Open Access Article
Open Access ArticleAFM evaluation of a humanized recombinant antibody affecting C. auris cell wall and stability†
Tania Vanzolini *a,
Tomas Di Mambrob,
Mauro Magnani
*a,
Tomas Di Mambrob,
Mauro Magnani a and
Michele Menotta
a and
Michele Menotta *a
*a
aDepartment of Biomolecular Sciences, University of Urbino Carlo Bo, Via Saffi 2, 61029 Urbino, Italy. E-mail: michele.menotta@uniurb.it; t.vanzolini@campus.uniurb.it
bDiatheva s.r.l., Via Sant'Anna 131/135, 61030, Cartoceto, Italy
First published on 20th February 2023
Abstract
Fungal infections are increasingly impacting on the health of the population and particularly on subjects with a compromised immune system. The resistance phenomenon and the rise of new species carrying sometimes intrinsic and multi-drug resistance to the most commonly used antifungal drugs are greatly concerning healthcare organizations. As a result of this situation, there is growing interest in the development of therapeutic agents against pathogenic fungi. In particular, the Candida genus is responsible for severe life-threatening infections and among its species, C. auris is considered an urgent threat by the Center for Disease Control and Prevention, and is one of the three leading causes of morbidity and mortality worldwide. H5K1 is a humanized monoclonal antibody (hmAb) that selectively binds to β-1,3-glucans, vital components of the fungal cell wall. It has been previously demonstrated that it is active against Candida species, especially against C. auris, reaching its greatest potential when combined with commercially available antifungal drugs. Here we used atomic force microscopy (AFM) to assess the effects of H5K1, alone and in combination with fluconazole, caspofungin and amphotericin B, on C. auris cells. Through an extensive exploration we found that H5K1 has a significant role in the perturbation and remodeling of the fungal cell wall that is reflected in the loss of whole cell integrity. Moreover, it contributes substantially to the alterations in terms of chemical composition, stiffness and roughness induced specifically by caspofungin and amphotericin B. In addition to this, we demonstrated that AFM is a valuable technique to evaluate drug–microorganism interaction.
Introduction
Fungi are slowly becoming a real plague amongst microbiological infections. They represent a great public health issue especially considering (I) the limited drug arsenal, (II) the rapid spread of resistance to commercially available antifungal drugs, (III) the impossibility to excessively increase the frequency or the number of the doses of the administered drugs to preserve patients' safety, (IV) the rise of new dangerous pathogens that sometimes carry intrinsic multi-drug resistance and (V) the broadening of the possible patients due to the current wide use of immunomodulators.1 In particular, candidemia is one of the most common systemic infections worldwide with an incidence of 7/100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 people per year in the US2 and 3.88/100
000 people per year in the US2 and 3.88/100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 in Europe.3 Invasive candidiasis is associated with a high mortality rate,4 and even if C. albicans is still the most common species, a mycological shift toward the non-albicans Candida (NAC) species has brought to light new risky species; among them C. auris was claimed as urgent global health threat by the Centers for Disease Control and Prevention.5 More than 90% of C. auris isolates have an innate resistance to one antifungal class, more than 30% to two classes and around 1% to all three major classes.6 These numbers must be also correlated to its rapid spread from 2009 in Japan to over 47 countries worldwide7–10 and with a mortality rate between 32 and 66%.9 In view of this alarming panorama, a high pressure is put on the study of new agents and strategies to be used in clinics and, among them, the employ of antibodies seems to be a great step forward. In this regard, H5K1 is a humanized monoclonal full-length antibody able to recognize and bind selectively.
000 in Europe.3 Invasive candidiasis is associated with a high mortality rate,4 and even if C. albicans is still the most common species, a mycological shift toward the non-albicans Candida (NAC) species has brought to light new risky species; among them C. auris was claimed as urgent global health threat by the Centers for Disease Control and Prevention.5 More than 90% of C. auris isolates have an innate resistance to one antifungal class, more than 30% to two classes and around 1% to all three major classes.6 These numbers must be also correlated to its rapid spread from 2009 in Japan to over 47 countries worldwide7–10 and with a mortality rate between 32 and 66%.9 In view of this alarming panorama, a high pressure is put on the study of new agents and strategies to be used in clinics and, among them, the employ of antibodies seems to be a great step forward. In this regard, H5K1 is a humanized monoclonal full-length antibody able to recognize and bind selectively.
β-1,3-Glucans which are fundamental and vital components of the cell wall of several pathogenic fungi.11 β-1,3-Glucans are recognized by the effectors of the immune system hence, they are often masked to limit their exposure.12 Indeed, C. auris cell wall composition is almost unique: compared to C. albicans, it consists of fewer cell wall proteins and enzymes involved in the cell wall remodeling but more structural lipids.13 In addition, they have similar levels of mannan and glucans, but chitin is more abundant in C. auris, thus explaining partially its higher natural low susceptibility to some antifungal classes.14 Nevertheless, it was previously demonstrated that H5K1 binding to the antigen on C. auris cell wall isn't negatively affected, furthermore H5K1 is effective against C. auris growth both alone and in combination with other commercially available antifungal drugs establishing an additivity with echinocandins and a synergy with amphotericin B.11 In this study we proved the versatility of AFM exploiting its ultrastructural analytical performances in the C. auris cell wall study. We investigated the morphological, mechanical, and chemical properties of C. auris cells and cell wall in normal condition and when treated with fluconazole, caspofungin, amphotericin B and H5K1 alone or in combination with the already mentioned antifungal drugs. We focused on the differences in the topographic and sub-topographic domains especially in terms of roughness, stiffness, elasticity, and molecule distribution.
Methods
Materials
C. auris DSM 21092 was purchased from Leibniz Institute DSMZ-German Collection of Microorganisms and Cell Cultures GmbH, Germany; the humanized monoclonal antibody H5K1 was produced by Takis s.r.l., Italy; caspofungin, fluconazole, amphotericin B, RPMI (powder) and MOPS were bought from Sigma-Aldrich, Germany, while paraformaldehyde 37% w/v was acquired from Carlo Erba reagents s.r.l., Italy.Sample preparation
From an overnight inoculum in RPMI + MOPS (0.165 M, pH 7), a C. auris suspension was washed and resuspended in phosphate buffered saline (PBS). 3 × 106 CFU were treated respectively with the hmAb H5K1, fluconazole, caspofungin and amphotericin B at sub-MIC concentrations (Fig. S1 in ESI†) and with the combinations of H5K1 with each of the mentioned antifungal drugs (1 h, 37 °C). PBS was used for the controls. Samples were washed three times with PBS before fixing with paraformaldehyde 4% (1 h, 4 °C). Finally, after washing again, the cells were resuspended in 18 MΩ water, layered on fresh cleaved muscovite mica and dried by nitrogen flow.AFM analyses
AFM analysis was carried out with an XE-100 Atomic Force microscope (PARK Systems Inc., Suwon, South Korea). The microscope was equipped with a 50 μm scanner controlled by the XEP 1.8.6 software. The XY stages and the Z scan were set in closed-loop manner and in high voltage mode.The speed scan was set between 0.2 Hz and 1.5 Hz. The cantilevers used in this study were Non-Contact High Resonant (NCHR) tips (spring constant between 35 and 44 N m−1) with a typical resonant frequency between 200 and 300 kHz. The instrument was set in true non-contact mode and topography, amplitude, error, and phase signals were acquired during the imaging.
