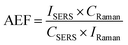Open Access Article
Open Access ArticleAg NP-filter paper based SERS sensor coupled with multivariate analysis for rapid identification of bacteria†
Rong Wang * and
Jiamin Luo
* and
Jiamin Luo
Chemical Engineering College, Sichuan University of Science and Engineering, Zigong, Sichuan 643000, China. E-mail: 29073964@qq.com
First published on 21st December 2022
Abstract
Rapid and accurate identification of bacteria is essential to ensure food safety and prevent pathogenic bacterial infection. In this study, a highly efficient method was established for accurately identifying bacterial species by applying Ag NP-filter paper based Surface enhanced Raman spectroscopy (SERS) analysis and Partial Least Squares-Discriminant Analysis (PLS-DA) statistical methods. The flexible Ag NP filter paper substrate with high sensitivity and uniformity was prepared by a facile and low-cost silver mirror reaction at room temperature, which exhibited desirable SERS activity in bacteria detection. Furthermore, PLS-DA was successfully employed to distinguish SERS spectra from S. aureus CMCC 26003, E. faecalis ATCC29212 and L. monocytogenes ATCC 19115 with a sensitivity of 93.3–100%, specificity of 96.7–97%, and overall predicting accuracy of 95.8%. This exploratory study demonstrates that a Ag NP-filter paper based SERS sensor coupled with PLS-DA has great potential for rapid and effective detection and identification of bacteria.
1. Introduction
In recent years, safety incidents caused by pathogenic bacterial pollution in food have occurred frequently.1,2 Pathogenic bacterial pollution has become a high-risk problem threatening global public health security.3 It is estimated that more than a quarter of deaths worldwide are caused by infectious diseases every year, which is second only to cardiovascular disease.4 Therefore, the accurate identification and discrimination of bacteria play a vital role in ensuring food safety and reducing pathogenic bacterial infection. Previously, several analytical methods have been used to detect bacteria, such as the traditional bacterial culture detection method, polymerase chain reaction (PCR), flow cytometry (FCM)5–7 and enzyme-linked immunosorbent assay (ELISAs).8 Although, these methods presented excellent accuracy and sensitivity in bacterial detection they are inherently time-consuming, laborious, expensive, and complex, and require well-trained personnel and cumbersome facilities, making them less attractive than low-cost and rapid detection applications. Thus, it is highly desired to develop a fast, reliable, cost-effective, high sensitivity and specificity method for identification.SERS technology has been widely applied in bacterial detection, and its high sensitivity and inherent specificity fingerprints provide the potential capability of bacterial identification and discrimination.9,10 Some studies have reported the successful detection and identification of bacteria by combining SERS detection with multivariate analysis.11
As we all know, SERS based bacteria identification heavily relys on the sensitivity and repeatability of individual SERS spectra. However, SERS based bacteria detection suffers from poor sensitivity as current available SERS substrates lack sufficient uniformity or interaction with bacterial cells.12 To address the above issues, a variety of SERS substrates, such as Au NRs,13 Fe3O4@Ag magnetic nanoparticle,14 lectin-modified BCNCs,15 Ag@TiO2 nanofibrous felts,16 Ag/Au NPs/SiNWs,17 have been developed for bacterial detection. Although significant progress has been made in the fabrication of SERS active substrates, some non-negligible problems still hinder further application in bacterial detection. For example, the traditional colloidal substrate undergoseasy aggregation and poor signal reproducibility,13 while the main drawbacks to the solid substrate fixed on the rigid support (such as glass and silicon) are either high production cost, poor uniformity, complex preparation process or low batch-to-batch reproducibility.18
Filter paper is emerging as an ideal scaffold for fabricating SERS substrate due to its quality of rich natural resources, affordable, 3D porous structure, portability, and flexibility.19,20 Filter paper substrate can continuously collect the samples via swabbing or capillary suction,21 making the bacteria more closely bound to the substrate, thus improving the sensitivity and reproducibility of bacterial testing.22,23 In addition, the 3D structure of filter paper fiber can not only provide a larger surface for assembling more nanoparticles, but also help to form more abundant “hot spots”, which is beneficial to generate stronger SERS enhancement.20,24
Herein, the performance of filter paper based SERS substrate is investigated by using R6G as probe molecules. The SERS performance of Ag NPs filter paper substrate was studied and optimized. Through capillary force suction bacteria sample, the optimized substrate was applied to detection of S. aureus CMCC 26003, E. faecalis ATCC29212 and L. monocytogenes ATCC 19115. In order to improve the efficiency of bacterial species identification, PLS-DA models was user to effectively identify the SERS of these bacterial species.
