A MALDI-MS-based impact assessment of ZnO nanoparticles, nanorods and quantum dots on the lipid profile of bacterial pathogens
Judy
Gopal
 a,
Pei-Yang
Hua
b,
Manikandan
Muthu
*a and
Hui-Fen
Wu
*bcde
a,
Pei-Yang
Hua
b,
Manikandan
Muthu
*a and
Hui-Fen
Wu
*bcde
aDivision of Research and Innovation, Department of Biotechnology, Saveetha School of Engineering, Saveetha Institute of Medical and Technical Sciences (SIMATS), Thandalam, Chennai 602105, Tamil Nadu, India. E-mail: bhagatmani@gmail.com
bDepartment of Chemistry, National Sun Yat Sen University, Kaohsiung, 804, Taiwan. E-mail: hwu@faculty.nsysu.edu.tw; Fax: +886-7-5253908; Tel: +886-7-5252000-3955
cSchool of Pharmacy, College of Pharmacy, Kaohsiung Medical University, Kaohsiung, 800, Taiwan
dCenter for Nanoscience and Nanotechnology, National Sun Yat-Sen University, Kaohsiung, 804, Taiwan
eDoctoral Degree Program in Marine Biotechnology, National Sun Yat-Sen University, Kaohsiung, 804, Taiwan
First published on 28th November 2022
Abstract
MALDI-MS was used for studying the impact of zinc oxide (ZnO) nanomaterials on Pseudomonas aeruginosa and Staphylococcus aureus. The growth patterns of both these bacterial pathogens in the presence of the ZnO nanomaterials and the subsequent lipidomic changes were assessed using an optimized simple, rapid MALDI-MS based methodology. All three nanostructures tested exhibited differential bactericidal activity unique to P. aeruginosa and S. aureus. The results indicated that the ZnO nanomaterials were highly inhibitory to S. aureus even at 70 mg L−1, while in the case of P. aeruginosa, the ZnO nanomaterials were compatible for up to 10 h and beyond 10 h only marginal growth inhibition was observed. The results proved that the shapes of the ZnO nanomaterials did not affect their toxicity properties. MALDI-MS was applied to study the lipidomic changes of P. aeruginosa and S. aureus after nanomaterial treatment, in order to throw light on the mechanism of growth inhibition. The results from the MALDI-MS studies showed that the ZnO nanostructures exhibited only marginal changes in the lipidomic profile both in the case of P. aeruginosa and S. aureus. These preliminary results indicate that the mechanism of growth inhibition by the ZnO nanomaterial is not through lipid-based interactions, but apparently more so via protein inhibitions.
Introduction
Materials properties of bulk substances of approximately ≥1 μm sizes have been studied thoroughly. Reducing the size of the material to a collection of a few atoms often provides materials with unique physical and chemical properties which absolutely depend on the size and shape of the particle. This phase transition occurs in particles sized between 1 and 100 nm and due to their nanometer sizes, these materials are called nanomaterials. At this scale, electron confinement and the lack of symmetry of materials offer new properties to the particles which are neither present for the individual constituents nor the bulk.1 Such properties of the nanomaterials render them useful in various fields including biomedicine, optics, electronics, magnetics, mechanics, catalysis, and energy science.2 Furthermore, they adequately commensurate with molecules of biological origin, making them attractive candidates for various microbiological, biomedical, and clinical applications.3,4Nanoparticles can become substitutes for traditional chemical drugs in the treatment of pathogenic bacteria since the development of drug resistance in pathogenic bacteria against chemical drugs has become a serious concern. Numerous nanoparticles have been used for bactericidal,5,6 fungicidal7–9 and anticancer applications.8–11 Still other nanoparticles such as TiO2 and NiO are used as dermatological protectants from ultraviolet radiation.12 It is also essential to protect the skin simultaneously from harmful bacterial pathogens along with UV damage. Chemically synthesized drugs may not comply with both the applications but in contrast, the nanoparticle-based approach could offer dual protection both from UV and dermatological pathogens. Zinc oxide (ZnO) is one of the metal oxides, which exhibits a wide band gap and large excitation binding energy at room temperature.13 Hence, zinc oxide nanomaterials have electronic and optical properties that are essential for various applications. The majority of ZnO-based nanoparticles are believed to be nontoxic, biocompatible, and safe and therefore used in various day-to-day applications such as cosmetic products, fillings in medical materials and drug carriers, etc.,14,15 Besides this, a few reports highlight these nanoparticles as bactericidal agents.15–17 The toxicity/biocompatibility of any nanoparticles relies on their particle morphology and size.18,19 ZnO nanoparticles have long been established as toxic to bacterial and biological systems,20–32 but so far the possible development of a biocompatible ZnO nanomaterial by tuning its size and shape has never been investigated.33
Mass spectrometry that combines ionizing molecules and a laser is called MALDI-MS. It is a soft ionization technique employed for the analysis of a wide variety of biomolecules and is considered to be a significant tool for understanding the molecular mechanisms of cells/organisms or at the tissue level. Furthermore, this technique serves as a rapid and reliable platform for the detection of pollutants in environmental samples34,35 and also in cancer detection.36–38 In addition to this, mass spectrometry (MS) has also been used for the analysis of microorganisms using intact cells. Through intact MALDI-MS analysis, it is possible to achieve the mass spectral fingerprints of microbes which can be used to identify the organisms from real samples.39 MALDI MS was also used for studying molecular interactions at micro and macromolecular levels.18,40,41 Furthermore, this technique is also used to study drug or nanoparticle–microbe interactions18 in which the molecular profiles obtained from the bacteria after nanoparticle treatment have been used for identifying the mode of interaction of nanoparticles with bacterial cells. However, these studies did not focus on one of the most significant molecules in bacteria, namely lipids, on which nanoparticles act during interactions.
Therefore, in the present paper, we report the morphological effects of different ZnO nanomaterials on Gram-positive and Gram-negative bacterial strains. We employed 3 different nanomaterials of ZnO such as quantum dots, nanoparticles and nanorods, and studied their interactions with S. aureus and P. aeruginosa. The bacterio-toxic/compatible properties of the 3 different sized ZnO nanoparticles were carefully investigated by studying their growth curves in the presence of various concentrations of ZnO nanomaterials. To identify the possible mode of interaction of the ZnO nanoparticles with bacterial cells, the lipid profiles were analyzed by extracting the total lipids of bacterial cells and analyzing the bacterial lipids using MALDI-MS. The analysis settings and the matrix used were all optimized with respect to the bacterial lipid samples. The MALDI-MS results throw light on the mechanism behind the killing of the pathogens and the role of lipid destruction behind the killing effect.