Amplitude signal images were used for topographic subdomains contrast enhancement while the phase images were adopted for surface chemical characterization.15,16
The Force Modulation Microscopy (FMM) was achieved by using a NSC14 tip (5 N m−1) and the local nanomechanical properties were tested by amplitude and phase signals.
For the force/distance spectroscopy analysis (F/D) a calibrated NCHR tip (39.7 N m−1) was employed. Briefly, at least 4 points were randomly chosen in the middle of each cell and F/D assay was performed. The tip moved forward the samples for 1 mm at 10 nm s−1, while the retrace velocity was set at 50 nm s−1 with a force limit of 3 mN. The stiffness of the sample was determined from the slope of the force–separation curve in the contact region and reported as nanoNewton.
Statistical analyses
Data inferred by Park XEI software (PARK Systems Inc., Suwon, South Korea) were analyzed by GraphPad Prism. The statistical Kruskal–Wallis and uncorrected Dunn's test for multiple comparisons was adopted.Results and discussion
C. auris is a new life-threatening pathogenic yeast that gained sad interest for the intrinsic multi-/drug resistance of most strains. This low susceptibility to antifungals is likely due to a unique cell wall composition that is enriched in chitin in order to cover components otherwise easily recognized by the immune system and in particular by myeloid cells.12,17 Among the “hot-components” there are β-1,3-glucans which represents, with chitin, the cell wall backbone hence, are vital for the fungal survival.18 For these reasons and for the advantage of a single yeast-like morphotype,19,20 C. auris was chosen as a model to determine the nano- and micrometrical effects of the humanized monoclonal antibody (hmAb) H5K1, able to bind selectively β-1,3-glucans. In previous studies, we demonstrated that there is a synergistic or additive effect of H5K1 in combination with commercially available antifungal drugs. Thereby, we investigated the activity our antibody H5K1 not just alone but also in combination with antifungals belonging to three different classes, fluconazole (FLC), caspofungin (CAS) and amphotericin B (AMB). Fluconazole is one of the major representatives of the azole family and works inhibiting the fungal Cytochrome P450-dependent enzyme lanosterol 14-α-demethylase. This enzyme converts lanosterol to ergosterol by demethylation and FLC prevents the reaction causing an accumulation in the cell membrane of methylated sterols that arrest the fungal growth.21 Caspofungin is an echinocandin that blocks the synthesis of β-1,3-glucans by inhibiting in a non-competitive manner the enzyme β-1,3-glucan synthase. The cease production of β-1,3-glucans heavily affects the cell wall synthesis reducing the fungal growth but also alters the integrity of the cell wall that inevitably leads to the loss of mechanical strength and inability to resist to the intracellular osmotic pressure.22 Finally amphotericin B belongs to the polyene class and its binding to ergosterol of the fungal membrane creates transmembrane channels which dramatically disrupt the membrane integrity and change the permeability allowing the deadly leakage of intracellular components. The agents tested were never studied in AFM on C. auris hence, this study represents an exquisite opportunity to get a first look of their effects on this specie. Sub-MIC concentrations were used since our aim was to observe the outcomes on living cells. Following the methodology found in literature and often applied for yeasts,23–27 after the treatment we fixed and air-dried C. auris cells. Several evidences28–32 demonstrated that the analysis of the surface topography is not affected by the fixation, as the data obtained with live cells without fixation are comparable to that obtained from fixed cells. Furthermore, the use of the internal control sample guarantees the correctness of the data as, even if the absolute values extrapolated from the analyses could be somehow affected by the sample preparation, the relative correlations remain the same. Other methods for samples preparation had to be avoided for different reasons like the dangerousness of the specie (that prevent the use of living cells in our facilities) and the optimal performing conditions for the analyses as the use of liquid medium (that otherwise would need a stable immobilization process that could negatively affect the samples) or the substrate-attaching techniques like that with polylysine (suitable for force measurements but considered not adequate for surface analyses since a lot of morphological information are lost31).Image-surface profile parameters chosen for topographic and sub-topographic analyses are the main descriptors used to delineate the cell morphology and the surface.33,34 Together with roughness parameters, they are sensitive indicators of cell's health state.35 They are suitable for monitoring the damages caused by specific agents on biological samples36 and, in the present work, the effects of antifungal compounds on C. auris cells. To have a complete sight of the observed phenomena, force modulation microscopy was adopted to probe the nanomechanical properties alongside the topographic data37 and finally the cellular stiffness was evaluated to investigate a larger structure stiffness and to gain insight of the integrity and of the mechanics of the cells.38
hmAb alone and in combination with drugs alters the surface morphology producing sharp and frequent vertical variations
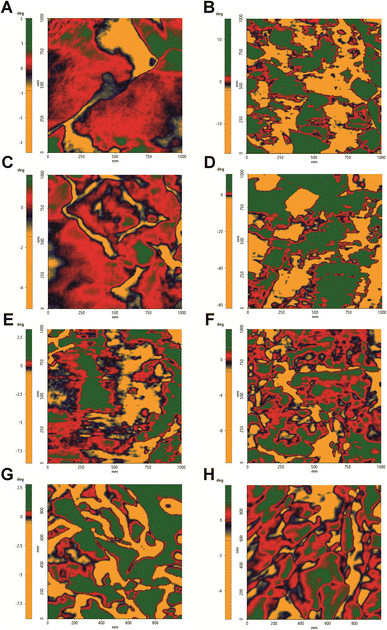