2. Experimental section
2.1 Chemicals and regents
Three different bacterial species used in the experiment were obtained from Beijing Microbiological Culture Collection Center. Silver nitrate (AgNO3, 99.7%) and glucose were purchased from Sinopharm Chemical Reagent Co, Ltd (Shanghai, China). Ammonium hydroxide were purchased from Aladdin's Reagent (Shanghai, China). Filter paper was purchased from Fushun filter paper factory. All reagents were not further purified.2.2 Apparatus
Field emission scanning electron microscope coupled with EDS-system (Hitachi S4800, Japan) was employed to elucidate the morphologies and composition of Ag NPs-filter paper substrate. The crystal phase of Ag NPs-filter paper substrates were measured on X-ray powder diffraction using Cu Kα radiation (λ = 1.5406 nm) (XRD, Bruker, D2 PHASER). The SERS spectra were measured using a DXR Raman microscope (Thermo Fisher Scientific, Inc. USA) equipped with CCD detector, excitation wavelength of 780 nm laser, and a 10× objective lens.2.3 In situ growth Ag NPs on filter paper
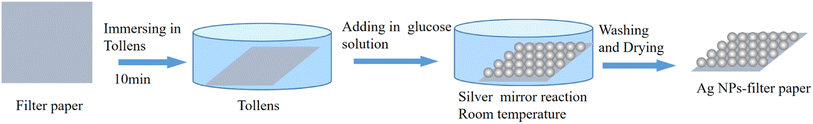
In order to prepare Ag NPs-filter paper substrate, the filter papers were cut into pieces of 5 cm × 5 cm, and immersed in 20 mL freshly prepared Tollens solution of 10 mM Tollens for 10 min. Then 1 mL 0.6 mol L−1 aqueous glucose solution was dropwise added with shaking. Subsequently, it was standing for a certain time at room temperature. Afterwards, the Ag-filter paper substrate were drawn from solution and washed successively with plenty of deionized water and ethanol to remove impurities and unbonded Ag NPs, and then, it was dried with nitrogen and stored in a brown vacuum desiccator for further use.2.4 SERS detection of small molecules
To demonstrate the performance of Ag NPs -filter paper substrate, R6G was used as probe molecules for testing. Firstly, the as-prepared Ag NPs filter paper substrates were cut into 3 × 4 mm small paper and immersed in R6G solutions with different concentrations for 20 min, then washed with deionized water, and put it on a clean glass slide. After the solution was evaporated and dried, SERS test was carry out by using a DXR Raman microscope. The excitation power was 5 mW with an exposure time of 5 s and accumulation of two times.2.5 Bacterial culture and SERS detection
All bacterial species used in this work, including S. aureus (CMCC 26003), E. faecalis (ATCC29212) and L. monocytogenes (ATCC 19115) were cultured in Luria Bertani (LB) broth medium at 37 °C for 24 h shaking at 180 rpm respectively. Following incubation, 5 mL bacteria suspension (∼107 CFU mL−1) were centrifuged at 6000 rpm for 5 min, and washed three times with sterilized deionized water to eliminate the interference of culture medium, and then resuspended in sterilized deionized water to obtain a final concentration of 102–107 CFU mL−1.The bacterial SERS spectra were collected on a DXR Raman microscope. The excitation laser with a power of 5 mW was focused on the bacteria through a 10× objective lens (3 μm focus diameter). The integration time was set to 10 s with accumulation of two times. Each sample was measured for at last 5 times and take the average spectral.
