Open Access Article
Open Access ArticleNano-biosensor for SARS-CoV-2/COVID-19 detection: methods, mechanism and interface design
Yansheng Liu
*ab,
Zhenle Qina,
Jin Zhoua,
Xiaobo Jiaa,
Hongli Lia,
Xiaohong Wanga,
Yating Chena,
Zijun Sun a,
Xiong Hea,
Hongda Li
a,
Xiong Hea,
Hongda Li ab,
Guofu Wang
*a and
Haixin Chang
ab,
Guofu Wang
*a and
Haixin Chang
 *b
*b
aSchool of Electronic Engineering, Guangxi University of Science and Technology, Liuzhou 545616, Guangxi, China
bQuantum-Nano Matter and Device Lab, State Key Laboratory of Material Processing and Die and Mould Technology, School of Materials Science and Engineering, Huazhong University of Science and Technology Wuhan, 430074, Hubei, China
First published on 13th June 2023
Abstract
The epidemic of coronavirus disease 2019 (COVID-19) was a huge disaster to human society. The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which led to COVID-19, has resulted in a large number of deaths. Even though the reverse transcription-polymerase chain reaction (RT-PCR) is the most efficient method for the detection of SARS-CoV-2, the disadvantages (such as long detection time, professional operators, expensive instruments, and laboratory equipment) limit its application. In this review, the different kinds of nano-biosensors based on surface-enhanced Raman scattering (SERS), surface plasmon resonance (SPR), field-effect transistor (FET), fluorescence methods, and electrochemical methods are summarized, starting with a concise description of their sensing mechanism. The different bioprobes (such as ACE2, S protein-antibody, IgG antibody, IgM antibody, and SARS-CoV-2 DNA probes) with different bio-principles are introduced. The key structural components of the biosensors are briefly introduced to give readers an understanding of the principles behind the testing methods. In particular, SARS-CoV-2-related RNA mutation detection and its challenges are also briefly described. We hope that this review will encourage readers with different research backgrounds to design SARS-CoV-2 nano-biosensors with high selectivity and sensitivity.
1. Introduction
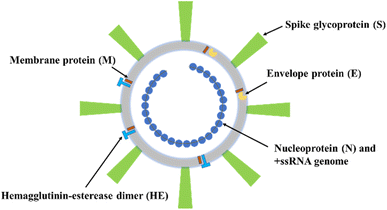
In December of 2019, the first unknown outbreak of a new type of pneumonia was discovered in the city of Wuhan, Hubei Province, in China. Further, numerous infected cases were diagnosed, and deaths were gradually reported. On January 20, 2020, the city of Wuhan was blocked to avoid the spread of this pneumonia-like illness that was later named COVID-19 disease. In the following months, the SARS-CoV-2 virus that caused the COVID-19 epidemic spread quicky throughout the world. In the following year, the SARS-CoV-2 vaccine1 was developed against the virus. Even though a lot of effort has been made by countries around the world to control and fight against the SARS-CoV-2 virus, it still is spreading and affecting both people who have been inoculated with vaccines or have not been inoculated. SARS-CoV-2 possesses the characteristics of strong transmission ability, strong infection ability, strong mutation ability, long incubation period, and long survival time.2 These properties make the global anti-epidemic work with great difficulty. It has been widely accepted that the main means of SARS-CoV-2 spreading are touching, viral aerosol, air contamination, the oral-fecal route, blood, from mothers to newborns, and animals to humans.3,4 Thus far, mutated viruses such as Omicron, Delta, and Alpha SARS-CoV-2 variants are widely observed and recognized.5A coronavirus (CoVs) is an enveloped virus that entraps non-segmented ribonucleic acid (ssRNA), positive sense, and single strand. The CoVs have a genome size of 26 to 32 kb, which is the largest RNA virus known today.6 SARS-CoV-2 is a member of the coronaviruses family and it belongs to β-coronaviruses.7,8 SARS-CoV-2 possesses a crown-shaped appearance with a diameter of 60–140 nm, which has been observed using transmission electron microscopy.9 The complete genome of SARS-CoV-2 was reported by Khailany et al.10 Fig. 1 illustrates the SARS-CoV-2 structure,8,11,12 which consists of a spike protein (S), an envelope protein (E), membrane protein (M), and nucleocapsid (N) proteins. The M and E proteins form the virus envelope, and the N protein responds by assembling the virus.13 The S protein is the key protein for viruses to invade susceptible cells, and it plays a crucial role in infection.14 The S protein is a homotrimer,15 and each monomer is folded and glycosylated by sialic acid distally.16 The host proteases cleave the S protein into two subunits, S1 and S2. The S2 subunit acts as an anchor to bind the viral membrane and it responds to the virus fusion with the host cell.15 The S1 subunit plays a role in cell recognition, cell invasion, and antibody neutralization.17 The invasion process of SARS-CoV-2 is mediated by the binding between the S1 subunit and angiotensin-converting enzyme 2 (ACE2), which acts as a receptor on the host cell cytomembrane.18–22
At present, two detection categories for detecting SARS-CoV-2 are widely applied, which are gold immunochromatography assay (GICA) and viral polymerase chain reaction (PCR).23–27 The humoral immune response generates SARS-CoV-2 antibodies approximately 1 week or more after infection.28 In older adults, children, and patients with immune deficiency disease, the antibody production capacity is lower in immune-compromised humans, which may lead to a potentially false categorization or delayed diagnosis.29 In the GICA, SARS-CoV-2 can be detected by detecting IgG or IgM antibodies. Since IgM antibodies in the blood can be detected after the infection of 3–6 days, and IgG antibodies can only be detected 8 days after infection, the antibody detection misses the window period of prevention of the SARS-CoV-2 epidemic. PCR can recognize virus-infected cases at an early stage with high confidence and detection limits of a few copies. PCR requires a high level of amplification of the oligonucleotide, in which the nucleic acid contaminants introduced in the sampling process or amplification steps may also serve as templates, resulting in inaccurate assay results.30 Moreover, the inhibitor used in the propagation of viral nucleic acid is a threat to the enzymatic amplification reaction. Usually, the product after propagation needs to be further purified, which makes the PCR complicated and cumbersome.31 Studies have shown that the reporting rate of the PCR “false negative” results is as high as 20% to 40% when re-examining suspected cases through medical images.23 This result indicates that the reliability of PCR, which is regarded as a “gold standard” in detecting SARS-CoV-2 in practice, is decreased due to its relatively complicated operation process. Therefore, it is of great significance to develop and research key technologies for the rapid, efficient, highly sensitive, and economical virus detection of SARS-CoV-2.
The basic principle of nano-biosensors is to convert the biometric information to optical or electrical signals, which can be directly observed. Its fast, sensitive, reliable, and easy-to-use characteristics are suitable for the detection of SARS-CoV-2. By combining the specific bio-probe (e.g., S protein, ACE2, antibody, and viral RNA sequence) with a sensitive physical and chemical sensor, the target biomolecules could be specifically detected. Until now, different kinds of nano-biosensors have been designed and fabricated in detecting bio-materials, such as proteins, oligonucleotides, and viruses. In this review, the nano-biosensors based on optical and electrical methods are collected and presented. The physical/chemical mechanisms are presented, as well as the brief fabrication process. A comparable discussion of each kind of biosensor is presented. The section discusses the possibility of detecting mutated SARS-CoV-2 has been individually given.