From the preliminary analysis of the 1 μm × 1 μm topographic images, there are evidence of the cell wall perturbation due to the presence of topographic alterations, leading to a more inhomogeneous surface in treated samples compared to the control cells (Fig. 1). The imaging was performed on single cells with altered morphology, and it revealed a smooth surface in the control in contrast to the bumpy aspect for all the treatments. The smooth and homogeneous surface of the control is consistent with the presence of the outer layer rich in mannans, mannoproteins and chitin. The irregular surfaces of the treated cells are characterized by deformations which are further accentuated in the hmAb H5K1 combinations. FLC causes minor irregularities (blebs) that we suggest are directly correlated with the accumulation of methylated sterols due to the inhibition of the lanosterol 14-α-demethylase. The presence of these molecules negatively affects not just the structure of the plasma membrane but also its functionality hence, as the cell volume and morphology are regulated by a fine-tuned ion balance, a disruption of this equilibrium can lead to changes such as cell swelling, as stated by Wonjong et al.21 who noticed a similar effect on C. albicans. CAS, both alone and in combination, produces the most severe and extensive effect. This is in agreement with its activity that, differently from the other two drugs, affects directly the fungal cell wall. The reorganization of the cell wall architecture and the cell swelling are the results of a compromised cell wall synthesis caused by a reduction of β-1,3-glucans production which normally have the critical role of maintaining cell shape, mechanical strength and resistance to osmotic pressure.39,40 Moreover, it is known that caspofungin triggers in Candida cells a cascade of intracellular responses in order to protect from further damages. Among these responses there are the remodeling of the cell wall and the redistribution of certain molecules between the plasma membrane and the cell wall. Badrane et al. and Rueda et al. demonstrated a mislocalization of septin and phosphatidylinositol-(4,5)-bisphosphate [PI(4,5)P2] in plasma membrane, the presence of cells wall proteins in the plasma membrane and increased chitin synthesis and deposition in both cell membrane and wall.22,41–43 These fundamental changes are likely to be responsible of not just the topographic deformation but also of the variation of the mechanical properties. AMB visibly perturbates C. auris surface despite it directly affects the plasma membrane and just indirectly the cell wall. When AMB interacts with lipids, preferably ergosterol but also other sterols and phospholipids, it produces pores that lead to the loss of the normal permeability.44 Kristanc et al. and Zemljič Jokhadar et al. observed the formation of blebs followed by their coalescence into “cell vesicles” as a result of nystatin treatment. Authors explained this behavior by a first cell swelling caused by the influx of water due to building up of the osmotic pressure in the intracellular environment (increase of Na+ and Cl− concentrations and decrease of that of K+), followed by a volume reduction at the reach of the critical membrane tension that leads to the non-selective extrusion of a portion of the cell content.45,46 We are likely to suppose that a similar effect can be attributed also to AMB, thus clearly explaining the formation of blebs on the fungal cell surface. Furthermore AMB triggers the formation of ROS which produce an oxidative damage at different levels: (i) the high intracellular production can arise an oxidative burst mediating the killing of fungal cells;47 (ii) ROS oxidize lipids of the membrane further contributing to the structural and functional destabilization;48 (iii) ROS can affect different vital functions such as the activity of membrane proteins, the pheromone signaling, the vacuole fusion, the endocytosis etc. giving additional destabilization to the cells.49–51 We believe that these primary and secondary effects noticed in different fungal and mammalian cells can be transferred also to C. auris cells allowing us to suggest a rational description of the biological and microbiological events occurring during treatments.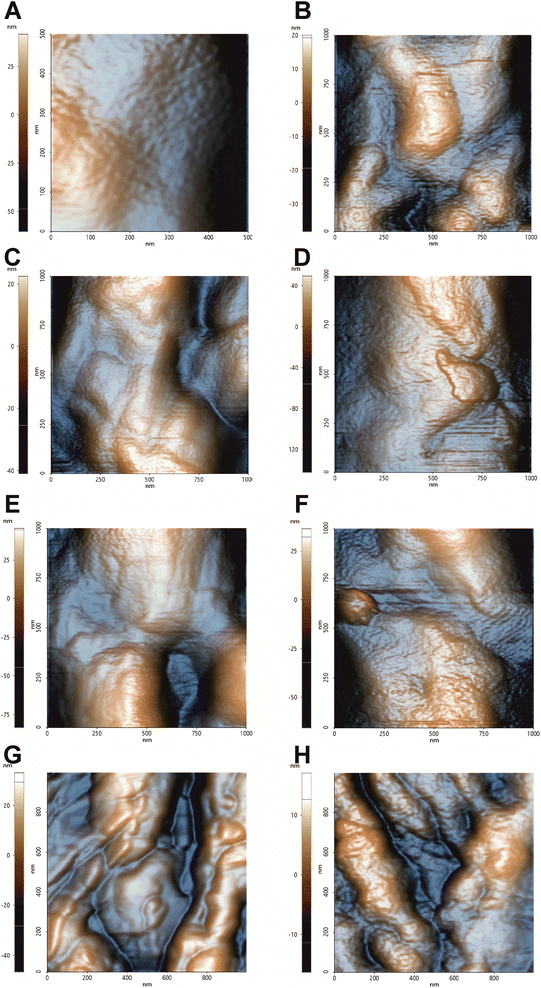 | ||
| Fig. 1 Topographic images by NCM AFM of C. auris cell surface. 1 μm × 1 μm topographic analysis conducted in air to better detect morphological changes on the surface. (A) Control, (B) treatment with hmAb H5K1, (C) treatment with fluconazole, (D) treatment with the combination of hmAb H5K1 and fluconazole, (E) treatment with amphotericin B, (F) treatment with the combination hmAb H5K1 and amphotericin B, (G) treatment with caspofungin, (H) treatment with the combination of hmAb H5K1 and caspofungin. Matching data are illustrated in Fig. 2. | ||
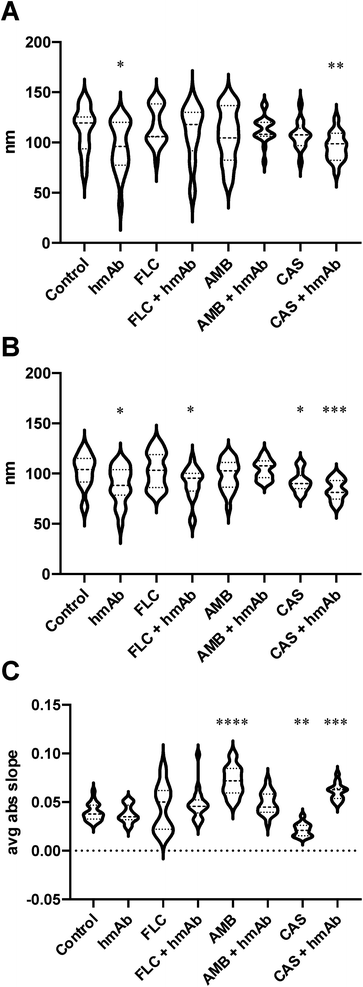
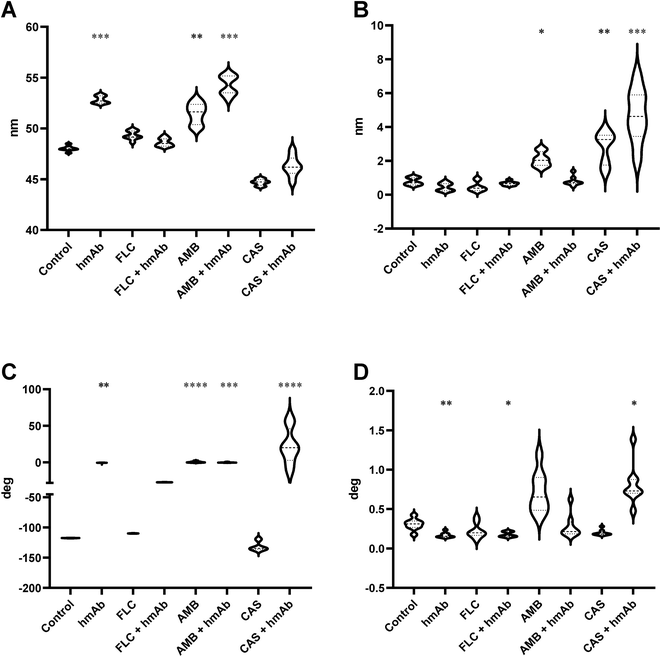
In order to numerically describe these alterations composed by crests and hollows, the mean spacing of profile irregularities (Sm) and the average wavelength of the profile (la) were evaluated as spatial parameters, while the average absolute slope (Da) was estimated as hybrid factor (Fig. 2). For what concerns the Sm (Fig. 2A), the calculated average spacing for the control was ∼113 nm (Table 1) while for hmAb and CAS + hmAb treated cells, the Sm values were lower and statistically significant, thus meaning a higher frequency of topographical changes. This outcome was also confirmed by using la parameter (Fig. 2B). In fact, hmAb treated sample and hmAb combined respectively with FLC and CAS produced a statistically significant decrement of la compared to the control (Table 1). The average absolute slope (Da) (Fig. 2C) was considered to characterize the magnitude of the topographical variations. By this parameter, statistical differences were noted in AMB, CAS and CAS + hmAb treated samples. Taken together these