymeric nanoparticles, have high antibacterial activity.36,37 Their antibacterial activity is relative to the controllability of their length and the charge of the polymer. Carbon-based nanomaterials (CNMs) with lower toxicity and higher safety can be endowed with antibacterial activities23 by regulating their lateral sizes, shapes, layer numbers, and surface charges.38–40 Therefore, many strategies can regulate the antibacterial activity of nanomaterials. The reported relevant reviews have focused on the mechanisms and molecular targets of the antimicrobial activity of metals,14 the biological action of antibacterial silver with different forms,16,27,41 the antibacterial efficiency of gold nanoparticles (AuNPs) with nonantibiotic or antibiotic molecules,42 the antibacterial properties and surface functionality of different NPs,15,43–46 and the role of designed nanomaterials in eradicating bacteria in biofilms.47 However, the universal relationship between the antibacterial activity and properties of different nanomaterials has not been understood systematically. The confirmation of clear design rules is very important to build customized antibacterial nanomaterials.
In this review, we categorize the antibacterial mechanisms of different nanomaterials and the key factors for controlling their antibacterial activity, providing selective strategies for regulating antibacterial nanomaterials (Fig. 1). To begin with, we discuss how the nanomaterials interact with the bacteria and destroy the bacterial structure to exert their antibacterial activity. Next, we mainly review six factors for regulating the antibacterial activity of nanomaterials, including their element composition, size dimension, charge type, surface topography, shape selection, and modification density. In addition, two other factors, namely stiffness and hydrophobicity, are listed briefly. We hope that this comprehensive review can help researchers precisely customize nanomaterials to meet the requirements for developing intelligent antibacterial agents.
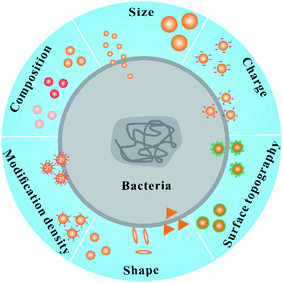 | ||
| Fig. 1 The main factors for regulating the antibacterial activity of nanomaterials: composition, size, charge, topography, shape and modification density. | ||
2. Antibacterial manipulation of nanomaterials
Based on their unique physiochemical properties, nanomaterials exert antibacterial activity according to three main bactericidal pathways (Fig. 2). The integrity of the cell membrane is destroyed with initial binding of the nanomaterials to the bacterial surface, causing possible leakage of intracellular molecules (1).48 The composition, charge type, surface topography, and modification density of the nanomaterials are of great significance to regulate the initial interaction of the nanomaterials and bacteria. After penetrating into the bacterial cell membrane, nanomaterials interact with various biomolecules in the cell, such as DNA, ribosomes and enzymes, disturbing the function of biomolecules, such as DNA replication, protein synthesis, and molecular catalysis (2).49,50 The size and shape of nanomaterials can affect their entry into bacterial cells. The inherent properties of nanomaterials with different compositions disturb the activity of biomolecules in the cells. In addition, with the addition of nanomaterials with different compositions and charges, oxidative stress and electrolyte imbalance are induced in the cell, causing death of the bacterial cell due to a lack of normal cellular behaviour (3).24 Therefore, the operation of bactericidal pathways depends on the core structure, size, shape, and surface status of the nanomaterials, which is of great significance to regulate their interaction with bacteria and biomolecules, obtaining broad-spectrum antibacterial activity and simultaneously reducing their toxicity against mammalian cells.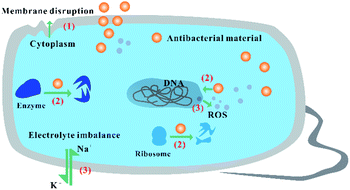 | ||
| Fig. 2 The fundamental antibacterial performance of nanomaterials according to different bactericidal pathways. | ||
3. Selective strategies for the antibacterial regulation of nanomaterials
3.1. Composition
Nanomaterials with different inherent compositions perform different antibacterial reactions. Metal-based NPs with different sizes in the range of 1–100 nm, such as silver, copper, gold, zinc and titanium, produce different physicochemical, electrical, optical, and biological properties. AgNPs release free Ag+ ions as active agents, destroying the bacterial membrane and disrupting electron transport and DNA function.51 The bactericidal effect appears in copper NPs (CuNPs) due to the formation of ROS via the function of copper-containing nanozymes.45,46 In addition to ROS-induced bactericidal effects, CuNPs can degrade into copper ions to enter the bacterial cell, destroying functions of DNA and RNA.52,53 Degradable Cu-doped phosphate-based glass (Cu-PBG) can simultaneously generate ROS and release copper ions, leading to excellent antibacterial effects (Fig. 3).54 Different from silver and copper, gold does not readily dissociate into ions due to its higher inertness and stability.55 It is necessary to endow AuNPs with antibacterial activity by grafting some compounds on their surface. Nanomaterials based on ZnO and TiO2 also cause cell membrane damage and generate ROS, killing the bacteria.15,43,56 In addition to metal-based NPs, using different synthesis methods, carbon atoms have been interconnected to form a variety of conventional CNMs, such as carbon dots, graphene quantum dots, carbon nanotubes (CNTs), and graphene oxide (GO). The CNMs can induce physical damage of the bacterial outer membrane or cell wall to exert antibacterial activity. For example, GO as a surface oxygen carrier can perform knife-like or lipid-extraction membrane destruction57,58 and oxidation reactions, leading to the death or inactivation of bacteria.59 CNTs can act as “nanodarts” to penetrate across the bacterial cell membrane.60–62 With the generation of ROS via electron transfer from their interaction with bacterial molecules, CNTs can inactivate proteins, lipids and nucleic acids to kill the bacteria.40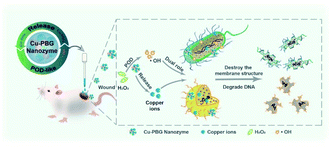 | ||
| Fig. 3 Cu-PBG-mediated antibacterial activity due to the formation of ROS and the release of copper ions.54 Reproduced from ref. 54 with permission from the American Chemical Society, Copyright© 2021. | ||
3.2. Size
The extraordinary properties of the materials are tailored via precise control on the molecular and atomic scales.63,64 The size-dependent characteristics of the nanomaterials contribute to their particular applications in surface-enhanced Raman scattering (SERS), green catalysts, and superior magnetic resonance imaging.65–68 More importantly, the regulation of size endows the nanomaterials with tunable physiological properties, including cell uptake, biodistribution, pharmacokinetics, cytotoxicity, and targeting efficiency.69–74 Especially focusing on the specific cytotoxicity of nanomaterials to bacteria, by decreasing the size of the materials from NPs to nanoclusters (NCs), the antimicrobial activity of broad-spectrum nanomaterials has been enhanced, effectively avoiding antimicrobial resistance.75,76 For example, ultrasmall gold NCs (AuNCs) readily interact with bacteria (Staphylococcus aureus as model bacteria), producing high antimicrobial activity with a wide spectrum. In the process, metabolic imbalance occurs after the internalization of AuNCs into bacterial cells. Subsequently, the increased ROS in the bacterial cell causes the death of bacteria (Fig. 4A).77 The inert and inactive AuNPs can be endowed with antibacterial activity by decreasing the size of the NPs.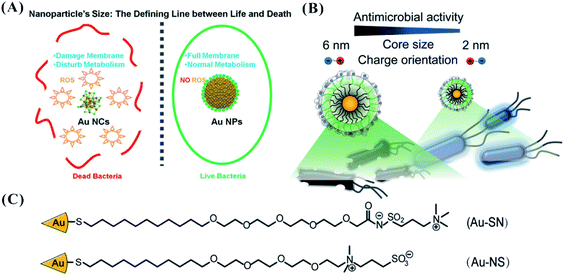 | ||
| Fig. 4 (A) The enhanced antibacterial activity of AuNCs relative to AuNPs due to the formation of ROS.77 Reproduced from ref. 77 with permission from the American Chemical Society, Copyright© 2017. (B) Controllability of the antibacterial activity of AuNPs by regulating the NP size.86 (C) The ligand structure for the synthesis of AuNPs.86 Reproduced from ref. 86 with permission from the American Chemical Society, Copyright© 2016. | ||
In addition to gold-based nanomaterials, the size of zinc oxide NPs (ZnO NPs) plays a significant role in the antibacterial activity of NPs, where smaller ZnO NPs have higher antibacterial efficiency.78,79 Similarly, the antibacterial activity of ZnO NPs is attributed to both ROS generation and NP accumulation in the bacteria. Due to their enhanced contact surface area, compared with larger NPs, smaller AgNPs ranging from 1 to 10 nm more readily bind to the surface of the cell membrane, disturbing the permeability and respiration of the cell.31,80 The increased reactive groups make the NPs produce higher toxicity.81–83 The AgNPs, after entry into the bacterial cell, interact with the thiol groups of proteins, preventing the synthesis of ribosomal subunits, the activity of cellular components,80 and the function of DNA replication.84,85
The precise size control of the nanomaterials depends on the ligand modification on the structural surface. The number and species of the ligands endow the nanomaterials with the defined size and stability. Cationic AuNPs with sizes of 6 nm (NP1) and 2 nm (NP2) displayed different cell lysis behaviours against bacteria.87 NP1 and NP2 were synthesized via place exchange of undecanethiol-capped AuNPs88 and pentanethiol-capped AuNPs, respectively.89 NP2 (2 nm) are more toxic than NP1 (6 nm) against Bacillus subtilis.87 Using one of two different zwitterionic ligands (SN or NS ligand), the effective antibacterial activity of AuNPs can be obtained through modulation of the NP size (2, 4, and 6 nm). Different from the higher antibacterial activity with smaller size, AuNPs stabilized by zwitterionic ligands (oligo(ethylene glycol)-functionalized interior) showed increased antimicrobial efficiency with increased size through bacterial membrane disruption (Fig. 4B and C).86 As the size changed from 2 to 6 nm, the MIC concentration of zwitterionic AuNPs against Pseudomonas aeruginosa could decrease from 8000 to 50 nM. Compared with smaller NPs (2 nm) with disorganized shells of ligands, larger NPs (>4.4 nm) with ordered “2D-like” surfaces more readily interact with the cell surface. In the design of nanomaterials, we should fully consider the action of the ligands when seeking a small size for improving the antibacterial activity.