2. Bio-probes for detecting SARS-CoV-2
The S protein is the most exposed viral protein on the SARS-CoV-2 virion surface. In the invasion process of SARS-CoV-2, the S protein is responsible for receptor recognition and membrane fusion.32 S1 subunits of the S protein have the receptor binding domain (RBD) that can interact directly with the ACE2 peptidase domain (PD).33 Meanwhile, the subunits S2 of the S protein play a role in membrane fusion. The cooperation of S1 and S2 with the assistance of ACE2 make the SARS-CoV-2 affect humans. So, ACE2 can be applied as a bio-probe in detecting SARS-CoV-2 through the ACE2-S protein pair. Except for the S protein, N protein detection as an outer structure of the SARS-CoV-2 virus also has been applied as an RBD, and the N protein antibody is the bio-probe. For each virus, the genome sequence is unique. The size of the genome sequence length of SARS-CoV-2 is around 30 kb, with a 5′-cap structure and 3′-poly-(A) tail enveloped by a complex of structural proteins.34 Due to this uniqueness of the genome sequence, the perfectly matched nucleic acid sequence is manually fabricated to selectively bind to the target RNA sequence of the SARS-CoV-2. IgG and IgM are processed by the human body when the SARS-CoV-2 invades healthy cells. IgG and IgM can specifically bind to the SARS-CoV-2 coronavirus antigen. So, IgG/IgM antibodies are good bioprobes in the detection of SARS-CoV-2.3. Surface-enhanced Raman scattering (SERS)
Raman spectroscopy is a very important, promising, and powerful technique in characterizing the structure of chemical materials or bio-material without any damage. The discovery of wavelength-shifted scattering occurred from 1922 to 1923.35 In 1928, C. V. Raman found “a new radiation”, which is now widely known as the Raman effect.36,37 In the process of Raman scattering, incident light excites the molecules, and most of the photons are scattered in an elastic way with energy maintained. The minority of photons are scattered inelastically with energy loss or gain. Raman scattering can provide vibrational information about molecules, but only 106 to 108 photons illuminated on the sample can undergo Raman scattering. The remaining photons experience competing pathways, such as fluorescence, Rayleigh elastic scattering, and heating.38 Surface-enhanced Raman scattering (SERS) as a surface sensing technique has been widely applied in the ultrasensitive and selective detection in biology and chemistry.39–41 By applying the proper SERS-enhanced substrate, the Raman signals could be largely enhanced.42 Until now, SERS is the only capable method for simultaneously detecting a single molecule and providing its chemical fingerprint.43–47 For SERS, two mechanisms are widely accepted, which are the chemical mechanism (CM) and the electromagnetic mechanism (EM).48,49 CM is formed by a charge transfer between the target molecule and the substrate. It usually has a minor enhancement factor of 10 to 100.44 CM is commonly referred to as the “first layer effect” because of the small distances needed for charge transfer. In some studies, the first monolayer molecules absorbed on the substrate often show a much larger SERS cross-section than the second layer.50 Such a reason necessitated a small uniform rough surface to obtain an improved chemical signal. The localized electromagnetic field caused by surface excitation plasmon (SPs) is the basis of the EM mechanism. SP is the coherent oscillation of conduction electrons under the condition of light illumination.51–53 The light-induced electromagnetic field results in a significant increment of the cross-section of the Raman scattering, and the Raman enhancement factor could reach up to 108 or more.50,54,55 To achieve higher SERS performance, easily fabricated metallic nanostructures are the most common and useful SERS-enhanced materials. The most used materials are Au and Ag, which can induce larger electromagnetic fields under light excitation. It has been proved that the higher SERS performance occurs at the edge of the metallic structures.48,56 Different kinds of metallic nanomaterials with verifying morphology have been designed and applied in SERS, which illustrate good performance such as nanodisks,57,58 nanopores,59–61 nanopillar,62,63 nanoparticles,64,65 nanoprime,66,67 nano spheres,68 nanostar,69 nanocubic,70 nanooctahedron71 and nanowires.72 Wang73 designed and fabricated an Au@Ag core–shell structure with a strong localized plasma resonance effect. By applying the 4-MBA (4-mercaptobenzoic acid) as reporter molecules, he could detect the prostate-specific antigen with a LOD of 0.94 fg mL−1. Ganesh74 designed a quantum organic semiconductor with a strong charge-transfer effect with bio-molecules, and applied it to detect DNA in the cancer cell. Due to the CM mechanism of SERS, the SERS signal was enormously enhanced and the LOD was 10−15 mol L−1. By analyzing the SERS spectra, Ganesh decoded the bases of the cancer genome, and revealed the expression of genomic DNA variation.74 Huang75 used disordered gold nanopores as SERS substrates, and successfully analyzed the order of bases in oligonucleotides through the SERS spectra. Owing to the high sensitivity and selectivity, SERS illustrates the great potential in the detection of SARS-CoV-2.Except for the morphology-dependent SERS performance, the gap between the coherent nanostructures also affects the enhanced SERS intensity. It has been widely accepted that the smaller gap results in larger SERS enhancement factors.80–82 Ojodomo J. Achadu76 reported that a 3-dimensional porous magnetic nanoparticle 3D mag-MoO3–PDA@Au NS (molybdenum trioxide–polydopamine-gold functionalized nanosphere) was designed as SERS “hot spots” in detecting SARS-CoV-2 (Fig. 2a). By applying an external magnetic field, the aggregated 3D mag-MoO3–PDA@Au NS showed an unprecedented SERS amplification. ACE2 was used as a bio-probe to specifically bind to the SARS-CoV-2 spike protein, which was applied as an analyte. During his experiments, the smaller nanogap between the coherent magnetic nanoparticles was magnetically induced by an external magnetic field, which resulted in high SERS performance. Meanwhile, the smaller gaps can generate the sandwich architecture that is the (NS-ACE2)-(S protein)-(ACE2-NS) structure. In this sandwich architecture, the SERS signal binding between the ACE2-(S protein) was largely enhanced with a detection limit of 4.5 fg mL−1.
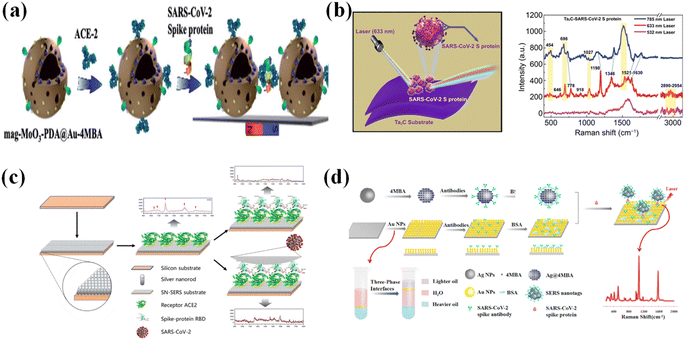 | ||
| Fig. 2 (a) Diagram of the 3D mag-MoO3-PDA@Au nanostructure modified with ACE2 acting as SERS material, and scheme of the sandwich structure after immunoreaction with a spike protein. Reprinted with permission from ref. 76. (b) Raman schematic diagram and the SERS spectra of the Ta2C nano-sheets (NSs) for the Raman scattering of the SARS-CoV-2 S protein with a concentration of 10−9 mol L−1 under the excitation laser at 532, 633, and 785 nm. Reproduced with permission from ref. 77. (c) State-of-the-art diagram of silver-nanorod SERS (SN-SERS) substrate functionalized with ACE2 for interrogating SARS-CoV-2. The ACE2@SN-SERS array generated strong SERS signals that were significantly quenched after the binding of the SARS-CoV-2 spike protein. Adapted with permission from ref. 78. (d) Schematic illustration of the SERS-based immunoassay. The 4-MBA labeled AgNPs-antibody and AuNPs-antibody layers were both stabilized by BSA, and they were applied to form a (4MBA-AgNPs-antibody)-(S protein)-(AuNPs-antibody) sandwich structure that illustrates good detecting sensitivity. Adapted with permission from ref. 79. | ||
SERS can directly identify the structural information of analytes. Yusi Peng77 used Nb2C and Ta2C MXenes as SERS substrates to directly identify the SARS-CoV-2 S protein with a LOD of 5 × 10−9 mol L−1 (Fig. 2b). However, direct identification methods are mainly applied in detecting pre-purified molecules. For complex compounds, the overlap of the characterized SERS peaks limits its application in detecting SARS-CoV-2 due to multi-component samples. The application of a bio-probe is more reliable in detecting SARS-CoV-2. ACE2 and the S protein antibody are important molecules that can bind to the S protein of SARS-CoV-2. The ACE2 possesses strong characteristic Raman peaks, and the intensities of some characteristic peaks are reduced when the interaction occurs between ACE2 and S protein78 (Fig. 2c). By analyzing the intensity change of the characteristic peaks (e.g., 1189 cm−1) of ACE2 and the appearance of a signal at 1182 cm−1 belonging to the SARS-CoV-2 virus, it was easy to confirm the existence of the SARS-CoV-2 virus. The SERS performance of biomolecules is relatively weak. To enhance the SERS intensities,83 the Raman-sensitive molecules are always selected as indicators, which were used to label the bioprobes. Zhang79 used 4-MBA (4-mercaptobenzoic acid)-labeled Ag NPs as SARS-CoV-2 spike antibody nano-tags (Fig. 2d). The few-layer Au NPs modified by the SARS-CoV-2 spike antibody were fabricated as SERS-immune substrates. This SERS-based sandwich biosensor can detect the SARS-CoV-2 spike protein with a LOD of 0.77 fg mL−1 in phosphate-buffered saline and 6.07 fg mL−1 in untreated saliva, respectively. In addition, this biosensor is able to detect the SARS-CoV-2 spike protein in serum and blood with the LOD of 7.60 fg mL−1 and 0.10 pg mL−1, respectively.
The testing of IgG and IgM antibodies is a good method for COVID-19 diagnosis. It has been reported that the anti-SARS-CoV-2 IgM/IgG antibodies are detectable within 0–5 days after symptom onset.86 Normally, lateral flow immunoassay (LFIA) methods are widely applied to detect anti-SARS-CoV-2 IgM/IgG antibodies. In a typical LFIA process, a liquid sample containing the target analytes flows along the strip.87 During the movement of the sample, the target analytes are labeled by metallic Au NPs and then flow to the test lines. The test lines are functionalized with probes that can specifically bind to the analyte and illustrate a color change.88,89 However, LFIA is based on colorimetric analysis, and it has the disadvantage of low sensitivity and poor quality. The SERS-LFIA method is a good candidate for diagnosing COVID-19 through strong, specific, and stable SERS signals.90–94 In a study by Haifeng Liu,84 the SARS-CoV-2 IgM/IgG antibodies in clinical samples were analyzed through a SERS-LFIA in 25 minutes (Fig. 3a–c). Shiliang Chen85 also performed similar work in which the IgM and IgG test lines were integrated on the same strip (Fig. 3d and e). The LOD for IgM and IgG were 1 and 0.1 ng mL−1, respectively.
