DOI:
10.1039/D2RA05192C
(Paper)
RSC Adv., 2022,
12, 27793-27808
Gravimetric and electrochemical statistical optimizations for improving copper corrosion resistance in hydrochloric acid using thiosemicarbazone-linked 3-acetylpyridine†
Received
19th August 2022
, Accepted 20th September 2022
First published on 29th September 2022
Abstract
Thiosemicarbazone-linked 3-acetylpyridine (T3AP), was synthesized and tested on copper strips in hydrochloric acid. Gravimetric measurements and electrochemical impedance spectroscopy were used to investigate the optimized inhibitory behavior of T3AP using the response surface methodology (RSM), with the optimized result obtained using a temperature of 42.90 °C, acid concentration of 2.38 M, inhibitor concentration of 3.80 mM, and time of 18.97 h, with inhibition efficiency up to approximately 93%. Validation of the experimental and predicted RSM showed that no significant difference in the inhibition efficiency with the confidence level value up to 97% was obtained. The isotherm study shows that T3AP obeys the Langmuir isotherm adsorption model, with physisorption and chemisorption adsorption mechanisms. The effectiveness of inhibitor performance of T3AP can be visually observed using scanning electron microscopy and X-ray photoelectron spectroscopy. The characterization revealed that the reactive S and N atoms in the T3AP inhibitor form strong chemical adsorption through N–Cu and Cu–S bonds on the copper surface. Computational analysis was also carried out, and we found that the stable energy gap between the occupied and unoccupied molecular orbitals (4.6891 eV) and high binding energy (540.962 kJ mol−1) adsorption from molecular dynamics were in agreement with the experimental findings.
1. Introduction
A variety of chemical processes rely on mineral acid solutions, such as hydrochloric and sulfuric acids, to remove scale and dirt from metal piping and other components.1,2 However, these acids aggressively attack metal surfaces, like copper, if there is no protective layer on the surface to prevent the metal from being harmed. Copper metals are particularly vulnerable to corrosion in acidic environments; hence, inhibitors are a critical component of corrosion prevention strategies.3,4 Researchers have studied numerous corrosion inhibitors to reduce the gap between the theoretical corrosion tests and actual application in industry. While corrosion inhibitors are still being developed, more research is needed to fully understand the mechanisms of effective inhibitor action. The remarkable adsorptive inhibitory activity of Schiff base compounds, owing to the presence of conjugated imine (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N) groups in the compounds, makes them attractive materials for inhibiting metal corrosion under acidic conditions.5 Schiff bases are used as corrosion inhibitors in commercial applications and they are preferred over other organic compounds because of their availability, cost-efficiency, simple synthesis, high purity, low toxicity, and environmental friendliness.6,7 The organic compounds, particularly Schiff bases, that contain 3-acetylpyridine and thiosemicarbazone8–10 can be effective corrosion inhibitors based on their ability to suppress hydrochloric acid solution corrosion. Hence, investigating new inhibitor platforms specifically for copper metal is a good start for this corrosion science study. On the other hand, having efficient inhibitors is not sufficient if their efficiencies are affected by the surrounding conditions, thus studying and optimizing the conditions is important to understand and ensure the maximum efficiency of the inhibitors.
N) groups in the compounds, makes them attractive materials for inhibiting metal corrosion under acidic conditions.5 Schiff bases are used as corrosion inhibitors in commercial applications and they are preferred over other organic compounds because of their availability, cost-efficiency, simple synthesis, high purity, low toxicity, and environmental friendliness.6,7 The organic compounds, particularly Schiff bases, that contain 3-acetylpyridine and thiosemicarbazone8–10 can be effective corrosion inhibitors based on their ability to suppress hydrochloric acid solution corrosion. Hence, investigating new inhibitor platforms specifically for copper metal is a good start for this corrosion science study. On the other hand, having efficient inhibitors is not sufficient if their efficiencies are affected by the surrounding conditions, thus studying and optimizing the conditions is important to understand and ensure the maximum efficiency of the inhibitors.
Traditional ways of studying optimization processes, such as one variable at a time, are no longer valid due to their demand on time and resources. Therefore, in this study, an experimental design with response surface methodology (RSM) was used to examine the inhibition efficiency of copper in an acidic medium using thiosemicarbazone-linked 3-acetylpyridine (T3AP). Analyzing the effects of variables on their own and in combination with one another is a valuable part of experimental design with RSM.11,12 Most reports in the literature have only used conventional methods to consider how inhibitors can mitigate corrosion. The optimization of corrosion inhibitors using RSM in the literature is minimal and less comprehensive. As a result, it is still necessary to identify the conditions within which an inhibitor will work optimally and provide the best response possible. To address this problem, RSM is can be used as a multi-objective analytical optimization method, along with the most comprehensive collection of mathematical and statistical methods. The primary advantage of RSM is that it reduces the number of experimental runs required to determine the optimum state.13,14
In the present study, T3AP has been synthesized to act as a corrosion inhibitor. The RSM method has been used to optimize the four parameters selected using central composite design (CCD), namely the temperature, acid concentration, inhibitor concentration, and immersion time, with two outputs, which were weight loss and impedance inhibition efficiency. Once the optimized conditions were obtained, the mechanism of the adsorption was characterized using an isotherm and transition state study. The surface analysis was conducted by scanning electron microscopy (SEM) and X-ray photoelectron spectroscopy (XPS) to visualize the morphology and binding interactions of the copper with T3AP. Furthermore, a computational study using density functional theory (DFT) quantum chemical calculations and Molecular Dynamic (MD) were carried out to determine the relationships between the electronic properties, the active sites and binding interaction of the T3AP inhibitor that function in inhibiting the copper from corrosion effect.
2. Materials and methods
2.1 Synthesis and characterization of the 3-acetylpyridine thiosemicarbazone
Equal amounts of 5 mmol of thiosemicarbazide [99%, Aldrich] (0.46 g) and a ketone 3-acetylpyridine [98%, Acros Organics] (0.61 g) were dissolved in 40 ml and 10 ml ethanol 97% solution [HmbG], respectively, using a condensation method. The solution containing the ketone was added dropwise into the thiosemicarbazide solution with slow heating and constant stirring. Then, five drops of sulfuric acid [95–97%, QReC] were added dropwise. The resultant solution was refluxed for 3 hours. The solution was allowed to cool at room temperature for the precipitate to form. Subsequently, the precipitate of T3AP was washed and filtered with cold ethanol and further dried in a desiccator. The yield of the reaction was weighed and kept under vacuum to avoid any moisture from the air. Then, the T3AP was characterized by spectroscopic analysis, elemental analysis of carbon, hydrogen, and sulfur, Fourier transform infrared spectroscopy (FTIR PerkinElmer, USA), and nuclear magnetic resonance [Bruker, USA] (1H NMR, 13C NMR 400 MHz, DMSO-d6). Fig. S1† depicts the general synthesis reaction for the T3AP inhibitor.
Yield: 78%; melting point: 195.3–197.4 °C; 1H-NMR (Fig. S2†) (DMSO-d6, 500 MHz, δ/ppm): 10.36 (s, 1H), 9.21 (s, 1H), 8.57 (d, 2H, J = 1.77 Hz), 8.36 (d, 2H, J = 1.6 Hz), 7.42 (t, 1H, J = 1.55 Hz), 8.11 (s, 1H), 8.34 (s, 1H), 2.34 (s, 3H). 13C-NMR (Fig. S3†) (DMSO-d6, 100 MHz δ/ppm) δ 179.52, 150.19, 148.24, 146.10, 133.67, 123.72, 14.25; CHNS elemental; 47.05, 5.57, 26.48, 16.22; FTIR (KBr, cm−1): 3385 (N–H), 3242 (N–H), 3166 (N–H), 1607 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1022, 856 (C
N), 1022, 856 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) S).
S).
2.2 Response surface methodology (RSM)
For this study, CCD was used to determine the optimal factors in inhibiting the copper metal with T3AP. RSM software (Design Expert 11) was used to model and analyze problems where responses are influenced by several variables. The ranges in this study were selected by the screening experiments to ensure the main parameters that can provide the optimization of corrosion inhibitors.
For the optimization of T3AP on a copper strip, the independent variables were selected as A: temperature (Xa), B: acid concentration (Xb), C: inhibitor concentration, (Xc), and D: immersion time (Xd). The responses (dependent variables) were the inhibition efficiency (%) of the gravimetric analysis (Y1) and the impedance response (Y2). The equation for the quadratic models is given as:
| | |
Y = X0 + XaA + XbB + XcC + XdD + XabAB + XacAC + XadAD + XbcBC + XbdBD + XcdCD + XaaA2 + XbbB2 + XccC2 + XddD2
| (1) |
CCD was performed with 30 experimental runs and six replications at the central points. Table 1 shows the set of 30 experiments based on CCD with their results. The determination set of experimental runs in this study was based on the CCD (30 = 2k + 2k + cp, where k is the number of independent variables and cp is the number of central points) with six replicated central points as proposed by the software. Additionally, the run order for these experiments was randomized to avoid systematic errors.