Although the resistance of bacteria to nanomaterials has a lower occurrence than their resistance to antibiotics, it remains present.47,90 Size regulation can prevent drug resistance. 4,6-Diamino-2-pyrimidine thiol (DAPT)-capped AuNPs (DAPT-AuNPs) with a positive charge readily absorb on the negatively charged surfaces of bacteria.51,91,92 After Escherichia coli was cultured 183 successive times using DAPT-AuNPs with increased concentrations, DAPT-AuNP-resistant E. coli were obtained. The bactericidal activities of DAPT-AuNPs to the resistant strain were recovered by tuning the size of the DAPT-AuNPs without introduction of new chemicals (Fig. 5).18 Size regulation can act as an alternative strategy for avoiding drug resistance of nanomaterials.
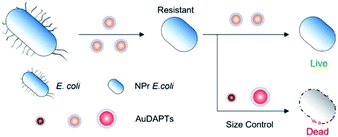 | ||
| Fig. 5 Schematic design of size-specific resistance of E. coli to DAPT-AuNPs.18 Reproduced from ref. 18 with permission from the American Chemical Society, Copyright© 2021. | ||
The size-dependent antibacterial activity of some nanomaterials is shown in Table 1. The MIC of nanomaterials with sizes of 1.8–12 nm changes from 50 nM to 7 mM, or several μg mL−1. However, an accurate antibacterial comparison between different size-dependent nanomaterials is difficult to perform due to the disturbance of different selected ligands and indicator bacteria.
| No. | Nanomaterial | Ligand | Size (nm) | Bacteria | MIC | Reference |
|---|---|---|---|---|---|---|
| 1 | AuNCs | 6-Mercaptohexanoic acid | <2 nm | S. aureus | 2.5 μM | 77 |
| 2 | ZnO NPs | Tetramethylammonium hydroxide | ∼12 nm | S. aureus | 4 to 7 mM | 79 |
| 3 | AgNPs | Glycol-thiol | 3 nm | E. coli | 6% survival rates at 2.2 mg L−1 | 28 |
| 4 | AgNPs | Gallic acid | 7 nm | E. coli | 6.25 μg mL−1 | 82 |
| 5 | AuNPs | Hexyl-substituted, ammonium-functionalized thiol | 2 nm | B. subtilis | 80% cell damage at 300 nM | 87 |
| 6 | AuNPs | Zwitterionic ligand | 6 nm | P. aeruginosa | 50 nM | 86 |
| 7 | AuNPs | 4,6-Diamino-2-pyrimidine thiol | 1.8 nm | E. coli | 3 μg mL−1 | 18 |
3.3. Charge
The adhesion events of nanomaterials and bacteria depend on force interactions (including static electricity, hydrophobicity and van der Waals) and receptor–ligand recognition.24 Ligands with positively charged, negatively charged, and/or zwitterionic moieties have been immobilized onto the surface of NPs, providing the basis for the adhesion action of nanomaterials and bacteria. Teichoic acids and lipopolysaccharide are characteristic components in the cell walls of Gram-positive and Gram-negative bacteria, respectively. Phosphate groups (also carboxylates in Gram-negative bacteria) in the structure endow bacterial surfaces with a negative charge.44 Usually, ligands with cationic groups are considered to be ideal candidates for designing and regulating antibacterial agents.93 Multiple cationic antimicrobials have been constructed by sufficiently utilizing the facile electrostatic attraction of a cationic agent and negative bacterial surface, such as peptides,94,95 polymers96 and NPs.42,87,97–99 By separately using the ligands 3-mercaptopropionic acid (MPA), 3-mercaptopropylamine (MPNH2), and poly(allylamine hydrochloride) (PAH), three types of AuNPs with different charge compositions were designed. Compared with anionic MPA-AuNPs, cationic PAH-AuNPs with more positive charge provided higher toxicity to bacteria (Fig. 6).99 Similarly, compared with anionic polymaleic anhydride-alt-1-octadecene (PMAO), quantum dots (QDs) coated with cationic polyethylenimine (PEI) exhibited higher inhibition against Pseudomonas stutzeri.100 The growth inhibition and death of the bacteria occurred after introducing these cationic agents according to the following steps. After binding initially with the cell wall, the antibacterial agents further penetrate and interact with lipids or proteins in the cell membrane, leading to membrane disorganization and leakage of small molecules in the cell. Finally, the gradual degradation of active biomolecules (such as proteins and nucleic acids) and lysis of bacteria cause the bacterial death.93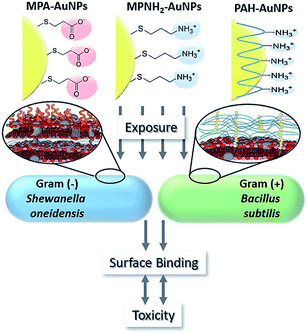 | ||
| Fig. 6 Antibacterial performance of AuNPs functionalized with anionic or cationic ligands.99 Reproduced from ref. 99 with permission. | ||
Quaternary ammonium (QA) compounds are the most frequently used compounds for producing antiseptics and disinfectants. The resin acids in QA can act as an active hydrophobic component to promote antibacterial activity.101 QA groups more readily interact with the cell membranes of bacteria, leading to disruption of the membranes to execute the antibacterial properties.102 Cationic poly(p-phenyleneethynylene)s (PPEs) with QA can bind advantageously to the surface of bacteria, indicating effective antibacterial activity against various bacteria.103–105 Using magnetic NPs functionalized with poly(quaternary ammonium) (PQA), 100% biocidal efficiency was still retained after performing the antibacterial cycle eight times.106 In addition, some polymeric QA compounds (polyquaternium-1) have good selective antimicrobial activity on bacterial and fungous microorganisms. They lead to high antimicrobial activity by destroying the spheroplasts of bacterial Serratia marcescens, but they show low inhibition against fungous Candida albicans due to the different microbial structures of bacteria and fungi.107
In addition to conventional QA as a cationic source, other ammonium-contained agents have been explored for obtaining good antibacterial activity. Positively charged cetyltrimethylammonium bromide (CTAB) was used to coat gold nanorods (AuNRs). CTAB-AuNRs bound with negatively charged teichoic acid in the cell wall of Gram-positive B. cereus and deposited on the bacterial surface due to the electrostatic interaction.108 Based on dimethyldecylammonium chitosan-graft-poly(ethylene glycol)methacrylate (DMDC-Q-g-EM) and poly(ethylene glycol)diacrylate, a polycationic hydrogel was built to achieve excellent antimicrobial efficacy. Like an ‘anion sponge’, the polycationic hydrogel drew anionic phospholipids out of the bacterial cell membrane into the gel pores. The positive charge density and pore size of the polycationic hydrogel determined the killing efficacy of the bacteria.109 However, positively charged NPs can produce substantial toxicity (e.g. hemolytic activity) to mammalian cells, resulting in difficulty of their application for killing pathogenic bacteria in infectious diseases.110,111 Neutral zwitterionic ligands integrated into cationic NPs can decrease their toxicity to mammalian cells while maintaining their antibacterial properties.112,113 The exquisite balance of zwitterionic and cationic ligands guarantees the stability as well as the antibacterial activity of the NPs.99,114,115 Using zwitterionic ligands with different charge orientations, several AuNPs were designed. The cationic charge in the outer layer is preferable to the charge inside the layer to perform antimicrobial activity (Fig. 4C).86
Although a cationic group is preferable, the origin of the antibacterial activity of nanomaterials is not confined to the modification of positive charge. Negatively charged carboxyl-modified single-walled carbon nanotubes can cause growth inhibition of Paracoccus denitrificans due to the influence of bacterial gene expression.116 In addition to the cationic ligands responsible for the combination of nanomaterials and the bacterial surface, anionic ligands with carboxylate headgroups may disrupt hydrogen bonding within the cell wall during their interaction, leading to cell lysis.117,118 Due to the different compositions of the cell wall in the two kinds of bacteria, tunable selectivity for two bacterial types was achieved by tailoring the surface chemistry of NPs. NPs functionalized with different ratios of charge indicated different antibacterial activities against Gram-positive and Gram-negative bacteria. The balance between polyvalent electrostatic and non-covalent interactions acts together to disrupt the bacterial cell. Using different ratios of positively charged N,N,N-trimethyl(11-mercaptoundecyl)ammonium chloride (TMA) and negatively charged 11-mercaptoundecanoic acid (MUA) ligands, NPs with different charge ratios were fabricated (Fig. 7A). Different charge ratios induced by the two ligands endowed the NPs with different surface charges (Fig. 7B). NPs with 48![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 52 and 80
52 and 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 TMA
20 TMA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) MUA could selectively kill Gram-positive and Gram-negative bacteria, respectively, at commensurate rates.119 However, the results did not clarify at the molecular level how NPs with mixed charge selectively interact with the cell wall in Gram-specific bacteria.
