Open Access Article
Open Access ArticleA system for artificial light signal transduction via molecular translocation in a lipid membrane†
Huiting
Yang
a,
Shengjie
Du
a,
Zhicheng
Ye
a,
Xuebin
Wang
a,
Zexin
Yan
a,
Cheng
Lian
 *a,
Chunyan
Bao
*a,
Chunyan
Bao
 *ab and
Linyong
Zhu
*ab and
Linyong
Zhu
 ab
ab
aKey Laboratory for Advanced Materials, Joint International Research Laboratory of Precision Chemistry and Molecular Engineering, Feringa Nobel Prize Scientist Joint Research Center, Frontiers Science Center for Materiobiology and Dynamic Chemistry, Institute of Fine Chemicals, School of Chemistry and Molecular Engineering, East China University of Science and Technology, Shanghai 200237, China. E-mail: liancheng@ecust.edu.cn; baochunyan@ecust.edu.cn
bShanghai Frontiers Science Center of Optogenetic Techniques for Cell Metabolism, East China University of Science and Technology, Shanghai 200237, China
First published on 4th February 2022
Abstract
Light signal transduction pathways are the central components of mechanisms that regulate plant development, in which photoreceptors receive light and participate in light signal transduction. Chemical systems can be designed to mimic these biological processes that have potential applications in smart sensing, drug delivery and synthetic biology. Here, we synthesized a series of simple photoresponsive molecules for use as photoreceptors in artificial light signal transduction. The hydrophobic structures of these molecules facilitate their insertion into vesicular lipid bilayers, and reversible photoisomerization initiates the reciprocating translocation of molecules in the membrane, thus activating or deactivating the hydrolysis reaction of a precatalyst in the transducer for an encapsulated substrate, resulting in a light controllable output signal. This study represents the first example of using simplified synthetic molecules to simulate light signal transduction performed by complex biomolecules.
Introduction
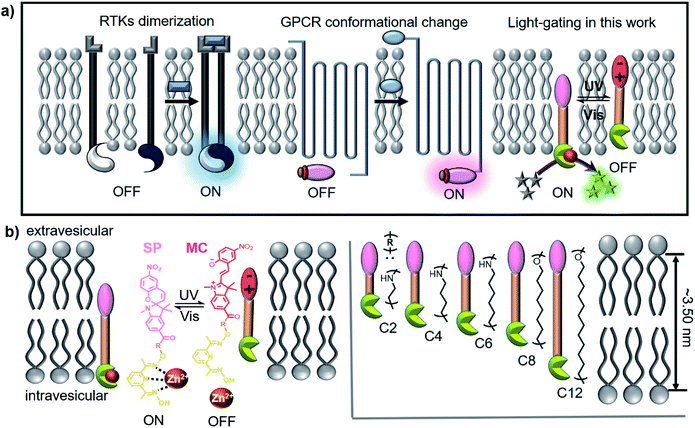
Signal transport and transduction are fundamental to cellular survival, where signals are transmitted into or out of the cell environment through an appropriate response of membrane-bound protein receptors to external stimuli.1–3 The complex structures of natural membrane protein receptors are easily inactivated away from the cell membrane; thus, synthetic systems simulating this process have aroused considerable interest and have been proven to have potential applications in drug delivery, sensing and disease treatment.4–6Generally, a signal transduction event involves three stages: (1) molecular recognition between a primary messenger and a membrane-bound protein receptor; (2) transmembrane signal transduction; (3) the release of a second messenger, which triggers a series of biochemical reactions and finally turns on gene expression and controls all cell functions.7 Receptor tyrosine kinases (RTKs) and the G protein-coupled receptor superfamily (GPCR)8 are prominent examples of receptors, where the binding of ligands induces dimerization or conformational changes of the receptors, thus transmitting signals and triggering a cascade of events inside cells (Scheme 1a). Several research groups have used minimal synthetic systems to mimic the mechanism of biological signal transduction.9–11 Some of these groups, such as the Clayden/Webb,12,13 Schrader14,15 and Hunter/Williams16,17 groups, have even utilized simple artificial mechanisms that do not exist in biological systems to achieve transmembrane signal transduction.