Table 1 The design layout based on the Design Expert for the experiment of T3AP
| Run |
Factor 1 |
Factor 2 |
Factor 3 |
Factor 4 |
Response 1 |
Response 2 |
| Temperature (°C) |
Acid (M) |
Inhibitor (mM) |
Time (h) |
WL IE% |
EIS IE% |
| 1 |
40 |
5 |
5 |
6 |
46.83 ± 1.37 |
67.97 ± 0.47 |
| 2 |
60 |
1 |
3 |
15 |
59.24 ± 0.19 |
73.31 ± 0.09 |
| 3 |
40 |
1 |
5 |
6 |
55.04 ± 1.34 |
77.76 ± 0.15 |
| 4 |
40 |
5 |
1 |
24 |
29.30 ± 0.93 |
60.37 ± 1.00 |
| 5 |
60 |
3 |
3 |
24 |
74.28 ± 0.19 |
84.01 ± 0.77 |
| 6 |
80 |
3 |
3 |
15 |
72.35 ± 0.29 |
76.20 ± 0.25 |
| 7 |
60 |
3 |
3 |
15 |
79.36 ± 0.47 |
86.66 ± 0.06 |
| 8 |
40 |
5 |
1 |
6 |
31.75 ± 1.37 |
56.04 ± 0.62 |
| 9 |
60 |
3 |
3 |
15 |
78.53 ± 0.64 |
87.08 ± 0.37 |
| 10 |
60 |
3 |
5 |
15 |
70.10 ± 0.13 |
80.13 ± 0.32 |
| 11 |
40 |
1 |
1 |
24 |
51.35 ± 0.30 |
54.38 ± 0.22 |
| 12 |
80 |
1 |
1 |
24 |
31.63 ± 0.13 |
59.14 ± 0.20 |
| 13 |
40 |
1 |
1 |
6 |
58.14 ± 2.33 |
48.58 ± 0.16 |
| 14 |
60 |
3 |
3 |
6 |
68.45 ± 1.03 |
74.37 ± 0.41 |
| 15 |
60 |
3 |
3 |
15 |
79.77 ± 0.71 |
86.17 ± 0.23 |
| 16 |
80 |
1 |
5 |
24 |
27.55 ± 0.26 |
53.37 ± 0.21 |
| 17 |
60 |
3 |
3 |
15 |
79.36 ± 0.78 |
85.74 ± 0.44 |
| 18 |
80 |
1 |
1 |
6 |
45.51 ± 0.60 |
40.69 ± 1.16 |
| 19 |
60 |
5 |
3 |
15 |
54.52 ± 0.75 |
75.41 ± 0.33 |
| 20 |
80 |
1 |
5 |
6 |
19.16 ± 0.60 |
35.87 ± 0.16 |
| 21 |
60 |
3 |
3 |
15 |
77.92 ± 0.47 |
87.49 ± 0.12 |
| 22 |
80 |
5 |
5 |
6 |
26.05 ± 0.66 |
33.08 ± 0.36 |
| 23 |
80 |
5 |
5 |
24 |
44.03 ± 0.36 |
45.29 ± 1.21 |
| 24 |
80 |
5 |
1 |
6 |
35.25 ± 0.66 |
54.26 ± 0.21 |
| 25 |
40 |
1 |
5 |
24 |
78.11 ± 0.12 |
79.89 ± 0.03 |
| 26 |
60 |
3 |
3 |
15 |
78.33 ± 0.62 |
86.76 ± 0.25 |
| 27 |
40 |
3 |
3 |
15 |
93.65 ± 0.39 |
92.19 ± 0.18 |
| 28 |
60 |
3 |
1 |
15 |
61.30 ± 0.62 |
76.48 ± 0.13 |
| 29 |
40 |
5 |
5 |
24 |
73.54 ± 0.45 |
68.03 ± 0.26 |
| 30 |
80 |
5 |
1 |
24 |
29.61 ± 0.19 |
69.58 ± 0.21 |
2.3 Materials and solution
Copper strip samples (99% purity) with dimensions of 2 × 2 × 0.08 cm were used in the experiments. Dilution with distilled water of 37% HCl [R&M] was used to create various concentrations of HCl acidic solution based on the selected parameters. Following that, different concentrations of 1, 3, and 5 mM of inhibitors were dissolved in 10 mL of ethanol to dissolve any leftover solid organic compounds that had formed. Then, the 100 mL test tube was used to immerse the copper strips in HCl solution with or without the addition of the inhibitors. The temperature and time for the immersion were controlled based on the parameter selected from the RSM.
2.4 Gravimetric analysis (first response for RSM)
The gravimetric analysis experiment was conducted to analyze the first response toward inhibition efficiency by T3AP on copper strips. A preliminary experiment was conducted to determine the RSM's recommended ranges for each factor. The copper strips were immersed in HCl solutions with different concentrations using solution volumes of 15 ml. The immersion time was recorded at three separate times based on the RSM range parameter selected, which were 6, 15, and 24 hours, respectively.
The weight of the copper strip was measured before and after immersion. The average decrease in weight was calculated when the experiments were conducted in triplicate simultaneously on each run. Using the CCD shown in Table 1, the temperatures, acid concentration, inhibitor concentrations, and time of the 30 sample runs were measured and recorded. Using eqn (2), the corrosion rate (r) was determined:15
| |
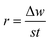 | (2) |
where Δ
w is the weight loss of the copper strip,
t is the immersion period, and
s is the entire surface area of the copper strip. This was followed by a calculation of inhibition efficiency (percentage) using
eqn (3) below:
15| |
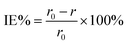 | (3) |
where
r0 and
r are the corrosion rates of copper strips without and with the inhibitor, respectively.
2.5 Electrochemical impedance spectroscopy (second response for RSM)
After gravimetric analysis, the sample was then used for the second response RSM of electrochemical impedance spectroscopy (EIS) inhibition efficiency. The same copper strip samples (99% purity) with dimensions of 2 × 2 × 0.08 cm square shape were used in the EIS experiments. All sample runs were carried out with the help of a three-electrode electrochemical cell, which was connected to an Auto-lab PGSTAT-302N potentiostat/galvanostat (AUTOLAB, USA). A copper strip was set as a working electrode, platinum rods as counter electrodes, and silver/silver chloride (Ag/AgCl) as a reference electrode. EIS analysis was performed in a potentiostatic condition with a 10 mV AC signal amplitude and a frequency limit of 0.01 Hz to 100 kHz at open circuit potential (OCP). Triplicate readings were recorded for every run of the experiment. The optimized EIS response was fitted using the Zview software version 3.4e. The Tafel plot curve for potentiodynamic polarization measurements was performed after the EIS optimization with same optimized condition where the potential was swept at a rate of 1 mV s−1 at OCP, primarily from −500 to −100 mV.
2.6 Scanning electron microscopy (SEM)
The surface morphologies of the copper strips were examined by SEM (EVO 50, USA) before and after being treated with T3AP immersed in HCl at the optimal conditions. The SEM images were generated with a scanning electron microscope at high vacuum using an electron high tension voltage of 10.0 kV and magnifications of 100 and 500×.
2.7 X-ray photoelectron spectroscopy (XPS)
After being immersed in HCl with and without the T3AP inhibitor at the optimal conditions, the copper strip samples were inserted into the XPS instrument to study the surface chemical characterization properties. The XPS experiments were conducted using the Kratos Axis ULTRA, UK spectrometer with a monochromatized Al Kα radiation source of 1486.6 eV and an X-ray beam diameter of around 100 μm. All the testing was carried out under ultrahigh vacuum, with a chamber base pressure greater than 1 × 10−9 torr. The XPS spectrum included both a wide and a narrow scan, depending on the type of analysis being performed. A narrow scan was performed in this study for characterizing the element of interest which were Cu 2p, Cl 2p, C 1s, N 1s, and S 2p. The XPS data spectra were first transformed into the standard VAMAS format and then deconvoluted using the commercial fitting program, CasaXPS.
2.8 DFT simulations
The B3LYP/DFT hybrid functional approach was used to optimize the ground states of the investigated compounds.16 A triple-ζ Pople-type basis set 6-311+G (d,p) was utilized, in which polarized and diffuse functions are included. Moreover, several electronic properties, such as the ionization potential (I), electronic affinity (A), energy gap, electronegativity (χ), global hardness (η), chemical softness (S), and highest occupied and lowest unoccupied molecular orbitals (HOMO and LUMO, respectively), were determined to describe the interactions of the copper metal with T3AP. Koopman's theorem was used to determine the electronic parameters A, I, χ, and η, where I = −EHOMO and A = −ELUMO. Thus, χ, η, and S were identified using the following equations:17,18| |
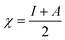 | (4) |
| |
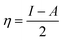 | (5) |
| |
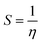 | (6) |
In addition, Mulliken atomic charges were used to discover which atoms were most likely to react with copper strips. The Gaussian 09 program was used for all of the theoretical computations, including ground state geometry optimization and frequency analysis.18
2.9 Molecular dynamics simulation
All of the molecular dynamics (MD) simulations were conducted in Material Studio. With the use of the COMPASS (Condensed Phase Optimized Molecular Potentials for Atomistic Simulation Studies) force field and a simulation box (25.55 by 22.55 by 34.86 Å) with periodic boundary conditions, we evaluated how the inhibitor T3AP interacts with the copper surface. The adsorption process was stimulated by using the most frequent and stable Cu (111) surface. Cu (111) has a low Miller index. Prior to using the 6 Å slab, the Cu crystal was imported and cleaved on the (111) plane. The smart minimizer generates a larger area for inhibitor interactions by lowering the energy of the copper (111) surface and extending it to a supercell of (10 × 10). A zero-thickness vacuum slab was then manufactured. In the end, an amorphous cell was built with the best possible configuration of T3AP inhibitors.
3. Results and discussion
The thiosemicarbazone-linked 3-acetylpyridine (T3AP) compound was synthesized by reacting the thiosemicarbazide and 3-acetylpyridine, as can be seen in Fig. S1.† Fig. S2 and S3† provide the 1H NMR and 13C NMR spectra used to elucidate the chemical structure of T3AP. Based on Fig. S2,† the proton located in the most downfield region, which showed as a singlet signal at a frequency of δ 10.36 ppm, corresponded to the N(3)–H found in the heterocyclic substituent. The proton signals for carbon [C(8)–H] that adjacent to the nitrogen atom in the pyridine ring can be observed at δ 9.12 ppm. The proton in C(7)–H, next to nitrogen in the pyridine ring, gives a double duplet signal at 8.57 ppm due to a different environment from the nitrogen and carbon C(6). The proton for amino [N(1)–H2] can be observed at two different peaks at δ 8.34 and 8.11 ppm. Multiplet signals can be seen occurring in the region of δ 7.42 ppm. These signals correspond to the heterocyclic substituent's proton bound to carbons C(6). The three protons bound to the carbon C(4) in the methyl group, known as an electron-withdrawing group, occur at the most shielded region in which the singlet signal emerged at a frequency of δ 2.34 ppm. In the 13C NMR spectrum, eight signals were observed within the δ 180 to 10 ppm (Fig. S3†). The signal that was the most downfield emerged at δ 179.52 ppm, corresponding to the carbon C(1) due to the electronegative atom sulfur of the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) S bond. The chemical shift of carbon C(3)
S bond. The chemical shift of carbon C(3)![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N can be assigned at δ 150.19 ppm. The carbon in the heterocyclic ring of pyridine C(4), C(5), C(6), C(7), and C(8) can be observed in the range of δ 148 to 123 ppm. The most shielded chemical shift belonged to the methyl group's C(2) atom, which appeared at δ 14.25 ppm.
N can be assigned at δ 150.19 ppm. The carbon in the heterocyclic ring of pyridine C(4), C(5), C(6), C(7), and C(8) can be observed in the range of δ 148 to 123 ppm. The most shielded chemical shift belonged to the methyl group's C(2) atom, which appeared at δ 14.25 ppm.