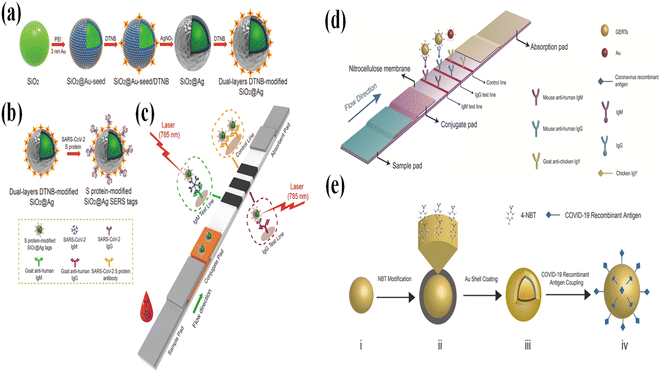 | ||
| Fig. 3 (a) Scheme of the preparation of the dual-layers DTNB-modified SiO2@Ag NPs, in which the DTBA acts as indicators. (b) Image of the preparation of the SARS-CoV-2 S protein-modified SiO2@Ag SERS tags. (c) Operating principle of the high-sensitivity and simultaneous analysis of anti-SARS-CoV-2 IgM/IgG via the SERS-LFIA strip. Adapted with permission from ref. 84. (d) Schematic diagram of the SERS-based strip for the detection of IgM and IgG. (e) Scheme of the preparation for the Au core@4-NBT@Au shell structure. Reproduced with permission from ref. 85. | ||
These methods, combined with artificial intelligence (AI), are a promising way to distinguish small changes in the optic spectra, and it is meaningful in SARS-CoV-2 detection. In principle, SERS can be directly applied in detecting SARS-CoV-2 without any probes or labels. However, unexpected organic compounds always coexist when taking samples from human bodies. The mixed sample shows a complex SERS spectrum, which makes it very difficult to distinguish the characteristic peaks of SARS-CoV-2. In such a case, deep learning could help.96 Deep learning methods combined with SERS have been widely applied in analyzing bio or chemical materials, such as cancers,97,98 bacterial lysate,99 honey varieties,100 and viruses.101 In the study of Jinglin Huang,95 the Ag NPs-coated Si was applied as a SERS substrate to detect the SARS-CoV-2 antigen (Fig. 4). In his study, a deep learning model was applied to train to distinguish the SERS detection spectrum of S proteins of SARS-CoV-2, SARS-CoV, and Middle East respiratory syndrome coronavirus (MERS-CoV), and the model illustrated an identification accuracyof 87.7%. C. Carlomagno102 also designed a deep learning model for detecting SARS-CoV-2 in saliva samples with an accuracy within the range of 89% to 92%. These methods, combined with artificial intelligence (AI), is a promising way to distinguish small changes of the optic spectra, and it is meaningful in SARS-CoV-2 detection.
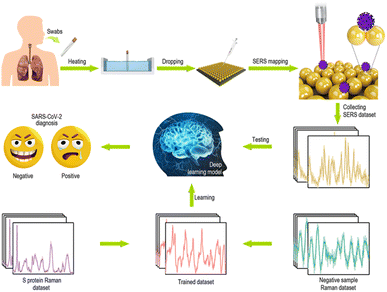 | ||
| Fig. 4 Schematic illustration of the detection process of the deep learning-based SERS technique. Adapted with permission from ref. 95. | ||
Breath volatile organic compounds (BVOCs) analysis is a non-invasive method that allows for analyzing cancer at an early stage through analyzing biomarkers in exhaled breath.104 Interesting research has been performed by Shi Xuan Leong,103 who came up with a strategy for directly identifying infection of SARS-CoV-2 by combining SERS with breath volatile organic compounds (BVOCs) (Fig. 5). SARS-CoV-2 is a virus that can cause pneumonia, which makes the BVOCs a method of detecting SARS-CoV-2 with an innate advantage. It has been reported that the coronavirus can induce immune responses and metabolic changes, which has been confirmed by analyzing the changing of aldehydes, ketones, and alcohols in exhaled breath.105,106 Normally, mass spectra are widely applied in detecting BVOCs107,108 with a sensitivity and specificity of around 80%. However, it is a time-consuming and money-consuming method. The combination of Raman spectra and BVOCs technique illustrates advantages, such as being simple, portable, and inexpensive.
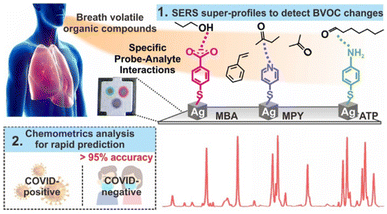 | ||
| Fig. 5 The mechanism of detecting breath volatile organic compounds (BVOCs) changes by using SERS with an accuracy of 95%. Adapted with permission from ref. 103. | ||
SERS sensors combine the detection of optical physical information and biological chemical information, and can achieve ultrahigh sensitivity with high selectivity. However, to achieve the best performance, the morphology of the SERS substrate formed by the noble metal needs to be optimized for best Raman signal enhancement. In addition, the repeatable utilization of the SERS sensor and the specially designed noble metal substrate are challenging for developing low-cost SARS-CoV-2 sensors. Finally, SERS needs a relatively complicated Raman system to support the collection of high-resolution Raman spectra for high-sensitivity detection. The commercially-available fiber-based Raman system has been limited by low resolution in Raman spectra. Therefore, the development of compact, high-resolution, convenient, reusable SERS systems and sensors is highly desirable.
4. Surface plasmon resonance (SPR)
Surface plasmon resonance (SPR) is a collection of oscillations of free electrons on a metal surface that is induced by incident light. The first application of the SPR biosensor was designed by Liebenberg and Nylander in 1982.109 Since then, the SPR biosensor as a surfacing sensing technique has been widely applied in the interdisciplinary study between chemistry, physics, and biology.110 Nowadays, SPR biosensors have been widely applied in fundamental biological studies, drug discovery, affinity of bimolecular binding, clinical diagnosis, and environmental science and agriculture. The surface plasmon resonance (SPR) sensor as a surface sensing detection method has been applied in bio-applications for a long time due to its advantages, such as the simple, low-cost, rapid, real-time monitoring, label-free detection, revealing the binding kinetics of patient antibodies.111,112 The SPR biosensing platform relies on changes in the localized refractive index (RI) of the sensing surface, which is caused by the binding between bio-probes and bio-analytes. In the SPR surface technique, the incident light can cause excitation of the charge density oscillation, which propagates along the interface between the metal–dielectric interface. To excite the SPR, the incident light has to satisfy the following condition:113
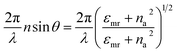 | (1) |
In comparison to a planar Au film, the metallic nanostructure illustrates a higher performance. The metallic nanostructures (such as Au and Ag NPs) possess plasmonic properties of strong and controllable optical signal enhancement, and manipulation capabilities.111 By proper design of the morphology of the metallic nanostructures, the SPR wavelength can be determined at a certain wavelength, which could improve the sensitivity of the SPR biosensors.131,132 In a study by Guangyu Qiu,127 the dual-functional plasmonic photothermal biosensor was applied in detecting the SARS-CoV-2 RNA sequence (Fig. 6a). Two-dimensional gold nanoscale islands (AuNIs) functionalized with DNA complementary receptors formed the sensing interface. The DNA reporter with specific DNA sequences can interact with SARS-CoV-2 RNA through nucleic acid hybridization. A microfluidic chip integrated with the sensing surface was designed as the detection chip. During the detection process, an external laser was applied to heat the sensing surface, which was induced by the plasmonic photothermal (PPT) effect of AuNIs and the calculated temperature of around 41 °C. Using two different angles of incidence, the plasmonic resonances of PPT and LSPR can be excited at two different wavelengths, which increased the stability, sensitivity, and reliability of the detection. By applying the PPT effect in SPR, the kinetics of DNA hybridization was largely increased, which decreased the binding time. The SPR biosensor responsible sensitivity also increased and the LOD was 0.22 pM.
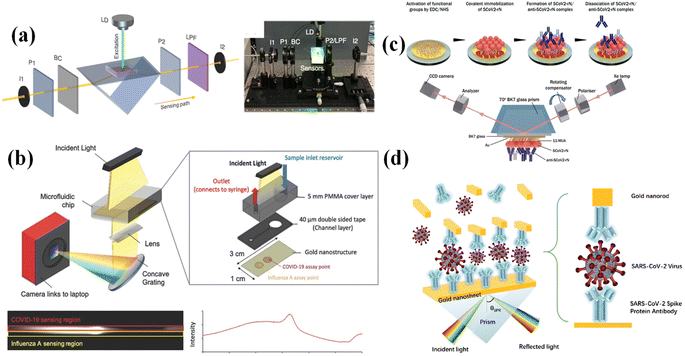 | ||
| Fig. 6 (a) Schematic and experimental setup of the dual-functional PPT-enhanced LSPR biosensing system. Reproduced with permission from ref. 127. (b) Scheme of the SPR spectral imaging platform integrated with a microfluidic chip. A typical spectral image is shown, and it was obtained from the CCD. Reproduced with permission from ref. 128. (c) Schematic representation of the gold-coated sensor disk (Au-disk) surface modified with antigen SCoV2-rN, which was applied as an interactive surface. Reproduced with permission from ref. 129. (d) The SPR biosensor was designed using a sandwich structure. Reproduced with permission from ref. 130. | ||
To improve the sensitivity, another way is to increase the “mass” of analytes by designing sandwich structures that largely change the RI of the sensing surface. Normally, the Au NPs modified with a bio-probe are applied as the top materials to bind to the analytes, which have been bound to the sensing surface. In the architecture of the sandwich structure, Au NPs play two functions: (1) the localized plasmon effects between Au NPs and the Au film can enlarge the changes of resonance wavelength. (2) The Au NPs bounded to the sensing surface can make a bigger mass change, resulting in a larger RI change. By applying the sandwich structure, the detection limits for detecting the SARS-CoV-2 RAN sequence128 (Fig. 6b), N protein129 (Fig. 6c), and S protein130 (Fig. 6d) are improved.