MUA could selectively kill Gram-positive and Gram-negative bacteria, respectively, at commensurate rates.119 However, the results did not clarify at the molecular level how NPs with mixed charge selectively interact with the cell wall in Gram-specific bacteria.
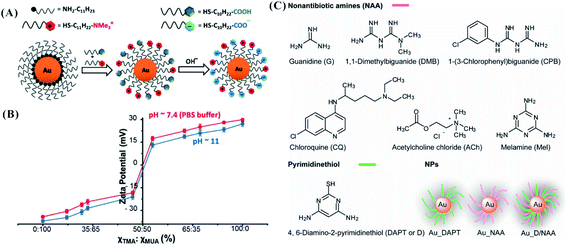 | ||
Fig. 7 The design of mixed-charge NPs. (A) The synthesis of mixed-charge NPs with different ratios of TMA/MUA. (B) The plotted curves of the NP charge polarities vs. the ratio of TMA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) MUA. The blue and red curves are the zeta potentials at pH 11 and pH 7.4, respectively. The MUAs were fully deprotonated at pH 11, while a few MUAs were protonated at pH 7.4 (PBS buffer).119 Reproduced from ref. 119 with permission from WILEY-VCH, Copyright© 2016. (C) The molecular structures of NAA and DAPT, and the synthesis of AuNPs using single or mixed ligands (including NAA and DAPT).120 Reproduced from ref. 120 with permission from the American Chemical Society, Copyright© 2013. MUA. The blue and red curves are the zeta potentials at pH 11 and pH 7.4, respectively. The MUAs were fully deprotonated at pH 11, while a few MUAs were protonated at pH 7.4 (PBS buffer).119 Reproduced from ref. 119 with permission from WILEY-VCH, Copyright© 2016. (C) The molecular structures of NAA and DAPT, and the synthesis of AuNPs using single or mixed ligands (including NAA and DAPT).120 Reproduced from ref. 120 with permission from the American Chemical Society, Copyright© 2013. | ||
In addition to the introduction of negative charge, drugs have been added to coat nanomaterials to enhance their antibacterial activity. AuNPs stabilized by DAPT (DAPT-AuNPs) only sufficiently inhibit Gram-negative bacteria. Synergistic effects were obtained when modifying non-antibiotic amines (NAA) and DAPT (D, pyrimidinethiol) as co-ligands on the surface of AuNPs via Au–N and Au–S interactions, forming Au_D/NAA NPs (Fig. 7C).120 The synergistic effects from the co-ligands of NAA and DAPT are attributed to the increased permeability of both the bacterial cell wall and membranes. The hydroxyl and amido groups from ampicillin were chelated with AgNPs to form an AgNP–ampicillin complex. The ampicillin-induced damage of the bacterial cell wall enhanced the penetration of AgNPs into the cell to bind with DNA, producing better antimicrobial effects.121
Most ligands on NPs undergo nonspecific interactions with bacterial surfaces. The selectivity of nanomaterials on bacteria is of great significance to specifically kill pathogenic bacteria. In addition to charge-dependent Gram selectivity, some ligands binding specifically with bacteria have been used to coat NPs, such as antibodies, antibiotics, and trehalose. Antibodies conjugated to NPs allow the targeting of specific microbes. For example, AuNPs linked with antibodies can specifically induce the death of Pseudomonas aeruginosa under near-infrared irradiation.122 Similarly, with the introduction of anti-protein A antibodies, AuNPs can selectively kill S. aureus under strong laser-induced overheating effects.123 A similar specific antibacterial reaction against S. aureus appeared in lysostaphin-antibody-conjugated NPs.124 In addition, vancomycin-conjugated AuNPs could specifically inhibit vancomycin-resistant and sensitive Enterococci.125 The conjugation of trehalose can lead silica NPs to selectively bind with Mycolicibacterium smegmatis on M. smegmatis-treated A549 cells, contributing to the development of targeted therapy for the corresponding infectious disease.126
The charge-dependent antibacterial activity of some nanomaterials is shown in Table 2. The MIC of nanomaterials with cationic charges from different ligands were obtained from 1 μg mL−1 to 1 mg mL−1, or from 16 nM to 112 μM.
| No. | Nanomaterial | Ligand | Charge | Bacteria | MIC | Reference |
|---|---|---|---|---|---|---|
| 1 | AuNPs | Poly(allylamine hydrochloride) | Cationic | Bacillus | 5 μg mL−1 | 99 |
| 2 | QD | Polyethylenimine | Cationic | P. stutzeri | IC50 at 7.25 ± 0.43 nM | 100 |
| 3 | Magnetic NPs | Quaternized poly(2-(dimethylamino)ethyl methacrylate) | Cationic | E. coli | 1 mg mL−1 | 106 |
| 4 | Hydrogel | DMDC-Q-g-EM and poly(ethylene glycol)diacrylate | Cationic | S. aureus | 49 μg mL−1 | 109 |
| 5 | AuNPs | Polythiophene | — | Listeria monocytogenes | 112 μM | 115 |
| 6 | AuNPs | Pentanethiol | Cationic and hydrophobic group | E. coli | 16 nM | 127 |
| 7 | AuNPs | 1,1-Dimethylbiguanide and DAPT | Cationic | E. coli | 1 μg mL−1 | 120 |
3.4. Surface topography
In nature, the introduction of topological features endows natural substances with elevated surface adhesion ability. In the pollination of pollen grains by insects and the infection of host cells by viruses, the pollen and cells provide rough surfaces for multivalent interactions. Similar to the interaction of pollen and the hairy legs of honey bees in nature,128 a hairy bacterial surface with pili129 can bind robustly with pollen-like nanomaterials. The surface roughness of nanomaterials is of great significance to regulate the interactions between the nanomaterials and bacteria.130 For example, designed nanoscale roughness on the surface of NPs is able to enhance attachment of bacteria to the nanomaterials.131,132 By surface topographical modification, compared to smooth mesoporous hollow silica (MHS) with hydrophilic hydroxyl groups, MHS NPs with surface roughness obtained by adding silica shell particles became more hydrophobic due to the repulsion of trapped air in the void and cavity domains. The rough MHS NPs facilitate not only the controllable load and release of antibiotic vancomycin but also the enhanced interaction with bacteria.133,134 Using three reagents, resorcinol (R), formaldehyde (F), and tetraethyl orthosilicate (TEOS), as the reactants through the Stöber method, the RF core was formed and subsequently underwent co-condensation with silica, producing rough mesoporous silica hollow spheres (RMSHSs) (named silica nanopollens) (Fig. 8A). The silica nanopollens with a spiky surface possess enhanced adhesion toward bacteria surfaces compared to their counterparts with smooth surfaces. Their accessible inner cavity can sufficiently load lysozymes, obtaining good antimicrobial activity toward E. coli (Fig. 8B).135 In the design of nanomaterials, nature-inspired concepts can be introduced to build a novel library of antibacterial nanomaterials.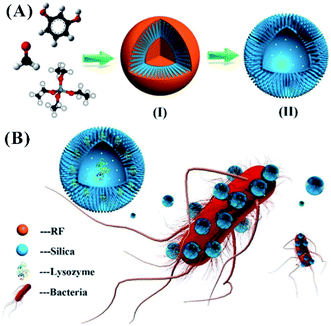 | ||
| Fig. 8 Silica nanopollens for antibacterial effects. (A) The synthesis of silica nanopollens. (B) The delivery of lysozymes due to the enhanced adhesive reaction of silica nanopollens and the bacterial surface.135 Reproduced from ref. 135 with permission from the American Chemical Society, Copyright© 2016. | ||
The surface topography is capable of affecting the affinity communication of a cell and the contact surface. The number and type of the bound cells depend on the topography of the used contact surface.136 Especially, the topography and nanoscale roughness of synthetic surfaces play an important role in bacterial responses. For example, when using polyurethane-coated glass plates, the number of adherent bacteria is related to the surface roughness. The influence of the surface nano-roughness on the adhesion of bacteria is relative to the fimbriae, flagella or polymeric substances excreted by the bacteria.137 The bacteria more readily aggregate on randomly ordered surfaces.138 Microorganisms with different sizes and shapes possess different retentions on titanium-coated silicon wafers with size-regulated pits.139 In addition, the surface topography can trigger and guide specific biological events. After treating bacteria with nanoscale metal substrates with a surface topography, the stress-related pathways of E. coli were activated by the nanorough surface. Different from the successful formation of bacterial fimbria on a flat surface (Fig. 9A), the synthesis of bacterial FimE protein on the nanorough surface is beneficial to convert the operon of the fimbrial promoter to the “OFF” status, inhibiting the transcription of all the fimbrial subunits and degrading the fimbrial structure (Fig. 9B).140 However, in the above finding, surface roughness does not directly influence the number of adherent E. coli. The selected marine bacteria produce stronger attachments to smooth glass surfaces, accompanied by the increase of secreted extracellular polymeric substances.141 Topography is only one factor for explaining the strong adhesion of bacteria. In addition to the surface topography, the adhesion between bacteria and the contact surface depends on the physicochemical properties of the substrates as well as the selected bacterial species and growing environment.