Materials and methods
Chemicals
Zinc acetate dihydrate and chloroform were purchased from Mallinckrodt Baker, Inc., Phillipsburg. Potassium hydroxide, ethylene glycol, and 2,5-dihydroxybenzoic acid were purchased from Riedel-de Haen (Seelze, Germany). Tetraethyl orthosilicate was purchased from Fluka (Buchs, Switzerland). All chemicals were of analytical grade and the bacterial culture media (nutrient broth and nutrient agar) used were obtained from Sigma-Aldrich. The water was purified by using a Milli-Q system (Millipore, Bedford, MA, USA).Preparation of ZnO nanoparticles
For the preparation of ZnO nanoparticles, we have followed the method described by Xia and Tang42 for ZnO nanoparticles. The ZnO nanoparticles were synthesized through the hydrolysis of zinc acetate by dissolving 10 g of Zn(CH3COO)2·2(H2O) in 50 mL of ethylene glycol (EG), followed by heating at 80 °C. Then 1.4 g of sodium dodecyl sulfate (SDS) and 50 mL of distilled water were blended together with constant mixing. The temperature was further increased to 95 °C for 4 h. This resulted in ZnO nanoparticles being separated by centrifugation; the nanoparticles were then washed many times with distilled water and finally resuspended in deionized water by ultrasonication for 5 min. The successful synthesis of the ZnO nanoparticles was verified by UV absorption spectrophotometery (U3501, Hitachi, Tokyo, Japan). The morphology of the ZnO nanoparticles was confirmed by analyzing the nanoparticles using a high resolution transmission electron microscope (TEM) (JEOL TEM-3010, Tokyo, Japan) at 75 keV.Preparation of ZnO quantum dots
We employed an alkaline reduction method for the synthesis of ZnO quantum dots. We prepared zinc acetate solution (0.55 g of Zn(CH3COO)2·2(H2O) in 25 mL) and KOH solution (2.8 g of potassium hydroxide in 25 mL) separately in methanol. This was then dissolved in 25 mL and 50 mL methanol separately by dissolving the respective salts with constant stirring. Furthermore, the KOH solution was added dropwise to zinc acetate solution at room temperature. Finally, the pH value of the solution was maintained at ∼10 with magnetic stirring for 1 h. 0.25 mL of tetraethylorthosilcate (TEOS) solution and 0.5 mL of distilled water were added into zinc acetate solution to achieve the ZnO quantum dots. The resulting quantum dots were washed using distilled water and methanol. The successful synthesis of the ZnO quantum dots was further verified using the UV absorption spectrum (U3501, Hitachi, Tokyo, Japan). The quantum effect of the ZnO quantum dots was also checked using a fluorescence spectrophotometer (Hitachi F2700, Japan). The morphology of the ZnO quantum dots was confirmed by analyzing the nanoparticles using a high resolution transmission electron microscope (TEM) (JEOL TEM-3010, Tokyo, Japan) at 75 keV.Preparation of ZnO nanorods
ZnO nanorods were prepared following the method previously reported.43 4.45 mmol of Zn(CH3COO)2·2(H2O) was dissolved in 42 mL of methanol with 250 μL of distilled water and 9.62 mmol of potassium hydroxide was dissolved in 23 mL of methanol. Then KOH solution (82% w/v) was added dropwise to zinc acetate solution. The mixture was maintained at 60 °C for 2 h and 15 min. After cooling at room temperature, 10 mL of nitrogen gas was passed into the mixture and the solution became clear again. The resulting mixture was heated to 60 °C for 12 h and washed with ethanol three times. The gel obtained from the reaction was centrifuged and washed with ethanol followed by dilution in 50 mL of 50% w/v trisodium citrate (Sigma-Aldrich). The successful synthesis of the ZnO nanorods was further verified using the UV absorption spectrum (U3501, Hitachi, Tokyo, Japan). The morphology of the ZnO nanorods was confirmed by analyzing the nanoparticles using a high-resolution transmission electron microscope (TEM) (JEOL TEM-3010, Tokyo, Japan) at 75 keV.The effect of the ZnO nanomaterials on bacterial growth
Standard cultures of Staphylococcus aureus (subsp. aureus BCRC no. 10451) and Pseudomonas aeruginosa (BCRC no. 10303) were purchased from the Bioresource Collection and Research Center (BCRC), Hsin-Chu, Taiwan. The cultures (procured as lyophilized powder) were retrieved by growing them on a sterile nutrient agar medium at 37 °C overnight. From the pure colonies, one loopful of bacterial colonies was transferred into sterile nutrient broth and incubated at 37 °C for 12 h. These bacterial suspensions containing approximately 109 cfu mL−1 of bacteria served as the mother cultures for all our further experiments. For studying the effect of different ZnO nanomaterials on the bacterial strains, 1 mL of bacterial cultures at 0.6 optical density (OD) were added into a fresh nutrient medium containing ZnO nanomaterials at concentrations such as 70, 185, and 700 mg L−1 and incubated at 37 °C for 24 hours at 120 rpm. The cultures were harvested from each treatment at different time intervals such as 2, 4, 6, 8, 10, and 12 h. At each time point, the OD was measured at 600 nm in a double-beam spectrophotometer using a UV/visible spectrophotometer (U3501, Hitachi, Tokyo, Japan). OD at 600 nm or short OD600, is the most common method to estimate bacterial growth. The method is based on absorbance detection mode and basically determines which portion of the light passes through a sample, more specifically through a suspension of microorganisms.44Extraction of lipids from bacterial culture
For lipid analysis, total lipid from control and ZnO nanomaterial treated bacterial cells was extracted by an earlier published method.45 The cultured bacterial cells were harvested by centrifugation at 7000 × g for 10 min at 4 °C. The bacterial pellets were re-suspended in 1 mL distilled water and to this suspension, 3.75 mL chloroform![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) methanol (1
methanol (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 (v/v)) was added and vortexed for 10 min. After that, 1.25 mL of chloroform was added and mixed well. Finally, 1.25 mL of distilled water was also added to the mixture, resulting in the separation of the aqueous and organic phases. The lipids from the organic layer were analyzed by MALDI MS using DHB and 9-amino acridine (9-AA) as matrices.
2 (v/v)) was added and vortexed for 10 min. After that, 1.25 mL of chloroform was added and mixed well. Finally, 1.25 mL of distilled water was also added to the mixture, resulting in the separation of the aqueous and organic phases. The lipids from the organic layer were analyzed by MALDI MS using DHB and 9-amino acridine (9-AA) as matrices.