In comparison with the most common ATR structure, the SPR biosensor based on fiber optic surface plasmon resonance (FO-SPR) is also widely applied in bio-detection. In the FO-SPR sensor, the light propagates along with the optic fiber through total internal reflection. For each reflection, the evanescent wave is generated, which can excite the Au film and generate the surface plasma to propagate along with the Au film. With changing the surface complexes on the Au film, the exciting light wavelength intensity is changing or the excitation light wavelength should be optimized. The exposure sensing surface of FO-SPR can be directly put into the analyte's solution with the out-coupling of microfluidics channels. Moreover, the light is restrained in the optic fiber, which reduces the complexity of the alignment of the optic path. These properties make the FO-SPR an easy-to-fabricate device with high stability and good sensitivities. Jia-Huan Qu112 designed a fiber-optic surface plasmon resonance (FO-SPR) biosensor in detecting anti-SARS-CoV-2 antibodies (Fig. 7a). This sensor greatly reduces the detection time-to-result of only 30 min compared to the conventional ELISA, and with enormous potential towards other applications in need of antibody quantification and kinetic profiling. Alireza Samavati133 used graphene oxide/Au/fiber Bragg grating (FBG) as a sensing interface in detecting the SARS-CoV-2 S protein in the saliva sample (Fig. 7b). This FBG probe can quickly detect COVID-19 within 10 s.
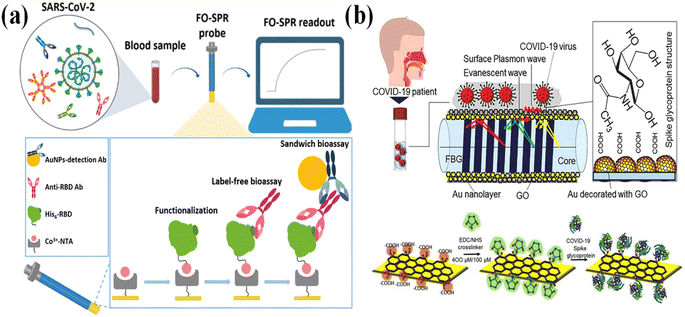 | ||
| Fig. 7 (a) Schematic illustration of the FO-SPR biosensor for the detection of anti-SARS-CoV-2 antibodies in label-free samples, and the detecting mechanism using sandwich formats. Reproduced with permission from ref. 112. (b) Sensing mechanism diagram of the interactive surface of FO-SPR biosensor and the process of binding the spike glycoprotein of COVID-19 virus with activated GO. Reproduced with permission from ref. 133. | ||
The SPR sensor has the great advantage of commercially-available systems and supporting facilities. However, for efficient SARS-CoV-2 detection, SPR faces several severe issues to resolve. First, the detection surrounding should be stable, especially for the temperature. Second, for the detection mechanism by SPR, interference of SARS-CoV-2 detection by SPR easily occurs by impurities in the solution, which reduces its specificity. The complexity of real samples needs to be minimized or eliminated by signal processing.
5. Field-effect transistors (FETs)
The FET biosensor is a label-free, highly sensitive, fast, and low-cost detection method.134 By combining it with kinds of bio-probe and advanced functional nanomaterials,135 FET biosensors have been applied in detecting various bio-molecules and diagnostic diseases.136 Through screening the readable and amplified electrical signals, which are generated through an interaction between target bio-molecules and bio-probe on the sensing interface, the FET-biosensor can real-time monitor the detection process.137,138Graphene is the most used channel material in FET. Graphene as an atomic thickness two-dimensional(2D) material has been regarded as the most renowned material.140–143 The near ballistic transport and high level of mobility143–145 of graphene make it an attractive platform for nanoelectronics applications,146,147 particularly for high-frequency applications. Due to the scalability of graphene, devices achieve 2D atomic dimensions.148,149 In addition, its optical and mechanical properties are ideal for micro/nano-mechanical systems, thin film transistors, transparent and conducting electrodes, and photonics.150,151 Graphene is a gapless semimetal with zero overlap between its conduction band and its valence band. At room temperature, graphene demonstrates a strong ambipolar electric field effect and possesses high carrier mobilities of ∼1 × 104 cm2 V−1 s−1.152 For the suspended graphene, its mobility can approach 2 × 105 cm2 V−1 s −1 at a low temperature.153 In the past decades, the graphene-based field-effect transistor (G-FET) biosensors were widely applied in the detection and diagnosis of diseases, such as Ebola virus,154 heart failure,155 human immunodeficiency virus (HIV),156 cancer,157 and viral exosomes in cells.158 Giwan Seo139 designed a graphene-based FET biosensor, and applied it in the real-time detection of SARS-CoV-2 (Fig. 8a) with a LOD of 242 copies/mL. This FET biosensor could detect the SARS-CoV-2 spike protein at concentrations of 1 fg mL−1 in phosphate-buffered saline and 100 fg mL−1 in clinical transport medium. Michael Taeyoung Hwang159 used a liquid top-gated graphene FET biosensor with SARS-CoV-2 antigens with a LOD of 1 aM. Jiahao Li31 applied reduced graphene oxide-FET in the detection of SARS-CoV-2 RAN with an LOD of 0.37 fM in 2 min. In the study of Jiahao Li,31 the RGO-FET was applied in detecting SARS-CoV-2 RAN. The RGO solution was drop-casted on the FET channel, and the AuNP functionalized with complementary phosphonodiamidite morpholino oligos (PMO) probe was decorated on RGO to form a detection surface (Fig. 8b). The RGO-FET was applied in detecting RNA-dependent RNA polymerase (RdRP) in PBS, throat swab, and serum with the LODs of 0.37, 2.29, and 3.99 fM, respectively. The RGO-FET also was applied in detecting clinical samples in 2 min. A similar study was also performed by Esteban Piccinini.160 He used the RGO-FET with a coplanar Ag/AgCl gate in the detection of the SARS-CoV-2 S protein with an LOD of 0.74 nM.
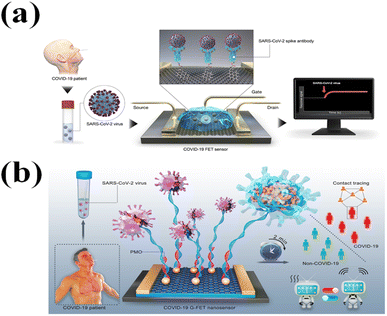 | ||
| Fig. 8 (a) Schematic diagram of the COVID-19 FET sensor operation procedure. Graphene is chosen as the sensing material, and the SARS-CoV-2 spike antibody is conjugated on the graphene sheet through the i-pyrenebutyric acid ester N-hydroxysuccinimide, which is an interfacing molecule as a probing linker. Reproduced with permission from ref. 139. (b) Schematic diagram of the PCR-free rapid direct identification of COVID-19 using the PMO-functionalized G-FET nanosensor. Reproduced with permission from ref. 31. | ||
Except for carbon-based 2D materials, one-dimensional carbon nanotubes (CNTs) based on FET have been widely studied in bio-detection. CNTs are molecular-scale wires with high mechanical stiffness and strength.163 An SWNT with different chirality illustrates different electrical properties. It can be metallic, semiconducting, or semimetallic.164 CNTs possess excellent electrical properties, good biocompatibility, and good size compatibility. These characteristics make it a good channel material to fabricate sensitive FET biosensors in detecting SARS-CoV-2.163,165 Mazin A. Zamzami161 designed a nanotube-based field-effect transistor in detecting SARS-CoV-2 S protein antigens. The single-wall carbon nanotubes (CNTS) with a length of 5–30 μm and a diameter of 1 to 2 nm were functionalized with the SARS-CoV-2 S protein antibody, which acts as a bio-probe to bind the SARS-CoV-2 S protein. The FET was fabricated on Si/SiO2 substrates and designed using a backdated structure. The CNTs-FET can respond to the SARS-CoV-2 S protein diluted from 0.1 to 5000 fg mL−1, and the LOD was 4.12 fg mL−1 of SARS-CoV-2 S protein in a 10 mM AA buffer solution (Fig. 9a). Wenting Shao162 designed an SWCNTs-FET, in which the top gated structure was constructed by using a silver/silver chloride reference electrode (Fig. 9b). The SWCNTs modified by anti-SARS-CoV-2 S protein antibody were applied as a sensing interface. The LODs for detecting the S protein antigen and N antigen (NAg) were 0.55 fg mL−1 and 0.016 fg mL−1, respectively. M. Thanihaichelvan166 used a top-gated CNT-FET in detecting the target sequence of the SARS-CoV-2 virus. The gate electrode is the Ag/AgCl electrode, and the CNT functionalized with probe oligonucleotide was used as the sending interface. The detection limit for the oligonucleotide is 10 fM.