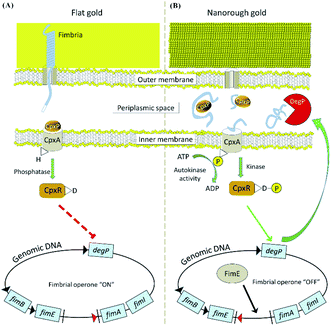 | ||
| Fig. 9 The different molecular mechanisms of E. coli on flat (A) and nanorough (B) gold substrates.140 The expression of type-1 fimbriae is active on the flat surface and inactive on the rough surface. Reproduced from ref. 140 with permission. | ||
3.5. Shape
The shape regulation of nanomaterials can control the antibacterial activity of the nanomaterials due to the effect of different contact killings. To investigate the correlation between shape and antimicrobial activity, using poly(N-vinyl-2-pyrrolidone) (PVP) as the stabilizer, nanosilvers with different shapes were designed, such as silver nanoplates (AgNPLs), silver nanorods (AgNRs) and AgNPs. The AgNPLs produced the best antibacterial activity against S. aureus and E. coli due to their best surface area for interacting with the bacterial surface.142 Similarly, compared to spherical and rod-shaped NPs, truncated triangular AgNPLs with more facets caused more membrane damage of bacteria, demonstrating strong antibacterial action,31 consistent with previous studies.85,143 Based on computer simulations, the anisotropy and initial orientation of the NPs affect the interaction between the NPs and the lipid bilayer in the translocation processes.144 Rod-like silica NPs (AR8) for releasing nitric oxide produced greater antibacterial action than their spherical counterparts (AR1) due to their higher aspect ratio.145A similar phenomenon occurs in the antibacterial operation of carbon nanomaterials.40 Sharp edges of these graphene nanomaterials are important to the acquisition of excellent antibacterial activities. Graphene films with different edge lengths and different angles of orientation exhibit different antibacterial efficiencies against P. aeruginosa and S. aureus, which is attributed to the formation of pores in the bacterial cell wall.146 In the graphene film, the presence of a smooth top side provides efficient inhibition for both types of bacteria, while the presence of a rough bottom side strongly kills only P. aeruginosa. Based on simulations, graphene sheets with corners or asperities along their irregular edges readily pierce and permeate into the cell membrane due to the existence of a low energy barrier.147 GO nanowalls with sharp edges result in bacterial inactivation.57 Needle-like single-walled carbon nanotubes and sharp-knife-like GO exhibit extremely strong antibacterial activity due to the destruction of the cell membrane.148 Therefore, the antibacterial activity of nanomaterials can be regulated by rationally designing their shape.
3.6. The density of surface modification
The surfaces of Gram-positive and Gram-negative bacteria possess different charge distributions. Gram-negative bacteria have more negative surface charges than Gram-positive bacteria.149 In addition to the limited selection of charge type, the charge density from surface ligands can regulate the antibacterial efficiency of nanomaterials. Using quaternized poly(vinylpyridine) chains grafted on glass surfaces, the charge density of cationic ligands in the organic layer can be regulated within the range of 1012 and 1016 amines per cm2.150 The biocidal efficiency can be improved by increasing the charge density. The antibacterial activity is attributed to the removal of divalent counterions from the bacteria during the interactions between the bacteria and surfaces, inducing disruption of the bacterial envelope. By initially immobilizing cationic NPs or poly-L-lysine (PLL) and subsequently backfilling the remaining areas with PEG (polyethylene glycol) brushes, the obtained surface starts to perform obvious bacteria adhesion above the threshold of the modification density. Two distinct thresholds are needed using different surface modifications due to their different protrusions, such as ∼180 NPs per μm2 and ∼1900 PLL per μm2. Two kinds of test surfaces with low density (280 NPs per μm2 and 3500 PLL per μm2) were designed, establishing relatively weak bacterial adhesion.151,152 The surfaces with NPs produced better killing efficiency of bacteria than those with PLL. Higher bactericidal efficiency was obtained on the surface with dense NPs.153 However, a loose surface density is beneficial to remove the bacteria by moderate shearing flow (Fig. 10A).154 The surface modification density using nanoparticles can regulate the adhesion and release of bacteria, affecting the antibacterial activity.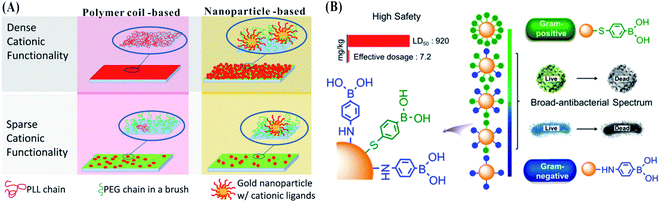 | ||
| Fig. 10 (A) Design of surfaces with different modification densities and substrates. Red color indicates a ligand with a cationic charge, while green color indicates a neutral PEG brush.154 Reproduced from ref. 154 with permission from the American Chemical Society, Copyright© 2014. (B) The design of AuNPs with different amounts and proportions of two ligands for obtaining bactericidal agents with tunable antibacterial spectra.156 Reproduced from ref. 156 with permission from the American Chemical Society, Copyright© 2020. | ||
Based on the mixed self-assembled monolayers containing positively and negatively charged thiols, the cellular uptake of NPs with unique stability at low and high pH was regulated. The presence of positively charged thiols allows for the uptake of particles with net negative charges.155 In addition to the tuned charge, the density of the ligands can regulate the antibacterial activity of NPs. By modification with different proportions of amine- and thiol-tethered phenylboronic acids via the binding affinities of Au–N and Au–S, including aminophenylboronic acid (ABA) and mercaptophenylboronic acid (MBA), the obtained AuNPs specifically bind to Gram-negative and Gram-positive bacteria. The AuNPs stabilized by ABA (A-AuNPs) bind to the lipopolysaccharide on Gram-negative bacteria, while the AuNPs stabilized by MBA (M-AuNPs) bind to the lipoteichoic acid on Gram-positive bacteria. The A-AuNPs and M-AuNPs show potent antibacterial effects on Gram-negative and Gram-positive bacteria, respectively. By co-modification of MBA and ABA on the AuNPs with varying ratios, the obtained A/M-AuNPs possess tunable antibacterial spectra with Gram selectivity (Fig. 10B).156 The density of the selective ligand can contribute to regulation of the Gram-selective antibacterial activities of the nanomaterials.
3.7. Other factors
In addition to the above six main factors, other physicochemical properties of nanomaterials can affect their antibacterial activity performance, such as their stiffness and hydrophobicity. It is well known that graphene possesses high stiffness. With the introduction of oxygen functional groups, the mechanical properties of the produced well-dispersed GO are severely decreased.157 GO with thin sheets may more readily wrap bacteria for performing antibacterial action.40 Compared with their flexible counterparts, long and stiff CNTs have shown higher toxicity due to their compression arising from lysosomal membranes, leading to cell death.158 The presence of polyacrylamide–gelatin–silver NPs or an N-acryloylsemicarbazide–gelatin scaffold can enhance the tensile and compression strength of gelatin-based ink, affording antibacterial properties.159,160 However, research on the direct link between the antibacterial activity and stiffness of nanomaterials remains rare.The hydrophobic interaction is a significant noncovalent reaction to control biomolecule absorption and cell adhesion in organisms.161–163 Hydrophobic moieties (alkanethiols and alkyl chains) were used to form AuNPs164,165 and silica NPs166,167 for enhancing drug loading and cellular delivery. AuNPs were synthesized with ligands containing groups with different chain lengths, nonaromatic characteristics, and aromatic characteristics via the varied R groups (Fig. 11A). The hydrophobicity of the ligands on the surface of NPs can affect the antibacterial activity of NPs. AuNPs stabilized by ligands with higher hydrophobic values of their end groups led to more effective activity against E. coli growth. NP3 with n-decane end groups produced an MIC of 32 nM against E. coli (Fig. 11B).127 The achievement of this antibacterial activity is attributed to the leakage of the cytoplasmic contents due to disruption of the bacterial membrane, eventually causing cell death. Even at 20 generations, E. coli remained susceptible to the original MIC of 16 nM. The design of NPs by regulating the types of modification groups can alleviate and avoid drug resistance. In addition, the increased hydrophobicity of AuNPs produced a linear increase in immune activity.168 In the practical application of nanomaterials in the medical field, hydrophobicity-dependent immune effects should be fully considered.