The stimulation of biological cells results in signal transduction, which provided cells with a lag time within which to determine an appropriate response. For example, RTK receptors are not always activated, and their activity can be reversibly changed through a phosphorylation–dephosphorylation process, whereby these receptors return to a deactivated state after completing a task and are ready for the next stimulation event. Light is the main natural energy source available and is often used as a stimulus switch for signal transduction, especially in many bacterial18 and plant19 systems. Photoreceptors enable organisms to use light for photosynthesis and photomorphogenesis. Simple artificial models can be used to simulate light signal transduction, whereby the underlying mechanisms can be elucidated and damaged transduction systems in nature can be replaced or salvaged. However, other than optogenetic systems,20 few successful synthetic cases are known. A team led by Clayden/Webb envisaged artificial light signal transduction using a light-responsive transmembrane helical switch in a 2016 Science paper.21
Hunter/Williams have reported to use pH change16 and ion recognition11 to trigger signal transduction by changing the position of receptors in lipid membranes. As is known, the spiropyran (SP) group can reversibly change its position in the hydrophobic lipid bilayers due to its photoisomerization between SP and merocyanine (MC) isomers.22 Therefore, we are inspired to use photoresponsive SP compounds that possess the necessary structural and functional elements to achieve light-switchable signal transduction on vesicular lipid bilayers. As shown in Scheme 1b, the transducers have hydrophobic molecular skeletons that comprise a sophisticated pyridine-oxime precatalyst and a photoresponsive SP moiety. The length of the entire molecule is regulated by an R spacer, which is designed to be not longer than the membrane thickness. After being inserted into the lipid membrane, the entire molecule can freely shuttle within the lipid membrane, enabling the precatalyst head to complex with metal ions in the vesicles and activate a catalytic reaction, resulting in a detectable output signal that starts signal transduction (activate/ON). Isomerization from SP to MC under UV light increases the hydrophilicity of the transducer to pull it towards the hydrophilic zone of the membrane and prevent the precatalyst head from complexing with the metal ions, which deactivates the catalytic reaction and stops signal transduction (deactivation/OFF). Therefore, reversible conversion between SP and MC under UV (365 nm) and visible light (530 nm) can realize repeatable switching between “ON” and “OFF” signal transduction states, which represents a crucial step toward designing artificial signalling systems that can respond to light stimulation.
Experimental section
Synthesis of compounds
See the details in the ESI.†The optimized molecular structures of transducers
The optimized structures of the transducers were obtained by ab initio calculations with the Gaussian09 package.23 The geometry optimization was performed at the B3LYP-D3/6-31G** level.24,25Molecular dynamics (MD) simulations
The behaviors of transducers in lipid bilayers were studied by molecular dynamics (MD) simulations. The systems were constructed including a transducer molecule C8, 70 egg yolk phosphatidylcholine (EYPC) molecules, 9 Zn2+ ions, 18 Cl− ions and 2265 water molecules to replicate the conditions of the experiment, respectively. The force field parameters were taken from the GROMOS force field.26 All MD simulations were performed with the GROMACS package.27 A cubic periodic boundary condition box and the SPC/E water model are used. The long-range electrostatic interactions were handled by the particle-mesh Ewald (PME) method28 with a cutoff radius of 1.4 nm. The LINCS algorithm29 constrains the bonds containing hydrogen atoms. The Verlet algorithm30 was used as an integrator of Newton's equation of motion. The time step is set as 2 fs for all the simulations. The systems were first subjected to energy minimization by the steepest descent and conjugate gradient method. Then, the systems were heated slowly to 310 K in 200 ps and equilibrated for 40 ns in an NPT ensemble with a v-rescale thermostat31 and Berendsen32 barostat. After that, the production MD run was performed for 20 ns in an NPT ensemble with a v-rescale thermostat and Parrinello–Rahman barostat,33 and the final 10 ns MD trajectories were used for sampling and analysis.Dynamic light scattering (DLS), calcein leakage experiment and membrane loading analysis using a liposome model
In a typical experiment, 1250 μL LUVs ⊃ calcein as prepared above was diluted with 1250 μL of HEPES buffer (10 mM HEPES, 100 mM KCl, and pH = 7.0) in a quartz cuvette for time dependent fluorescence tracking (λem = 518 nm) at an excitation wavelength λex = 497 nm. Then, transducers with a final concentration of 10.0 μM (10 μL stock solution in DMSO) was added at t = 1 min, and 60 μL of 5% Triton X-100 aqueous solution was added at t = 3 min. The temperature was kept at 25 °C with a stirrer and a temperature controller throughout the experiment.