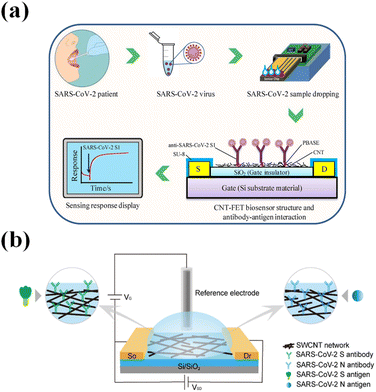 | ||
| Fig. 9 (a) Schematic diagram of the CNT-FET biosensor for detecting the SARS-CoV-2 S protein. Reproduced with permission from ref. 161. (b) Schematic illustration of a liquid-gated SWCNT FET for the detection of SARS-CoV-2 SAg and NAg. Reproduced with permission from ref. 162. | ||
Except for graphene, 2D semiconducting materials hold great promise in the detection of biomolecules with higher sensitivity due to their tunable bandgap and the high surface-to-volume ratio.169 In the 2D materials domain, transition metal dichalcogenides (TMDs) attracted much attention in designing TMDs FET-biosensor due to their direct bandgap, biocompatibility, and high mobility.170 Molybdenum disulfide (MoS2) as a new type of 2D layered transition metal dichalcogenides has been widely applied in FET. MoS2 possesses lots of unique electronic, optical, and mechanical properties. Unlike the electric properties of graphene,171,172 MoS2 is a semiconductor with both the top and bottom surface of the Mo atoms layer bonded by the sulfur atoms to generate a covalent Mo–S band with the polarity in the vertical direction to the surface.173,174 Junqing Wei167 designed a MoS2-FET in detecting antibodies using the SARS-CoV-2 S protein receptor in vaccinated serum specimens (Fig. 10a). The MoS2-FET can quantitively detect the IgG within the range of 10−3 to 10−9 mg mL−1.
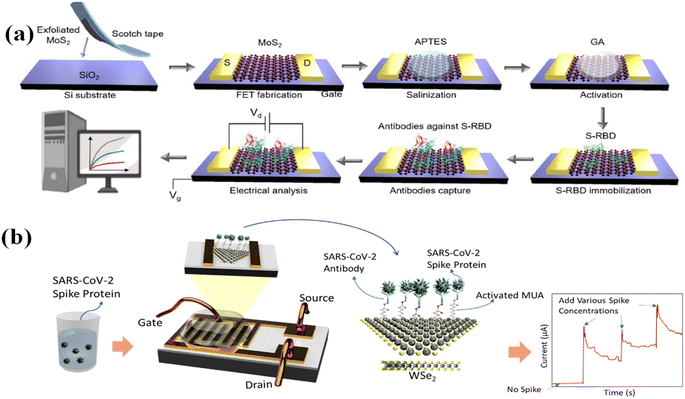 | ||
| Fig. 10 (a) The schematic of the MoS2-FET fabrication, functionalization, and its application for antibodies against S protein detection. Reproduced with permission from ref. 167. (b) Schematic diagram of the fabricated monolayer WSe2-based SARS-CoV-2 FET biosensor. Reproduced with permission from ref. 168. | ||
Like single-layer MoS2, WSe2 as a TMD material is a direct bandgap semiconductor with bandgaps (Eg) between 1.2–1.8 eV.175 WSe2 films have pristine surfaces free of dangling bonds that can reduce surface roughness scattering, and lead to high mobility.176 The free dangling bonds also reduce interface traps, and result in a low density of interface states on the semiconductor–dielectric interface.176 Parvin Fathi-Hafshejani168 designed and fabricated a WSe2-based FET, and it was applied to detect the SARS-CoV-2 virus with a LOD of 25 fg μL−1 (Fig. 10b).
The FET detection of SARS-CoV-2 can achieve high sensitivity, but the selectivity is relatively low. The challenge for the FET SARS-CoV-2 sensor is to achieve highly selective recognition. For FETs, the electrical signal changes induced by bio-interfaces need to be screened by stable control samples, and some fake electrical signals should be carefully identified. Another problem is the re-utilization of the FET SARS-CoV-2 sensor. The regeneration of SARS-CoV-2 bio-interfaces may be complicated for FETs construction. The reliable regeneration of the FET SARS-CoV-2 nano-biosensor needs more investigation.
6. Fluorescence
Fluorescence spectroscopy is a powerful identification technique and images the emitters with ultra-high sensitivity, and its LOD reaches the single molecule level.177 Fluorescence-based sensing techniques possess advantages, such as high speed, easy operation, good sensitivity, real-time motoring, good biocompatibility, and noninvasive characteristics.178,179 Fluorescence biosensors are the most popular optic devices, and they have been widely studied and used in bio-detection and disease diagnosis.180–182 Fluorescence biosensors can directly provide the readout of bio-analytes or bio-interaction information that is located or occurs in different biological organisms and living cells.183The most common fluorescence biosensors are based on metal-enhanced fluorescence or plasmon-enhanced fluorescence. Metallic nanostructures can generate localized surface plasmon, which can excite the fluorophore near the metallic nanostructures more efficiently and increase the decay rate, resulting in stronger fluorescence.188,189 The enhancement of the fluorescence can be affected by several factors, such as the morphology of metallic NPs, the distance between the fluorophore and the NPs, the quantum yield of fluorophores, as well as the spectral overlap between SPR and excitation/emission spectra of the fluorophores.189 A grating-coupled fluorescent plasmonic (GC-FP) was studied in detecting SARS-CoV-2 by Nathaniel C. Cady184 (Fig. 11a). The sensing interface was a gold-coated nanoscale grating, and the SARS-CoV-2 antigens or control proteins were sprinted with an array of 400 μm diameter spots. The Alexa Fluor 647 tagged anti-human IgG was applied to enhance the detection fluorescence intensity. This fluorescence-enhanced biosensor can simultaneously detect the antibody levels for multiple antigens using one sample, and the maximum measuring time was 27 min. The GC-FP biosensor possesses a selectivity of 100%. In comparison with the fluorescence biosensors based on a single signal change, the radiometric fluorescence biosensors are more attractive. Förster resonance energy transfer (FRET) is a good choice to design a radiometric fluorescence biosensor. FRET is a non-radiative physical process, in which the donor is excited by extra energy, and then transfers its energy to an acceptor proximal to the ground state via long-range dipole–dipole interactions.190 The molecular chromophore that offers the energy is regarded as a donor (D), and the molecular chromophore that accepts the energy is considered an acceptor (A). The donor chromophore, originally in the ground state, is transferred by absorbing an external photon into an excited state. Its energy is transferred to the acceptor through the nonradiative process of energy transfer from dipole to dipole, thus being transferred back to the ground state. There is also a competing process that exists. The excited photons in acceptors and donors both can lose energy to the ground state by emitting fluorescence. During the FRET process, several requirements must be satisfied. Firstly, adequate spectral overlap between the excitation of the acceptor and the emission of the donor should exist. Secondly, the FRET process is efficiently affected by the distance between donor and acceptor. The FRET efficiency is inversely associated with the sixth power of the distance between the donor and the acceptor. For such reason, the distance between the donor and acceptor should be within a range of 10 nm.191–195 Thirdly, for the dipole–dipole interaction, at least one donor and acceptor should possess a certain degree of rapid rotational freedom to obtain favorable energy transfer orientation. When all of these conditions are satisfied, the excited electrons in the donor have a possible competing pathway to transfer their energy to the acceptor, instead of emitting energy as fluorescence. For the fluorescence biosensors, two elements are required, which are the recognition element and a reporter.196 The recognition element is responsible for selectively binding to the analyte, while the reporter can generate the signal. When the recognition element combines with the analyte, the fluorescence signal that comes from the reporter could be quenched197 or enhanced198 fluorescence. Byunghoon Kang185 developed an ACE2 mimic peptide beacon (COVID-19-PEB) for the simple detection of SARS-CoV-2 using a FRET system (Fig. 11b). The COVID-19-PEB can specifically bind to the S protein RBD of the SARS-CoV-2. In the absence of the SARS-CoV-2, the COVID-19-PEB is a form of a hairpin structure, in which the fluorescence signal is minimum due to the FRET process between the BHQ2 (as acceptor) and Cy3 (as donor). Under the condition of the existence of ARS-CoV-2, the hairpin structure of COVID-19-PEB is open, resulting in an increase in the fluorescence signal within a short period. The LOD of this method is 52.5 pM. Ghasem Rezanejade Bardajee186 designed a FRET pair, where the BHQ2 was applied as an acceptor and Cy3 was applied as a donor in detecting the specific RAN sequence of the SARS-CoV-2 (Fig. 11c). The complementary sequence of the target SARS-CoV-2 sequence was divided into two parts. One part is modified with Cy3 organic dye, and another part was functionalized with BHQ2. In the absence of a complementary sequence, the FRET occurred. In the presence of the complementary sequence, the efficiency of the FRET was blocked, resulting in the fluorescence intensity decrease. Through such a design, the LOD is 0.0995 μM. He187 used CdTe/ZnS QDs as a donor and BHQ2 as an acceptor (Fig. 11d). By properly designing the CdTe/ZnS-DNA and BHQ2-DNA structures, the sandwich structure (CdTe/ZnS-DNA)-(target SARS-CoV-2 RNA)-(sequence-DNA) was formed, in which the FRET between the CdTe/ZnS and BHQ2 occurred. By adjusting the on/off of the FRET process, the target SARS-CoV-2 RNA sequence was detected with an LOD of 0.000823 μM.