MALDI-MS detection
The lipid samples for MALDI MS analysis were prepared by directly spotting 1 μL lipid extracts extracted from bacteria on hydrophobic MALDI target plates (MTP AnchorChip 384, Bruker inc., Germany) and overlaying them with 1 μL (equal quantity) of 9-AA matrix (10 mg 9-AA in 1 mL of isopropanol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) acetonitrile (3
acetonitrile (3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2)). The DHB matrix using positive ion mode was also attempted (same proportions as above), but since the 9-AA matrix gave better results and showed lower interference, 9-AA was chosen as the ideal matrix for the bacterial lipdomic studies. All the mass spectra were obtained from a MALDI-TOF MS (Microflex, Daltonics Bruker, Bremen Germany) equipped with a nitrogen laser (337 nm), and the accelerating voltages were acquired with the following parameters set on the MALDI-MS: IS1, 19.0 kV; IS2, 16.15 kV; lens, 9.35 kV and a reflector at 20.0 kV in a negative ion mode. The laser energy was adjusted to be slightly above the threshold to obtain good resolution and signal-to-noise ratios at 60 Hz and 200 laser shots were applied for each spectrum. Fig. 1 gives the schematic workflow of this investigation.
2)). The DHB matrix using positive ion mode was also attempted (same proportions as above), but since the 9-AA matrix gave better results and showed lower interference, 9-AA was chosen as the ideal matrix for the bacterial lipdomic studies. All the mass spectra were obtained from a MALDI-TOF MS (Microflex, Daltonics Bruker, Bremen Germany) equipped with a nitrogen laser (337 nm), and the accelerating voltages were acquired with the following parameters set on the MALDI-MS: IS1, 19.0 kV; IS2, 16.15 kV; lens, 9.35 kV and a reflector at 20.0 kV in a negative ion mode. The laser energy was adjusted to be slightly above the threshold to obtain good resolution and signal-to-noise ratios at 60 Hz and 200 laser shots were applied for each spectrum. Fig. 1 gives the schematic workflow of this investigation.
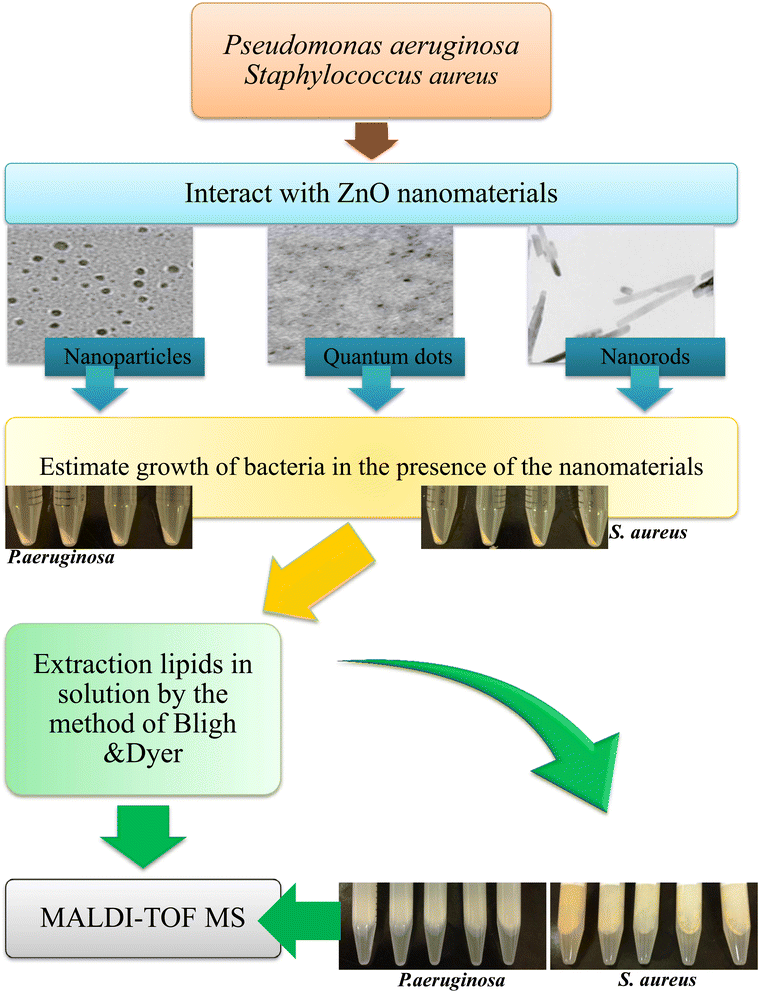 | ||
| Fig. 1 Schematic showing nanomaterial interaction with bacteria and changes in the lipidomic profile. | ||
Results and discussion
Characterization of ZnO nanomaterials by TEM and FT-IR
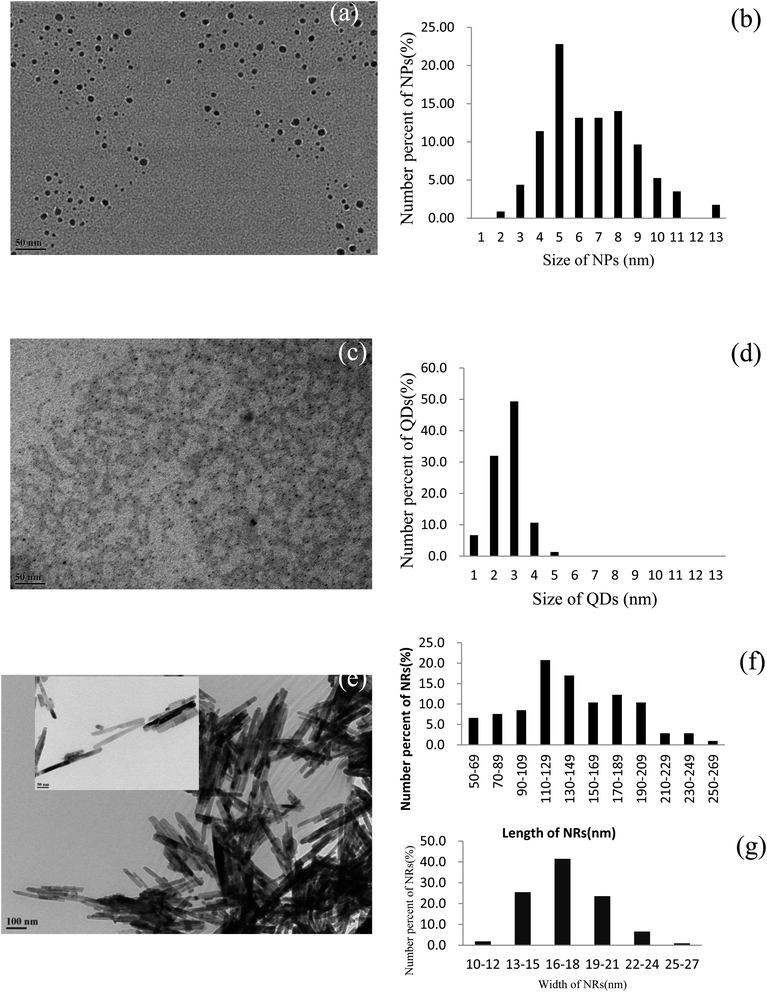
Three morphologically different nanomaterials of zinc oxide origin were synthesized inhouse and characterized using TEM (Fig. 2). From the TEM analysis, we observed that the different methodologies applied for preparing ZnO nanomaterials resulted in the same nanomaterial consisting of three distinct morphologies (Fig. 2a, c and e). Fig. 1a shows the morphology of ZnO nanoparticles and it is evident from the micrograph that all the nanoparticles were spherical irrespective of their sizes. The particle sizes of the indigenously synthesized ZnO nanoparticles existed in the regime of 2 to 13 nm, in which the particles with sizes from 5 to 8 nm predominated followed by the 6 and 7 nm sized particles (Fig. 2b). The successful synthesis of ZnO quantum dots was also confirmed and the TEM micrograph of the spherical shaped ZnO quantum dots is observed in Fig. 2c. The size distribution of ZnO quantum dots was found to be extremely small (between 1 and 5 nm) with a uniform distribution, in which 3 nm particles predominated (Fig. 2d). The ZnO nanorods are shown in Fig. 2e. The length of the synthesized ZnO nanorods was between 50 and 269 nm whereas the breadth was between 10 and 27 nm. Amidst these sizes, nanorods with 110–129 nm length and 16–18 nm width were found to occur predominantly (Fig. 2f and g).Evaluating the toxicity/biocompatibility of ZnO nanomaterials on P. aeruginosa and S. aureus