Preparation of giant unilamellar vesicles (GUVs)
A solution containing 400 μL EYPC (EYPC, 25 mg mL−1, and 10 mg) in deacidified chloroform was dried with a N2 flow to form a thin lipid film. The resulting film was subsequently dried under vacuum for 12 h and hydrated at 37 °C overnight with 2 mL sucrose solution (0.2 M). Then, 250 μL of above-prepared GUVs was transferred to a glass bottle followed by addition of 1.0 μL C8 containing DMSO solution (10 mM) and the obtained solution was irradiated with 365 nm LED light with an intensity of 2 mW cm−2 for 10 s, which was subjected to microscopic observation. Fluorescence imaging was performed on a Nikon A1R Laser Scanning Confocal microscope; the bright field images were obtained in the monochromatic mode and the fluorescence images were taken at an excitation wavelength of 514 nm.Signal transduction assays
| Normalized FL intensity = (I − I0)/(I∞ − I0) |
| I = 1 − e−kobst |
Results and discussion
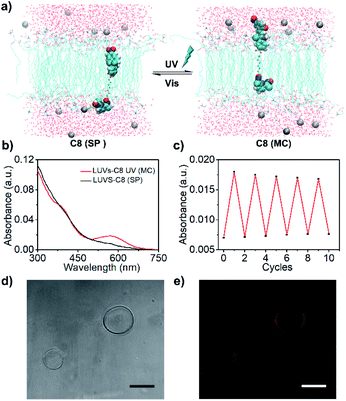
A series of photosensitive transducers C2, C4, C6, C8 and C12 were synthesized, all of which exhibited relative hydrophobicity (Table 1, Clog![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) p, 4.5–10.8), which is a prerequisite for effective insertion into a hydrophobic phospholipid bilayer membrane. Calculated according to the Corey–Pauling–Koltun (CPK) model, the optimized molecular structures (Fig. S1†) show molecular lengths of approximately 2.59 nm (C2), 2.77 nm (C4), 3.01 nm (C6), 3.26 nm (C8) and 3.71 nm (C12), which are shorter (C2–C6) than or similar (C8 and C12) to the hydrophobic thickness of a lipid bilayer membrane (∼3.50 nm). The photoisomerization of the transducers produced sensitive and reversible switches between the hydrophobic SP and hydrophilic MC states (Fig. S2†). The disappearance of the UV absorption peak at ∼550 nm shows that MC isomers can be efficiently transformed into SP isomers with a high yield. C8 was used as an example to study the photoresponsive behavior of a transducer in lipid bilayers by molecular dynamics (MD) simulations, and snapshots of the systems before and after photoisomerization were obtained by using VMD software.34Fig. 1a shows that when a photoresponsive group is in the SP form, the precatalyst group of C8 is closer to the water layer than the hydrophobic SP group, and a coordination bond between the N atom and Zn2+ ion is formed. After photoisomerization, the hydrophilic MC group is closer to the water layer than the precatalyst group, so as to prevent the coordination with Zn2+ ions by pulling the precatalyst group into the membrane. A similar result was obtained by calculating the radial distribution function (rdf) between the N atom of the precatalyst group and Zn2+; that is, a clear peak appeared at 0.8 nm for the SP isomer but not for the MC isomer (Fig. S3†). These theoretical results are consistent with previous studies35 and clearly show that the change of polarity by photoisomerization can trigger the relocation of transducers in the lipid membrane, so that the precatalyst head tends to complex/decomplex with Zn2+ in the aqueous solution. This provides a theoretical basis for light signal transduction by changing the position of transducers in the membrane. The decomplexing can be supported by the high hydration energy of Zn2+ (∼1955 kJ mol−1)36 and the poor binding constant between the precatalyst and Zn2+ (k = 1562 M−1 and ΔG = −18.2 kJ mol−1, Fig. S4†).