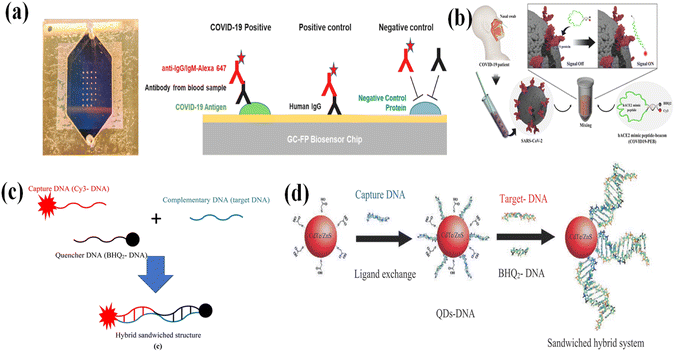 | ||
| Fig. 11 (a) Immunosensing mechanism of S-IgG detection using an optofluidic pointing-of-care testing platform (POCT) and a typical fluorescence signal trace for S-IgG detection. Reproduced with permission from ref. 184. (b) Schematic illustration of the simple detection method of SARS-CoV-2 using the hACE2 mimic peptide-based molecular beacon (COVID 19-PEB). Reproduced with permission from ref. 185. (c) A schematic representation for pairing different DNA single strands (containing Cy3-DNA, BHQ 2-DNA, and complementary DNA) to form a hybrid sandwiched structure. Reproduced with permission from ref. 186. (d) A schematic of the DNA-conjugated CdTe/ZnS QDs nanoprobe for the detection of complementary (target-DNA) derived from the COVID-19 virus genome. Reproduced with permission from ref. 187. | ||
Fluorescence methods have the advantage of direct low-cost identification with highly commercially-available detection systems. However, fluorescent SARS-CoV-2 sensors are affected by the detection conditions. The modification of the labeled bioprobes is complicated, and the fluorescence intensity can be affected by several factors. Therefore, the fabrication of reliable and easily fabricated fluorophore label bioprobes needs to be further studied.
7. Electrochemistry
Electrochemical transducers possess the characteristics of cost efficiency, simplicity, high selectivity, and high sensitivity.199 The electrochemical sensors have been applied in detecting various viruses, such as Hepatitis B virus,200 Ebola virus,201 HIV,202 Hantavirus,203 and Epstein–Barr virus204 with excellent sensitivities. Electrochemical biosensors are a combination of electroanalytical methods and bio-interaction, which occurs between the bio-probes and the target analytes. The bio-probes are responsible for identifying and binding to the analytes, and the electroanalytical event plays a role in generating electrochemical signals, which is monitored by a transducer that is related to the analyte concentration. There are mainly two kinds of electrochemical biosensors that are biocatalytic devices and affinity sensors.205 For biocatalytic devices, enzyme electrodes are applied to identify target analytes such as glucose, lactose, and xanthine and produce electroactive species. For the affinity sensors, the electrodes functionalized with the bio-probes are utilized to selectively bind to the analyte, such as an antibody, nucleic acid, or receptors. The most used affinity sensors in biology are based on immunosensors, where the antigen or hapten are detected by using antibodies as bio-probes. The electrochemical transducer transfers the binding event into a readable electrical signal. The RNA/DNA affinity sensors also play an important role in medical diagnostics, cancers detections, viral infections, and genetic diseases, where the complementary nucleic acid sequence is applied as a probe to identify the target nucleic acid sequence.In the electrochemical techniques, the current, potential, and impedance are the main measurable parameters, in which the voltammetry technique, impedance technique, conductometry technique, and potentiometry technique are applied.209 The voltammetric technique measures the current changes between the working electrode reference electrode and reference electrode by applying a potential. Electrolysis is the result of the current using an electrochemical reduction or oxidation at the working electrode. Electrolysis currents are limited by the mass transport rate of the molecules toward the electrode. The current is a reflection of the electrochemical reduction or oxidation at the working electrode. When a scanning voltage is applied, the current response is a spike that is proportional to the analyte concentration. Voltametric methods include linear sweep voltammetry (LSV), cyclic voltammetry (CV), differential pulse voltammetry (DPV) and square-wave voltammetry (SWV).210 Lokman Liv206 used a CV platform to detect SARS-CoV-2 spike antibodies in saliva and oropharyngeal swab samples (Fig. 12a). The glassy carbon electrode (GCE) was applied as the working electrode, and the BSA/S-gene/GluAl/CysAm/Au/GCE (BSA: bovine serum albumin, S-gene: SARS-CoV-2 antigen, GluAl: glutaraldehyde, CysAm: cysteamine) complex was applied as a sensing surface. With the presence of the SARS-CoV-2 spike antibody sample, the scan peak height decreased due to the large size of the immuno-complex to block the electron transfer. The proposed method illustrated a linear response to the SARS-CoV-2 spike antibody in synthetic media and saliva/oropharyngeal swab samples varied from 0.1 to 1000 ag mL−1, and the LOD was 0.01 ag mL−1. Differential pulse voltammetry (DVP) is a very sensitive technique owing to its low capacitive current.211,212 In DVP, short pulses with small amplitude are superimposed on a linear ramp, and enhanced discrimination of faradaic currents is measured before and after applying the pulse.213 By analyzing the relationship between the potentials and the current difference of each pulse, the potentials of the envisaged oxidation or reduction reaction can be calculated.214 Hui Zhao207 applied a super sandwich-type recognition strategy in detecting RNA of SARS-CoV-2 by using portable electrochemical smartphone equipment with differential pulse voltammetry (DPV) (Fig. 12b). The probe nucleic acid sequence was functionalized with Au@Fe3O4, and it was applied to bind the target ssDNA. The total structure of nucleic acid@Au@Fe3O4 was named premix A. The premix B was a host–guest complex that was Au@SCX8-TB-RGO-(labeled signal obe) acting as a bioconjugate. The premix A, premix B, and SARS-CoV-2 target sequence form a sandwich structure, which illustrates a strong DPV signal with a LOD of 200 copies mL. Leila Farzin208 developed an early diagnosis of RNA of SARS-CoV-2 by using DPV methods (Fig. 12c). The CPE–HT18C6(Ag)/chitosan/SiQDs@PAMAM/probe (CPE: carbon paste electrode, HT18C6: hexathia-18-crown-6, SiQDs: Si quantum dots) sequence was applied in the detection of SARS-CoV-2 RdRP (RdRP: RNA-dependent RNA polymerase) sequence. The proposed nanosensor exhibited a wide linear range of detection, which was 1.0 pM–8.0 nM.
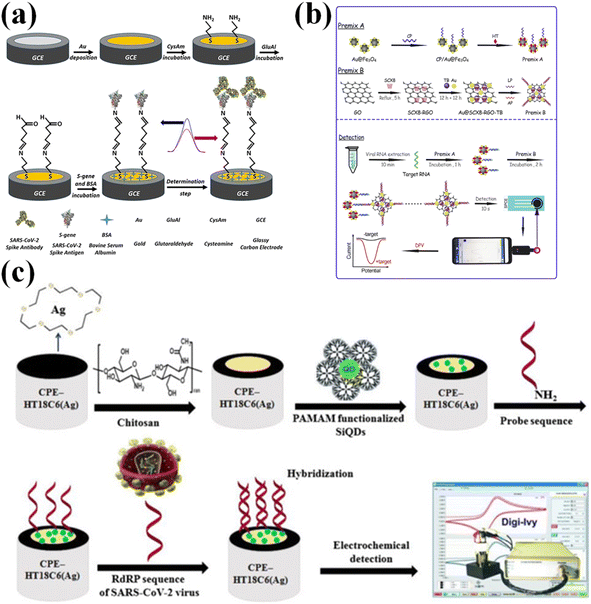 | ||
| Fig. 12 (a) Schematic representation of SARS-CoV-2 detection using the electrochemical biosensor. Schematic illustration of the fabrication process of an electrochemical genosensor. Adapted with permission from ref. 206. (b) Preparation of a premixed A and B, and the detecting process by using electrochemical methods equipped with a smartphone. Adapted with permission from ref. 207. (c) Schematic illustration of the fabrication process of an electrochemical genosensor. Adapted with permission from ref. 208. | ||
In electrochemical impedance spectroscopy (EIS) measurements, a small amplitude sinusoidal AC signal with different frequencies are applied at working electrodes. Upon changing the frequency of the AC, the ratio of the voltage to the current of the AC signal changes. A small change in the electrode surface can change the magnitude of the system impedance. Juan Carlos Abrego-Martinez215 used EIS to detect the S protein of the SARS-CoV-2. The LOD was 66 pg mL−1 in 40 min. Jie Zeng216 applied an impedimetric biosensor in detecting COVID-19 antibodies in serum. The gold micro-interdigitated electrodes were photolithographed on SiO2. The SARS-CoV-2 S protein was functionalized between the Au electrodes with a channel length of 4 μm. With the assistance of AuNPs modified by antibodies, the LOD of detecting purified anti-S protein antibodies was 200 ng mL−1. With the assistance of dielectrophoretic (DEP), the detection time was shortened to 30 min by sacrificing the LOD, which was 2 μg mL−1.