2.6 Data analysis
The pretreatment and multivariate statistical analysis for bacteria SERS spectra were performed on MATLAB R2014a (MathWorks, USA). Prior to multivariate statistical analysis, all of bacteria SERS spectra were preprocessed by baseline correction, Savitzky–Golay smoothing and area normalization in the range of 300–2000 cm−1. The preprocessed bacteria SERS spectral data was analyzed by using PLS Toolbox in the Matlab software. The SERS spectral date of bacterial were randomly divided into 2/3 training data set and 1/3 test data set. The PLS-DA model was established on a training data set along with venetian blinds cross-validation, and then the testing data set was used as a blind sample to external verify the accuracy of the PLS-DA model.3. Results and discussion
3.1 Preparation and characterization of Ag NPs-filter paper substrate
Here, we introduce a facile, low-cost and eco-friendly strategy for in situ synthesis of Ag NPs on filter paper with high uniformity and density. The fabrication processing was illustrated in Fig. 1.First, a new filter paper with the size of 5 × 5 cm2 w was soaked in the Tollens for 10 min, which is easy to attach positively charged Ag(NH3)2+ through capillary attraction and electrostatic attraction.25 Then aqueous glucose solution was dropwise added and allowed to stand for a certain amount of time at room temperature, which caused a slow silver mirror reaction between Tollens and glucose, resulting in the in situ growth of Ag NPs on the filter paper26 based on reaction (1-1).
| CH2OH(CHOH)4CHO + 2Ag(NH3)2OH → CH2OH(CHOH)4COONH4 + 2Ag + 3NH3 + 2H2O | (1-1) |
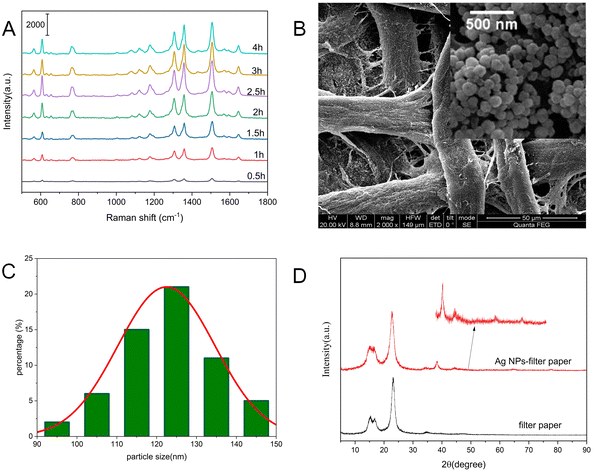
It is well known that the SERS substrate performance is highly associated with the particle size and aggregates morphology of Ag NPs, which is well controlled by adjusting the reaction time. The sensitivity of Ag NPs-filter paper substrates prepared at different reaction times were investigated by using R6G as probe. As shown in Fig. 2A, the SERS intensities of R6G increased with the increase of reaction time ranging from 0.5 h to 2.5 h, and reached a maximum at 2.5 h. Interestingly, the SERS intensity of R6G decreased slowly with further prolonging the reaction time. As we all know, the sensitivity of SERS depends mainly on the size, shape and neighboring particle interaction. The results reported in this work showed that the size, shape of neighboring particle of silver nanostructures obtained at 3 h provide the highest Raman signal enhancement.