p, 4.5–10.8), which is a prerequisite for effective insertion into a hydrophobic phospholipid bilayer membrane. Calculated according to the Corey–Pauling–Koltun (CPK) model, the optimized molecular structures (Fig. S1†) show molecular lengths of approximately 2.59 nm (C2), 2.77 nm (C4), 3.01 nm (C6), 3.26 nm (C8) and 3.71 nm (C12), which are shorter (C2–C6) than or similar (C8 and C12) to the hydrophobic thickness of a lipid bilayer membrane (∼3.50 nm). The photoisomerization of the transducers produced sensitive and reversible switches between the hydrophobic SP and hydrophilic MC states (Fig. S2†). The disappearance of the UV absorption peak at ∼550 nm shows that MC isomers can be efficiently transformed into SP isomers with a high yield. C8 was used as an example to study the photoresponsive behavior of a transducer in lipid bilayers by molecular dynamics (MD) simulations, and snapshots of the systems before and after photoisomerization were obtained by using VMD software.34Fig. 1a shows that when a photoresponsive group is in the SP form, the precatalyst group of C8 is closer to the water layer than the hydrophobic SP group, and a coordination bond between the N atom and Zn2+ ion is formed. After photoisomerization, the hydrophilic MC group is closer to the water layer than the precatalyst group, so as to prevent the coordination with Zn2+ ions by pulling the precatalyst group into the membrane. A similar result was obtained by calculating the radial distribution function (rdf) between the N atom of the precatalyst group and Zn2+; that is, a clear peak appeared at 0.8 nm for the SP isomer but not for the MC isomer (Fig. S3†). These theoretical results are consistent with previous studies35 and clearly show that the change of polarity by photoisomerization can trigger the relocation of transducers in the lipid membrane, so that the precatalyst head tends to complex/decomplex with Zn2+ in the aqueous solution. This provides a theoretical basis for light signal transduction by changing the position of transducers in the membrane. The decomplexing can be supported by the high hydration energy of Zn2+ (∼1955 kJ mol−1)36 and the poor binding constant between the precatalyst and Zn2+ (k = 1562 M−1 and ΔG = −18.2 kJ mol−1, Fig. S4†).
| Transducer | Clog![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) pa pa |
Molecular lengthb (nm) | Membrane loadingc (mol%) | I | k obs (10−5 s−1) |
|---|---|---|---|---|---|
a Calculated log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) p, the estimated lipophilicity, obtained using ALOGPS 2.1.
b Molecular length based on the optimized structures.
c Membrane loading of transducers, defined as mol% relative to the lipid.
d The normalized fluorescence intensity after 24 h.
e The first order rate constant of the transduction curves of transducers at 10 μM (2 mol% relative to the lipid). p, the estimated lipophilicity, obtained using ALOGPS 2.1.
b Molecular length based on the optimized structures.
c Membrane loading of transducers, defined as mol% relative to the lipid.
d The normalized fluorescence intensity after 24 h.
e The first order rate constant of the transduction curves of transducers at 10 μM (2 mol% relative to the lipid).
|
|||||
| C2 | 4.50 | 2.59 | 0.57 | 0.50 | 0.76 |
| C4 | 4.40 | 2.77 | 0.94 | 0.33 | 0.43 |
| C6 | 5.47 | 3.01 | 1.24 | 0.23 | 0.30 |
| C8 | 8.75 | 3.26 | 1.48 | 1.00 | 2.06 |
| C12 | 10.80 | 3.71 | 1.34 | 0.80 | 1.37 |
To achieve signal transduction, transducers must be able to insert into a lipid membrane and form an effective transmembrane structure. First, a dynamic light scattering (DLS) analysis of large unilamellar vesicles (LUVs) was performed. It confirms that the addition of the transducer and subsequent light irradiation would not compromise the membrane integrity even after 24 h storage (Fig. S5 and S6†). The results of a calcein leakage experiment also verify that the insertion of transducers does not induce the leakage of the encapsulated molecules (Fig. S7 and S8†). In order to achieve high signal transduction activity, the effective insertion of transducers in the lipid membrane is very important. Then, the membrane loading ratio was explored by mini-column centrifugation and determined by HPLC analysis (Fig. S9†). The results as summarized in Table 1 suggest that the more hydrophobic (Clog![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) p) the molecule is, the higher the LUV membrane loading ratio is. For example, a 1.48 mol% (relative to lipid) membrane loading with C8 is obtained using only 2 mol% C8. However, when the hydrophobicity of the molecules is increased further, the membrane loading ratio decreases, as shown by the results for C12, which can be attributed to the competitive precipitation of molecules in aqueous solution. Finally, in order to explore whether the transducers in the lipid membrane can undergo reversible photoisomerization as in solution, the C8 loaded LUVs purified by removing the extravesicular compounds (LUVs–C8) were analyzed by UV-Vis absorption spectroscopy. As illustrated in Fig. 1b and c, C8 in the lipid membranes can also undergo reversible isomerization, and the conversion from SP to MC does not remove molecules from the lipids (Fig. S10†), indicating in situ photoisomerization of C8 in the lipid membrane.