Fig. 3 and 4 show the growth responses of P. aeruginosa (Fig. 3a–c) and S. aureus (Fig. 4a–c) to the ZnO nanomaterials with 3 distinct morphologies. To assess the effect of ZnO nanomaterials on the growth of P. aeruginosa, the organism was grown in the presence of the respective (three different) nanomaterials with 3 different concentrations such as 70, 185 and 700 mg L−1. As observed from Fig. 3a–c, P. aeruginosa entered the exponential phase after 2 hours of nanomaterial treatments which is very much similar to the untreated control (Fig. 3a–c). Up to 10 h, as seen in Fig. 3a–c, all three ZnO nanomaterials apparently did not bring about any inhibition of bacterial growth. It is beyond 10 h that we see a decrease in bacterial growth. This trend was observed irrespective of the different concentrations. In the case of ZnO nanoparticles, the growth of the test organism at 70 mg L−1 treatment was 0.282 whereas on 185 mg L−1 treatment and 700 mg L−1 treatment the growth showed a slight decrease (0.271 and 0.268 respectively). However, it was observed that the ZnO quantum dot interaction with P. aeruginosa led to a gradual increase in OD from 0.108 (70 mg L−1) to 0.254 (185 mg L−1) and 0.254 for 700 mg L−1 quantum dot concentrations. ZnO nanorods showed an effect similar to that observed in the case of the ZnO nanoparticles where a marginal decrease in growth was observed beyond 10 h. The growth trend was OD (0.289) for 70 mg L−1, (0.190) for 185 mg L−1 and (0.241) for 700 mg L−1. Thus, with respect to P. aeruginosa, in the case of the ZnO nanoparticles and nanorods, irrespective of the concentration, be it 70 mg L−1, 185 mg L−1 or 700 mg L−1 no growth inhibition was evident up to 7 h of incubation time. Beyond 7 h an abrupt drop in the growth curve could be seen in the case of all three nanomaterial interactions at all concentrations, except in the case of ZnO quantum dots at the lowest concentration of 70 mg L−1. This concentration did not lead to any inhibition or toxicity. These results are interesting since so far ZnO nanoparticles have been deemed to be highly toxic to bacterial cells; however from our results we see that the nanomaterials prepared inhouse could be biocompatible for up to 10 h periods even at considerably high concentrations.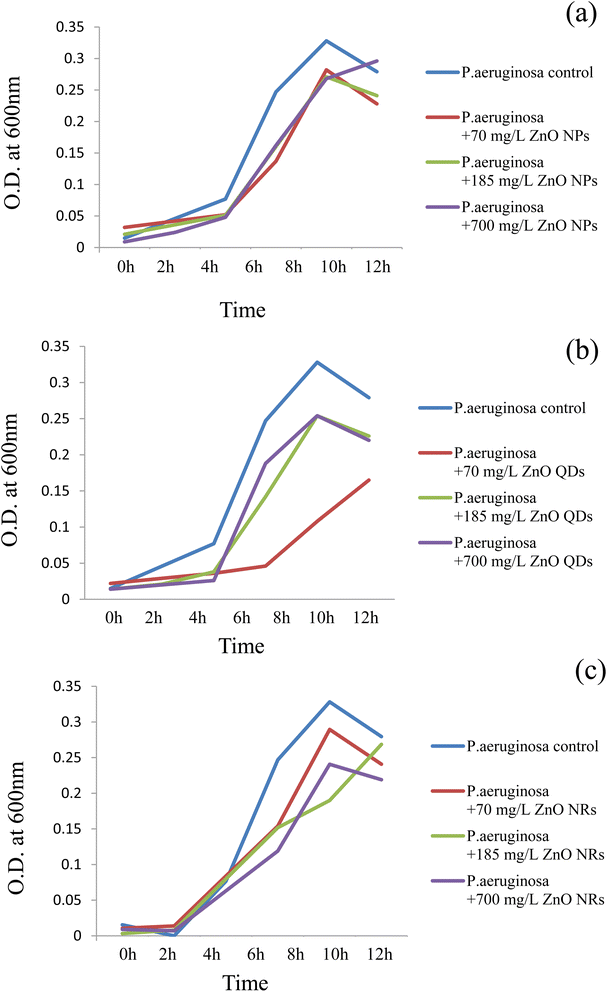 | ||
| Fig. 3 Growth responses of P. aeruginosa at different concentrations of ZnO nanomaterials. (a) ZnO nanoparticles; (b) ZnO quantum dots; (c) ZnO nanorods. | ||
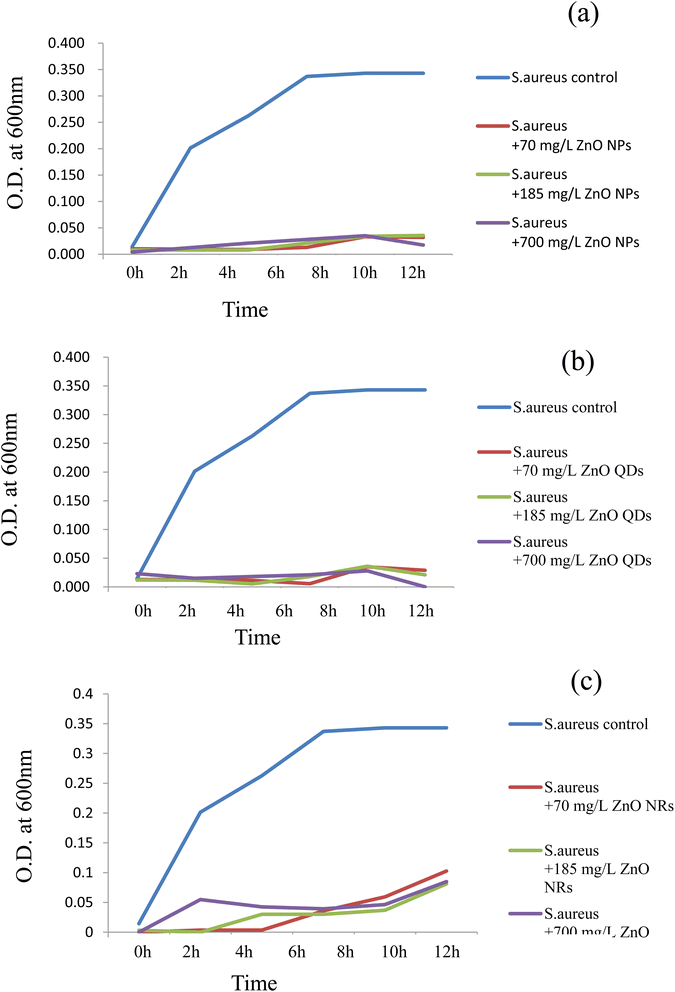 | ||
| Fig. 4 Growth responses of S. aureus at different concentrations of ZnO nanomaterials. (a) ZnO nanoparticles; (b) ZnO quantum dots; (c) ZnO nanorods. | ||