To improve the uniformity of the substrate, a strategy of in situ synthesis of Ag NPs was successfully conducted by reducing silver ammonia solution with glucose at room temperature. In comparison with silver nanoparticles prepared at 60–80 °C, the superior uniformity of the AgNPs distributed on the filter paper was attributed to the slower nucleation and growth rate of Ag NPs at room temperature.26 According to the typical SEM image of Ag NPs-filter paper substrate obtained with a reaction time of 3 h (Fig. 2B), the surface of filter paper was covered by homogeneous and dense Ag NPs with an average particle size around 120 nm (Fig. 2C), by which a large number of “hot spots” were able to form between neighboring Ag NPs, thus providing a high-activity and reliable SERS response. Fig. 2D shows that the XRD patterns of the as-prepared Ag NPs-filter paper substrate exhibit not only the diffraction peaks of Ag NPs, but also strong diffraction peaks of filter paper.26 Among them, the diffraction peaks at 2θ values of 16.6°, 22.7° and 34.2° belong to filter paper. The diffraction peaks at 2θ values of 38.2°, 44.3°, 64.6° and 77.5° can be assigned to the reflections of (111), (200), (220) and (311) planes of face centered cubic (FCC) silver (JCPDF, no. 04-0783), respectively,27,28 demonstrating the pure and well crystallized nature of the produced Ag NPs.
3.2 Evaluation of SERS substrate
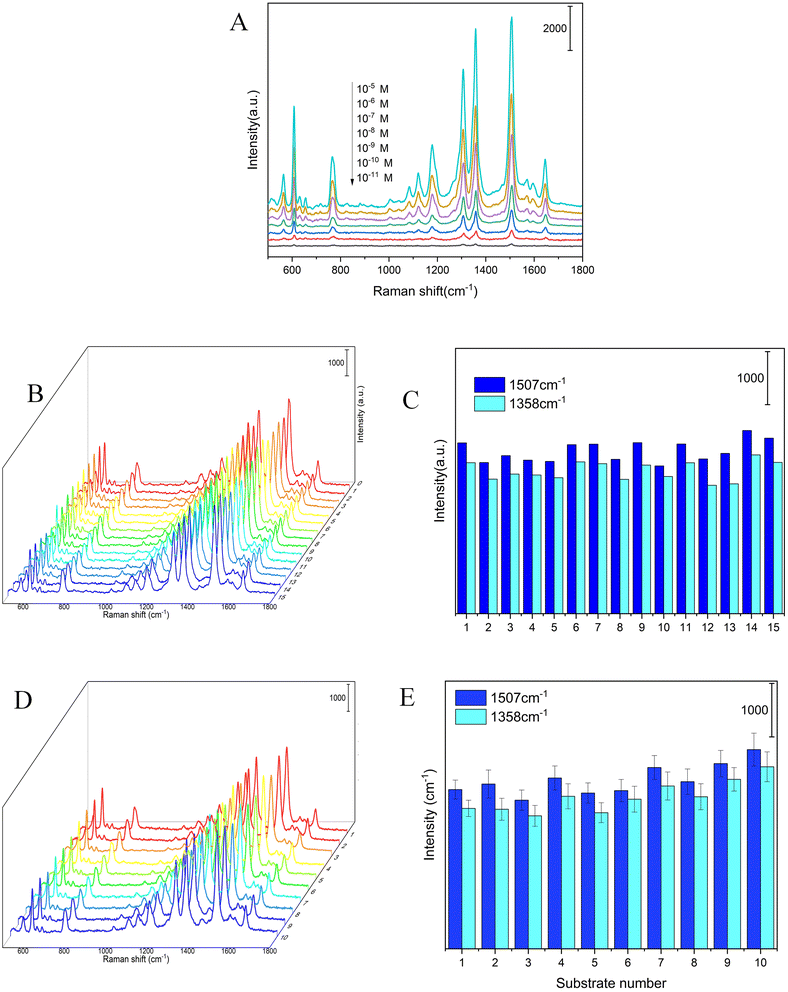
The main advantage of the as-prepared Ag NPs-filter paper was possessing high activity and reproducibility of the SERS spectra. The SERS performances of the as-prepared Ag NPs-filter paper substrate was preliminarily evaluated by using R6G as probe molecules. The SERS spectra of R6G of different concentrations (10−5–10 −11 M) were collected on the optimal Ag NPs-filter paper substrates, as shown in Fig. 3A. The prominent characteristic peak of R6G was observed even at a concentration as low as 10−11 M, demonstrating a high sensitivity of the SERS substrate. To further quantitatively evaluate the enhancement ability of the as-prepared SERS substrate, the analytical enhancement factor (AEF) for R6G was calculated based on the following equation:29,30where CSERS and ISERS are R6G concentration (1 × 10−11 M) and SERS intensity collected on the as-prepared SERS substrate, respectively. CRaman and IRaman are R6G concentration (1 × 10−2 M) and Raman intensity collected on a gold wafer (Fig. S1†), respectively. Consequently, the AEF of the as-prepared SERS substrate was as high as 4 × 109 and 2.9 × 109 for R6G at 1358 cm−1 and 1507 cm−1, respectively, which indicates that it has better SERS activity.