data suggests that changes in surface roughness were induced by hmAb H5K1 alone or in combination with other drugs. In the AMB treated sample the Da distribution showed higher values probably indicating the presence of sharper vertical variations possibly caused by the capacity of amphotericin B's aggregates to produce digging after their embedding in surface rich in ergosterol.52 This digging appears to be just the prelude to the typical pores produced by amphotericin B.52 Moreover, pores dimensions and depth, which affect Sm, Da and la, are influenced by different factors such as the number of AMB monomers that aggregate which ranges between 4 and 12 (ref. 53) giving holes from 1, 6 to 16 nm diameter,52,54 the presence of zones sterol-rich called rafts with affect the size of pores55 or in case of phospholipids, the length of the chains of the fatty acyl groups that control the depth,56 and the thickness of the membrane57 that can change not just among fungal species but also among strains. The combination of hmAb with CAS produces a similar trend, hence we are likely to suppose that the enhancement of hmAb H5K1 on caspofungin can lead to an effect comparable to that of amphotericin B on the surface or to an indirect effect that should be better understood. Of note the statistically significant decrease of Da in presence of caspofungin alone, that we speculate could be due to the unmasking activity of caspofungin24,39,58 to the simultaneous decrease of β-glucans content and increase of chitin and mannans or to both of effects already seen in other Candida species or other yeasts.40,59 We suppose that the formation of blebs and the correlated increase of roughness can be considered as adaptive responses of the cells to the drugs which are progressively disintegrating the cell wall. The concomitant presence of hmAb perhaps impedes this mechanism forcing a cellular reaction that reflects in the alteration of the whole cell.
| Control | hmAb | FLC | FLC + hmAb | AMB | AMB + hmAb | CAS | CAS + hmAb | ||
|---|---|---|---|---|---|---|---|---|---|
| Sm | Mean | 113 | 95.35 | 116.6 | 109.1 | 107.4 | 113.1 | 107.4 | 95.57 |
| SEM | 4.231 | 5.703 | 4.42 | 6.562 | 6.017 | 3.011 | 3.893 | 3.561 | |
| λa | Mean | 102.4 | 88.28 | 103.8 | 89.8 | 98.21 | 104.3 | 91.87 | 83.9 |
| SEM | 3.86 | 4.187 | 4.013 | 3.955 | 3.442 | 2.214 | 2.863 | 2.669 | |
| Δa | Mean | 0.03957 | 0.03766 | 0.04411 | 0.04804 | 0.07142 | 0.0491 | 0.02152 | 0.06089 |
| SEM | 0.002013 | 0.002065 | 0.005429 | 0.003777 | 0.003403 | 0.002667 | 0.001888 | 0.002214 | |
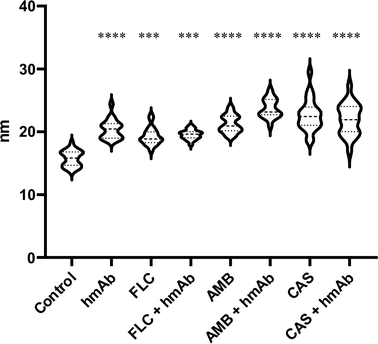
| Amp. λa | Mean | 15.76 | 20.41 | 19.28 | 19.55 | 21.24 | 23.72 | 22.68 | 22 |
| SEM | 0.226 | 0.3403 | 0.3008 | 0.1623 | 0.2981 | 0.3503 | 0.5463 | 0.5393 | |
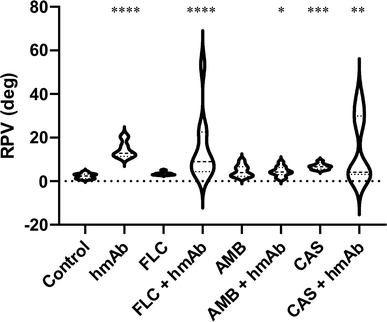
| Phase (deg) | Mean | 2.351 | 14.47 | 3.398 | 14.96 | 4.678 | 4.518 | 6.706 | 14.66 |
| SEM | 0.3692 | 1.141 | 0.2260 | 4.825 | 0.8359 | 0.7139 | 0.4113 | 4.757 | |
| FMM amp. (nm) median | Mean | 48.02 | 52.83 | 49.33 | 48.62 | 51.47 | 54.25 | 44.70 | 46.29 |
| SEM | 0.09034 | 0.1310 | 0.1500 | 0.1401 | 0.3634 | 0.3379 | 0.1075 | 0.3530 | |
| RPV amp. (nm) FMM | Mean | 0.7805 | 0.4575 | 0.4768 | 0.6619 | 2.087 | 0.8000 | 2.793 | 4.668 |
| SEM | 0.07615 | 0.08226 | 0.1023 | 0.04104 | 0.1489 | 0.09742 | 0.3344 | 0.5578 | |
| FMM phase (deg) median | Mean | −117.5 | −0.7914 | −109.8 | −27.14 | 0.5188 | −0.1118 | −131.3 | 22.06 |
| SEM | 0.06237 | 0.3164 | 0.05930 | 0.02724 | 0.3150 | 0.1230 | 2.678 | 8.552 | |
| RPV phase (deg) FMM | Mean | 0.3060 | 0.1678 | 0.2251 | 0.1769 | 0.7158 | 0.2786 | 0.1989 | 0.8104 |
| SEM | 0.02552 | 0.01144 | 0.03402 | 0.01159 | 0.09190 | 0.05332 | 0.01286 | 009![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 302 302 |
|
| Stiffness (nN) | Mean | 93.91 | 90.40 | 64.27 | 77.73 | 65.45 | 74.38 | 94.21 | 87.09 |
| SEM | 1.905 | 1.015 | 2.445 | 1.204 | 2.666 | 1.664 | 1.011 | 0.7766 |
To further confirm the changes in the surface topography, the images obtained by tip amplitude signal were used to highlight sub-nanometric structures (Fig. 3). As illustrated in Fig. 3, the filtered image obtained by this technique showed a dense net of “fibers”. In the control, these segmentations were denser while in all the treated samples, domains underwent alterations that became even more slacken in the presence of hmAb. Again, by measuring the average wavelength of the profile belonging to the amplitude signal, all the treated samples differed from the control with increased la of the amplitude signal (amp. la in Table 1) (Fig. 4). This evidence suggests that there are differences in the molecule distribution on the surface of the cells as we previously hypothesized looking at the topographic images and at the literature, and, in this context, hmAb further affects the canonical ultrastructural organization. As reported in literature and stated before, CAS mislocalizes and increases some components both in plasma membrane and cell wall41,43 while AMB seems to initiate an ergosterol redistribution and produces a relocation of other components changing the lipid environment and increasing the distance between molecules.60 This can explain the slacken aspects of the sub-topographic domains and, not by chance, the fact that at this level, the higher visible effect is observed for AMB which affect primarily the plasma membrane and not for CAS that, to the other hand, firstly act on the cell wall (Fig. 3). Since the λa increased in all treated samples (Table 1), it is possible to suppose that the cell wall components were displaced away by the treatments and, considering the high influence of hmAb both alone and in combination, we are confident stating that among the components, β-1,3-glucans are for sure affected.