3.1 Statistical analysis and modelling using RSM
The study analysis was conducted using four independent variables: temperature (A), acid concentration (B), inhibitor concentration (C), and immersion time (D). Weight loss and EIS inhibition efficiency are two responses that were selected as outputs. It is shown in Table 1 the input and output results for both responses. Eqn (7) and (8) describe the weight loss and impedance IE regression equation derived from the analysis of variance (ANOVA) results, respectively.| | |
WL IE = 78.546 + −10.365 × A + −3.04722 × B + 3.69833 × C + 2.95667 × D + 4.51938 × AB + −6.76187 × AC + −2.10562 × AD + 4.45687 × BC + 1.61312 × BD + 6.55688 × CD + 4.7864 × A2 + −21.3336 × B2 + −12.5136 × C2 + −6.8486 × D2
| (7) |
| | |
EIS IE = 86.5912 + −7.65167 × A + 0.391111 × B + 1.215 × C + 4.74667 × D + 1.33375 × AB + −8.14625 × AC + 3.1975 × AD + −4.37375 × BC + −0.7475 × BD + −0.75 × CD + −2.33746 × A2 + −12.1725 × B2 + −8.22746 × C2 + −7.34246 × D2
| (8) |
3.1.1 Normal probability plots. It is possible to use the normal probability plot to visualize the data and determine if it has a normal distribution. In this sort of figure, data are plotted against a theoretical normal distribution, and a straight-line graph shows the agreement between the predicted and actual data. If the values deviate from this line, it reflects deviations from the normality of the data. Plots for both responses are shown in Fig. S4,† revealing the normal probability plot for the WL and EIS IE responses against residuals. Design Expert software was used to create this graph. As seen in Fig. S4,† the ANOVA projected value has a very high degree of accuracy since the points are extremely close to the line, exhibiting low dispersion.
3.2 ANOVA results
Table 2 shows the complete summary of the quadratic model results for inhibitor efficiency with ANOVA regression analysis regarding the weight loss efficiency. The significance of the RSM model depends on the P-value obtained. The P-value is considered to be significant when it is lower than 0.05.19 Each of the regression equations for the response inhibition efficiency is significant for all factors. Table 3 shows that the WL IE's R2 adj value is 99.64%. Therefore, the model can accurately anticipate the response output. The F-value (1226.07) and T-value (−35.02) indicate that the temperature shows the most significant effect on corrosion efficiency.
Table 2 Quadratic model of T3AP WL IE
| Source |
Sum of squares |
df |
Mean square |
F-value |
p-value |
T-value |
| Model |
12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 749.50 749.50 |
14 |
910.68 |
577.39 |
<0.0001 |
— |
| A |
1933.80 |
1 |
1933.80 |
1226.07 |
<0.0001 |
−35.02 |
| B |
167.14 |
1 |
167.14 |
105.97 |
<0.0001 |
−10.29 |
| C |
246.20 |
1 |
246.20 |
156.10 |
<0.0001 |
12.49 |
| D |
157.35 |
1 |
157.35 |
99.77 |
<0.0001 |
9.99 |
| AB |
326.80 |
1 |
326.80 |
207.20 |
<0.0001 |
14.39 |
| AC |
731.57 |
1 |
731.57 |
463.83 |
<0.0001 |
−21.54 |
| AD |
70.94 |
1 |
70.94 |
44.98 |
<0.0001 |
−6.71 |
| BC |
317.82 |
1 |
317.82 |
201.50 |
<0.0001 |
14.20 |
| BD |
41.63 |
1 |
41.63 |
26.40 |
0.0001 |
5.14 |
| CD |
687.88 |
1 |
687.88 |
436.13 |
<0.0001 |
20.88 |
| A2 |
59.36 |
1 |
59.36 |
37.63 |
<0.0001 |
6.13 |
| B2 |
1179.18 |
1 |
1179.18 |
747.63 |
<0.0001 |
−27.34 |
| C2 |
405.71 |
1 |
405.71 |
257.23 |
<0.0001 |
−16.04 |
| D2 |
121.52 |
1 |
121.52 |
77.05 |
<0.0001 |
−8.78 |
| Lack of fit |
21.06 |
10 |
2.11 |
4.05 |
0.0680 |
— |
Table 3 Fit statistics of the model for T3AP
| Terms |
Response |
| WL IE |
EIS IE |
| Standard deviation |
1.26 |
0.6910 |
| Mean |
57.00 |
68.54 |
| R2 |
0.9981 |
0.9991 |
| Adjusted R2 |
0.9964 |
0.9983 |
| Predicted R2 |
0.9871 |
0.9955 |
| Adequate precision |
85.8217 |
120.8982 |
| Coefficient of variation (CV) % |
2.20 |
1.01 |
Tables 3 and 4 show the results of an ANOVA regression analysis of the RSM models for EIS inhibition efficiency, as well as the conclusions of the RSM models for inhibitor efficiency. The results of the ANOVA (Table 4), suggest that each expression of the regression equations for the EIS IE response is statistically significant as the P-value of the model was <0.0001. The R2 adj for the EIS IE response was 99.83% which indicated that the model can accurately predict the output. Table 3 indicates that the WL and EIS IE R2 assessments are 99.81 and 99.91%, respectively. Thus, the model predicts the response output with great accuracy. Similar to the gravimetric results, the EIS study also shows that temperature has the most significant effect on corrosion efficiency based on the F-value (2207.11) and T-value (−46.98) obtained.
Table 4 Quadratic model of T3AP EIS IE
| Source |
Sum of squares |
df |
Mean square |
F-value |
p-value |
T-value |
| Model |
8322.00 |
14 |
594.43 |
1244.91 |
<0.0001 |
— |
| A |
1053.86 |
1 |
1053.86 |
2207.11 |
<0.0001 |
−46.98 |
| B |
2.75 |
1 |
2.75 |
5.77 |
0.0297 |
2.40 |
| C |
26.57 |
1 |
26.57 |
55.65 |
<0.0001 |
7.46 |
| D |
405.56 |
1 |
405.56 |
849.35 |
<0.0001 |
29.14 |
| AB |
28.46 |
1 |
28.46 |
59.61 |
<0.0001 |
7.72 |
| AC |
1061.78 |
1 |
1061.78 |
2223.69 |
<0.0001 |
−47.16 |
| AD |
163.58 |
1 |
163.58 |
342.59 |
<0.0001 |
18.51 |
| BC |
306.08 |
1 |
306.08 |
641.01 |
<0.0001 |
−25.32 |
| BD |
8.94 |
1 |
8.94 |
18.72 |
0.0006 |
−4.33 |
| CD |
9.00 |
1 |
9.00 |
18.85 |
0.0006 |
−4.34 |
| A2 |
14.16 |
1 |
14.16 |
29.65 |
<0.0001 |
−5.44 |
| B2 |
383.89 |
1 |
383.89 |
803.98 |
<0.0001 |
−28.35 |
| C2 |
175.38 |
1 |
175.38 |
367.30 |
<0.0001 |
−19.17 |
| D2 |
139.68 |
1 |
139.68 |
292.53 |
<0.0001 |
−17.10 |
| Lack of fit |
5.20 |
10 |
0.5201 |
1.33 |
0.3980 |
— |
3.3 Multi objective optimization using 3-D surface plot
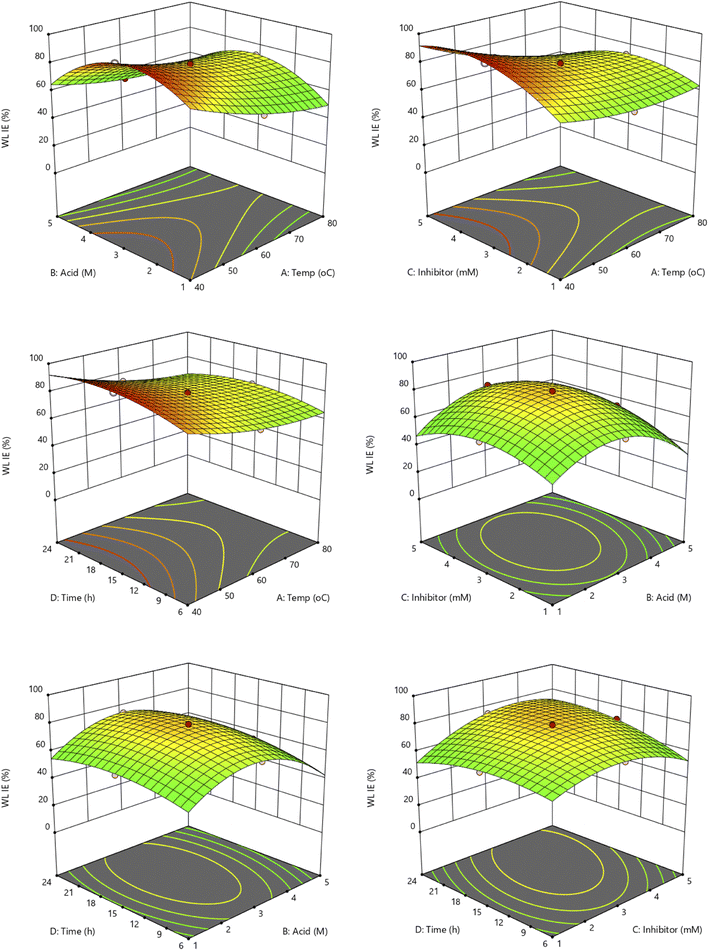
The relationships among the four independent variables (temperatures, acid concentration, inhibitor concentration, and time) were investigated further using a 3-D surface plot. The 3-D plot can show the trends and relationships between two factors and the optimized output results. The copper strip's inhibitory efficiency properties in harsh conditions can be improved by optimizing these four factors. The two output responses from the experimental design are weight loss and EIS inhibition efficiency.
3.3.1 Interaction effect of the independent variables on the weight loss inhibition efficiency. Fig. 1 represents the influence of four independent variables on the weight loss inhibition efficiency. It can be observed from the 3D surface plot when the temperature is increasing the efficiency is decreasing. High temperature enhances the corrosion rate effect of the copper metal. For acid concentration and inhibitor concentration, both show the same trend of shape where it increased and decreased after reaching the optimal conditions. Inhibitors should not be used excessively because they can cause drawback effects by inhibiting the corrosion since they will react with the acid solutions. Meanwhile, for immersion time, the inhibition increased as the time increased, since the calculation involving the rate of corrosion efficiency where the longer the time, the smaller the rate of the corrosion. It can be concluded from the surface plots that the temperature was the best at a lower temperature (40–45 °C). For acid and inhibitor concentrations best range around 3.0 M and 3.0 mM, respectively. The immersion time gives the best result at 15–19 h for the weight loss experiments. However, if the time is prolonged further above 24 h, the inhibitor would lose its adsorption on the copper surface because the dissolution from corrosion still proceeds.20
 |
| | Fig. 1 3D plots for evaluating weight loss inhibition efficiency response of T3AP. | |
3.3.2 Interaction effect of the independent variables on the impedance inhibition efficiency. The second response study of impedance inhibition efficiency (EIS IE) 3D surface plot can be seen in Fig. S5.† It can be observed that the result of EIS IE gives more significance in terms of their inhibition efficiency because it is a study on the immediate reaction and electrochemical study. However, the trend of the interaction is almost similar to the weight loss response study. For the temperature factor, the inhibition seems to be decreased as the temperature increases. Notably, when the temperature of the electrolyte medium is raised, the adsorption tendency of the organic compounds naturally decreases.21,22 This is because an increase in kinetic energy due to high temperature has a deleterious effect on the intermolecular attraction between the inhibitor and metal surface.23 This phenomenon can be illustrated in the schematic diagram in Fig. 2. In addition, a higher temperature can cause the catalyzation of the fragmentation or rearrangement of the organic compounds particularly heterocycles.21,23 The acid concentration reached the optimal conditions at the middle of the study range of 1–5 M, around 3 M. Acid concentration played an important role in the electrochemical study because it affected the number of ions in the solutions of the electrochemical system, where the higher the number of ions, the higher the current needed. The aggressiveness of the HCl solution will reduce the efficiency of the inhibitor's protection on the metal surface as some previous study which used the high HCl solution concentration.24,25 Furthermore, the concentration of inhibitor gives the same trend as acid concentration with a concave shape, where it reached the maximum efficiency at 3.0 mM. Same with acid factor, inhibitors also give significant results to EIS study since these inhibitors are the ones that blocked the copper surface from the HCl ions attacked. The effects of the time trend were found to be the best at around 15–18 h result. From these interaction results, all the factors show a significant effect on the inhibition efficiency for both responses as in line with the ANOVA result obtained.