As we all know, signal reproducibility is also a crucial factor, affecting its performance and reliability for SERS substrate used in the practical application. The homogeneity and reproducibility of the as-prepared Ag NPs-filter paper substrate were evaluated by measuring 15 points that were randomly selected on the same substrate loaded with 10−7 M R6G, as shown in Fig. 3B and C. The relative standard deviation (RSD) values of spot-to-spot SERS peak intensity at 1358 and 1507 cm−1 are 6.6% and 6.5%, respectively, indicating good reproducibility, which may result from the uniform distribution of Ag NPs on the surface of filter paper. For another, the reproducibility of the SERS spectra between different batches is also a key factor affecting the application. The reproducibility of Ag NPs-filter paper substrate from batch to batch were also investigated, and the results are shown in Fig. 3D and E. The RSD of the SERS spectra intensity at 1358 and 1507 cm−1 from different batches were 10.4% and 9.2%, respectively, revealing high batch-to-batch reproducibility. Therefore, this method has the potential for mass production.
3.3 SERS detection of bacteria
The SERS substrate is provided with high SERS sensitivity and signal reproducibility which lays a foundation for the detection of bacteria.31 The S. aureus was employed to evaluate the detection ability of the as-prepared Ag NPs-filter paper substrate to bacteria. The S. aureus was diluted to bacterial densities in the range 102–106 CFU mL−1 with steriled deionized water, and then, 5 mL of the S. aureus solution was centrifugated. Subsequently, the bacteria was collected on the flexible Ag NPs-filter paper substrate by swabbing the bottom of the centrifuge tube. After that, The SERS spectra of S. aureus were captured by focusing the laser spot with a microscope. As shown in Fig. S2.† The SERS spectrum of S. aureus was significantly enhanced on the Ag NPs-filter paper substrate. As the concentration of S. aureus decreased, its spectral shape was very stable and its intensity gradually decreased. The limit of detection of S. aureus was at the level of 102 CFU mL−1.3.4 SERS classification of bacterial species
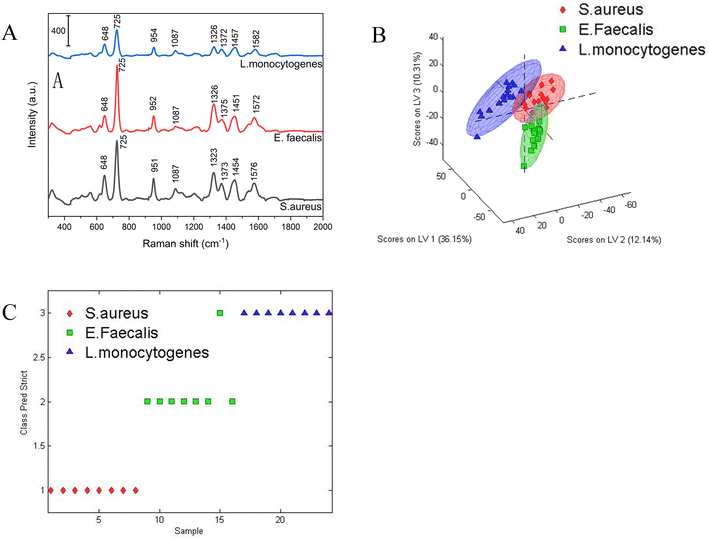
To verify the identification capability for bacterial species, three different bacterial species (S. aureus, E. faecalis and L. monocytogenes) were investigated based on the Ag NPs-filter paper substrate combined with multivariate statistical method. For each of the bacterial species, 5 mL pure bacteria solution (∼107 CFU mL−1) were centrifuged at 6000 rpm for 5 min, and then resuspended in 50 μL sterilized deionized water. Subsequently, 10 μL bacterial solution was dripped onto the Ag-filter paper substrate and used for SERS detection after dried. The average SERS spectra belonging to S. aureus (n = 23), E. faecalis (n = 23) and L. monocytogenes (n = 26) were shown in Fig. 4A. As can be seen clearly, the characteristic SERS peaks from different bacteria species showed strong similarities and the positions of the characteristic peaks are very close, because of the similar component of cell wall, membrane and nucleic acid of different bacteria species. The characteristic peaks of these three bacterial species mainly around at ∼648, ∼725 ∼950, ∼1087, ∼1326, ∼1375, ∼1457, 1576 cm−1. According to some literature on SERS peak assignment, the assignment of these peaks was shown in Table 1.