The effect of ZnO nanomaterials on the growth of S. aureus was assessed by growing the organism in the presence of all three nanomaterials at 3 different concentrations such as 70, 185 and 700 mg L−1 (Fig. 4a–c). It was rather interesting to observe that the same three nanomaterials were significantly toxic and totally suppressed the growth of S. aureus cells. Irrespective of the concentration used the ZnO nanoparticles and ZnO quantum dots proved to exhibit strong bactericidal properties. The ZnO nanorods also showed bactericidal activity but to a lesser degree compared to that of quantum dots and nanoparticles. Thus, these results indicate that the toxicity/bacterial compatibility of the ZnO nanoparticles was not a universal rule, but instead it differed with respect to the bacterial species. P. aeruginosa belongs to Gram-negative bacteria whereas S. aureus is a Gram-positive bacterium. Our results show that the Gram-positive pathogen S. aureus was more susceptible to ZnO toxicity compared to its Gram-negative counterpart. These preliminary results suggest that a species related variation with respect to ZnO nanomaterials was evident in this case. Such significant differences in the impact of antibacterial agents/photocatalytic activity/nanomaterials such as CdS quantum dots on Gram-negative and Gram-positive bacteria have been observed previously too.7,9,46,47
MALDI-MS based lipid analysis of P. aeruginosa and S. aureus
MALDI-MS was used as a rapid analytical tool for the lipidomic profiling of the test bacterial cells upon interaction with different sized/shaped ZnO nanomaterials, so that the mechanism of growth inhibition could be determined. The cultures were grown in the presence of different concentrations of the three different shapes of nanomaterials. Equal numbers of bacterial cells were obtained after the respective nanomaterial treatment, by measuring the optical density and adjusting the OD values to 1.0 OD, and then the cells were harvested and the total lipids were extracted. We studied the influence of two different matrices such as DHB and 9-AA on lipids from untreated control cells of P. aeruginosa (Fig. 5A) and S. aureus (Fig. 5B). It is observed that the 9-AA matrix exhibited more number of MALDI MS peaks with good intensity and fewer matrix interferences (Fig. 5A(a) and B(a)) than the DHB matrix (Fig. 5A(b) and B(b)). Therefore, we have used the 9-AA matrix for the subsequent studies.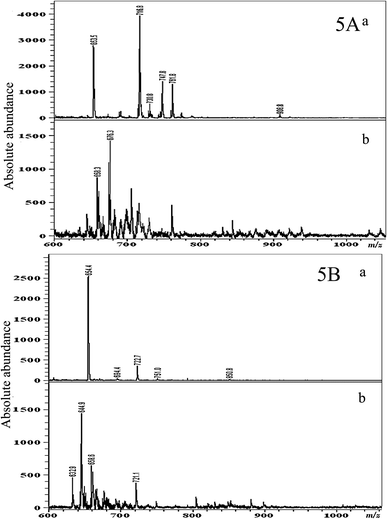 | ||
| Fig. 5 MALDI MS based total lipid analysis of lipid extracts obtained from the control cells of P. aeruginosa (A) and S. aureus (B). (a) DHB as the matrix and (b) 9-amino acridine as the matrix. | ||
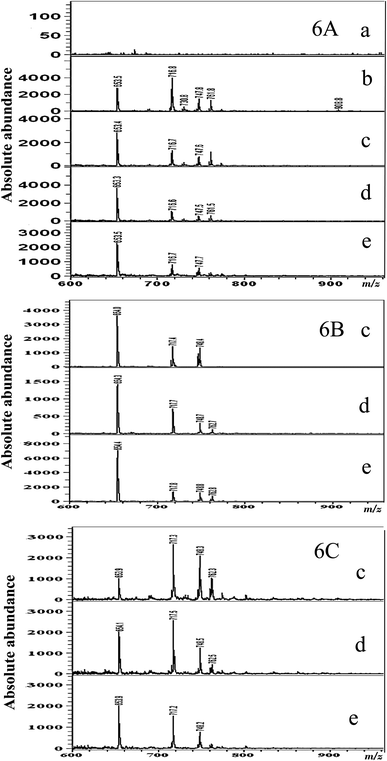
The effect of ZnO nanomaterials on the lipid profile of P. aeruginosa from the MALDI MS analysis is given in Fig. 6A–C. Fig. 6A displays the results of the MALDI-MS analysis conducted with P. aeruginosa lipid samples extracted after treatment with different concentrations of ZnO nanoparticles after an incubation time of 12 h. As can be observed in Fig. 6A(a)–(e), compared to those of the untreated control P. aeruginosa cells (Fig. 6A(b)), very trivial differences in lipid peaks were observed on exposure to ZnO nanoparticles. Marginal inhibition of MALDI MS peaks at m/z 908.8, 1403.9, and 1425.7 was observed at all 3 concentrations of ZnO nanoparticle treatments (Fig. 6A(c)–(e)). Although in terms of bacterial growth, as discussed in the previous section, the ZnO nanomaterial could bring about growth inhibition, yet in terms of the lipid profile not many changes (as shown by the MALDI-MS profiling) were observed.