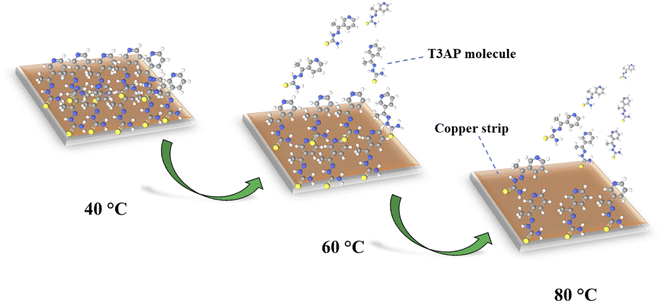 |
| | Fig. 2 The schematic diagram of temperature effect on the desorption process of T3AP molecules. | |
3.4 Method validation
The model of the RSM outcomes was validated using both responses based on the optimum parameters suggested by the RSM. The predicted values for weight loss and EIS inhibition efficiency were obtained from the eqn (7) and (8), respectively. The weight loss predicted value from RSM is 93.93%, and the optimum response obtained from the average gravimetric result was 91.22 ± 0.58% as shown in Table 5. This indicated that the predicted and experimental weight loss responses exhibit 97.03% agreement. For EIS inhibition efficiency response, the predicted value is 92.41% and the experimental result for EIS obtained is 94.34% (Table 6). Thus, the confidence level between both results is 97.95%. It is best to conclude the correlation between the predicted value from the RSM and experiments from both responses shows very close agreement which means that the accuracy of the RSM model is accepted. Then, the optimum parameters are applicable to identify the best inhibition efficiency for gravimetric and impedance experiments for copper metal with T3AP.
Table 5 Validation of the optimal values of temperature, acid concentration, inhibitor concentration, and time for weight loss inhibition efficiency T3AP
| |
Predicted RSM |
Experimental data |
Confident level |
| Weight loss inhibition efficiency (%) |
93.93 |
91.22 ± 0.58 |
97.03 |
Table 6 Validation of the impedance data for copper strip in the absence and presence of T3AP at optimum condition
| Inhibitor |
Rs (Ω cm2) |
Rct (Ω cm2) |
η (%) |
Predicted RSM |
Confident level |
| Blank (untreated) |
1.176 |
8.042 |
— |
— |
— |
| T3AP (treated) |
1.130 |
142.200 |
94.34 ± 0.05 |
92.41 |
97.95 |
3.5 Electrochemical inhibition tests
3.5.1 Electrochemical impedance spectroscopy (EIS). EIS tests were used to look for T3AP inhibition efficiency for the second response. For the validation of the optimum result, the copper strips were immersed in HCl solution at RSM's optimal conditions, and the resulting Nyquist plots are shown in Fig. 4. Copper immersed in the HCl solution without inhibitor had the lowest impedance values. The copper strip's impedance module values grew dramatically reaching a maximum value of inhibition efficiency as the concentration of 3.80 mM T3AP inhibitor at the temperature, acid concentration, and time of 42.90 °C, 2.38 M, and 18.97 h, respectively.
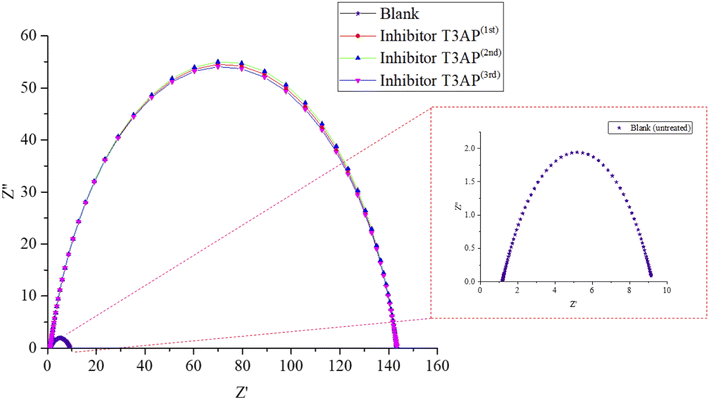 |
| | Fig. 4 Nyquist diagrams of copper strips with and without T3AP at optimum condition. | |
Fig. 3 shows the equivalent circuit model that was used to fit all EIS spectra to get a quantitative description of the corrosion system.20,26,27 Based on the equivalent circuit (Fig. 3), Rs denotes the solution resistance, CPE stands for constant phase element, Rct stands for the charge transfer resistance related to the corrosion reaction. T3AP inhibitors had a considerable impact on the corrosion activity on the copper surface, based on the varied Rct values obtained in the absence and presence of the compounds. The Rct value of the copper strip immersed in HCl without T3AP obtained 8.042 cm2, which is quite low. In contrast, it was shown that the Rct levels increased significantly when T3AP inhibitors were used. Using the 3.80 mM T3AP solution, the Rct readings reached their highest value of 142.20 Ω cm2 (Fig. 4) at the optimum condition, which was almost 18 times greater than the untreated sample under the same conditions. The inhibition efficiency calculated based on the Rct result matches the predicted RSM value by 94.34%. T3AP molecules are effectively adsorbing onto the copper metal as the Rct grows significantly, which means that less copper is exposed to the HCl electrolyte, and this slows down the corrosion process.20,28
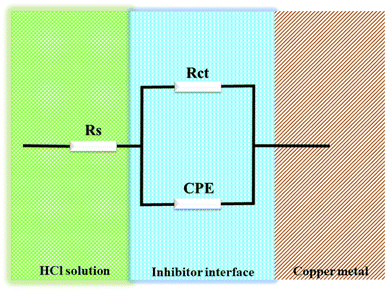 |
| | Fig. 3 Model of equivalent circuit of EIS for this study. | |
3.5.2 Polarization and linear polarization resistance measurements. The optimal concentration used for this test was 3.80 mM with optimal temperature and acid concentration conditions. Fig. 5 depicts the typical polarization curves for the copper in 2.38 M HCl with and without T3AP inhibitors. Table 7 lists all the important corrosion parameters obtained from the polarization resistance measurements. It is clear from these results that when an inhibitor is applied, current densities decrease. This could imply that the T3AP molecules are adsorbing to the copper surface. Copper corrosion rates in inhibited solutions are significantly lower than in untreated solutions. Both anodic and cathodic processes are affected by the addition of 3.80 mM T3AP to the HCl, as shown in Fig. 5 polarization curves. This means that the cathodic hydrogen reaction mechanism has been altered and the copper surface has been blocked from the active sites.29,30 An increase in the number of T3AP inhibitors adsorbing on the metal-solution interface can be seen in the shift of the Ecorr to the more negative direction when T3AP is present. These findings indicate that T3AP is a mixed-type inhibitor. The polarization measurement results show that the inhibition efficiency obtained was nearly the same as the EIS efficiency result of 94%. The absolute corrosion rate can be determined using this method in millimeters per year (mm per year). Thus, corrosion rates obtained with T3AP inhibitors (3.5542 mm per year) were far lower than uninhibited solutions (47.050 mm per year). It is clear from these results that T3AP inhibitors have a significant impact on copper dissolution. In terms of corrosion inhibition, these results are in agreement with the gravimetric and EIS measurements.
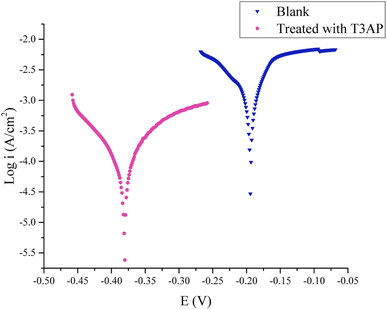 |
| | Fig. 5 Polarization curves for copper strips in HCl at optimal conditions with and without the addition of T3AP. | |
Table 7 Parameters of potentiodynamic polarization of copper strip in HCl with and without T3AP
| Inhibitor |
Ecorr (mV) |
Icorr (mA cm−2) |
bc (mv dec−1) |
ba (mv dec−1) |
η (%) |
CR (mm per year) |
| Blank (untreated) |
−195.18 |
4.0491 |
184.17 |
141.51 |
— |
47.050 |
| T3AP (treated) |
−381.44 |
0.3059 |
432.65 |
117.05 |
92.45 |
3.5542 |
3.6 Adsorption isotherm
The result obtained from the optimized RSM result was then matched with the adsorption isotherm study to determine the type of adsorption of the T3AP and copper metal reaction. Various adsorption isotherm was analyzed, and the result shows Langmuir isotherm fit the best result as shown in Fig. 6. The data can be expressed using eqn (9):| |
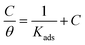 | (9) |
where θ is inhibitor coverage surface area, Cinh is T3AP concentration and Kads is the equilibrium constant for adsorption. The slope obtained from the plotted graph of C/θ vs. C is 1.0429, which is almost equal to 1, and R2 is equal to 0.9956. The purpose of finding Kads is to calculate their adsorption standard free energy (ΔGads), which can be obtained from eqn (10):| |
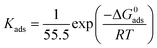 | (10) |
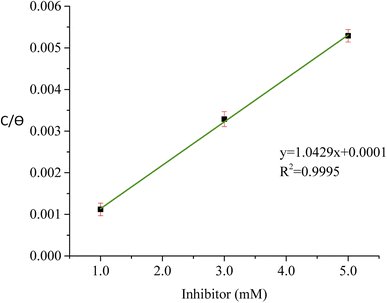 |
| | Fig. 6 Langmuir graph of T3AP. | |
The type of adsorption mechanism of metal-inhibitor can be characterized based on the magnitude of ΔGads. If the ΔGads value of −40 kJ mol−1 or more is negative, then the chemisorption process is confirmed. However, if the ΔGads value is between −20 kJ mol−1 and −40 kJ mol−1, then physio chemisorption is confirmed.31–33 From the result obtained, the ΔGads for copper-3APT was −34.76 kJ mol−1 (Table 8) which is between −20 and −40 kJ mol−1, thus the reaction involves physisorption and chemisorption mechanisms. Spontaneous adsorption and stability of the adsorbent layer on the copper metal externality can be inferred from the negative values of the ΔGads parameter.32,34
Table 8 Equilibrium constant and standard free energy of adsorption of copper strips with T3AP
| Inhibitor |
Kads (M−1) |
ΔGads (kJ mol−1) |
| T3AP |
1.00 × 104 |
−34.76 |
3.7 Activation thermodynamic parameters
The thermodynamic properties of the activation such as activation energy (Ea), enthalpy (ΔHa), and entropy (ΔSa) were determined via the following relationships.35,36| |
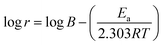 | (11) |
| |
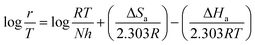 | (12) |
where B is the pre-exponential factor, N is Avogadro's number, and h is the Planck constant.