p) the molecule is, the higher the LUV membrane loading ratio is. For example, a 1.48 mol% (relative to lipid) membrane loading with C8 is obtained using only 2 mol% C8. However, when the hydrophobicity of the molecules is increased further, the membrane loading ratio decreases, as shown by the results for C12, which can be attributed to the competitive precipitation of molecules in aqueous solution. Finally, in order to explore whether the transducers in the lipid membrane can undergo reversible photoisomerization as in solution, the C8 loaded LUVs purified by removing the extravesicular compounds (LUVs–C8) were analyzed by UV-Vis absorption spectroscopy. As illustrated in Fig. 1b and c, C8 in the lipid membranes can also undergo reversible isomerization, and the conversion from SP to MC does not remove molecules from the lipids (Fig. S10†), indicating in situ photoisomerization of C8 in the lipid membrane.
In order to realize light controllable signal transduction, the photostability of transducer isomers in the lipid membrane is an important parameter. Taking LUVs–C8 as an example, MC isomers are thermally unstable in lipid membranes compared to SP isomers. However, the recovery form MC to SP in the dark is quite slow, and only 15% of MC isomers recover to SP isomers after 10 min of storage in the dark (Fig. S11†), which provides a basis for the subsequent light controlled signal transduction strategy. In addition, as the MC isomer is fluorescent, the insertion of transducers into lipid bilayers can be visualized by performing fluorescence microscopy on giant unilamellar vesicles (GUVs). As illustrated in Fig. 1d and e, the addition of C8 does not collapse the GUVs, and significant fluorescence is exhibited around the GUV wall after UV irradiation. All these results strongly support the successful integration of our transducers into lipid bilayers and in situ photoisomerization.
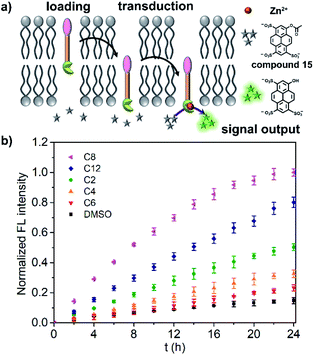
Before investigating the light signal transduction of transducers, the catalytic hydrolysis activity of transducers in aqueous solution was investigated and compared (Fig. S12†). It suggests that the linker length has a great effect on the catalytic activity with the order of C8 > C12 > C2 ≈ C4 > C6, and the photoisomerization doesn't affect the catalytic activity. Then, the transduction activity of the transducers in liposomes was explored by using LUVs encapsulated with zinc chloride and substrate molecules 15 (LUVs ⊃ 15). As shown in Fig. 2a, a transducer in the SP form with a correct orientation can initiate the hydrolysis of 15 in vesicles by complexation of pyridine-oxime and Zn2+, thus producing a fluorescent signal. The results for the transduction activity are presented in Fig. 2b, showing that a control of DMSO alone hydrolyzes the substrate at a slow background rate over a period of 24 h. However, the addition of transducers induces a significant increase in the fluorescence, where C8 exhibits the highest transduction activity (I ≈ 1.00 at t = 24 h) and the highest rate constant (kobs, Fig. S13†) at a given concentration (10.0 μM), which is consistent with its highest membrane loading ratio (Table 1). After comparison, it can be concluded that the transduction activity of transducers on liposomes is basically in the same order as its catalytic activity in aqueous solution (C8 > C12 > C2 ≈ C4 > C6), although the structure–activity relationship is very complicated on liposomes.37 Based on the working mechanism of the transducers on liposomes, the transduction activity is mainly affected by the membrane loading, the accessibility of the catalytic head group to the inner compartment (effective insertion) and the inherent catalytic activity. With the consideration of the membrane loading of transducers, the transduction activity of the vesicle bound compounds shows an order of C8 ≈ C2 > C12 > C4 > C6 (kobs/membrane loading), in which membrane incorporated C2 is as effective as that of C8. For transducer C8, high catalytic activity endows it with high transduction activity. However, for transducer C2, the effective insertion of transducers in the membrane may play a key role. It is presumably due to the small molecular structure of C2, which facilitates molecular shuttling across the membrane, thus increasing the accessibility of the catalytic head to the inner compartment and initiating the hydrolysis of 15 inside the vesicles.