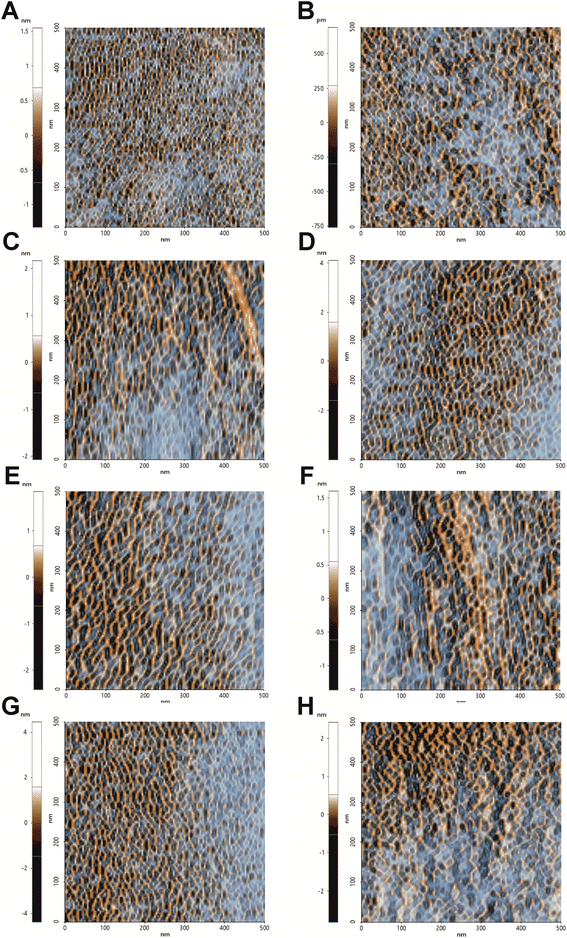 | ||
| Fig. 3 Sub-nanometric structures of C. auris cell surface. 0.5 μm × 0.5 μm analysis of the surface sub-domains. Images were obtained from FFT filtered amplitude signal analysis of images performed in NCM. This signal processing allows to highlight and contrast the topography-dependent subdomains. (A) Control, (B) treatment with hmAb H5K1, (C) treatment with fluconazole, (D) treatment with the combination of hmAb H5K1 and fluconazole, (E) treatment with amphotericin B, (F) treatment with the combination hmAb H5K1 and amphotericin B, (G) treatment with caspofungin, (H) treatment with the combination of hmAb H5K1 and caspofungin. Matching data are illustrated in Fig. 4. | ||
The phase roughness is significantly increased by hmAb and its combination with the drugs
The surface composition alteration was also assayed by non-contact mode (NCM) phase imaging (Fig. 5). By this technique it is possible to monitor surface chemical properties as the roughness of the phase signal was demonstrated to be a parameter dependent on the chemical and physical properties.61 Control sample showed a homogeneous distribution of the phase signal whereas all treated cells exhibited a variegate phase signal distribution, quantitatively measured as peak-to-valley mean-height roughness (RPV roughness) (Fig. 6 and Table 1). hmAb H5K1 provided the major contribution to the roughness, and this is highlighted by the statistically significant difference in RPV of hmAb monotreatment and of its combinations compared to the control. Fluconazole and amphotericin B don't cause any alteration on the surface chemistry while caspofungin was statistically different from the control and this is consistent with the results obtained by Hasim et al. that reported an increase of roughness in C. albicans treated with caspofungin probably due to the β-1,3-glucans unmasking which produce greater frequencies of interaction39,58 or by the great exposure induced by CAS of the adhesion protein Als1 which affects several biophysical properties such as hydrophobicity, adhesion, elasticity and conformation and extension of the surface proteins.59 Interesting is the different distribution of the RPV values: those of control and of the antifungals' monotreatments are more homogeneous while in the presence of hmAb H5K1 alone and in combination with fluconazole or caspofungin the phase signal roughness is notably increased, indicating an extremely widely distribution of variegate molecules on the surface. These data indicates that fluconazole and caspofungin alter the surface composition and unravel a reorganization of the cell wall especially when hmAb is present. AMB produces pores on the surface, hence in the combination AMB + hmAb, we consider the possibility that hmAb may wedge into the pores resulting less influential in altering the chemical distribution. In fact, the displacement of the combination is not considerably different from AMB monotreatment meaning that perhaps, in this case, hmAb effect was not probed to the core. In addition to that, we believe that in control sample the molecules examined are mainly those more exposed in the outer layers i.e. chitin, mannans, glucans and cell wall proteins, while in the samples with hmAb, besides these entities, the hmAb itself and the molecules exposed as effect of the perturbation of the antibody were registered as well. This can also explain the wider and varied molecule distribution on the surface.hmAb affects the nanomechanical properties of the cell wall altering the local stiffness and elasticity and their respective variations
As the chemical distribution influences the mechanics of the cell wall, the surface alterations induced by the treatments were also assayed by a physical perspective. The AFM force modulation mode (FMM) was employed to investigate the nanomechanical properties by the FMM amplitude and the FMM phase signals to obtain information about the superficial and local stiffness and elasticity (Fig. 7). The middle amplitude values are dependent on analyzed regions and are little informative. Anyway, the median of the amplitude signal was higher in the presence of AMB and hmAb H5K1 alone and in combination with AMB (Fig. 8A) indicating stiffer surfaces compared to the control. As it is difficult to fully understand how fungal cell stiffness is regulated, we can just agree with Kyung Sook et al. saying that probably is due to the progressive cell death caused by the fungal agent/s.62 No differences were recorded in the other treated samples. Samples treated with CAS and CAS + hmAb underwent a decrease of the FMM amplitude signal but without resulting statistically different compared to the control. This data is consistent with similar analyses performed of different Candida species such as in C. albicans where the treatment with caspofungin caused a softening not only of the cell wall but of the entire cell because of the decrease of the intracellular turgor pressure (in agreement with the cell swelling seen in topographic analyses) and the formation of osmotically fragile cells.59 The FMM amplitude roughness revealed that nanomechanical proprieties dependent on molecules arrangement were altered in samples treated with AMB, CAS and CAS + hmAb (Fig. 8B) meaning the presence of regions extremely hard