Table 9 lists the values of Ea determined from the slope and intercept of the graphs ln![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) r vs. 1/T depicted in Fig. 7(a). A comparison of the Ea values in the presence and absence of T3AP shows that the creation of an energy barrier makes metal corrosion more difficult in the presence of the T3AP.1 Meanwhile, the enthalpy (ΔHa) and entropy (ΔSa) of the reaction system were calculated from the slope and intercept of ln(r/T) vs. 1/T in Fig. 7(b). Positive values of ΔHa (Table 9) are a strong indicator that endothermic processes are involved in copper dissolution.37 The fact that the ΔSa value (Table 9) was higher in the treated solution with T3AP compared to the untreated solution suggests that there was a greater degree of disorder in the system.36
r vs. 1/T depicted in Fig. 7(a). A comparison of the Ea values in the presence and absence of T3AP shows that the creation of an energy barrier makes metal corrosion more difficult in the presence of the T3AP.1 Meanwhile, the enthalpy (ΔHa) and entropy (ΔSa) of the reaction system were calculated from the slope and intercept of ln(r/T) vs. 1/T in Fig. 7(b). Positive values of ΔHa (Table 9) are a strong indicator that endothermic processes are involved in copper dissolution.37 The fact that the ΔSa value (Table 9) was higher in the treated solution with T3AP compared to the untreated solution suggests that there was a greater degree of disorder in the system.36
Table 9 Data for thermodynamic corrosion of copper strip in HCl solution with absence and presence of T3AP
| Inhibitor |
Ea (kJ mol−1) |
ΔHa (kJ mol−1) |
ΔSa (kJ mol−1 K−1) |
| Blank |
40.99 |
38.22 |
−195.92 |
| T3AP |
70.72 |
67.95 |
−117.17 |
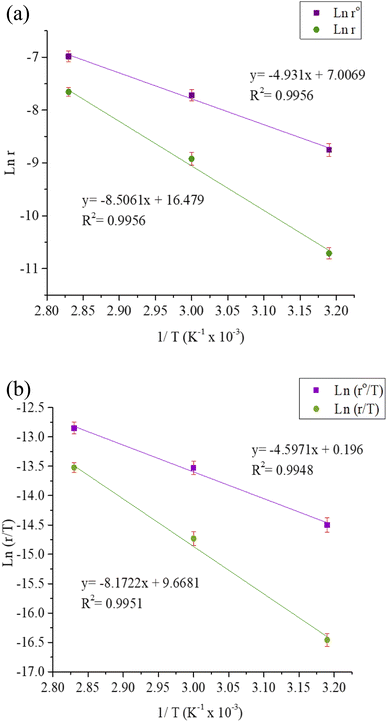 |
| | Fig. 7 (a) Arrhenius plots for copper strip in the absence and presence of T3AP. (b) Transition state plots for copper strip in the absence and presence of T3AP. | |
3.8 Surface analysis
3.8.1 Scanning electron microscopy (SEM) analysis. The result of SEM visualized the morphology surface of the copper strip with two magnification when immersed in HCl solution at the optimum conditions in presence and absence of T3AP were obviously different. It can be seen in Fig. 8(a and b) that the copper surface show relatively rough with deep holes and cracks. This indicated copper strips excessively corroded without the inhibitor due to the metal dissolution by HCl. In comparison, the copper surface with the addition of the inhibitor T3AP in Fig. 8(c and d) at the optimum conditions, was smooth and not affected much compared with the blank.
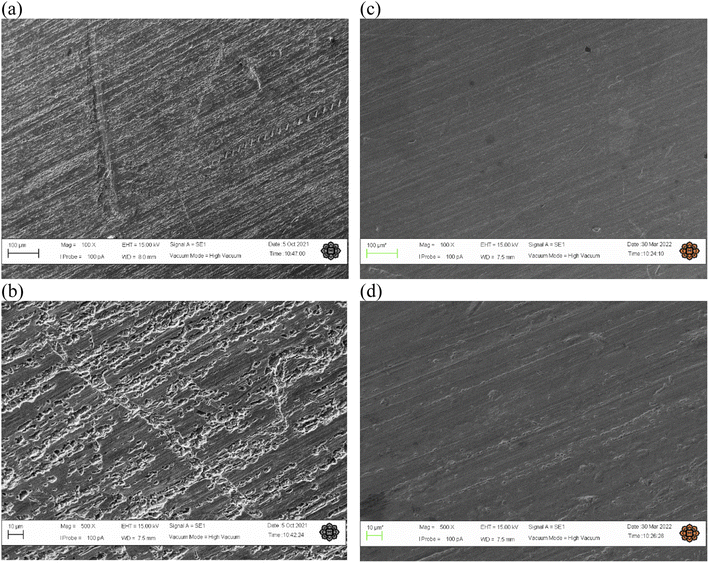 |
| | Fig. 8 The SEM result for untreated copper samples at 100× magnification (a) and 500× magnification (b), and treated (T3AP) copper samples at 100× magnification (c) and 500× magnification (d). | |
3.8.2 X-ray photoelectron spectroscopy (XPS). The XPS analysis of copper metal in the absence and presence of T3AP was investigated for the surface chemical characterization bonding of the adsorption between copper metal and the inhibitor which was shown in Fig. S6 until S11.† Since the corrosion inhibitor got the maximum inhibitory efficiency from the RSM optimization, the T3AP at its optimal concentration of 3.80 mM was chosen for XPS investigation. The elements of interest (Cu 2p, Cl 2p, C 1s, N 1s, and S 2p) in samples with and without inhibitor T3AP were obtained from narrow scans and resolved using a procedure called spectrum deconvolution fitting. Based on the results, the untreated sample revealed two elements of interest, namely chlorine and copper. The first element is represented by deconvoluted peaks of Cl 2p, as shown in Fig. S6,† and the assignment peaks are presented in Table S1.† It was discovered that 198.91 and 200.56 eV denoted for Cl 2p3/2 and Cl 2p1/2, respectively. These results originate from the chlorine solution that coated the surface of the copper metal. The peak on Cl 2p3/2 was assigned to be the bonding of H–Cl which represent the solution used for the copper corrosion.38,39 The Cl 2p1/2 was observed for the bonding of CuCl2 (ref. 40) because chloride ions are electrostatically attracted to the positively charged copper ions on the interface between the metal and the solution's layer, creating the adsorption surface complex Cu–Cl2. Thus, the HCl solution caused corrosion on the surface of the copper strips study. Fig. S7 and S8† show the two doublets Cu 2p3/2 and Cu 2p1/2 that are present in the Cu 2p narrow scan spectra for copper metal-solution interfaces without and with T3AP protection. Only two peaks (Fig. S7†) were obtained by the deconvolution of the high-resolution Cu 2p3/2 XPS spectra when the untreated sample was used, but three peaks (Fig. S8†) were produced when T3AP was used. The peaks of Cu(I) can be seen in Fig. S8,† and they are located at 932.53 and 932.20 eV. The Cu(I) may have reacted with oxygen and chlorine ions in the solution to formed Cu2O and CuCl. This indicated that the copper surface has oxidized to Cu(I). This can be explained by the mechanism as illustrated below:| | |
2Cu + H2O → Cu2O + 2H+ + 2e
| (13) |
| | |
Cu + HCl → CuCl + H+ + e
| (14) |
The Cu2O can be observed due to the existence of an adsorbed water film on the copper substrate which is most likely the result of the physisorption of water vapor from the surrounding air at the contact between the metal and the solution.41 However, the solution protected with T3AP inhibitor produced peaks for Cu–S, Cu–N, and CuCl. The Cu–S and Cu–N can be observed at binding energy at 932.02 and 933.23 eV, respectively. This bond was possible to be observed since the sulfur and nitrogen atoms present in the structure of the T3AP inhibitor clarify the successful adsorption of the inhibitor with the copper metal. These helped to improve the corrosion resistance of copper in HCl solutions because they were formed up of a firmly cohesive layer, which contributed to insulating and conserving the copper strip from the reaction that triggered corrosion. The CuCl (932.26 eV) is still present since there are still Cl atoms from the HCl solution on the exterior copper surface.