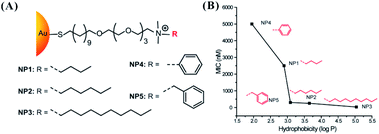 | ||
Fig. 11 (A) Molecular structures of functional ligands with different R groups for modifying AuNPs.127 (B) MIC determination of AuNPs with different hydrophobic ligands against E. coli DH5R. Log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P is the calculated hydrophobic values of the end groups.127 Reproduced from ref. 127 with permission from the American Chemical Society, Copyright© 2014. P is the calculated hydrophobic values of the end groups.127 Reproduced from ref. 127 with permission from the American Chemical Society, Copyright© 2014. | ||
4. Conclusions and outlook
The regulation of nanomaterials for obtaining customizable bactericidal agents is of great significance to inhibit the growth of pathogenic bacteria, providing more alternative strategies for treating infectious diseases. However, the reported literature has not systematically summarized the correlations between antibacterial activity and nanomaterials with various structures, leading to confusion of researchers regarding the rational design of antibacterial nanomaterials. To address these deficiencies, different from reported reviews that focus on the biological action of different antimicrobial NPs, this review summarized the universal rules determining the antibacterial activity of nanomaterials in six main aspects, including composition, size, charge, surface topography, shape and modification density. The clear description and discussion of every factor can provide precise guidance for constructing antibacterial nanomaterials.The component selection of nanomaterials is the first factor for regulating their antibacterial activity. In addition to the innate antibacterial activity of AgNPs and CuNPs, NPs based on gold, zinc and titanium must be modified with ligands to exert their bactericidal effects by destroying cell integrity and forming ROS. Smaller size facilitates entry of nanomaterials into bacterial cells to perform their intracellular reactions, such as enzyme inactivation, DNA/RNA structure damage, and ROS formation. However, the action of the ligand should be considered in the size regulation. In the performance of antibacterial reactions, the binding of the nanomaterial and bacterial surface is the first step of the entry of the nanomaterial into bacterial cells. Due to the negatively charged properties of the bacterial surface, the introduction of cationic ligands is beneficial to enhance the antibacterial activity of nanomaterials. Interestingly, the introduction of neutral zwitterionic ligands can decrease the toxicity of nanomaterials with positive charge to mammalian cells. In addition, nanomaterials with a mixed ratio of cationic and anionic ligands can selectively and specifically kill Gram-positive or Gram-negative bacteria.
In addition to the charge effect, the surface topography of nanomaterials can regulate the adhesion of bacteria on the contact surface. Compared with a smooth surface, a rough surface is beneficial to enhance the interaction of bacteria and nanomaterials, leading to increased antibacterial activity. The selected bacterial species and growing environment may affect the adhesion of bacteria on the contact surface. The shape is another factor for regulating the antibacterial activity of nanomaterials. A shape with a high surface area is preferable to obtain good antibacterial activity. For example, silver nanoplates have higher antibacterial efficiency than silver nanorods and AgNPs. Rod-like NPs have higher activity than their spherical counterparts. The modification density of the surface of nanomaterials can regulate the interaction of nanomaterials and bacteria. The structure with dense modification is beneficial to bind with bacteria, producing excellent antibacterial activity. The density of the ligands can selectively inhibit Gram-positive and Gram-negative bacteria. In the real design of antibacterial nanomaterials, the involved factors should not be singly be considered due to the comprehensive antibacterial effect of nanomaterials.
Although antimicrobial nanomaterials are promising to become alternatives to antibiotics, their potential toxicity to human health remains uncertain. Due to nonspecific interactions with the bacterial surface, for most antimicrobial nanomaterials, it is difficult to specifically differentiate between microbial and human cells. More specific ligands for binding with bacteria should be found. The immune responses of nanomaterials as drugs should not be neglected. It is necessary to balance the death of bacteria and inflammatory reactions in the practical application of antimicrobial nanomaterials. Therefore, the direct replacement of common antibiotics with antimicrobial nanomaterials still has many large challenges. In the future, two significant tasks should receive more emphasis. Firstly, the effects of antimicrobial nanomaterials on health concerns and ecosystems should be investigated. Secondly, to promote the application of nanomaterials as antimicrobial agents, unified standards must be constructed to compare the antimicrobial effects of different nanomaterials.
In sum, this review provides alternative rules for designing a variety of antibacterial nanomaterials. By having a good understanding of the corresponding correlation between antibacterial activity and nanomaterials, researchers can follow the rules for precisely designing the properties of nanomaterials, finally achieving nanomaterials with maximized antibacterial activity in complex biological media and minimizing the cytotoxicity to host cells. The facile regulation of nanomaterials can provide alternative strategies for eradicating drug resistance and developing effective therapeutic next-generation materials. The achievement of effective antibacterial nanomaterials requires interdisciplinary collaborations of many researchers in the fields of chemistry, biology, medicine and engineering. The treatment strategies based on the regulated nanomaterials are promising to provide an alternative to antibiotics for intractable infections, alleviating the challenges of the post-antibiotic era.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was financially supported by National Natural Science Foundation of China (NSFC Grants 21804089).Notes and references
- K. E. Jones, N. G. Patel, M. A. Levy, A. Storeygard, D. Balk, J. L. Gittleman and P. Daszak, Nature, 2008, 451, 990–993 CrossRef CAS PubMed.
- A. P. Anisimov and K. K. Amoako, J. Med. Microbiol., 2006, 55, 1461–1475 CrossRef CAS PubMed.
- C. J. Leaver and J. Edelman, Nature, 1965, 207, 1000–1001 CrossRef CAS.
- K. Kümmerer, Chemosphere, 2009, 75, 417–434 CrossRef PubMed.
- Q. Zhang, G. Lambert, D. Liao, H. Kim, K. Robin, C. K. Tung, N. Pourmand and R. H. Austin, Science, 2011, 333, 1764–1767 CrossRef CAS PubMed.
- H. C. Neu, Sci. Justice, 1992, 257, 1064–1073 CrossRef CAS PubMed.
- L. L. Ling, T. Schneider, A. J. Peoples, A. L. Spoering, I. Engels, B. P. Conlon, A. Mueller, T. F. Schäberle, D. E. Hughes, S. Epstein, M. Jones, L. Lazarides, V. A. Steadman, D. R. Cohen, C. R. Felix, K. A. Fetterman, W. P. Millett, A. G. Nitti, A. M. Zullo, C. Chen and K. Lewis, Nature, 2015, 517, 455–459 CrossRef CAS PubMed.
- S. Reardon, Nature, 2015, 521, 402–403 CrossRef CAS PubMed.
- K. M. G. O. Connell, J. T. Hodgkinson, H. F. Sore, M. Welch, G. P. C. Salmond and D. R. Spring, Angew. Chem., Int. Ed., 2013, 52, 10706–10733 CrossRef PubMed.
- I. Yosef, M. Manor, R. Kiro and U. Qimron, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, 7267–7272 CrossRef CAS.
- R. J. Citorik, M. Mimee and T. K. Lu, Nat. Biotechnol., 2014, 32, 1141–1145 CrossRef CAS PubMed.
- R. J. Krom, P. Bhargava, M. A. Lobritz and J. Collins, Nano Lett., 2015, 15, 4808–4813 CrossRef CAS PubMed.
- S. Galdiero, A. Falanga, R. Berisio, P. Grieco, G. Morelli and M. Galdiero, Curr. Med. Chem., 2015, 22, 1665–1677 CrossRef CAS PubMed.
- J. A. Lemire, J. J. Harrison and R. J. Turner, Nat. Rev. Microbiol., 2013, 11, 371–384 CrossRef CAS PubMed.
- K. P. Miller, L. Wang, B. C. Benicewicz and A. W. Decho, Chem. Soc. Rev., 2015, 44, 7787–7807 RSC.
- S. Chernousova and M. Epple, Angew. Chem., Int. Ed., 2013, 52, 1636–1653 CrossRef CAS PubMed.
- K. Malzahn, W. D. Jamieson, M. Dröge, V. Mailänder, A. Jenkins, C. K. Weiss and K. Landfester, J. Mater. Chem. B, 2014, 2, 2175–2183 RSC.
- W. Zheng, Y. Jia, Y. Zhao, J. Zhang and X. Jiang, Nano Lett., 2021, 21, 1992–2000 CrossRef CAS PubMed.
- U. Bogdanovic, V. Lazic, V. Vodnik, M. Budimir, Z. Markovic and S. Dimitrijevic, Mater. Lett., 2014, 128, 75–78 CrossRef CAS.
- H. Yuan, Z. Liu, L. Liu, F. Lv, Y. Wang and S. Wang, Adv. Mater., 2014, 26, 4333–4338 CrossRef CAS PubMed.
- Y. Wang, H. Chen, M. Li, R. Hu, F. Lv, L. Liu and S. Wang, Polym. Chem., 2016, 7, 6699–6702 RSC.
- H. Bai, H. Zhang, R. Hu, H. Chen, F. Lv, L. Liu and S. Wang, Langmuir, 2017, 33, 1116–1120 CrossRef CAS PubMed.
- Q. Xin, H. Shah, A. Nawaz, W. Xie, M. Z. Akram, A. Batool, L. Tian, S. U. Jan, R. Boddula, B. Guo, Q. Liu and J. R. Gong, Adv. Mater., 2019, 31, 1804838 CrossRef CAS PubMed.