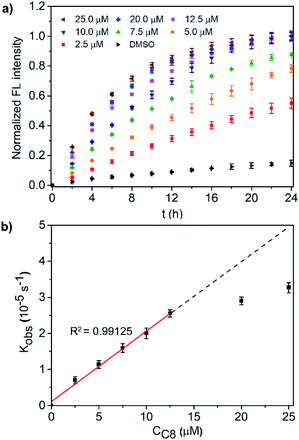
A detailed study of the transduction activity of C8 was carried out by varying the C8 concentrations in LUVs ⊃ 15 and concentration-dependent transduction activity is shown in Fig. 3a. By fitting the fluorescence kinetic curves with a first-order exponential decay equation, the observed first-order rate constants kobs of the C8 transducer at different concentrations were obtained (Fig. S14†). Further plotting kobs against concentration (Fig. 3b), it is found that the Kobs of transducer C8 exhibits a concentration-dependent linear relationship (R2 = 0.99125) when the applied concentration is less than 12.5 μM and then tends to equilibrate at higher concentrations. This indicates that a unimolecular active structure of C8 is involved in transduction. To verify that the output fluorescent signal comes from the hydrolysis of compound 15 catalyzed by the binding of the precatalyst head and intravesicular Zn2+, two control experiments in the absence of Zn2+ or the precatalyst head were performed (Fig. S15†). The ineffective transduction confirms our hypothesis shown in Fig. 2a; that is, the transducers in the SP form can activate the catalyst to hydrolyze compound 15 through membrane insertion, translocation and pyridine-oxime coordination with Zn2+ inside the vesicles to produce an output signal. It should be mentioned that the direction of C8 molecules in the lipid membrane is random, and only those molecules in the direction shown in Fig. 2a can exhibit signal transduction function. In terms of probability, up to 50% of molecules can work. Therefore, limiting the insertion direction of these transducers will be a promising strategy to further improve the transduction activity.
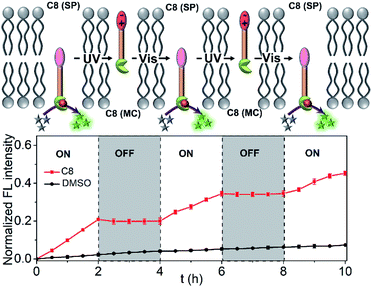
Due to the most effective signal transduction ability, C8 is selected to explore the light signal transduction process. Before the experiment, the illumination conditions were explored and optimized. To avoid photobleaching of all organic molecules, the intensity of the 365 nm LED light and the 540 nm LED light used for alternating irradiation was set as 2 mW cm−2. Considering the difference of the thermal stability of SP and MC isomers, an intermittent UV irradiation mode of 10 seconds every 5 minutes was used to ensure the maximum existence of MC isomers of C8. Due to the excellent thermal stability of SP isomers, only 2 min of visible light irradiation was required for the conversion from MC to SP in the lipid membrane. As is known, the hydrolysis of substrate 15 is not affected by C8 isomerization in aqueous solution (Fig. S12†). However, when C8 is added to the LUVs ⊃ 15 system, the situation is completely opposite. Fig. 4 shows the cycles of the light signal transduction process. Due to the rapid photoresponsiveness of the spiropyran, there is no delay in light regulated signal transduction. When C8 is added in the SP form, the precatalyst is activated by binding to zinc inside the vesicles, which induces the increase of fluorescence, and transduction occurs in the “ON” state. UV light irradiation changes the molecular position of C8 in the membrane, which deactivates the precatalyst and turns the signalling process off. The fluorescence does not increase further in this “OFF” state. Finally, alternating irradiation with 365 nm UV and 530 nm visible light induces the reciprocating translocation of C8 across the lipid bilayers to turn the transduction process on or off. This result demonstrates that the signaling process can be controlled by light, thus realizing artificial light signal transduction.