A very similar trend continued in the case of MALDI-MS spectra acquired from lipid samples extracted from P. aeruginosa incubated with different concentrations of ZnO quantum dots (Fig. 6B(c)–(e)). MALDI MS peaks at m/z 908.8, 1403.9, and 1425.7 at all 3 concentration of ZnO quantum dot treatments (Fig. 6B(c)–(e)) showed minimal suppression. In the case of P. aeruginosa with ZnO NR treatment, the lowest concentration (70 mg L−1) appeared to have no effect as shown by the presence of peaks at m/z 1403.9 and 1425.7; however the peak at m/z 908.8 was suppressed (Fig. 6C(c)). Furthermore, at the increasing concentrations of ZnO NR treatments, peaks at m/z 908.8, 1403.9, and 1425.7 were absent (Fig. 6B(d) and (e)). However, the changes in the lipid profile were not significant enough to explain the growth inhibition mechanism by the ZnO nanomaterials. The strong lipid peak continued to stay unaltered following ZnO exposure. Gopal et al., 2010![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 48 have reported the predominant disappearance of protein peaks of E. coli, a Gram-negative pathogen, on exposure to ZnO nanoparticles. Such a distinct inhibitory pattern was not observed with lipids in this case. Thus, these results indicate that the mode of action of the ZnO nanomaterial may not be via lipid attack but could have been through disruption of the protein moieties of the bacterial cells.
48 have reported the predominant disappearance of protein peaks of E. coli, a Gram-negative pathogen, on exposure to ZnO nanoparticles. Such a distinct inhibitory pattern was not observed with lipids in this case. Thus, these results indicate that the mode of action of the ZnO nanomaterial may not be via lipid attack but could have been through disruption of the protein moieties of the bacterial cells.
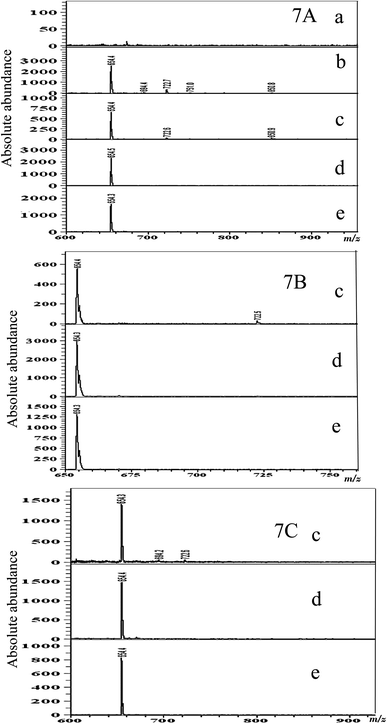
The lipid profile changes of S. aureus during ZnO nanomaterial treatment as shown by the MALDI-MS analysis are given in Fig. 7A–C. The results of the MALDI-MS analysis conducted with S. aureus lipid samples extracted after treatment with different concentrations of ZnO nanoparticles are given in Fig. 7A. As can be observed in Fig. 7A(a)–(e), untreated control S. aureus cells (Fig. 7A(b)) exhibited very few dominant lipid peaks compared to the Gram-negative P. aeruginosa lipid profile. Inhibition of the low intensity MALDI MS peaks at m/z 694.4, 722.7, 751.0 and 850.8 was observed at 185 and 700 mg mL−1 ZnO nanoparticle treatments (Fig. 7A(d) and (e)); however, the stronger peak at m/z 654 was unaffected at all concentrations with respect to all three ZnO nanomaterial morphologies (nanoparticles, nanorods, and quantum dots). In the case of ZnO quantum dots and nanorods, the suppressing effect on S. aureus lipids was also not very pronounced (Fig. 7B(c) and C(c)); a few of the smaller peaks had disappeared, but on the whole, it was observed that the ZnO nanomaterial did not affect the lipid profile of the bacteria as much as it affected the growth of the bacterium. The most noteworthy point is that the ZnO nanoparticles, quantum dots and nanorods exhibited high toxicity and inhibitory activity to the growth of S. aureus at all concentrations tested, but none of these nanomaterials, at all the concentrations tested, brought about significant changes in the bacterial lipids. Thus, using MALDI-MS we were able to conclude that the inhibitory action of ZnO nanoparticles, nanorods and quantum dots on S. aureus had nothing to do with its lipids. This study shows that only marginal inhibition of lipids was observed. As shown by one of our previous studies, it is probable that the ZnO nanoparticle attack is targeted toward bacterial proteins.48 Also, the study confirmed that the shape of the ZnO nanomaterials used in this study did not significantly affect their toxicity/biocompatibility properties, and on the other hand the species of bacteria, as well as the type (Gram-positive/Gram-negative) of bacteria, did significantly impact the nanomaterial interaction. More fundamental mechanistic studies are required to correlate these observations, for a proper understanding of the nanomaterial/bacterial interactions.
Conclusion
The study showed that irrespective of the shape of the ZnO nanomaterials, their interactions with the bacteria were similar. However, ZnO nanoparticles, quantum dots and nanorods used in this study were observed to show the least inhibitory activity on P. aeruginosa even up to an incubation time of nearly 10 h, and only beyond this time was marginal toxicity observed. In the case of S. aureus right from 1 h of incubation all three ZnO nanomaterials showed extremely high toxicity by total inhibition of S. aureus growth. MALDI-MS analysis of the damaged cells showed no significant change in the lipid profile. This study proves that the growth inhibition/toxicity is not by lipid attack; rather a protein related attack or other biomolecular inhibition based antibacterial activity is suspected. This preliminary study suggests that the bacterial toxicity of ZnO nanomaterials is not universal but is highly specific to individual genera of bacteria. More studies are needed to specifically highlight the mode of action leading to inhibition of bacteria. MALDI-MS analysis is no doubt a rapid, simple and straightforward tool leading to a basic understanding of underlying bacterial/nanomaterial interactions.Author contributions
J. G., M. M, conceptualization, investigation, writing-original draft, review and editing; P-Y. H., data curation; H-F. W., supervision, funding.Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
We acknowledge the Ministry of Science and Technology for financial support.References
- S. Eustis and M. A. El-Sayed, Why gold nanoparticles are more precious than pretty gold: noble metal surface plasmon resonance and its enhancement of the radiative and nonradiative properties of nanocrystals of different shapes, Chem. Soc. Rev., 2006, 35, 209–217 RSC.