- L. Wang, H. Chen and L. Shao, Int. J. Nanomed., 2017, 12, 1227–1249 CrossRef CAS PubMed.
- M. E. Quadros and L. C. Marr, Environ. Sci. Technol., 2011, 45, 10713–10719 CrossRef CAS PubMed.
- J. S. Kim, E. Kuk, K. N. Yu, J. H. Kim, S. J. Park, J. L. Hu, S. H. Kim, Y. K. Park, H. P. Yong and C. Y. Hwang, Nanomedicine, 2007, 3, 95–101 CrossRef CAS PubMed.
- L. Rizzello and P. P. Pompa, Chem. Soc. Rev., 2014, 43, 1501–1518 RSC.
- Z. Xiu, Q. Zhang, H. L. Puppala, V. L. Colvin and P. J. J. Alvarez, Nano Lett., 2012, 12, 4271–4275 CrossRef CAS PubMed.
- O. Choi and Z. Hu, Environ. Sci. Technol., 2008, 42, 4583–4588 CrossRef CAS PubMed.
- A. E. Badawy, R. G. Silva, B. Morris, K. G. Scheckel, M. T. Suidan and T. M. Tolaymat, Environ. Sci. Technol., 2010, 45, 283–287 CrossRef PubMed.
- S. Pal, K. T. Yu and J. M. Song, Appl. Environ. Microbiol., 2007, 73, 1712–1720 CrossRef CAS PubMed.
- Y. Inoue, M. Hoshino, H. Takahashi, T. Noguchi, T. Murata, Y. Kanzaki, H. Hamashima and M. Sasatsu, J. Inorg. Biochem., 2002, 92, 37–42 CrossRef CAS PubMed.
- R. Kumar and H. Münstedt, Biomaterials, 2005, 26, 2081–2088 CrossRef CAS PubMed.
- M. Lv, S. Su, Y. He, Q. Huang, W. Hu, D. Li, C. Fan and S. T. Lee, Adv. Mater., 2010, 22, 5463–5467 CrossRef CAS PubMed.
- K. Zheng and J. Xie, ACS Nano, 2020, 14, 11533–11541 CrossRef CAS PubMed.
- B. Hisey, P. J. Ragogna and E. R. Gillies, Biomacromolecules, 2017, 18, 914–923 CrossRef CAS PubMed.
- T. Gao, H. Fan, X. Wang, Y. Gao and Y. J. Wang, ACS Appl. Mater. Interfaces, 2017, 9, 25738–25746 CrossRef CAS PubMed.
- H. E. Karahan, C. Wiraja, C. Xu, J. Wei and C. Yuan, Adv. Healthcare Mater., 2018, 7, 1701406 CrossRef PubMed.
- A. J. Ahmed, A. Surjith, B. Kateryna and J. Mohan, Mater. Corros., 2017, 10, 1066 Search PubMed.
- X. Zou, Z. Li, Z. Wang and Y. Luo, J. Am. Chem. Soc., 2016, 138, 2064–2077 CrossRef CAS PubMed.
- X. Chen and H. J. Schluesener, Toxicol. Lett., 2008, 176, 1–12 CrossRef CAS PubMed.
- Y. Zhao and X. Jiang, Nanoscale, 2013, 5, 8340–8350 RSC.
- M. J. Hajipour, K. M. Fromm, A. A. Ashkarran, D. D. Aberasturi, I. Larramendi, T. Rojo, V. Serpooshan, W. J. Parak and M. Mahmoudi, Trends Biotechnol., 2012, 20, 499–511 CrossRef PubMed.
- A. Gupta, S. Mumtaz, C. H. Li, I. Hussain and V. M. Rotello, Chem. Soc. Rev., 2019, 48, 415–427 RSC.
- A. F. Halbus, T. S. Horozov and V. N. Paunov, Adv. Colloid Interface Sci., 2017, 249, 134–148 CrossRef CAS PubMed.
- A. P. Ingle, N. Duran and M. Rai, Appl. Microbiol. Biotechnol., 2014, 98, 1001–1009 CrossRef CAS PubMed.
- J. Makabenta, A. Nabawy, C. H. Li, S. Schmidt-Malan and V. M. Rotello, Nat. Rev. Microbiol., 2021, 19, 23–36 CrossRef CAS PubMed.
- A. Gupta, R. F. Landis and V. M. Rotello, F1000Research, 2016, 5, 364 Search PubMed.
- S. Shahzadi, Z. Noshin, R. Saira, S. Rehana, N. Jawad and N. Shahzad, Nanomaterials, 2016, 6, 1–10 Search PubMed.
- A. K. Chatterjee, R. Chakraborty and T. BaSu, Nanotechnology, 2014, 25, 135101 CrossRef PubMed.
- Y. Zhao, Y. Tian, C. Yan, W. Liu, W. Ma and X. Jiang, J. Am. Chem. Soc., 2010, 132, 12349–12356 CrossRef CAS PubMed.
- M. Vincent, R. E. Duval, P. Hartemann and M. Engels-Deutsch, J. Appl. Microbiol., 2017, 124, 1032–1046 CrossRef PubMed.
- G. Grass, C. Rensing and M. Solioz, Appl. Environ. Microbiol., 2011, 77, 1541–1547 CrossRef CAS PubMed.
- Y. Liu, N. Nie, H. Tang, C. Zhang, K. Chen, W. Wang and J. Liu, ACS Appl. Mater. Interfaces, 2021, 13, 11631–11645 CrossRef CAS.
- B. Hammer and J. K. Norskov, Nature, 1995, 376, 238–240 CrossRef CAS.
- A. Sirelkhatim, S. Mahmud, A. Seeni, N. Kaus, C. A. Ling, S. Bakhori, H. Hasan and D. Mohamad, Nano-Micro Lett., 2015, 7, 219–242 CrossRef CAS PubMed.
- O. Akhavan and E. Ghaderi, ACS Nano, 2010, 4, 5731–5736 CrossRef CAS PubMed.
- Y. Tu, M. Lv, P. Xiu, T. Huynh, M. Zhang, M. Castelli, Z. Liu, Q. Huang, C. Fan and H. Fang, Nat. Nanotechnol., 2013, 8, 594–601 CrossRef CAS PubMed.
- M. Lee, S. Yang, K. J. Kim, S. Kim and H. Lee, J. Phys. Chem. C, 2014, 118, 1142–1147 CrossRef CAS.
- S. Kang, M. Herzberg, D. F. Rodrigues and M. Elimelech, Langmuir, 2008, 24, 6409–6413 CrossRef CAS PubMed.
- S. Liu, L. Wei, L. Hao, N. Fang and Y. Chen, ACS Nano, 2009, 3, 3891–3902 CrossRef CAS PubMed.
- S. Kang, M. Pinault, L. D. Pfefferle and M. Elimelech, Langmuir, 2007, 23, 8670–8673 CrossRef CAS PubMed.
- M. C. Daniel and D. Astruc, Chem. Rev., 2004, 104, 293–346 CrossRef CAS PubMed.
- A. M. Alkilany, X. Huang, C. J. Murphy, M. A. El-Sayed and E. C. Dreaden, Chem. Soc. Rev., 2012, 41, 2740 RSC.
- J. Kneipp, H. Kneippa and K. Kneippa, Chem. Soc. Rev., 2008, 37, 1052–1060 RSC.
- X. Zhou, W. Xu, G. Liu, D. Panda and P. Chen, J. Am. Chem. Soc., 2010, 132, 138–146 CrossRef CAS PubMed.
- D. Alloyeau, C. Ricolleau, C. Mottet, T. Oikawa, C. Langlois, Y. L. Bouar, N. Braidy and A. Loiseau, Nat. Mater., 2009, 8, 940–946 CrossRef CAS PubMed.
- B. L. Li, M. I. Setyawati, H. L. Zou, J. X. Dong, H. Q. Luo, N. B. Li and D. T. Leong, Small, 2017, 13, 1700527 CrossRef PubMed.
- A. Albanese, P. S. Tang and W. Chan, Annu. Rev. Biomed. Eng., 2012, 14, 1–16 CrossRef CAS PubMed.
- W. Jiang, B. Kim, J. T. Rutka and W. Chan, Nat. Nanotechnol., 2008, 3, 145–150 CrossRef CAS PubMed.
- C. He, Y. Hu, L. Yin, C. Tang and C. Yin, Biomaterials, 2010, 31, 3657–3666 CrossRef CAS PubMed.
- P. Yu, S. Neuss, A. Leifert, M. Fischler and W. Jahnen-Dechent, Small, 2007, 3, 1941–1949 CrossRef PubMed.
- M. I. Setyawati, C. Y. Tay, B. H. Bay and D. T. Leong, ACS Nano, 2017, 11, 5020–5030 CrossRef CAS PubMed.
- M. I. Setyawati and D. T. Leong, ACS Appl. Mater. Interfaces, 2017, 9, 6690–6703 CrossRef CAS PubMed.
- K. Zheng, M. I. Setyawati, T. P. Lim, D. T. Leong and J. Xie, ACS Nano, 2016, 10, 7934–7942 CrossRef CAS PubMed.