| Peak position cm−1 | Assignments | ||
|---|---|---|---|
| S. aureus CMCC 26003 | E. faecalis ATCC29212 | L. monocytogenes ATCC 19115 | |
| 648 | 648 | 648 | Ring vibration of guanine |
| 725 | 725 | 725 | Ring vibration of adenine |
| 951 | 952 | 954 | Polysaccharide |
| 1087 | 1087 | 1087 | Aliphatic esters |
| 1323 | 1326 | 1326 | Ring vibration of adenine |
| 1373 | 1375 | 1372 | Amino acids, protein |
| 1454 | 1451 | 1457 | Saturated lipids |
| 1576 | 1572 | 1582 | Protein amide II |
It's confirmed that different bacterial species present unique fingerprint SERS spectra, however, it is difficult to distinguish bacterial species by visual observation as the SERS spectra of different bacterial species are similar due to biochemical composition among different species.
To distinguish the SERS spectra of different bacterial species more effectively, a PLS-DA based model was developed as a differentiation tool to analyze the bacterial SERS spectra obtained from 23 S. aureus CMCC 26003, 23 E. faecalis ATCC29212 and 26 L. monocytogenes ATCC 19115, in which the 72 bacterial SERS spectra were randomly divided into 48 training date set (15 S. aureus CMCC 26003, 15 E. faecalis ATCC29212 and 18 L. monocytogenes ATCC 19115) and 24 testing date set (8 S. aureus CMCC 26003, 8 E. faecalis ATCC29212 and 8 L. monocytogenes ATCC 19115). Prior to multivariate statistical analysis, all raw SERS spectra were pre-processed by baseline correction, Savitzky–Golay smoothing and area normalization in the range of 300–2000 cm−1 using MATLAB R2014a software to eliminate the influence of measurement conditions on bacterial classification. The three-dimensional scores plot distribution of bacterial SERS spectra on the first three latent variables were shown in Fig. 4B. It can be seen that the spectra of three different bacterial species were separated from each other, while the spectra of the same type of bacteria were well clustered together. The PLS-DA model displayed a high sensitivity of 93.3%, 100%, 94.4% and a specificity of 97%, 97%, 96.7% for S. aureus CMCC 26003, E. faecalis ATCC29212 and L. monocytogenes ATCC 19115, respectively. To further evidence the performance of PLS-DA model, the ROC curve with an AUC was also made in Fig. S3.† The AUC value of S. aureus CMCC 26003, E. faecalis ATCC29212 and L. monocytogenes ATCC 19115 were 0.97, 1.00 and 0.99, respectively, which further demonstrated that the PLS-DA-based bacterial SERS spectral classification method has the potential capability for species identification.