The C 1s spectrum for copper metal with T3AP in HCl solution is ascribed to four primary photoelectron peaks. This suggests that four kinds of distinct C atoms are present on the copper metal-solution interface, as illustrated in Fig. S9.† As can be seen in Table S1,† the molecule T3AP is chemisorbed via an interaction involving a covalent bond π to π* represented by C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C (284.74 eV) of the pyridine ring, C
C (284.74 eV) of the pyridine ring, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N or C–N (286.97 eV), as well as C
N or C–N (286.97 eV), as well as C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) S at 287.89 eV. The presence of these conjugated double bonds C
S at 287.89 eV. The presence of these conjugated double bonds C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C, C
C, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N, and C
N, and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) S in the T3AP inhibitor increased adsorption on the copper surface's outermost layer by providing a barrier to shield the underlying copper metal–solution interface from further oxidation by an acidic HCl solution during an extended immersion duration. The C–C bond (282.48 eV) also appeared in the spectrum since the chemical structure of T3AP contained C–CH3 bond. There are four primary synthetic photoelectron peaks displayed in Fig. S10† by fitting the deconvolution of N 1s spectra for the adsorbed film on copper when T3AP is present. The peaks were for N–H, N–N/C
S in the T3AP inhibitor increased adsorption on the copper surface's outermost layer by providing a barrier to shield the underlying copper metal–solution interface from further oxidation by an acidic HCl solution during an extended immersion duration. The C–C bond (282.48 eV) also appeared in the spectrum since the chemical structure of T3AP contained C–CH3 bond. There are four primary synthetic photoelectron peaks displayed in Fig. S10† by fitting the deconvolution of N 1s spectra for the adsorbed film on copper when T3AP is present. The peaks were for N–H, N–N/C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N, N–Cu, and N–O which appeared at 399.17, 399.11, 398.55, and 397.522 eV, respectively. The T3AP molecule is responsible for the formation of the N–H bond as well as the N–N/C
N, N–Cu, and N–O which appeared at 399.17, 399.11, 398.55, and 397.522 eV, respectively. The T3AP molecule is responsible for the formation of the N–H bond as well as the N–N/C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N bond. The N–Cu bond results from the combination of copper and N in the T3AP molecule and the N–O bond is caused by the oxide layer on the copper strip.41 The formation of the N–Cu bond is accomplished by transferring a lone pair electron from a nitrogen atom into an unoccupied d orbital of a copper atom. This process also leads to the production of protective coating chemical bonds on the surface of the copper atom. Fig. S11† demonstrates that there were two obvious S 2p peaks (S 2p1/2 and S 2p3/2) on the copper surface. The binding energy at 162.40 eV, 163.25 eV, and 167.52 eV were assigned for Cu–S, C–S, and S–O bonds, respectively. The interaction of Cu ions with the lone pair of S atoms in T3AP was the main reason for the adsorption of the S 2p spectrum, followed by C
N bond. The N–Cu bond results from the combination of copper and N in the T3AP molecule and the N–O bond is caused by the oxide layer on the copper strip.41 The formation of the N–Cu bond is accomplished by transferring a lone pair electron from a nitrogen atom into an unoccupied d orbital of a copper atom. This process also leads to the production of protective coating chemical bonds on the surface of the copper atom. Fig. S11† demonstrates that there were two obvious S 2p peaks (S 2p1/2 and S 2p3/2) on the copper surface. The binding energy at 162.40 eV, 163.25 eV, and 167.52 eV were assigned for Cu–S, C–S, and S–O bonds, respectively. The interaction of Cu ions with the lone pair of S atoms in T3AP was the main reason for the adsorption of the S 2p spectrum, followed by C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) S (C–S) which indicated the T3AP molecules.41,42 Meanwhile, the S–O bonds were indicated for the oxidized S atom on the copper surface. Therefore, from the findings of the XPS study, it may be inferred that the T3AP molecules and copper are reacting chemically. N–Cu bonds and Cu–S bonds suggest that T3AP molecules are firmly attached to the copper surface by chemical bonding, preventing copper corrosion and making the surface more resistant to corrosion.
S (C–S) which indicated the T3AP molecules.41,42 Meanwhile, the S–O bonds were indicated for the oxidized S atom on the copper surface. Therefore, from the findings of the XPS study, it may be inferred that the T3AP molecules and copper are reacting chemically. N–Cu bonds and Cu–S bonds suggest that T3AP molecules are firmly attached to the copper surface by chemical bonding, preventing copper corrosion and making the surface more resistant to corrosion.
3.9 DFT calculation
The chemical properties of the T3AP were further characterized using the DFT approach. The quantitative characteristics of T3AP molecules were optimized and calculated using Gaussian software. The optimized T3AP stable molecular configuration is shown in Fig. 9. Prior to performing any additional calculations, it was necessary to verify the consistency of the experimentally obtained and optimized theoretical structural value, as shown in ESI Fig. S12.† The HOMO and LUMO electron cloud distribution diagram orbitals were illustrated in Fig. 10. HOMO and LUMO orbitals of T3AP electron clouds are essentially uniformly dispersed. This demonstrates that T3AP can be adsorbed in a parallel pattern on the copper surface. T3AP has a HOMO (ionization potential) and LUMO (electron affinity) values of −6.3664 eV and −1.6773 eV, respectively. The organic molecules HOMO and LUMO orbitals correlate to the molecule's electron supply and gain capacities, respectively.43–45 The ΔE value of a compound energy gap serves as a general indicator of a molecule's.46–48 The HOMO–LUMO band gap energy is a key measure for comparing the reactivity of the substances studied.49 It is easier for the corrosion inhibitor molecules to adsorb on copper surfaces with a highly stable energy gap value, resulting in a superior anti-corrosion performance.44 A good anti-corrosion property of T3AP is demonstrated by its ΔE value of 4.6891 eV. In addition, the global hardness (η) and chemical softness (S) calculated with other parameters are summarized in Table 10. Concerning inhibitor molecular activity and stability, these characteristics are critical. Based on hard and soft acids bases (HSAB) theory, the reaction that takes place involves hard acids reacting with hard bases and soft acids reacting with soft bases.6,7 Copper performed the role of a soft base by reacting with the softer molecule form of T3AP while a corrosion reaction was taking place. As shown in Table 10, the protonated T3AP is considerably soft in terms of chemical softness (0.4265 eV−1) in comparison to the neutral form, which indicates the effectiveness of T3AP in protecting copper strips. The fact that T3AP has a low global hardness and a high chemical softness suggests that it is an excellent corrosion inhibitor, which aligns with HSAB theory. Additionally, a molecule's ability to be drawn to an electron is measured by its electronegativity (χ). Electrons tend to be drawn from the copper surface as the molecule's electronegativity increases. In contrast, the low electronegativity (−4.0190 eV) of protonated T3AP is linked to the protonated N atom, which may have an effect on the electronegativity of inhibitors. This is consistent with the absorption mechanisms observed in the isotherm study, where the inhibitors are not fully reacted chemically with the copper surface but had strong physical adsorption on the surface to protect the copper.
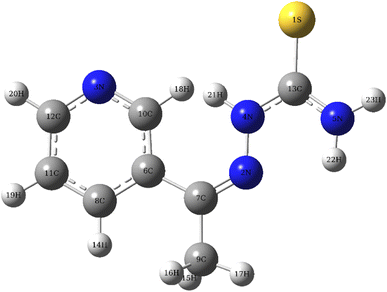 |
| | Fig. 9 The optimised T3AP molecular structure. | |
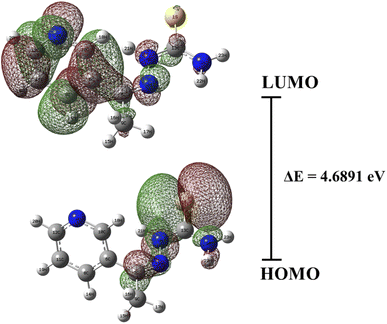 |
| | Fig. 10 The HOMO–LUMO of T3AP. | |
Table 10 Electronic and structural parameters for T3AP molecule employing DFT approach
| Parameters |
Value |
| Electron affinity (A) |
−1.6773 eV |
| Ionization potential (I) |
−6.3664 eV |
| Electronegativity (χ) |
−4.0190 eV |
| Global hardness (η) |
2.3446 eV |
| Chemical softness (S) |
0.4265 eV−1 |
3.9.1 MEP. The color in the electrostatic potential diagram of the T3AP molecule (Fig. 11) indicate the behavior of the reactive site of the compounds.50 The red-orange-yellow regions are electrophilic, whereas the green-blue regions are nucleophilic.18 N and S atoms dominate the red-orange-yellow regions. The electrophilic characteristic of copper atoms allows them to establish coordination bonds with N and S atoms, resulting in stable chemical adsorption.
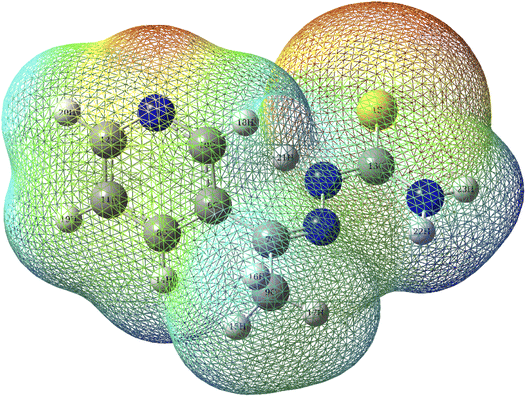 |
| | Fig. 11 Molecular electrostatic potential diagram of T3AP. | |
3.10 Molecular dynamics simulations
Further investigation into the adsorption process of the T3AP corrosion inhibitor molecule on a copper surface was carried out with the use of molecular dynamics modelling. Fig. 12 illustrates the stable adsorption configuration of T3AP molecules on the surface of Cu (111) when the temperature is optimized to 316.05 K. It can be deduced from the side view of the equilibrium configuration shown in Fig. 12(a) and the top view shown in Fig. 12(b) of the equilibrium configuration that the pyridine ring and conjugated thiosemicarbazone Scaffold of the T3AP molecule and the Cu (1 1 1) surface are virtually parallel to one another. As a matter of fact, it has been hypothesized that the T3AP molecule on the copper surface displays a parallel adsorption mode based on the results of the quantum chemistry experiment describing the geometrically flat adsorption of T3AP with planar conjugated structure of the pyridine ring.4 The surface area of interaction between the copper and the corrosion inhibitor molecule can be increased due to the T3AP presence of conjugated pyridine ring, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) S and C
S and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N bond parallel adsorption mechanism. As a result of this, the Cu surface is protected from the corrosive HCl medium since there is less surface area for the corrosive particles to contact the Cu. Table 11 also shows the relative kinetic parameters that were determined from the molecular dynamic simulation. Adsorption of T3AP onto a Cu (111) surface has a binding energy that can be calculated using the following formula (15) and (16).