- X. Yuan, M. I. Setyawati, A. S. Tan, C. N. Ong, D. T. Leong and J. Xie, NPG Asia Mater., 2013, 5, e39 CrossRef CAS.
- K. Zheng, M. I. Setyawati, D. T. L. Orcid and J. Xie, ACS Nano, 2017, 11, 6904–6910 CrossRef CAS PubMed.
- O. Yamamoto, Int. J. Inorg. Mater., 2001, 3, 643–646 CrossRef CAS.
- K. R. Raghupathi, R. T. Koodali and A. C. Manna, Langmuir, 2011, 27, 4020–4028 CrossRef CAS PubMed.
- M. Yamanaka, K. Hara and J. Kudo, Appl. Environ. Microbiol., 2005, 71, 7589–7593 CrossRef CAS PubMed.
- C. Carlson, S. M. Hussain, A. M. Schrand, L. K. Braydich-Stolle, K. L. Hess, R. L. Jones and J. J. Schlager, J. Phys. Chem. B, 2008, 112, 13608–13619 CrossRef CAS PubMed.
- G. A. Martínez-Castaón, N. Nio-Martínez, F. Martínez-Gutierrez, J. Martínez-Mendoza and F. Ruiz, J. Nanopart. Res., 2008, 10, 1343–1348 CrossRef.
- A. Ivask, O. Bondarenko, N. Jepihhina and A. Kahru, Anal. Bioanal. Chem., 2010, 398, 701–716 CrossRef CAS PubMed.
- Q. L. Feng, J. Wu, G. Q. Chen, F. Z. Cui and J. O. Kim, J. Biomed. Mater. Res., 2000, 52, 662–668 CrossRef CAS.
- J. R. Morones, J. L. Elechiguerra, A. Camacho, K. Holt, J. B. Kouri, J. Ramírez and M. J. Yacaman, Nanotechnology, 2005, 16, 2346–2353 CrossRef CAS PubMed.
- S. Huo, Y. Jiang, A. Gupta, Z. Jiang, R. F. Landis, S. Hou, X. Liang and V. M. Rotello, ACS Nano, 2016, 10, 8732–8737 CrossRef CAS PubMed.
- S. C. Hayden, G. Zhao, K. Saha, R. L. Phillips, X. Li, O. R. Miranda, V. M. Rotello, M. A. El-Sayed, I. Schmidt-Krey and U. H. F. Bunz, J. Am. Chem. Soc., 2012, 134, 6920–6923 CrossRef CAS PubMed.
- M. J. Hostetler, A. C. Templeton and R. W. Murray, Langmuir, 1999, 15, 3782–3789 CrossRef CAS.
- M. Brust, M. Walker, D. Bethell, D. J. Schiffrin and R. Whyman, Chem. Commun., 1994, 801–802 RSC.
- A. Panáček, L. Kvítek, M. Smékalová, R. Večeřová, M. Kolář, M. Röderová, F. Dyčka, M. Šebela, R. Prucek, O. Tomanec and R. Zbořil, Nat. Nanotechnol., 2018, 13, 65–71 CrossRef PubMed.
- W. Zheng, Y. Jia, W. Chen, G. Wang, X. Guo and X. Jiang, ACS Appl. Mater. Interfaces, 2017, 9, 21181–21189 CrossRef CAS PubMed.
- C. Yan, Y. Zhao, Y. Tian, Z. Wei, X. Lü and X. Jiang, Biomaterials, 2012, 33, 2327–2333 CrossRef PubMed.
- A. M. Carmona-Ribeiro and L. D. de Melo Carrasco, Int. J. Mol. Sci., 2013, 14, 9906–9946 CrossRef PubMed.
- R. Hancock and H. G. Sahl, Nat. Biotechnol., 2006, 24, 1551–1557 CrossRef CAS PubMed.
- K. A. Brogden, Nat. Rev. Microbiol., 2005, 3, 238–250 CrossRef CAS PubMed.
- V. Sambhy, B. R. Peterson and A. Sen, Angew. Chem., Int. Ed., 2008, 47, 1250–1254 CrossRef CAS PubMed.
- L. Liu, K. Xu, H. Wang, P. J. Tan, W. Fan, S. S. Venkatraman, L. Li and Y. Y. Yang, Nat. Nanotechnol., 2009, 4, 457–463 CrossRef CAS PubMed.
- F. Nederberg, Y. Zhang, J. Tan, K. Xu, H. Wang, C. Yang, S. Gao, X. D. Guo, K. Fukushima and L. Li, Nat. Chem., 2011, 3, 409–414 CrossRef CAS PubMed.
- Z. V. Feng, L. L. Gunsolus, T. A. Qiu, K. R. Hurley, L. H. Nyberg, H. Frew, K. P. Johnson, A. M. Vartanian, L. M. Jacob, S. E. Lohse, M. D. Torelli, R. J. Hamers, C. J. Murphyc and C. L. Haynes, Chem. Sci., 2015, 6, 5186–5196 RSC.
- Y. Yu, H. Zhu, V. L. Colvin and P. J. Alvarez, Environ. Sci. Technol., 2011, 45, 4988–4994 CrossRef PubMed.
- J. Wang, Y. P. Chen, K. Yao, P. A. Wilbon, W. Zhang, L. Ren, J. Zhou, M. Nagarkatti, C. Wang and F. Chu, Chem. Commun., 2011, 48, 916–918 RSC.
- S. Imazato, M. Torii, Y. Tsuchitani, J. F. Mccabe and R. Russell, J. Dent. Res., 1994, 73, 1437–1443 CrossRef CAS PubMed.
- S. Chemburu, T. S. Corbitt, L. K. Ista, E. Ji, J. Fulghum, G. P. Lopez, K. Ogawa, K. S. Schanze and D. G. Whitten, Langmuir, 2008, 24, 11053–11062 CrossRef CAS PubMed.
- L. Lu, F. H. Rininsland, S. K. Wittenburg, K. E. Achyuthan, D. W. Mcbranch and D. G. Whitten, Langmuir, 2005, 21, 10154–10159 CrossRef CAS PubMed.
- T. S. Corbitt, J. R. Sommer, S. Chemburu, K. Ogawa, L. K. Ista, G. P. Lopez, D. G. Whitten and K. S. Schanze, ACS Appl. Mater. Interfaces, 2009, 1, 48–52 CrossRef CAS PubMed.
- H. Dong, J. Huang, R. R. Koepsel, P. Ye, A. J. Russell and K. Matyjaszewski, Biomacromolecules, 2011, 12, 1305–1311 CrossRef CAS PubMed.
- C. E. Codling, M. Jean-Yves and R. A. Denver, J. Antimicrob. Chemother., 2003, 1153–1158 CrossRef CAS PubMed.
- V. Berry, A. Gole, S. Kundu, C. J. Murphy and R. F. Saraf, J. Am. Chem. Soc., 2005, 127, 17600–17601 CrossRef CAS PubMed.
- P. Li, Y. F. Poon, W. Li, H. Y. Zhu, S. H. Yeap, Y. Cao, X. Qi, C. Zhou, M. Lamrani and R. W. Beuerman, Nat. Mater., 2011, 10, 149–156 CrossRef CAS PubMed.
- C. M. oodman, C. D. McCusker, T. Yilmaz and V. M. Rotello, Bioconjugate Chem., 2004, 15, 897–900 CrossRef PubMed.
- Z. Zhang, Z. Min, S. Chen, T. A. Horbett, B. D. Ratner and S. Jiang, Biomaterials, 2008, 29, 4285–4291 CrossRef CAS PubMed.
- S. Jiang and Z. Cao, Adv. Mater., 2010, 22, 920–932 CrossRef CAS PubMed.
- G. Cheng, Z. Zhang, S. Chen, J. D. Bryers and S. Jiang, Biomater., Artif. Cells, Artif. Organs, 2007, 28, 4192–4199 CAS.
- J. Bresee, K. E. Maier, A. E. Boncella, C. Melander and D. L. Feldheim, Small, 2011, 7, 2027–2031 CrossRef CAS PubMed.
- M. Adhikari, S. Goswami, B. R. Panda, A. Chattopadhyay and A. Ramesh, Adv. Healthcare Mater., 2013, 2, 599–606 CrossRef CAS PubMed.
- X. Zheng, Y. Su, Y. Chen, R. Wan, M. Li, Y. Wei and H. Huang, Sci. Rep., 2014, 4, 5653 CrossRef CAS PubMed.
- R. Crh and C. Whitfield, Annu. Rev. Biochem., 2002, 71, 635–700 CrossRef PubMed.
- J. van Heijenoort, Glycobiology, 2001, 11, 25R–36R CrossRef CAS PubMed.
- P. P. Pillai, B. Kowalczyk, K. Kandere-Grzybowska, M. Borkowska and B. A. Grzybowski, Angew. Chem., Int. Ed., 2016, 55, 8610–8614 CrossRef CAS PubMed.
- Y. Zhao, Z. Chen, Y. Chen, J. Xu, J. Li and X. Jiang, J. Am. Chem. Soc., 2013, 135, 12940–12943 CrossRef CAS PubMed.
- A. M. Fayaz, K. Balaji, M. Girilal, R. Yadav, P. T. Kalaichelvan and R. Venketesan, Nanomedicine, 2010, 6, 103–109 CrossRef CAS PubMed.