and extremely soft (for AMB we can suppose it is due to the sterol-rich rafts). The wide variation reflects the alterations induced on the molecules on the surface thus explaining also the huge difference between CAS and CAS + hmAb compared to FLC and FLC + hmAb. For what concerns AMB + hmAb, the variation is reduced probably because the antibody binds to its antigen producing a general stiffening on the cell wall especially in proportion to AMB monotreatment. At the same time, elasticity properties were computed by the analysis of the phase signal and while AMB and AMB + hmAb treated cells are coherent with the structural rigidity (a stiffer region can also be less elastic), hmAb induces a great increase in the cell wall elasticity both alone and in combination with FLC and CAS. In particular, the effect of the combination with CAS is consistent with the inhibition of the β-(1,3)-glucan synthase by caspofungin that reduces the number of antigens for hmAb H5K1 and the relative amount of cross-linking. Again, the absolute measures depend on the zone analyzed, therefore the FMM phase roughness was observed to evaluate the spatial variation of the FMM phase signal medians. HmAb treated sample is less variegate and more homogeneous and the same occurs for its combination with FLC and AMB when compared to the antifungal drugs used alone. For CAS the effect is the opposite, but this is exquisitely in accordance with the previous data (Fig. 8B and C) in which the reduced presence of β-(1,3)-glucan limits the bonds of H5K1 hence decreases its stiffening effect.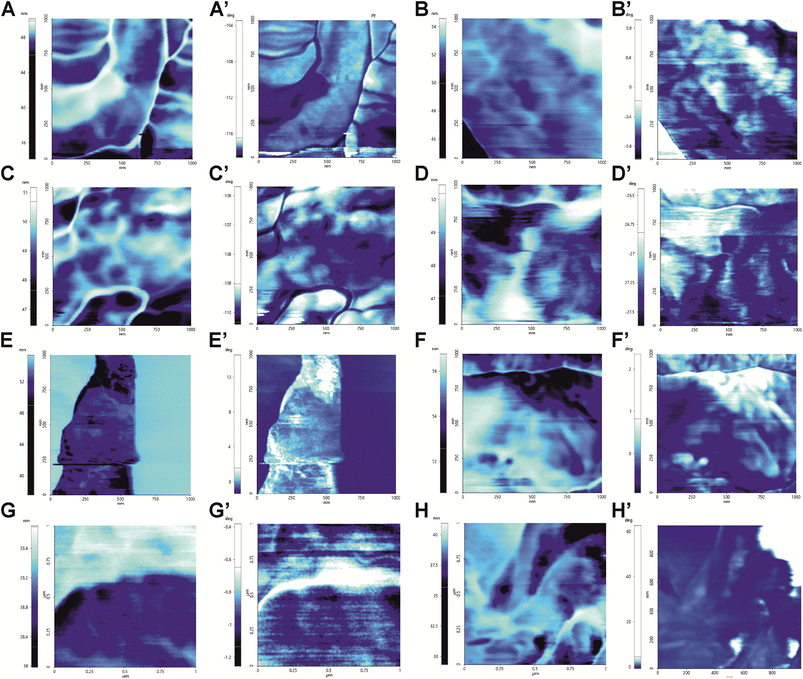 | ||
| Fig. 7 Force Modulation Mode (FMM) images obtained by amplitude (letters) and phase signals of the same wave function (marked letters). Images provide information about the local nanomechanics proprieties of C. auris surface. FMM amplitude describes the stiffness of the area: in bright the stiffest zones and in dark the softest zones. FMM Phase describes the elasticity of the analyzed area: in bright the most elastic zones while in dark the least elastic. (A/A′) Control, (B/B′) treatment with hmAb H5K1, (C/C′) treatment with fluconazole, (D/D′) treatment with the combination of hmAb H5K1 and fluconazole, (E/E′) treatment with amphotericin B, (F/F′) treatment with the combination hmAb H5K1 and amphotericin B, (G/G′) treatment with caspofungin, (H/H′) treatment with the combination of hmAb H5K1 and caspofungin. Signal analyses are reported in Fig. 8. | ||
hmAb in combination causes a global stiffening that reflects the frailty of the whole cells
In order to assess the effects of the treatments not only considering the local mechanical features but also a larger region of the cells, force–distance curves were performed randomly in different superficial spots of several cell. By force–distance curves gained through a physical deformation, a deeper cellular stiffness depending on the whole intracellular scaffold can be acquired. In line with what has been seen locally, fluconazole, amphotericin B, and the combination of hmAb with all the antifungal drugs presented decremented stiffness values compared to the control sample (Table 1) meaning that the sub surface structural integrity of the cell wall was altered. Easy-deformable zones on the surface reflect the weakening and the frailty of the cell (Fig. 9).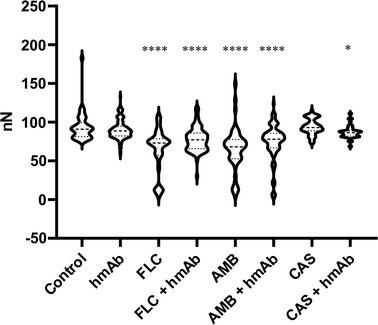 | ||
| Fig. 9 Cell Stiffness expressed an nN obtained by F/D curves analysis. Almost all treatments were able to reduce the cell strength. | ||
Conclusions
The drug resistance phenomenon and the lack of new therapeutic agents highlight the urgency in finding novel effective strategies for the treatment of fungal infections. The humanized monoclonal antibody H5K1 is an exquisite example of a step forward towards the application of biotechnological entities to the microbiological world.Even without the presence of the immune system apparatus, hmAb H5K1, binding selectively β-1,3-glucans of the fungal cell wall,11 can affect the cell integrity resulting as an enhancer especially for drugs like caspofungin and amphotericin B.
Summarizing our results, we demonstrated that: (I) H5K1 disturbs C. auris topography producing irregularities and bumpy aspects especially when combined with caspofungin and that all the treatments cause a substantial increase of the topographic roughness. (II) The perturbation extends also to the subdomains which appear more slacken and altered in terms of structural organization. (III) hmAb alone but also caspofungin, amphotericin B and their combination with the antibody cause changes in the chemical distribution of the molecules on the surface. (IV) The presence of hmAb alters the local stiffness that results inhomogeneous compared to the control. (V) hmAb combined with the drugs affects both the local and the global nanomechanical properties influencing the structural integrity of the cell that showed weak and frail when probed with force–distance curves.
In conclusion, this study wants to show the perturbating effects produced on C. auris by hmAb H5K1 alone and in combination with commercially available antifungal drugs but also to demonstrate that AFM is a valuable tool for the investigation of microorganisms and of their morphological, structural, and biochemical changes induced by drug treatments.