The prediction accuracy of the PLS-DA model for bacterial classification was further evaluated by blind predicting 24 spectra originating from three different bacterial samples. The prediction results were shown in Fig. 4C. It can be seen that except for one E. faecalis ATCC29212 was wrongly identified as L. monocytogenes ATCC 19115, the vast majority of spectra were correctly classified into their respective classes with an overall accuracy of 95.8%. These results fully proved the identification ability of this method in bacterial species.
4. Conclusions
In summary, a method for rapid discriminating of bacteria species was developed by combining Ag NPs filter paper based SERS detection with PLS-DA multivariate analysis. The advantages of the filter paper substrate allows silver nanoparticles to uniformly in situ deposited on filter paper at room temperature. This facile preparation process is not only cost-saving but also has the potential of massive production, which provides the possibility for the practical application of SERS technology. The as-prepared Ag-filter paper substrate showed excellent SERS performance, which can be used for highly sensitive detection of bacteria. Furthermore, a PLS-DA model for SERS spectra analysis was developed and successfully employed for bacteria identification with an overall accuracy rate of 95.8%. In future work, we will apply this new SERS-PLS-DA approach to study more bacteria species.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was financially supported by Key project of Science and Technology Department of Sichuan Province (2020YFS0347) and University Student Innovation Fund (cx2021037).References
- K. Tsougeni, G. Kaprou, C. M. Loukas, G. Papadakis, A. Hamiot, M. Eck, D. Rabus, G. Kokkoris, S. Chatzandroulis, V. Papadopoulos, B. Dupuy, G. Jobst, E. Gizeli, A. Tserepi and E. Gogolides, Sens. Actuators, B, 2020, 320, 128345 CrossRef CAS.
- A. C. Soares, J. C. Soares, V. C. Rodrigues, O. N. Oliveira and L. H. Capparelli Mattoso, Analyst, 2020, 145, 6014–6023 RSC.
- M. E. Berry, H. Kearns, D. Graham and K. Faulds, Analyst, 2021, 146, 6084–6101 RSC.
- E. O. F. S. de Siqueira, A. M. da Silva, M. T. T. Pacheco, H. E. Giana and L. Silveira, Jr., Lasers Med. Sci., 2021, 36, 289–302 CrossRef PubMed.
- S. He, X. Hong, M. Zhang, L. Wu and X. Yan, Anal. Chem., 2020, 92, 2393–2400 CrossRef CAS PubMed.
- S. He, X. Hong, T. Huang, W. Zhang, Y. Zhou, L. Wu and X. Yan, Methods Appl. Fluoresc., 2017, 5, 024002 CrossRef PubMed.
- L. Lv, L. Dong, J. Zheng, T. Maermaer, X. Huang, X. Fan, H. Zhang and T. Shen, Methods Appl. Fluoresc., 2021, 9, 8 Search PubMed.
- Y. Zhao, D. Zeng, C. Yan, W. Chen, J. Ren, Y. Jiang, L. Jiang, F. Xue, D. Ji, F. Tang, M. Zhou and J. Dai, Analyst, 2020, 145, 3106–3115 RSC.
- E. Yang, D. Li, P. Yin, Q. Xie, Y. Li, Q. Lin and Y. Duan, Biosens. Bioelectron., 2021, 172, 112758 CrossRef CAS PubMed.
- B. Hua, H. Pu and D.-W. Sun, Trends Food Sci. Technol., 2021, 110, 304–320 CrossRef.
- E. Witkowska, K. Niciński, D. Korsak, B. Dominiak, J. Waluk and A. Kamińska, J. Biophotonics, 2020, 13, e201960227 CrossRef PubMed.
- L. F. Tadesse, C. S. Ho, D. H. Chen, H. Arami, N. Banaei, S. S. Gambhir, S. S. Jeffrey, A. A. E. Saleh and J. Dionne, Nano Lett., 2020, 20, 7655–7661 CrossRef CAS PubMed.