N bond parallel adsorption mechanism. As a result of this, the Cu surface is protected from the corrosive HCl medium since there is less surface area for the corrosive particles to contact the Cu. Table 11 also shows the relative kinetic parameters that were determined from the molecular dynamic simulation. Adsorption of T3AP onto a Cu (111) surface has a binding energy that can be calculated using the following formula (15) and (16).| | |
Einteract = Etot − (Esubs + Einh)
| (15) |
| | |
Ebinding = −Einteract
| (16) |
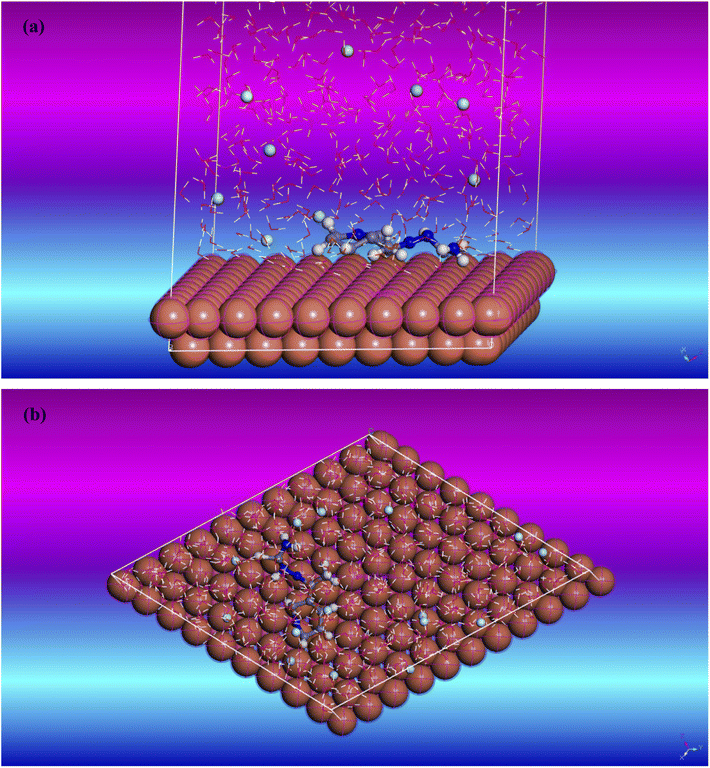 |
| | Fig. 12 Copper (111) surface with T3AP inhibitor simulation adsorption diagram (a) side view (b) top view. | |
Table 11 Molecular dynamics simulation relative parameters of T3AP at optimized temperature of 316.05 K
| Parameters |
Values (kJ mol−1) |
| Etot |
−41187.208 |
| Esubs |
−40664.911 |
| Einh |
18.665 |
| Einteract |
−540.962 |
| Ebinding |
540.962 |
According to the calculated binding energy, the T3AP molecule has a significant amount of inhibition on copper corrosion in HCl medium with binding energy (Ebinding) of 540.962 kJ mol−1. This is because a high binding energy indicates that the corrosion inhibitor molecules readily adsorb onto the metal surface and exert a strong binding effect, resulting in superior corrosion prevention efficacy.3,4
4. Conclusion
In this study, the synthesis of a novel Schiff base, 3-acetylpyridine linked with thiosemicarbazone, exhibited effective corrosion inhibition of copper strips in HCl solution, optimized using the RSM. The synthesized compound, T3AP, can effectively inhibit the corrosion up to 93% based on the response of gravimetric and EIS analysis. The optimal conditions for the four selected parameters were temperature of 42.90 °C, acid concentration of 2.38 M, inhibitor concentration of 3.80 mM, and time of 18.97 h. Furthermore, T3AP obeyed the Langmuir isotherm. Based on the calculated ΔGads, the molecule undergoes physisorption and chemisorption on the copper surface. The polarization study also indicated the T3AP was a mixed-type inhibitor. Treated copper with T3AP showed that its surface morphology was smooth and without deep holes and cracks, as analyzed using SEM. In addition, the presence of metal to inhibitor bonding, such as N–Cu and Cu–S bonds, revealed by XPS indicated the successful adsorption of T3AP onto copper strips. Moreover, DFT simulations were used to evaluate the correlation between T3AP's corrosion inhibition efficiency and its electronic characteristics. The efficacy of inhibitory signalling improves as the ionization potential and electronic affinity grow and are supported by the stability of the molecular energy gap (ΔE value = 4.6891 eV). The molecular dynamic adsorption with parallel configuration of T3AP molecule with copper surface exhibit high binding energy of 540.962 kJ mol−1 have justified the strong adsorption that become the reason of good corrosion inhibition obtained.
Author contributions
Muhammad Ammar: methodology and taking experiment part of inhibitors synthesized and tested, writing – original draft preparation. Mohammad Norazmi: supervision, software, resources, validation. Izan Izwan: supervision, resources, software. Erna Normaya: supervision, conceptualization, investigation, software, validation, review and editing article.
Conflicts of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
This paper is a part of a project that was funded by the Ministry of Higher Education, Malaysia (FRGS/1/2019/STG01/UIAM/02/2).
References
- H. M. A. El-lateef, Z. A. Abdallah and M. S. Mohamed, J. Mol. Liq., 2019, 296, 111800 CrossRef CAS.
- H. M. A. El-lateef, Corros. Sci., 2014, 92, 104–117 CrossRef.
- E. Gutiérrez, J. A. Rodríguez, J. Cruz-borbolla, J. G. Alvarado-rodríguez and P. Thangarasu, Eval. Progr. Plann., 2016, 108, 23–35 CrossRef.
- L. Luo, S. Zhang, Y. Qiang, N. Chen, S. Xu and S. Chen, Int. J. Electrochem. Sci., 2016, 11, 8192 Search PubMed.
- A. El Aatiaoui, M. Koudad, T. Chelfi, S. Erkac, M. Azzouzi, A. Aouniti, K. Savaş, M. Kaddouri, N. Benchat and A. Oussaid, J. Mol. Struct., 2020, 1226, 129372 CrossRef.
- X. L. Li, B. Xie, J. S. Feng, C. Lai, X. X. Bai, T. Li, D. L. Zhang, W. Y. Mou, L. Wen and Y. T. Gu, J. Mol. Liq., 2022, 345, 117032 CrossRef CAS.
- F. Tezcan, G. Yerlikaya, A. Mahmood and G. Kardaş, J. Mol. Liq., 2018, 269, 398–406 CrossRef CAS.
- H. H. Zhang and Y. Chen, J. Mol. Struct., 2019, 1177, 90–100 CrossRef CAS.
- Q. Yuan, R. Cheng, S. Zou, C. Ding, H. Liu, Y. Wang, D. Yang, X. Xiao, Q. Jiang, R. Tang and J. Chen, J. Mater. Res. Technol., 2020, 9, 11935–11947 CrossRef CAS.
- A. Fawzy, T. A. Farghaly, H. A. El-Ghamry and T. M. Bawazeer, J. Mol. Struct., 2020, 1203, 127447 CrossRef CAS.
- A. Ataei, A. Mehrizad and K. Zare, J. Mol. Liq., 2021, 328, 115476 CrossRef CAS.
- M. A. Bezerraa, R. E. Santelli, E. P. Oliveira, L. S. Villar and L. A. lia Escaleira, Talanta, 2008, 76, 965–977 CrossRef.
- N. S. Mohan Kumar, R. Ramasamy and H. K. Manonmani, Ind. Crops Prod., 2013, 43, 150–158 CrossRef CAS.
- F. Asadzadeh, M. Maleki-Kaklar, N. Soiltanalinejad and F. Shabani, Sci. Rep., 2018, 8, 1–8 CAS.
- P. Mourya, S. Banerjee, R. B. Rastogi and M. M. Singh, Ind. Eng. Chem. Res., 2013, 52, 12733–12747 CrossRef CAS.
- E. Normaya, M. N. Ahmad and Y. Farina, J. Braz. Chem. Soc., 2018, 29, 2197–2206 CAS.
- M. Talebian, K. Raeissi, M. Atapour, Z. Salarvand, S. Meghdadi, M. Amirnasr and R. M. Souto, Appl. Surf. Sci., 2018, 447, 852–865 CrossRef CAS.
- N. Z. N. Hashim, E. H. Anouar, K. Kassim, H. M. Zaki, A. I. Alharthi and Z. Embong, Appl. Surf. Sci., 2019, 476, 861–877 CrossRef CAS.
- M. N. Ahmad, N. U. Karim and E. Normaya, Sci. Rep., 2020, 10, 1–14 CrossRef PubMed.
- Y. Wang, J. Wang, L. Ma, C. Ren, D. Zhang, L. Ma and M. Sun, Appl. Surf. Sci., 2021, 568, 1–10 Search PubMed.
- N. I. Kairi and J. Kassim, Int. J. Electrochem.
Sci., 2013, 8, 7138–7155 CAS.
- A. Zarrouk, I. Warad, B. Hammouti, A. Dafali, S. S. Al-Deyab and N. Benchat, Int. J. Electrochem. Sci., 2010, 5, 1516–1526 CAS.
- A. Mishra, J. Aslam, C. Verma, M. A. Quraishi and E. E. Ebenso, J. Taiwan Inst. Chem. Eng., 2020, 114, 341–358 CrossRef CAS.
- I. B. Obot, N. K. Ankah, A. A. Sorour, Z. M. Gasem and K. Haruna, Sustainable Mater. Technol., 2017, 14, 1–10 CrossRef CAS.
- M. Yadav, L. Gope, N. Kumari and P. Yadav, J. Mol. Liq., 2015, 216, 78–86 CrossRef.
- L. Ma, J. Wang, C. Ren, P. Ju, Y. Huang, F. Zhang, F. Zhao, Z. Zhang and D. Zhang, Sens. Actuators, B, 2020, 321, 128617 CrossRef CAS.
- L. B. Coelho, D. Cossement and M. G. Olivier, Corros. Sci., 2018, 130, 177–189 CrossRef CAS.
- M. L. Zheludkevich, K. A. Yasakau, S. K. Poznyak and M. G. S. Ferreira, Corros. Sci., 2005, 47, 3368–3383 CrossRef CAS.
- M. Bobina, A. Kellenberger, J. P. Millet, C. Muntean and N. Vaszilcsin, Corros. Sci., 2013, 69, 389–395 CrossRef CAS.
- R. Solmaz, Corros. Sci., 2010, 52, 3321–3330 CrossRef CAS.
- P. Singh, D. S. Chauhan, S. S. Chauhan, G. Singh and M. A. Quraishi, J. Mol. Liq., 2019, 286, 110903 CrossRef CAS.
- A. S. Fouda, M. A. Ismail, A. A. Al-Khamri and A. S. Abousalem, J. Mol. Liq., 2019, 290, 111178 CrossRef CAS.
- I. Ahamad and M. A. Quraishi, Corros. Sci., 2010, 52, 651–656 CrossRef CAS.
- E. A. Flores, O. Olivares, N. V. Likhanova, M. A. Domínguez-Aguilar, N. Nava, D. Guzman-Lucero and M. Corrales, Corros. Sci., 2011, 53, 3899–3913 CrossRef CAS.
- W. Zhang, H. Li, M. Wang, L. Wang, A. Zhang and Y. Wu, J. Mol. Struct., 2019, 43, 413–426 CAS.
- M. El, F. Benhiba, H. About, Y. Kerroum and A. Guenbour, J. Colloid Interface Sci., 2020, 576, 330–344 CrossRef.
- P. Mourya, P. Singh, A. K. Tewari, R. B. Rastogi and M. M. Singh, Corros. Sci., 2015, 95, 71–87 CrossRef CAS.
- M. Mobin, S. Zehra and M. Parveen, J. Mol. Liq., 2016, 216, 598–607 CrossRef CAS.
- C. Verma, I. B. Obot, I. Bahadur, E. S. M. Sherif and E. E. Ebenso, Appl. Surf. Sci., 2018, 457, 134–149 CrossRef CAS.
- J. F. Moulder, W. F. Stickle, P. E. Sobol and K. D. Bomben, Handbook X-Ray Photoelectron Spectroscopy, Perkin Elmer Corporation, Minnesota, USA, 1992 Search PubMed.
- F. H. Zaidon, K. Kassim, H. Mohd Zaki, Z. Embong, E. H. Anouar and N. Z. Nor Hashim, J. Mol. Liq., 2021, 329, 115553 CrossRef CAS.
- R. A. Walton, Coord. Chem. Rev., 1980, 31, 183–220 CrossRef CAS.