Author contributions
Conceptualization: Tania Vanzolini, Tomas Di Mambro, Mauro Magnani, Michele Menotta. Data curation: Tania Vanzolini, Michele Menotta. Formal analysis: Tania Vanzolini, Michele Menotta. Funding acquisition: Mauro Magnani. Investigation: Tania Vanzolini, Michele Menotta. Methodology: Tania Vanzolini, Tomas Di Mambro, Michele Menotta. Project administration: Mauro Magnani. Resources: Mauro Magnani. Supervision: Mauro Magnani. Validation: Tania Vanzolini, Michele Menotta. Visualization: Tania Vanzolini. Writing – original draft: Tania Vanzolini. Writing – review & editing: Tania Vanzolini, Tomas Di Mambro, Mauro Magnani, Michele Menotta.Conflicts of interest
Diatheva s.r.l. supported the study but was not involved in carrying out or managing the investigation, nor was it involved in the analysis and interpretation of data, and in the preparation of the manuscript. T. D. M. is an employee of Diatheva s.r.l., M. Magnani holds shares in Diatheva s.r.l. This does not alter our adherence to RSC policies on sharing data and materials. All the other authors do not have any conflict of interest.Acknowledgements
We acknowledge for the support DIATHEVA s.r.l., ARS01_00876, “BIO-D” (Cod. ARS01_00876 dal titolo “BIO-D – Sviluppo di biomarcatori diagnostici per la medicina di precisione e la terapia personalizzata” – CUP B32F20000270005 – RNA-COR 3200158 finanziato nell'ambito del programma PON ≪R&I≫ 2014–2020 – Azione II – OS 1.b) and European Union POR MARCHE FESR 2014/2020. Asse 1, OS 2, Azione 2.1 - Intervento 2.1.1 – Sostegno allo sviluppo di una piattaforma di ricerca collaborativa negli ambiti della specializzazione intelligente. Thematic Area: "Medicina personalizzata, farmaci e nuovi approcci terapeutici". Project acronym: Marche BioBank www.marchebiobank.it. The content of the paper is the sole responsibility of the authors and can under no circumstances be regarded as reflecting the position of the European Union and/or Marche Region authorities.References
- D. Z. P. Friedman and I. S. Schwartz, J. Fungi, 2019, 5(3), 67–85 CrossRef CAS PubMed.
- P. Koehler, M. Stecher, O. A. Cornely, D. Koehler, M. J. G. T. Vehreschild, J. Bohlius, H. Wisplinghoff and J. J. Vehreschild, Clin. Microbiol. Infect., 2019, 25, 1200–1212 CrossRef CAS PubMed.
- P. G. Pappas, M. S. Lionakis, M. C. Arendrup, L. Ostrosky-Zeichner and B. J. Kullberg, Nat. Rev. Dis. Primer, 2018, 4, 18026 CrossRef PubMed.
- K. Kainz, M. A. Bauer, F. Madeo and D. Carmona-Gutierrez, Microb. Cell, 2020, 7, 143–145 CrossRef PubMed.
- Candida auris|Candida auris|Fungal Diseases|CDC, https://www.cdc.gov/fungal/candida-auris/index.html, accessed October 31, 2021 Search PubMed.
- CDC, Antibiotic-resistant Germs, https://www.cdc.gov/drugresistance/biggest-threats.html, accessed October 27, 2021 Search PubMed.
- K. Satoh, K. Makimura, Y. Hasumi, Y. Nishiyama, K. Uchida and H. Yamaguchi, Microbiol. Immunol., 2009, 53, 41–44 CrossRef CAS PubMed.
- Tracking Candida auris|Candida auris|Fungal Diseases|CDC, https://www.cdc.gov/fungal/candida-auris/tracking-c-auris.html, accessed October 10, 2021 Search PubMed.
- A. W. de Jong and F. Hagen, Mycopathologia, 2019, 184, 353–365 CrossRef PubMed.
- H. Heaney, J. Laing, L. Paterson, A. Walker, N. Gow, E. Johnson, D. MacCallum and A. Brown, Med. Mycol., 2020, 58(6), 744–755 CrossRef CAS PubMed.
- T. Di Mambro, T. Vanzolini, P. Bruscolini, S. Perez-Gaviro, E. Marra, G. Roscilli, M. Bianchi, A. Fraternale, G. F. Schiavano, B. Canonico and M. Magnani, Sci. Rep., 2021, 11, 19500 CrossRef CAS PubMed.
- N. A. R. Gow, F. L. van de Veerdonk, A. J. P. Brown and M. G. Netea, Nat. Rev. Microbiol., 2011, 10, 112–122 CrossRef PubMed.
- D. Zamith-Miranda, H. M. Heyman, L. G. Cleare, S. P. Couvillion, G. C. Clair, E. L. Bredeweg, A. Gacser, L. Nimrichter, E. S. Nakayasu and J. D. Nosanchuk, mSystems, 2019, 4, 00257 CrossRef PubMed.
- M. J. Navarro-Arias, M. J. Hernández-Chávez, L. C. García-Carnero, D. G. Amezcua-Hernández, N. E. Lozoya-Pérez, E. Estrada-Mata, I. Martínez-Duncker, B. Franco and H. M. Mora-Montes, Infect. Drug Resist., 2019, 12, 783–794 CrossRef CAS PubMed.
- H. Schönherr, V. Paraschiv, S. Zapotoczny, M. Crego-Calama, P. Timmerman, C. W. Frank, G. J. Vancso and D. N. Reinhoudt, Proc. Natl. Acad. Sci., 2002, 99, 5024–5027 CrossRef PubMed.
- R. D. Bellis, M. P. Piacentini, M. A. Meli, M. Mattioli, M. Menotta, M. Mari, L. Valentini, L. Palomba, D. Desideri and L. Chiarantini, PLoS One, 2019, 14, e0218734 CrossRef PubMed.
- G. D. Brown, Nat. Rev. Immunol., 2006, 6, 33–43 CrossRef CAS PubMed.
- A. Yoshimi, K. Miyazawa and K. Abe, J. Fungi, 2017, 3(4), 63–83, DOI:10.3390/jof3040063.
- G. Bravo Ruiz, Z. K. Ross, N. A. R. Gow and A. Lorenz, mSphere, 2020, 5(2) DOI:10.1128/mSphere.00151-20.
- X. Wang, J. Bing, Q. Zheng, F. Zhang, J. Liu, H. Yue, L. Tao, H. Du, Y. Wang, H. Wang and G. Huang, Emerging Microbes Infect., 2018, 7, 93 Search PubMed.
- W. Lee and D. G. Lee, Microbiology, 2018, 164, 194–204 CrossRef CAS PubMed.
- V. Letscher-Bru and R. Herbrecht, J. Antimicrob. Chemother., 2003, 51, 513–521 CrossRef CAS PubMed.
- E. A. Corbin, F. Kong, C. T. Lim, W. P. King and R. Bashir, Lab Chip, 2015, 15, 839–847 RSC.
- F. Quilès, I. Accoceberry, C. Couzigou, G. Francius, T. Noël and S. El-Kirat-Chatel, Nanoscale, 2017, 9, 13731–13738 RSC.
- S. Proa-Coronado, C. Séverac, A. Martinez-Rivas and E. Dague, Nanoscale Horiz., 2019, 5, 131–138 RSC.