For the architecture of electrochemical biosensors, a three-electrode system or a two-electrode system is widely applied, where the working electrode plays a pivotal role.219,220 A typical three-electrode electrochemical cell is made up of a working electrode, a reference electrode, and an auxiliary electrode. Normally, the semiconducting and conducting materials can be used as working electrodes, such as carbon or platinum, that can be in various sizes ranging from bulk materials and nanostructures, which could affect the performance selectivity and sensitivity.221 Normally, the reference electrodes and the auxiliary electrodes are Ag/AgCl and platinum wire, respectively. The two-electrode system consists of the working electrode and reference electrode under the condition of low current density. Normally, the two-electrode system is generally applied in disposable sensors because of their lower cost, and the long-time stability of the reference is not needed. In comparison with a two-electrode system, charge from electrolysis flows through the auxiliary electrode instead of the reference electrode, which protects the reference one from changing its half cell potential in a three-electrode system. Abdulhadee Yakoh217 designed an electrochemical biosensor fabricated by using paper applied in the detection of SARS-CoV-2 antibodies and antigens (Fig. 13b). The test sheet consisted of 3 folding layers: a working ePAD, a counter ePAD, and a closing ePAD (Fig. 13a). The three-dimensional wax barrier was a printed pattern to isolate the hydrophilic center of each zone in which the solution could flow through to the working ePAD (Fig. 13c). The counter electrode (CE) and the reference electrode (RE) were printed and integrated on the counter ePAD. Through such architecture, a three-electrode system was established. The SARS-CoV-2 spike protein was modified on the test zone of the working ePAD. The paper-based electrochemical biosensor can detect the SARS-CoV-2 IgG and IgM in the range from 1 to 1000 ng mL−1 with LODs of 0.96 and 0.14 ng mL−1, respectively. Razieh Salahandish218 designed a dual working-electrodes immune-biosensor in detecting SARS-CoV-2 N-protein in spiked samples, in which the N protein antibodies are applied as bio-probes (Fig. 13d). The two working electrodes can simultaneously give the detecting results, which can corroborate each other (Fig. 13e). This biosensor can reach the LOD of 56 fg mL−1 and the detection time is 1.5 min.
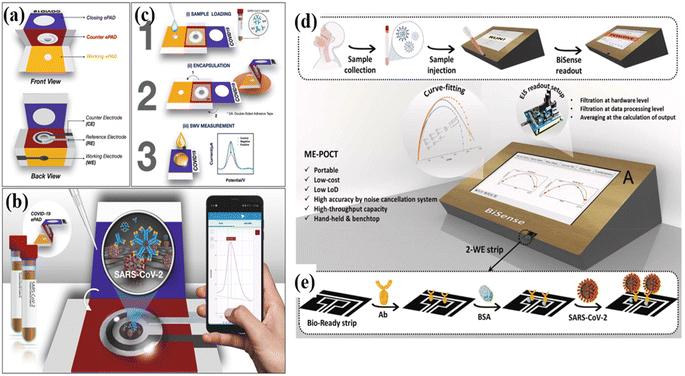 | ||
| Fig. 13 Schematic illustration of the (a) device components, (b) detection principle, and (c) detection procedure of the COVID-19 ePAD. Adapted with permission from ref. 217. (d) Schematic illustration of detecting SARS-CoV-2 by using the proposed two-working electrode electrochemistry biosensors. (e) Two-working electrode strip with a carboxyl-rich surface ready to be immobilized for direct antibody (Ab) immobilization, followed by surface blockage using bovine serum albumin (BSA) for ultimate SARS-CoV-2 virus conjugation in clinical specimens. Adapted with permission from ref. 218. | ||
For the electrochemical biosensors, the bio-probes play important roles in biosensing. Genxi Li and his research team have made great progress in designing electrochemical biosensors combined with kinds of bio-probes that have been applied in detecting SARS-CoV-2 viruses. In his study, signal amplification was applied, and it proves that the terminal deoxynucleotidyl transferase catalyzes the hairpin assembly (CHA) with amplificated electrochemical signals. By using this method, the LOD for detecting SARS-CoV-2 RNA,222 the N genes and the RdRp genes223 were 26 fM and 57 fM, respectively. He also proposed an electrochemical biosensor for the detection of SARS-CoV-2 based on the aid of the target-triggered cascade signal amplification strategy. This method possessed a LOD of 45 fM in detecting ssDNA of SARS-CoV-2, with a broad linear range between 0.1 pM and 3000 pM.224 Except for the common RNA or antibody detection, he came up with a new method that used papain-like cysteine protease (PLpro) as a biomarker in detecting SARS-CoV-2, which can be used in real surroundings, including blood and saliva with a LOD of 27.18 fM.225
Electrochemical SARS-CoV-2 sensors have the advantage of low-cost and commercially available systems. They can also achieve high sensitivity by using carefully designed sensing bio-interfaces. The challenges for electrochemical SARS-CoV-2 sensors remain in the repeatable quantified detection. For electrochemical detection, the detection process needs to be done in a three-electrode system in the electrolyte. Many environmental impurities may influence the electrochemical detection system, including potential, bio-interfaces or reference electrode stability. Therefore, a stable and robust electrochemical detection system should be designed to achieve repeatable quantified detection with high sensitivity and selectivity at the same time.
8. CRISPR-Cas and Argonaute
CRISPR-Cas226–228 and Argonaute229–231 systems are deemed as the next-generation diagnostic technology, as excellent gene editing tools. In particular, CRISPR-Cas12a and CRISPR-Cas13a have attracted interest due to their highly accurate gene-editing capabilities and crosscutting activities.232 CRISPR-Cas-based and Argonaute-based SARS-CoV-2 detection methods are more specific, convenient, and reliable. Cas12 and Cas13 as RNA-directed CRISPR-related proteins can be programmed to detect specific DNA and RNA sequences from pathogens with single-base pair specificity, respectively.226,233,234 Ma et al. designed the CRISPR-Cas12a powered SERS bioassay for SARS-CoV-2 detection with a LOD of 1.9 copies per mL.235 Ashwin Ramachandran et al. constructed a selective ionic focusing technique assistant CRISPR assay method in detecting SARS-CoV-2 RNA in 35 min. The assay was performed in a microfluidic channel, in which the electric field control was used to accelerate the CRISPR assay by cofocusing Cas12–gRNA re-porters, and targets.228Argonaute is a nucleic acid-oriented endonuclease that prefers to be guided by short 5′-phosphorylated single-stranded DNA and cuts DNA substrates in the absence of a proto-spatial adjacent motifs (PAM). The selection of target DNA cleavage sites is not subject to specific sequence restrictions. Wang et al. proposed a method called PLCR (Pyrococcus furiosus Argonaute coupled with modified Ligase Chain Reaction), and applied this technique to nucleic acid detection. In this method, the PfAgo (Pyrococcus furiosus Argonaute) was applied to specifically cleave DNA and LCR to precisely distinguish single-base mismatch, where the PfAgo can only cleave target DNA guided by phosphorylated DNA guides longer than 14-mer. By utilizing this technique in other coronaviruses and multiplexed detection, it effectively reduced the detection time to less than 70 minutes.230 Similar work also has been done by Wang et al., in which PfAgo mediated nucleic acid detection method can significantly reduce the time of detecting COVID-19.236 Ye et al. developed a multiplex Argonaute (Ago)-based nucleic acid detection system (MULAN), which was applied in RNA detection. The advantage of MULAN is that it possessed a detection resolution of a single base, which illustrated good performance in detecting mutant genotyping with a LOD of five copies per reaction.231
In summary, biosensing based on CRISPR-Cas and Argonaute proteins represents a conceptually novel paradigm for virus detection. Due to their unique biocharacters, they can be independently used to detect viruses with excellent sensitivity. The more important aspect is that they can be integrated with some existing methods to suit diverse needs, such as being coupled to fluorimeters,232 FETS,237 SERS,235 and electrochemical biosensors.226 These combinations have attracted researchers' interest, and have great potential in nucleic acid detection with high specificity and sensitivity.