- G. Schmid, Nanoparticles: from Theory to Application, John Wiley & Sons, 2011 Search PubMed.
- A. S. Ball, S. Patil and S. Soni, Introduction into nanotechnology and microbiology, Methods in Microbiology, Elsevier, 2019, pp. 1–18 Search PubMed.
- G. Grasso, D. Zane and R. Dragone, Microbial nanotechnology: challenges and prospects for green biocatalytic synthesis of nanoscale materials for sensoristic and biomedical applications, Nanomaterials, 2019, 10, 11 CrossRef PubMed.
- S. Sharmin, M. M. Rahaman, C. Sarkar, O. Atolani, M. T. Islam and O. S. Adeyemi, Nanoparticles as antimicrobial and antiviral agents: a literature-based perspective study, Heliyon, 2021, 7, e06456 CrossRef CAS PubMed.
- N. Tran, A. Mir, D. Mallik, A. Sinha, S. Nayar and T. J. Webster, Bactericidal effect of iron oxide nanoparticles on Staphylococcus aureus, Int. J. Nanomed., 2010, 5, 277 CAS.
- M. Ali, X. Wang, U. Haroon, H. J. Chaudhary, A. Kamal, Q. Ali, M. H. Saleem, K. Usman, A. Alatawi and S. Ali, Antifungal activity of Zinc nitrate derived nano Zno fungicide synthesized from Trachyspermum ammi to control fruit rot disease of grapefruit, Ecotoxicol. Environ. Saf., 2022, 233, 113311 CrossRef CAS PubMed.
- S. S. Hosseini, H. Joshaghani, T. Shokohi, A. Ahmadi and Z. Mehrbakhsh, Antifungal activity of ZnO nanoparticles and nystatin and downregulation of SAP1-3 genes expression in fluconazole-resistant Candida albicans isolates from vulvovaginal candidiasis, Infect. Drug Resist., 2020, 13, 385 CrossRef CAS PubMed.
- M. Manikandan and H.-F. Wu, Probing the fungicidal property of CdS quantum dots on Saccharomyces cerevisiae and Candida utilis using MALDI-MS, J. Nanopart. Res., 2013, 15, 1–13 CrossRef.
- M. Magro, A. Venerando, A. Macone, G. Canettieri, E. Agostinelli and F. Vianello, Nanotechnology-based strategies to develop new anticancer therapies, Biomolecules, 2020, 10, 735 CrossRef CAS PubMed.
- R. Tanino, Y. Amano, X. Tong, R. Sun, Y. Tsubata, M. Harada, Y. Fujita and T. Isobe, Anticancer activity of ZnO nanoparticles against human small-cell lung cancer in an orthotopic mouse model, Mol. Cancer Ther., 2020, 19, 502–512 CrossRef CAS PubMed.
- K. Niska, E. Zielinska, M. W. Radomski and I. Inkielewicz-Stepniak, Metal nanoparticles in dermatology and cosmetology: Interactions with human skin cells, Chem.-Biol. Interact., 2018, 295, 38–51 CrossRef CAS PubMed.
- N. Kamarulzaman, M. F. Kasim and R. Rusdi, Band gap narrowing and widening of ZnO nanostructures and doped materials, Nanoscale Res. Lett., 2015, 10, 1–12 CrossRef CAS PubMed.
- S. Gupta, R. Bansal, S. Gupta, N. Jindal and A. Jindal, Nanocarriers and nanoparticles for skin care and dermatological treatments, Indian Dermatol. Online J., 2013, 4, 267 CrossRef PubMed.
- Y. H. Mohammed, A. Holmes, I. N. Haridass, W. Y. Sanchez, H. Studier, J. E. Grice, H. A. Benson and M. S. Roberts, Support for the safe use of zinc oxide nanoparticle sunscreens: lack of skin penetration or cellular toxicity after repeated application in volunteers, J. Invest. Dermatol., 2019, 139, 308–315 CrossRef CAS PubMed.
- S. Jiang, K. Lin and M. Cai, ZnO nanomaterials: current advancements in antibacterial mechanisms and applications, Front. Chem., 2020, 8, 580 CrossRef CAS PubMed.
- L.-E. Shi, Z.-H. Li, W. Zheng, Y.-F. Zhao, Y.-F. Jin and Z.-X. Tang, Synthesis, antibacterial activity, antibacterial mechanism and food applications of ZnO nanoparticles: a review, Food Addit. Contam., Part A, 2014, 31, 173–186 CrossRef CAS PubMed.
- J. Gopal, N. Hasan, M. Manikandan and H.-F. Wu, Bacterial toxicity/compatibility of platinum nanospheres, nanocuboids and nanoflowers, Sci. Rep., 2013, 3, 1–8 Search PubMed.
- M. Manikandan, N. Hasan and H.-F. Wu, Platinum nanoparticles for the photothermal treatment of Neuro 2A cancer cells, Biomaterials, 2013, 34, 5833–5842 CrossRef CAS PubMed.
- M. Li, L. Zhu and D. Lin, Toxicity of ZnO Nanoparticles to Escherichia coli: Mechanism and the Influence of Medium Components, Environ. Sci. Technol., 2011, 45(5), 1977–1983 CrossRef CAS PubMed.
- H. Ma, P. L. Williams and S. A. Diamond, Ecotoxicity of manufactured ZnO nanoparticles: a review, Environ. Pollut., 2013, 172, 76–85, DOI:10.1016/j.envpol.2012.08.011.
- K. S. Siddiqi, A. Ur Rahman and A. Tajuddin Husen, Properties of zinc oxide nanoparticles and their activity against microbes, Nanoscale Res. Lett., 2018, 13, 141, DOI:10.1186/s11671-018-2532-3.
- J. Jiang, J. Pi and J. Cai, The advancing of zinc oxide nanoparticles for biomedical applications, Bioinorg. Chem. Appl., 2018, 2018, 1062562, DOI:10.1155/2018/1062562.
- C. Coll, D. Notter, F. Gottschalk, T. Sun, C. Som and B. Nowack, Probabilistic environmental risk assessment of five nanomaterials (nano-TiO2, nano-Ag, nano-ZnO, CNT, and fullerenes), Nanotoxicology, 2016, 10, 436–444, DOI:10.3109/17435390.2015.1073812.
- K. B. Kim, Y. W. Kim, S. K. Lim, T. H. Roh, D. Y. Bang and S. M. Choi, et al., Risk assessment of zinc oxide, a cosmetic ingredient used as a UV filter of sunscreens, J. Toxicol. Environ. Health, Part B, 2017, 20, 155–182, DOI:10.1080/10937404.2017.1290516.