- M. Jalal, M. A. Ansari, A. K. Shukla, S. G. Ali, H. M. Khan, R. Pal, J. Alam and S. S. Cameotra, RSC Adv., 2016, 6, 107577–107590 RSC.
- M. Takenaka, Y. Miyachi, J. Ishii, C. Ogino and A. Kondo, Nanoscale, 2015, 7, 4956–4963 RSC.
- E. Gibbs, J. Hsu, K. Barth and J. W. Goss, Yeast, 2021, 38, 480–492 CrossRef CAS PubMed.
- M. Osumi, M. Sato, S. A. Ishijima, M. Konomi, T. Takagi and H. Yaguchi, Fungal Genet. Biol., 1998, 24, 178–206 CrossRef CAS PubMed.
- M. Sipiczki, in Yeast Cytokinesis: Methods and Protocols, ed. A. Sanchez-Diaz and P. Perez, Springer, New York, NY, 2016, pp. 97–111 Search PubMed.
- A. V. Bolshakova, O. I. Kiselyova and I. V. Yaminsky, Biotechnol. Prog., 2004, 20, 1615–1622 CrossRef CAS PubMed.
- I. Yu. Sokolov, M. Firtel and G. S. Henderson, J. Vac. Sci. Technol., A, 1996, 14, 674–678 CrossRef CAS.
- N. Gavara, Microsc. Res. Tech., 2017, 80, 75–84 CrossRef PubMed.
- M. Raposo, Q. Ferreira and P. Ribeiro, Mod. Res. Educ. Top Microsc., 2007, 1, 1 Search PubMed.
- P. D. Antonio, M. Lasalvia, G. Perna and V. Capozzi, Biochim. Biophys. Acta, 2012, 1818, 3141–3148 CrossRef PubMed.
- A. da Silva Junior and O. Teschke, World J. Microbiol. Biotechnol., 2005, 21, 1103–1110 CrossRef CAS.
- W. Arnold, in Encyclopedia of Nanotechnology, ed. B. Bhushan, Springer Netherlands, Dordrecht, 2014, pp. 1–11 Search PubMed.
- K. Hayashi and M. Iwata, J. Mech. Behav. Biomed. Mater., 2015, 49, 105–111 CrossRef PubMed.
- S. Hasim, D. P. Allison, S. T. Retterer, A. Hopke, R. T. Wheeler, M. J. Doktycz and T. B. Reynolds, Infect. Immun., 2016, 85(1) DOI:10.1128/IAI.00601-16.
- C. Formosa, M. Schiavone, H. Martin-Yken, J. M. François, R. E. Duval and E. Dague, Antimicrob. Agents Chemother., 2013, 57, 3498–3506 CrossRef CAS PubMed.
- H. Badrane, M. H. Nguyen, J. R. Blankenship, S. Cheng, B. Hao, A. P. Mitchell and C. J. Clancy, Antimicrob. Agents Chemother., 2012, 56, 4614–4624 CrossRef CAS PubMed.
- H. Badrane, M. H. Nguyen and C. J. Clancy, Antimicrob. Agents Chemother., 2016, 60, 3591–3600 CrossRef CAS PubMed.
- C. Rueda, M. Cuenca-Estrella and O. Zaragoza, Antimicrob. Agents Chemother., 2014, 58, 1071–1083 CrossRef PubMed.
- D. M. Kamiński, Eur. Biophys. J., 2014, 43, 453–467 CrossRef PubMed.
- L. Kristanc, B. Božič, Š. Z. Jokhadar, M. S. Dolenc and G. Gomišček, Biochim. Biophys. Acta, Biomembr., 2019, 1861, 418–430 CrossRef CAS PubMed.
- Š. Z. Jokhadar, B. Božič, L. Kristanc and G. Gomišček, PLoS One, 2016, 11, e0165098 CrossRef PubMed.
- F. Sangalli-Leite, L. Scorzoni, A. C. Mesa-Arango, C. Casas, E. Herrero, M. J. S. M. Gianinni, J. L. Rodríguez-Tudela, M. Cuenca-Estrella and O. Zaragoza, Microbes Infect, 2011, 13, 457–467 CrossRef CAS PubMed.
- M. T. Lamy-Freund, V. F. Ferreira and S. Schreier, J. Antibiot., 1985, 38, 753–757 CrossRef CAS PubMed.
- Y. M. te Welscher, H. H. ten Napel, M. M. Balagué, C. M. Souza, H. Riezman, B. de Kruijff and E. Breukink, J. Biol. Chem., 2008, 283, 6393–6401 CrossRef CAS PubMed.
- K. C. Gray, D. S. Palacios, I. Dailey, M. M. Endo, B. E. Uno, B. C. Wilcock and M. D. Burke, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 2234–2239 CrossRef CAS PubMed.
- D. S. Palacios, I. Dailey, D. M. Siebert, B. C. Wilcock and M. D. Burke, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 6733–6738 CrossRef CAS PubMed.
- J. Milhaud, V. Ponsinet, M. Takashi and B. Michels, Biochim. Biophys. Acta, Rev. Biomembr., 2002, 1558, 95–108 CrossRef CAS PubMed.
- W. I. Gruszecki, M. Gagoś, M. Hereć and P. Kernen, Cell. Mol. Biol. Lett., 2003, 8, 161–170 CAS.
- T.-S. Yang, K.-L. Ou, P.-W. Peng, B.-C. Liou, W.-T. Wang, Y.-C. Huang, C.-M. Tsai and C.-H. Su, Biochim. Biophys. Acta, Rev. Biomembr., 2013, 1828, 1794–1801 CrossRef CAS PubMed.
- I. Fournier, J. Barwicz, M. Auger and P. Tancrède, Chem. Phys. Lipids, 2008, 151, 41–50 CrossRef CAS PubMed.
- O. S. Ostroumova, S. S. Efimova and L. V. Schagina, PLoS One, 2012, 7, e30261 CrossRef CAS PubMed.
- S. Matsuoka, H. Ikeuchi, N. Matsumori and M. Murata, Biochemistry, 2005, 44, 704–710 CrossRef CAS PubMed.
- R. T. Wheeler, D. Kombe, S. D. Agarwala and G. R. Fink, PLoS Pathog., 2008, 4, e1000227 CrossRef PubMed.
- S. El-Kirat-Chatel, A. Beaussart, D. Alsteens, D. N. Jackson, P. N. Lipke and Y. F. Dufrêne, Nanoscale, 2013, 5, 1105–1115 RSC.
- Y. Umegawa, N. Matsumori, T. Oishi and M. Murata, Biochemistry, 2008, 47, 13463–13469 CrossRef CAS PubMed.
- G. Pascual, B. Kim, M. Hong, K. Lee, Y. V. Li and D. A. Hendrickson, Microsc. Today, 2016, 24, 32–37 CrossRef CAS.
- K. S. Kim, Y.-S. Kim, I. Han, M.-H. Kim, M. H. Jung and H.-K. Park, PLoS ONE, 2011, 6, e28176 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2ra07217c |
| This journal is © The Royal Society of Chemistry 2023 |