Conclusions
In summary, we have presented an example of artificial light signal transduction using simple synthetic molecules. The hydrophobic structures of the transducer molecules facilitate their insertion into lipid bilayers. The significant conformational difference between the SP and MC isomers of the transducers enables the use of light to regulate the position of the transducers in the lipid membranes. Molecular photoisomerization induces reversible translocation of the transducers in lipid bilayers, which enables the activation and deactivation of a precatalyst in the transducer by controlling binding events in the vesicles and leads to the opening and closing of the transduction channel.Such an artificial light signal transduction system may find novel applications in numerous fields. For example, the light controllable catalytic hydrolysis can be used to construct intelligent carrier for drug release; the light induced molecular shuttling can be used to trigger cascade reactions in nanoreactors; the signaling sites of the transducers can be changed for the detection of bioanalytes.
Data availability
All relevant data is presented in the paper and ESI.†Author contributions
C. Bao proposed and supervised the project. H. Yang carried out the synthesis, characterization and data collection. S. Du, X. Wang, and Z. Yan performed the LUV based fluorescence experiments. Z. Ye and C. Lian carried out the MD theoretical analysis and also prepared the text for that section. C. Bao and L. Zhu prepared the paper and programmed the data from all authors. All the authors discussed the results and commented on the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Natural Science Foundation of China (NSFC, 22171085), Shanghai Sci. Tech. Comm. (21ZR1415500) and Shanghai Frontier Science Research Base of Optogenetic Techniques for Cell Metabolism (Shanghai Municipal Education Commission, grant 2021 Sci & Tech 03-28). We thank the Research Centre of Analysis and Test of East China University of Science and Technology for the help with the characterization.Notes and references
- M. A. Lemmon and J. Schlessinger, Cell, 2010, 141, 1117–1134 CrossRef CAS PubMed.
- I. Rey-Suarez, B. A. Wheatley, P. Koo, A. Bhanja, Z. Shu, S. Mochrie, W. Song, H. Shroff and A. Upadhyaya, Nat. Commun., 2020, 11, 439 CrossRef CAS PubMed.
- V. Katritch, V. Cherezov and R. C. Stevens, Annu. Rev. Pharmacol. Toxicol., 2013, 53, 531–556 CrossRef CAS PubMed.
- Z. Quinn, W. Mao, Y. Xia, R. John and Y. Wan, Bioact. Mater., 2021, 6, 749–756 CrossRef CAS PubMed.
- D. E. Clapham, Nature, 2003, 426, 517–524 CrossRef CAS PubMed.
- M.-C. Jonathan, S.-H. Adrián and A. Gonzalo, Mol. Aspects Med., 2021, 78, 100940 CrossRef CAS PubMed.
- P. Tompa, Chem. Soc. Rev., 2016, 45, 4252–4284 RSC.
- M. I. Simon, M. P. Strathmann and N. Gautam, Science, 1991, 252, 802 CrossRef CAS PubMed.
- J. Morstein, M. A. Dacheux, D. D. Norman, A. Shemet, P. C. Donthamsetti, M. Citir, J. A. Frank, C. Schultz, E. Y. Isacoff, A. L. Parrill, G. J. Tigyi and D. Trauner, J. Am. Chem. Soc., 2020, 142, 10612–10616 CrossRef CAS PubMed.
- S. Peng, A. Barba-Bon, Y.-C. Pan, W. M. Nau, D.-S. Guo and A. Hennig, Angew. Chem., Int. Ed., 2017, 56, 15742–15745 CrossRef CAS PubMed.
- M. J. Langton, N. H. Williams and C. A. Hunter, J. Am. Chem. Soc., 2017, 139, 6461–6466 CrossRef CAS PubMed.
- F. G. A. Lister, B. A. F. Le Bailly, S. J. Webb and J. Clayden, Nat. Chem., 2017, 9, 420–425 CrossRef CAS.
- F. G. A. Lister, N. Eccles, S. J. Pike, R. A. Brown, G. F. S. Whitehead, J. Raftery, S. J. Webb and J. Clayden, Chem. Sci., 2018, 9, 6860–6870 RSC.