- S. Liu, H. Li, M. M. Hassan, J. Zhu, A. Wang, Q. Ouyang, M. Zareef and Q. Chen, Int. J. Food Microbiol., 2019, 304, 58–67 CrossRef CAS PubMed.
- C. Wang, B. Gu, Q. Liu, Y. Pang, R. Xiao and S. Wang, Int. J. Nanomed., 2018, 13, 1159–1178 CrossRef CAS PubMed.
- A. Rahman, S. Kang, W. Wang, Q. Huang, I. Kim and P. J. Vikesland, ACS Appl. Nano Mater., 2022, 5, 259–268 CrossRef CAS.
- Y. Yang, Z. Zhang, Y. He, Z. Wang, Y. Zhao and L. Sun, Sens. Actuators, B, 2018, 273, 600–609 CrossRef CAS.
- O. Žukovskaja, S. Agafilushkina, V. Sivakov, K. Weber, D. Cialla-May, L. Osminkina and J. Popp, Talanta, 2019, 202, 171–177 CrossRef PubMed.
- S. K. Srivastava, H. B. Hamo, A. Kushmaro, R. S. Marks, C. Gruner, B. Rauschenbach and I. Abdulhalim, Analyst, 2015, 140, 3201–3209 RSC.
- Y. Xu, P. Man, Y. Huo, T. Ning, C. Li, B. Man and C. Yang, Sens. Actuators, B, 2018, 265, 302–309 CrossRef CAS.
- M. L. Mekonnen, Y. A. Workie, W.-N. Su and B. J. Hwang, Sens. Actuators, B, 2021, 345, 130401 CrossRef CAS.
- D. Zhang, H. Pu, L. Huang and D.-W. Sun, Trends Food Sci. Technol., 2021, 109, 690–701 CrossRef CAS.
- A. Zhua, S. A. Ali, Y. Xu and Q. O. yang, Spectrochim. Acta, Part A, 2022, 270, 120814 CrossRef PubMed.
- F. Zeng, W. Duan, B. Zhu, T. Mu, L. Zhu, J. Guo and X. Ma, Anal. Chem., 2019, 91, 1064–1070 CrossRef CAS PubMed.
- Y. H. Ngo, D. Li, G. P. Simon and G. Garnier, Langmuir, 2012, 28, 8782–8790 CrossRef CAS PubMed.
- https://doi.org/10.1016/j.talanta.2014.04.066.
- M. Verma, T. K. Naqvi, S. K. Tripathi, M. M. Kulkarni and P. K. Dwivedi, Environ. Technol. Innovation, 2021, 24, 102033 CrossRef CAS.
- W. Wei and Q. Huang, Spectrochim. Acta, Part A, 2017, 179, 211–215 CrossRef CAS PubMed.
- A. Zhu, T. Jiao, S. Ali, Y. Xu, Q. Ouyang and Q. Chen, Anal. Chem., 2021, 93, 9788–9796 CrossRef CAS PubMed.
- R. Wang, L. Zhang, S. Zou and H. Zhang, Microchem. J., 2019, 150, 104127 CrossRef CAS.
- P. Butmee, A. Samphao and G. Tumcharern, J. Hazard. Mater., 2022, 437, 129344 CrossRef CAS PubMed.
- A. Sivanesan, E. Witkowska, W. Adamkiewicz, L. Dziewit, A. Kaminska and J. Waluk, Analyst, 2014, 139, 1037–1043 RSC.
- L. Cui, D. Zhang, K. Yang, X. Zhang and Y. G. Zhu, Anal. Chem., 2019, 91, 15345–15354 CrossRef CAS PubMed.
- O. Prakash, S. Sil, T. Verma and S. Umapathy, J. Phys. Chem. C, 2019, 124, 861–869 CrossRef.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2ra05715h |
| This journal is © The Royal Society of Chemistry 2023 |