- J. Hou, Y. Wu, X. Li, B. Wei, S. Li and X. Wang, Toxic effects of different types of zinc oxide nanoparticles on algae, plants, invertebrates, vertebrates and microorganisms, Chemosphere, 2018, 193, 852–860, DOI:10.1016/j.chemosphere.2017.11.077.
- B. de Berardis, G. Civitelli, M. Condello, P. Lista, R. Pozzi and G. Arancia, et al., Exposure to ZnO nanoparticles induces oxidative stress and cytotoxicity in human colon carcinoma cells, Toxicol. Appl. Pharmacol., 2010, 246, 116–127, DOI:10.1016/j.taap.2010.04.012.
- J. Park, S. Kim, J. Yoo, J. S. Lee, J. W. Park and J. Jung, Effect of salinity on acute copper and zinc toxicity to Tigriopus japonicus: the difference between metal ions and nanoparticles, Mar. Pollut. Bull., 2014, 85, 526–531, DOI:10.1016/j.marpolbul.2014.04.038.
- H. C. Chuang, H. T. Juan, C. N. Chang, Y. H. Yan, T. H. Yuan and J. S. Wang, et al., Cardiopulmonary toxicity of pulmonary exposure to occupationally relevant zinc oxide nanoparticles, Nanotoxicology, 2014, 8, 593–604, DOI:10.3109/17435390.2013.809809.
- M. R. Cooper, G. H. West, L. G. Burrelli, D. Dresser, K. N. Griffin and A. M. Segrave, et al., Inhalation exposure during spray application and subsequent sanding of a wood sealant containing zinc oxide nanoparticles, J. Occup. Environ. Hyg., 2017, 14, 510–522, DOI:10.1080/15459624.2017.1296237.
- C. Monsé, O. Hagemeyer, M. Raulf, B. Jettkant, V. van Kampen and B. Kendzia, et al., Concentration-dependent systemic response after inhalation of nano-sized zinc oxide particles in human volunteers, Part. Fibre Toxicol., 2018, 15, 8, DOI:10.1186/s12989-018-0246-4.
- T. Gordon, L. C. Chen, J. M. Fine, R. B. Schlesinger, W. Y. Su and T. A. Kimmel, et al., Pulmonary effects of inhaled zinc oxide in human subjects, guinea pigs, rats, and rabbits, Am. Ind. Hyg. Assoc. J., 1992, 53, 503–509, DOI:10.1080/15298669291360030.
- M. Lagarrigue, R. M. Caprioli and C. Pineau, Potential of MALDI imaging for the toxicological evaluation of environmental pollutants, J. Proteomics, 2016, 144, 133–139 CrossRef CAS PubMed.
- S. Wang, H. Niu, T. Zeng, X. Zhang, D. Cao and Y. Cai, Rapid determination of small molecule pollutants using metal-organic frameworks as adsorbent and matrix of MALDI-TOF-MS, Microporous Mesoporous Mater., 2017, 239, 390–395 CrossRef CAS.
- M. Manikandan and H.-F. Wu, Bio-mimicked gold nanoparticles with complex fetal bovine serum as sensors for single cell MALDI MS of cancer cell and cancer stem cell, Sens. Actuators, B, 2016, 231, 154–165 CrossRef CAS.
- M. Pietrowska and P. Widłak, MALDI-MS-based profiling of serum proteome: detection of changes related to progression of cancer and response to anticancer treatment, Int. J. Proteomics, 2012, 2012, 926427 Search PubMed.
- P.-Y. Hua, M. Manikandan, H. N. Abdelhamid and H.-F. Wu, Graphene nanoflakes as an efficient ionizing matrix for MALDI-MS based lipidomics of cancer cells and cancer stem cells, J. Mater. Chem. B, 2014, 2, 7334–7343 RSC.
- M. Manikandan, P.-Y. Hua and H.-F. Wu, Rapid endophytic bacterial detection by enzyme incorporated MALDI MS, RSC Adv., 2014, 4, 50233–50240 RSC.
- M. Manikandan, H.-F. Wu and N. Hasan, Cell population based mass spectrometry using platinum nanodots for algal and fungal studies, Biosens. Bioelectron., 2012, 35, 493–497 CrossRef CAS PubMed.
- M. Giampà and E. Sgobba, Insight to Functional Conformation and Noncovalent Interactions of Protein-Protein Assembly Using MALDI Mass Spectrometry, Molecules, 2020, 25(21), 4979, DOI:10.3390/molecules25214979.
- G. Chen, M. Fan, Y. Liu, B. Sun, M. Liu, J. Wu, N. Li and M. Guo, Advances in MS Based Strategies for Probing Ligand-Target Interactions: Focus on Soft Ionization Mass Spectrometric Techniques, Front. Chem., 2019, 7(703), 1–17, DOI:10.3389/fchem.2019.00703.
- H.-L. Xia and F.-Q. Tang, Surface synthesis of zinc oxide nanoparticles on silica spheres: preparation and characterization, J. Phys. Chem. B, 2003, 107, 9175–9178 CrossRef CAS.
- W.-Q. Zhang, Y. Lu, T.-K. Zhang, W. Xu, M. Zhang and S.-H. Yu, Controlled synthesis and biocompatibility of water-soluble ZnO nanorods/Au nanocomposites with tunable UV and visible emission intensity, J. Phys. Chem. C, 2008, 112, 19872–19877 CrossRef CAS.
- J. A. Myers, B. S. Curtis and W. R. Curtis, Improving accuracy of cell and chromophore concentration measurements using optical density, BMC Biophys., 2013, 6, 4 CrossRef PubMed.
- E. G. Bligh and W. J. Dyer, A rapid method of total lipid extraction and purification, Can. J. Biochem. Physiol., 1959, 37, 911–917 CrossRef CAS PubMed.
- Z. Breijyeh, B. Jubeh and R. Karaman, Resistance of Gram-Negative Bacteria to Current Antibacterial Agents and Approaches to Resolve It, Molecules, 2020, 25(6), 1340, DOI:10.3390/molecules25061340 , PMID: 32187986; PMCID: PMC7144564..
- J. Gopal, R. P. George, P. Muraleedharan and H. S. Khatak, Photocatalytic Inhibition of Microbial Adhesion by Anodized Titanium, Biofouling, 2004, 20(3), 167–175, DOI:10.1080/08927010400008563.
- J. Gopal, H.-F. Wu and Y.-H. Lee, Matrix-assisted laser desorption ionization-time-of-flight mass spectrometry as a rapid and reliable technique for directly evaluating bactericidal activity: Probing the critical concentration of Zno nanoparticles as affinity probes, Anal. Chem., 2010, 82, 9617–9621 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2023 |