- K. Bernitzki and T. Schrader, Angew. Chem., Int. Ed., 2009, 48, 8001–8005 CrossRef CAS PubMed.
- S. Dutt, C. Wilch and T. Schrader, Chem. Commun., 2011, 47, 5376–5383 RSC.
- M. J. Langton, F. Keymeulen, M. Ciaccia, N. H. Williams and C. A. Hunter, Nat. Chem., 2017, 9, 426–430 CrossRef CAS PubMed.
- Y. Ding, N. H. Williams and C. A. Hunter, J. Am. Chem. Soc., 2019, 141, 17847–17853 CrossRef CAS PubMed.
- X. Li, C. Zhang, X. Xu, J. Miao, J. Yao, R. Liu, Y. Zhao, X. Chen and Y. Yang, Nucleic Acids Res., 2020, 48, e33 CrossRef CAS PubMed.
- Y. Jiao, O. S. Lau and X. W. Deng, Nat. Rev. Genet., 2007, 8, 217–230 CrossRef CAS PubMed.
- L. Klewer and Y.-W. Wu, Chem.–Eur. J., 2019, 25, 12452–12463 CrossRef CAS PubMed.
- M. De Poli, W. Zawodny, O. Quinonero, M. Lorch, S. J. Webb and J. Clayden, Science, 2016, 352, 575 CrossRef CAS PubMed.
- C. J. Wohl, M. A. Helms, J. O. Chung and D. Kuciauskas, J. Phys. Chem. B, 2006, 110, 22796–22803 CrossRef CAS PubMed.
- M. J. Frisch, G. W. Trucks, H. B. Schlegel, et al., Gaussian 09, Gaussian, Inc., Wallingford CT, 2009 Search PubMed.
- P. J. Stephens, F. J. Devlin, C. F. Chabalowski and M. J. Frisch, J. Chem. Phys., 1994, 98, 11623–11627 CrossRef CAS.
- R. Krishnan, J. S. Binkley, R. Seeger and J. A. Pople, J. Chem. Phys., 1980, 72, 650–654 CrossRef CAS.
- N. Schmid, A. P. Eichenberger, A. Choutko, S. Riniker, M. Winger, A. E. Mark and W. F. van Gunsteren, Eur. Biophys. J., 2011, 40, 843 CrossRef CAS PubMed.
- M. J. Abraham, T. Murtola, R. Schulz, S. Páll, J. C. Smith, B. Hess and E. Lindahl, SoftwareX, 2015, 1–2, 19–25 CrossRef.
- U. Essmann, L. Perera, M. L. Berkowitz, T. Darden, H. Lee and L. G. Pedersen, J. Chem. Phys., 1995, 103, 8577–8593 CrossRef CAS.
- B. Hess, J. Chem. Theory Comput., 2008, 4, 116–122 CrossRef CAS PubMed.
- H. Grubmüller, H. Heller, A. Windemuth and K. Schulten, Mol. Simul., 1991, 6, 121–142 CrossRef.
- G. Bussi, D. Donadio and M. Parrinello, J. Chem. Phys., 2007, 126, 014101 CrossRef PubMed.
- H. J. C. Berendsen, J. P. M. Postma, W. F. van Gunsteren, A. DiNola and J. R. Haak, J. Chem. Phys., 1984, 81, 3684–3690 CrossRef CAS.
- M. Parrinello and A. Rahman, J. Appl. Phys., 1981, 52, 7182–7190 CrossRef CAS.
- W. Humphrey, A. Dalke and K. Schulten, J. Mol. Graph., 1996, 14(33–38), 27–38 Search PubMed.
- F. Jonsson, T. Beke-Somfai, J. Andreasson and B. Norden, Langmuir, 2013, 29, 2099–2103 CrossRef CAS PubMed.
- Y. Marcus, J. Chem. Soc., Faraday Trans., 1991, 87, 2992–2999 Search PubMed.
- I. Kocsis, Y. Ding, N. H. Williams and C. A. Hunter, Chem. Sci., 2021, 12, 12377–12382 RSC.
Footnote |
| † Electronic supplementary information (ESI) available: Synthesis, supplementary data and procedures: Fig. S1–S12; appendix of NMR and mass spectra of new compounds. See DOI: 10.1039/d1sc06671d |
| This journal is © The Royal Society of Chemistry 2022 |