- R. S. Norman, J. W. Stone, A. Gole, C. J. Murphy and T. L. Sabo-Attwood, Nano Lett., 2008, 8, 302–306 CrossRef CAS PubMed.
- V. P. Zharov, K. E. Mercer, E. N. Galitovskaya and M. S. Smeltzer, Biophys. J., 2006, 90, 619–627 CrossRef CAS PubMed.
- R. Satishkumar and A. A. Vertegel, Nanotechnology, 2011, 22, 505103 CrossRef CAS PubMed.
- H. Gu, P. L. Ho, E. Tong, L. Wang and B. Xu, Nano Lett., 2003, 3, 1261–1263 CrossRef CAS.
- K. W. Jayawardana, H. S. N. Jayawardena, S. A. Wijesundera, T. D. Zoysa, M. Sundhoroa and M. Yan, Chem. Commun., 2015, 51, 12028–12031 RSC.
- X. Li, S. M. Robinson, A. Gupta, K. Saha and V. M. Rotello, ACS Nano, 2014, 8, 10682–10686 CrossRef CAS PubMed.
- R. W. Thorp, Plant Syst. Evol., 2000, 222, 211–223 CrossRef.
- L. Craig, M. E. Pique and J. A. Tainer, Nat. Rev. Microbiol., 2004, 2, 363–378 CrossRef CAS PubMed.
- C. Meng, Q. Yu and H. Sun, Int. J. Mol. Sci., 2013, 14, 18488–18501 CrossRef PubMed.
- E. P. Ivanova, V. K. Truong, J. Y. Wang, C. C. Berndt, R. T. Jones, I. I. Yusuf, I. Peake, H. W. Schmidt, C. Fluke, D. Barnes and R. J. Crawford, Langmuir, 2010, 26, 1973–1982 CrossRef CAS PubMed.
- V. K. Truong, R. Lapovok, Y. S. Estrin, S. Rundell, J. Y. Wang, C. J. Fluke, R. J. Crawford and E. P. Ivanova, Biomaterials, 2010, 31, 3674–3683 CrossRef CAS PubMed.
- Y. A. Nor, Y. Niu, S. Karmakar, L. Zhou, C. Xu, J. Zhang, H. Zhang, M. Yu, D. Mahony, N. Mitter, M. A. Cooper and C. Yu, ACS Cent. Sci., 2015, 1, 328–334 CrossRef PubMed.
- X. Wu, Y. Tian, Y. Cui, L. Wei, Q. Wang and Y. Chen, J. Phys. Chem. C, 2007, 27, 9704–9708 CrossRef.
- S. Hao, Y. A. Nor, M. Yu, Y. Yang and C. Yu, J. Am. Chem. Soc., 2016, 138, 6455–6462 CrossRef PubMed.
- E. Yim and K. W. Leong, Nanomedicine, 2005, 1, 10–21 CrossRef CAS PubMed.
- D. P. Bakker, H. J. Busscher, J. v. Zanten, J. d. Vries, J. W. Klijnstra and H. C. v. d. Mei, Microbiology, 2004, 150, 1779–1784 CrossRef CAS PubMed.
- C. Díaz, P. L. Schilardi, R. C. Salvarezza and M. F. de Mele, Langmuir, 2007, 23, 11206–11210 CrossRef PubMed.
- K. A. Whitehead, J. Colligon and J. Verran, Colloids Surf., B, 2005, 41, 129–138 CrossRef CAS PubMed.
- L. Rizzello, A. Galeone, G. Vecchio, V. Brunetti, S. Sabella and P. P. Pompa, Nanoscale Res. Lett., 2012, 7, 575 CrossRef PubMed.
- N. Mitik-Dineva, J. Wang, V. K. Truong, P. R. Stoddart, F. Malherbe, R. J. Crawford and E. P. Ivanova, Biofouling, 2009, 25, 621–631 CrossRef CAS PubMed.
- B. Sadeghi, F. S. Garmaroudi, M. Hashemi, H. R. Nezhad, A. Nasrollahi, S. Ardalan and S. Ardalan, Adv. Powder Technol., 2012, 23, 22–26 CrossRef CAS.
- I. Sondi and B. Salopek-Sondi, J. Colloid Interface Sci., 2004, 275, 117–182 Search PubMed.
- K. Yang and Y. Q. Ma, Nat. Nanotechnol., 2010, 5, 579–583 CrossRef CAS PubMed.
- D. L. Slomberg, Y. Lu, A. D. Broadnax, R. A. Hunter and M. H. Schoenfisch, ACS Appl. Mater. Interfaces, 2013, 5, 9322–9329 CrossRef CAS PubMed.
- V. Pham, V. K. Truong, M. Quinn, S. M. Notley, Y. Guo, V. A. Baulin, M. A. Kobaisi, R. J. Crawford and E. P. Ivanova, ACS Nano, 2015, 9, 8458–8467 CrossRef CAS PubMed.
- Y. Li, H. Yuan, D. Von, M. Creighton, R. H. Hurt, A. B. Kane and H. Gao, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 12295–12300 CrossRef CAS PubMed.
- X. Wang, X. Liu and H. Han, Colloids Surf., B, 2013, 103, 136–142 CrossRef CAS PubMed.
- Y. Shai, Biopolymers, 2002, 66, 236–248 CrossRef CAS PubMed.
- R. Kügler, O. Bouloussa and F. Rondelez, Microbiology, 2005, 151, 1341–1348 CrossRef PubMed.
- B. Fang, S. Gon, M. Park, K. N. Kumar, V. M. Rotello, K. Nusslein and M. M. Santore, Colloids Surf., B, 2011, 87, 109–115 CrossRef CAS PubMed.
- S. Gon, K. N. Kumar, K. Nüsslein and M. M. Santore, Macromolecules, 2012, 45, 8373–8381 CrossRef CAS PubMed.
- B. Fang, Y. Jiang, K. Nüsslein, V. M. Rotello and M. M. Santore, Colloids Surf., B, 2015, 125, 255–263 CrossRef CAS PubMed.
- B. Fang, Y. Jiang, V. M. Rotello, K. Nüsslein and M. M. Santore, ACS Nano, 2014, 8, 1180–1190 CrossRef CAS PubMed.
- P. P. Pillai, S. Huda, B. Kowalczyk and B. A. Grzybowski, J. Am. Chem. Soc., 2013, 135, 6392–6395 CrossRef CAS PubMed.
- L. Wang, S. Li, J. Yin, J. Yang, Q. Li, W. Zheng, S. Liu and X. Jiang, Nano Lett., 2020, 20, 5036–5042 CrossRef CAS PubMed.
- A. J. Clancy, D. B. Anthony and F. D. Luca, ACS Appl. Mater. Interfaces, 2020, 12, 15955–15975 CrossRef CAS PubMed.
- W. Zhu, V. Annette, X. Yi, Y. Qiu, Z. Wang, P. Weston, R. H. Hurt, A. B. Kane and H. Gao, Proc. Natl. Acad. Sci. U. S. A., 2016, 113, 12374–12379 CrossRef CAS PubMed.
- Y. Liu, J. Huang, Z. Xu, S. Li, Y. Jiang, G. w. Qu, Z. Li, Y. Zhao, X. Wu and J. Ren, Int. J. Biol. Macromol., 2021, 186, 396–404 CrossRef CAS PubMed.
- R. Yang, C. Fan, Y. Dou, X. Zhang, Z. Xu, Q. Zhang, Y. Sun, Q. Yang and W. Liu, Appl. Mater. Today, 2021, 24, 101089 CrossRef.
- T. Cedervall, I. Lynch, S. Lindman, T. Berggard, E. Thulin, H. Nilsson, K. A. Dawson and S. Linse, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 2050–2055 CrossRef CAS PubMed.
- A. E. Nel, L. Mädler, D. Velegol, T. Xia, E. M. V. Hoek, P. Somasundaran, F. Klaessig, V. Castranova and M. Thompson, Nat. Mater., 2009, 8, 543–557 CrossRef CAS PubMed.
- M. B. Salerno, B. E. Logan and D. Velegol, Langmuir, 2004, 20, 10625–10629 CrossRef CAS PubMed.
- A. Sethuraman, M. Han, R. S. Kane and G. Belfort, Langmuir, 2004, 20, 7779–7788 CrossRef CAS PubMed.
- M. R. Rasch, E. Rossinyol, J. L. Hueso, B. W. Goodfellow, J. Arbiol and B. A. Korgel, Nano Lett., 2010, 10, 3733–3739 CrossRef CAS PubMed.
- S. S. Bale, S. J. Kwon, D. A. Shah, A. Banerjee and R. S. Kane, ACS Nano, 2010, 4, 1493–1500 CrossRef CAS PubMed.
- J. Zhang, S. Karmakar, M. Yu, N. Mitter, J. Zou and C. Yu, Small, 2015, 10, 5068–5076 Search PubMed.
- D. F. Moyano, M. Goldsmith, D. J. Solfiell, D. Landesman-Milo, O. R. Miranda, P. Dan and V. M. Rotello, J. Am. Chem. Soc., 2012, 134, 3965–3967 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2022 |