- M. T. Majd, M. Ramezanzadeh, B. Ramezanzadeh and G. Bahlakeh, J. Mol. Liq., 2020, 382, 112750 CrossRef.
- J. He, Q. Li, X. Li, J. An, G. Chen, L. Zhao and W. Li, J. Mol. Liq., 2020, 320, 114494 CrossRef CAS.
- B. Tan, S. Zhang, Y. Qiang, L. Feng, C. Liao, Y. Xu and S. Chen, J. Mol. Liq., 2017, 248, 902–910 CrossRef CAS.
- M. T. majd, M. Ramezanzadeh, G. Bahlakeh and B. Ramezanzadeh, J. Mol. Liq., 2020, 304, 112750 CrossRef.
- Z. Sanaei, G. Bahlakeh, B. Ramezanzadeh and M. Ramezanzadeh, J. Mol. Liq., 2019, 290, 111176 CrossRef CAS.
- A. Dehghani, A. H. Mostafatabar, G. Bahlakeh, B. Ramezanzadeh and M. Ramezanzadeh, J. Mol. Liq., 2020, 309, 113035 CrossRef CAS.
- M. A. Mohammad Alwi, E. Normaya, H. Ismail, A. Iqbal, B. Mat Piah, M. A. Abu Samah and M. N. Ahmad, ACS Omega, 2021, 6, 25179–25192 CrossRef CAS.
- M. Drissi, N. Benhalima, Y. Megrouss, R. Rachida, A. Chouaih and F. Hamzaoui, Molecules, 2015, 20, 4042–4054 CrossRef CAS PubMed.
|
| This journal is © The Royal Society of Chemistry 2022 |
Click here to see how this site uses Cookies. View our privacy policy here.  Open Access Article
Open Access Article a,
Mohammad Norazmi Ahmad
a,
Mohammad Norazmi Ahmad a,
Izan Izwan Misnon
a,
Izan Izwan Misnon b,
Hariy Pauzic and
Erna Normaya
b,
Hariy Pauzic and
Erna Normaya *a
*a
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N) groups in the compounds, makes them attractive materials for inhibiting metal corrosion under acidic conditions.5 Schiff bases are used as corrosion inhibitors in commercial applications and they are preferred over other organic compounds because of their availability, cost-efficiency, simple synthesis, high purity, low toxicity, and environmental friendliness.6,7 The organic compounds, particularly Schiff bases, that contain 3-acetylpyridine and thiosemicarbazone8–10 can be effective corrosion inhibitors based on their ability to suppress hydrochloric acid solution corrosion. Hence, investigating new inhibitor platforms specifically for copper metal is a good start for this corrosion science study. On the other hand, having efficient inhibitors is not sufficient if their efficiencies are affected by the surrounding conditions, thus studying and optimizing the conditions is important to understand and ensure the maximum efficiency of the inhibitors.
N) groups in the compounds, makes them attractive materials for inhibiting metal corrosion under acidic conditions.5 Schiff bases are used as corrosion inhibitors in commercial applications and they are preferred over other organic compounds because of their availability, cost-efficiency, simple synthesis, high purity, low toxicity, and environmental friendliness.6,7 The organic compounds, particularly Schiff bases, that contain 3-acetylpyridine and thiosemicarbazone8–10 can be effective corrosion inhibitors based on their ability to suppress hydrochloric acid solution corrosion. Hence, investigating new inhibitor platforms specifically for copper metal is a good start for this corrosion science study. On the other hand, having efficient inhibitors is not sufficient if their efficiencies are affected by the surrounding conditions, thus studying and optimizing the conditions is important to understand and ensure the maximum efficiency of the inhibitors.
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1022, 856 (C
N), 1022, 856 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) S).
S).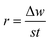
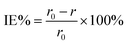
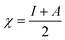
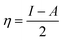
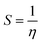
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) S bond. The chemical shift of carbon C(3)
S bond. The chemical shift of carbon C(3)![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N can be assigned at δ 150.19 ppm. The carbon in the heterocyclic ring of pyridine C(4), C(5), C(6), C(7), and C(8) can be observed in the range of δ 148 to 123 ppm. The most shielded chemical shift belonged to the methyl group's C(2) atom, which appeared at δ 14.25 ppm.
N can be assigned at δ 150.19 ppm. The carbon in the heterocyclic ring of pyridine C(4), C(5), C(6), C(7), and C(8) can be observed in the range of δ 148 to 123 ppm. The most shielded chemical shift belonged to the methyl group's C(2) atom, which appeared at δ 14.25 ppm.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 749.50
749.50
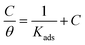
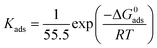
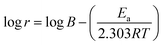
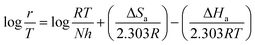
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) r vs. 1/T depicted in Fig. 7(a). A comparison of the Ea values in the presence and absence of T3AP shows that the creation of an energy barrier makes metal corrosion more difficult in the presence of the T3AP.1 Meanwhile, the enthalpy (ΔHa) and entropy (ΔSa) of the reaction system were calculated from the slope and intercept of ln(r/T) vs. 1/T in Fig. 7(b). Positive values of ΔHa (Table 9) are a strong indicator that endothermic processes are involved in copper dissolution.37 The fact that the ΔSa value (Table 9) was higher in the treated solution with T3AP compared to the untreated solution suggests that there was a greater degree of disorder in the system.36
r vs. 1/T depicted in Fig. 7(a). A comparison of the Ea values in the presence and absence of T3AP shows that the creation of an energy barrier makes metal corrosion more difficult in the presence of the T3AP.1 Meanwhile, the enthalpy (ΔHa) and entropy (ΔSa) of the reaction system were calculated from the slope and intercept of ln(r/T) vs. 1/T in Fig. 7(b). Positive values of ΔHa (Table 9) are a strong indicator that endothermic processes are involved in copper dissolution.37 The fact that the ΔSa value (Table 9) was higher in the treated solution with T3AP compared to the untreated solution suggests that there was a greater degree of disorder in the system.36
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C (284.74 eV) of the pyridine ring, C
C (284.74 eV) of the pyridine ring, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N or C–N (286.97 eV), as well as C
N or C–N (286.97 eV), as well as C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) S at 287.89 eV. The presence of these conjugated double bonds C
S at 287.89 eV. The presence of these conjugated double bonds C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C, C
C, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N, and C
N, and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) S in the T3AP inhibitor increased adsorption on the copper surface's outermost layer by providing a barrier to shield the underlying copper metal–solution interface from further oxidation by an acidic HCl solution during an extended immersion duration. The C–C bond (282.48 eV) also appeared in the spectrum since the chemical structure of T3AP contained C–CH3 bond. There are four primary synthetic photoelectron peaks displayed in Fig. S10† by fitting the deconvolution of N 1s spectra for the adsorbed film on copper when T3AP is present. The peaks were for N–H, N–N/C
S in the T3AP inhibitor increased adsorption on the copper surface's outermost layer by providing a barrier to shield the underlying copper metal–solution interface from further oxidation by an acidic HCl solution during an extended immersion duration. The C–C bond (282.48 eV) also appeared in the spectrum since the chemical structure of T3AP contained C–CH3 bond. There are four primary synthetic photoelectron peaks displayed in Fig. S10† by fitting the deconvolution of N 1s spectra for the adsorbed film on copper when T3AP is present. The peaks were for N–H, N–N/C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N, N–Cu, and N–O which appeared at 399.17, 399.11, 398.55, and 397.522 eV, respectively. The T3AP molecule is responsible for the formation of the N–H bond as well as the N–N/C
N, N–Cu, and N–O which appeared at 399.17, 399.11, 398.55, and 397.522 eV, respectively. The T3AP molecule is responsible for the formation of the N–H bond as well as the N–N/C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N bond. The N–Cu bond results from the combination of copper and N in the T3AP molecule and the N–O bond is caused by the oxide layer on the copper strip.41 The formation of the N–Cu bond is accomplished by transferring a lone pair electron from a nitrogen atom into an unoccupied d orbital of a copper atom. This process also leads to the production of protective coating chemical bonds on the surface of the copper atom. Fig. S11† demonstrates that there were two obvious S 2p peaks (S 2p1/2 and S 2p3/2) on the copper surface. The binding energy at 162.40 eV, 163.25 eV, and 167.52 eV were assigned for Cu–S, C–S, and S–O bonds, respectively. The interaction of Cu ions with the lone pair of S atoms in T3AP was the main reason for the adsorption of the S 2p spectrum, followed by C
N bond. The N–Cu bond results from the combination of copper and N in the T3AP molecule and the N–O bond is caused by the oxide layer on the copper strip.41 The formation of the N–Cu bond is accomplished by transferring a lone pair electron from a nitrogen atom into an unoccupied d orbital of a copper atom. This process also leads to the production of protective coating chemical bonds on the surface of the copper atom. Fig. S11† demonstrates that there were two obvious S 2p peaks (S 2p1/2 and S 2p3/2) on the copper surface. The binding energy at 162.40 eV, 163.25 eV, and 167.52 eV were assigned for Cu–S, C–S, and S–O bonds, respectively. The interaction of Cu ions with the lone pair of S atoms in T3AP was the main reason for the adsorption of the S 2p spectrum, followed by C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) S (C–S) which indicated the T3AP molecules.41,42 Meanwhile, the S–O bonds were indicated for the oxidized S atom on the copper surface. Therefore, from the findings of the XPS study, it may be inferred that the T3AP molecules and copper are reacting chemically. N–Cu bonds and Cu–S bonds suggest that T3AP molecules are firmly attached to the copper surface by chemical bonding, preventing copper corrosion and making the surface more resistant to corrosion.
S (C–S) which indicated the T3AP molecules.41,42 Meanwhile, the S–O bonds were indicated for the oxidized S atom on the copper surface. Therefore, from the findings of the XPS study, it may be inferred that the T3AP molecules and copper are reacting chemically. N–Cu bonds and Cu–S bonds suggest that T3AP molecules are firmly attached to the copper surface by chemical bonding, preventing copper corrosion and making the surface more resistant to corrosion.![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) S and C
S and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N bond parallel adsorption mechanism. As a result of this, the Cu surface is protected from the corrosive HCl medium since there is less surface area for the corrosive particles to contact the Cu. Table 11 also shows the relative kinetic parameters that were determined from the molecular dynamic simulation. Adsorption of T3AP onto a Cu (111) surface has a binding energy that can be calculated using the following formula (15) and (16).
N bond parallel adsorption mechanism. As a result of this, the Cu surface is protected from the corrosive HCl medium since there is less surface area for the corrosive particles to contact the Cu. Table 11 also shows the relative kinetic parameters that were determined from the molecular dynamic simulation. Adsorption of T3AP onto a Cu (111) surface has a binding energy that can be calculated using the following formula (15) and (16).