9. Colorimetric biosensor
The colorimetric biosensor is an excellent inspection technology because it is sensitive, simple, rapid, and inexpensive. Colorimetric biosensors can rapidly detect SARS-CoV-2, especially in resource-limited regions, where external devices and reagents are not needed.238 The colorimetric detection technique is capable of providing results that are visible to the human eye. The miniature devices based on the colorimetric technique are popular because they are user-friendly, and they can capture images via electronic benchtops, as well as wearable devices. By applying nanomaterials, it can dramatically amplify the signal intensity of colorimetric biosensors and improve the sensitivity of biological target molecules.239 Gold nanoparticles (AuNPs) are widely used in biomedical colorimetric assays because they are simple to produce and physically stable. AuNPs exhibit a unique optoelectronic feature where the chromogenic effect is caused by the aggregation of the AuNPs.240 Karakus et al. developed a simple and selective AuNP-based biosensing platform that enables the colorimetric detection of the SARS-CoV-2 spike antigen at the level of 1 ng mL−1. Karakus et al. used the color change phenomenon caused by the rapid and irreversible aggregation of AuNPs when the antibody interacts with the antigen. Through this method, the color change can be readily observed by eye, and its LOD was 48 ng mL−1.241 Some performance comparisons of the colorimetric biosensors are summarized in Table 1.| Nanomaterial | Biological recognition element | Target | LOD | Ref. |
|---|---|---|---|---|
| AuNPs | ssDNA | Gene N | 10 copies per μL | 242 |
| Fe3O4/Au core–shell | Anti-spike protein antibodies | Protein spike | 1200 PFU mL−1 | 243 |
| AuNPs | Antibody monoclonal (mAb) | Protein spike | 48 ng mL−1 | 241 |
| AuNPs | Antigen | Protein N | 150 ng mL−1 | 244 |
| AuNPs | Antibody rabbit IgG | Protein N | 3 ng mL−1 | 245 |
| AuNPs-ACE2 | ACE2 receptor | Protein spike | 1.54 × 10−4 ng mL−1 | 246 |
| AuNPs-N | protein N-antigen | IgG, IgM eIgA | 247 | |
| Polyurethane polydiacetylene | Anti-N protein | Protein spike | 248 |
10. Mutation detection
SARS-CoV-2 has shown a strong capability for mutation, and various mutations dominate the epidemic at different periods in different countries. For detecting the mutation of SARS-CoV-2, the best way is to detect the RNA sequence of the SARS-CoV-2 virus. By identifying the existence of the mutated RNA sequences, the mutated SARS-CoV-2 can be detected. For the SERS, the base information located on the specific position of RNA can be detected, but it requires a special SERS substrate or special bio-probes. Lei Wu249 designed a stem-loop structure that can specifically hybridize a perfectly matched DNA sequence and the mutated DNA sequence (Fig. 14a). The closed loop does show a SERS signal, while the open loop does not have a SERS signal. This design has been applied in identifying the mutated DNA sequence of cancer cells. Jian-An Huang250 reported an approach to control the residence time of the DNA sequence through an Au hole (Fig. 14b). The DNA is absorbed in a single nanoparticle and passes through the Au hole, and the four DNA bases can be distinguished by analyzing the characteristic Raman peaks of the bases. Another method based on Raman spectroscopy in detecting the mutation of oligonucleotides is tip-enhanced Raman scattering (TERS). For TERS, the signals are mainly enhanced by the nanoscale gap between a silver tip and a metal substrate. Due to the small size of the tip, the TERS can resolve a single-stranded DNA/RNA molecule with a spatial resolution below 1 nm (Fig. 14c).251,255,256 This extremely high resolution provides a possibility of figuring out the type of mutated base and its position. For detection of mutation by using SPR127,128 (Fig. 15a–c), fluorescence methods252,257 (Fig. 15d), FET253 (Fig. 15e), and electrochemistry methods254 (Fig. 15f), the kinetic of the hybridization can reveal the existence of a mutation. Because of the existence of mutated bases, the kinetic rate is changed, which can be monitored by measuring the hybridization signal intensity vs. hybridization time. However, these methods cannot specifically indicate where the mutated base is located.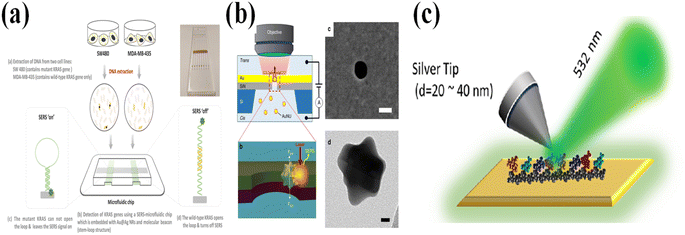 | ||
| Fig. 14 (a) The microfluidic chip for SERS analysis of cellular DNA mutations. The NDA and mutated DNA pass through the microfluidic channel coated with Au@Ag NRs. The mutant KRAS sequences maintain the stem-loop structure, resulting in the ‘SERS-on’ state, while the KRAS is disrupted due to the hybridization of target sequences with the molecular beacon resulting in the ‘SERS-off’ state. Adapted with permission from ref. 249. (b) The diagram of trapping the single DNA passing through the Au nanohole. By controlling the resident time, the specific base information of DNA can be extracted by plasmon effects induced by the Au and nanoparticles. Adapted with permission from ref. 250. (c) Diagram of identifying the base in the DNA/RNA sequence by using TESR. Adapted with permission from ref. 251. | ||
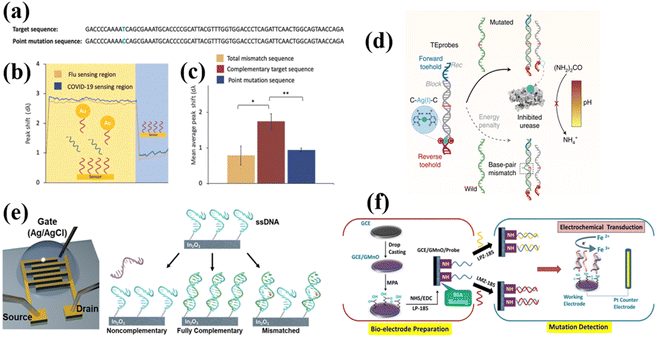 | ||
| Fig. 15 (a) The target and single basepoint mutation mimicking sequences. (b) The result of detecting a target and a single base mutated COVID-19 sequence (100 fM) by using SPR methods. (c) The wavelength signal shifts in both sensing regions in response to the working solution. Adapted with permission from ref. 128. (d) To precisely control the net thermodynamic energy of the strand displacement reaction induced by input RNAs, a double stranded DNA probe is designed to tune its forward and reverse toehold terminal sequences. The difference between mutant and wild-type viral RNAs is the strand displacement reaction based on the thermodynamic energy penalty derived from a single base pair mismatch. Recognition of viral RNAs leads to the release of a urease inhibitor metal ion, Ag(I), and thus controls the cleavage of urea and the level of NH4+. We identify key mutation markers of SARS-CoV-2 variants, such as alpha, beta, gamma and delta, and visualize them using pH indicators. Adapted with permission from ref. 252. (e) The structure of the FET in detecting mutated oligonucleotide, and FET functionalized with ssDNA that is exposed to non-complementary, fully complementary, or mismatched sequences. Adapted with permission from ref. 253. (f) Schematic representation of the fabrication of the electrochemical sensing platform, and the response principle of detecting mutated target sequence. Adapted with permission from ref. 254. | ||
11. Perspective
Table 2 illustrates the LOD and the detection time of biosensors applied in detecting SARS-CoV-2. All these biosensors have been applied to detect the real sample of the SARS-CoV-2 virus, and show good sensitivity and short detection time. The bio-probes such as ACE2, S protein antibody, and specific DNA sequences are selected to bind to the specific component of SARS-CoV-2. Among these biosensors, most of them rely on single optic or electric signal changes caused by the state changes on the sensing surface and the state changes, owing to bio-interaction between the probe and the target analyte. Consequently, the robust sample is always changing in identifying the SARS-CoV-2.| Methods | Analyte | LOD | Detection time | R2 for the linear fit | Selectivity | Ref. |
|---|---|---|---|---|---|---|
| SERS | SARS-CoV-2 spike protein | 1 fg mL−1 | 15 min | 0.9201 | 76 | |
| SARS-CoV-2 spike protein | 5 × 10−9 mol L−1 | — | 77 | |||
| Breath volatile organic compounds (BVOCs) | — | 5 min | 96.2% | 103 | ||
| SARS-CoV-2 virus in Crude water | 255 copies per L | 10 min | 0.9688 | 78 | ||
| SARS-CoV-2 spike protein | 0.77 fg mL−1 | 4 h | 0.99 | 79 | ||
| SARS-CoV-2-specific IgM/IgG antibodies | 100 fg mL−1 | 30 min | 90 | |||
| IgM/IgG | 1 pg mL−1 | 25 minutes | 0.99 | 84 | ||
| IgM/IgG | 0.1 nm mL−1 | 15 min | 85 | |||
| SPR | COVID-19 cDNA sequence | 1 pM | 3 min | 127 | ||
| COVID-19 NA sequence | 100 aM | Real-time | 128 | |||
| SARS-CoV-2 N protein | 85 fM | Real-time | 258 | |||
| Anti-SARS-CoV-2 antibodies in whole blood samples | 100 ng mL−1 | 30 min | 0.96 | 112 | ||
| SARS-CoV-2 in saliva | 1.6 × 103 copies per mL | 10 s | 133 | |||
| FET | SARS-CoV-2 antigen protein | 1 fg mL−1 in buff | Real-time | 139 | ||
| SARS-CoV-2 virus from clinical samples | 242 copy per mL | |||||
| SARS-CoV-2 RAN | 0.37 fM | Within 2 min | 0.997 | 31 | ||
| SARS-CoV-2 S protein antigens | 4.12 fg mL−1 | Real-time | 92% | 161 | ||
| N antigen of SARS-CoV-2 | 0.016 fg mL−1 | Within 5 min | 162 | |||
| RNA sequence of the SARS-CoV-2 virus | 10 fM | Real-time | 166 | |||
| SARS-CoV-2 S protein receptor | 10−3 mg mL−1 | — | 167 | |||
| SARS-CoV-2 virus | 25 fg μL−1 | Real-time | 168 | |||
| Fluorescence | SARS-CoV-2 IgG antibodies | 12.5 ng mL−1 | Within 7 min | 259 | ||
| SARS-CoV-2 virus in serum | — | ∼30 min | 0.998 | 100% | 184 | |
| SARS-CoV-2 virus | 52.5 pM | 1 h | 185 | |||
| RAN sequence of the SARS-CoV-2 | 0.0995 μM | 20 min | 0.9945 | 186 | ||
| SARS-CoV-2 RNA | 0.000823 μM | 25 min | 0.9825 | 187 | ||
| Electrochemical | SARS-CoV-2 spike antibodies in saliva/oropharyngeal swab | 0.01 ag mL−1 | 30 min | 0.98 | 206 | |
| RNA of SARS-CoV-2 | 200 copies per mL | 3 h | 0.999 | High | 207 | |
| RNA of SARS-CoV-2 | 1.0 pM | 25 min | High | 208 | ||
| S Protein of the SARS-CoV-2 | 66 pg mL−1 | 40 min | 0.9954 | 42.5% | 215 | |
| SARS-CoV-2 S protein | 200 ng mL−1 | 30 min | 0.999 | 216 | ||
| SARS-CoV-2 IgG and IgM | 0.96 ng mL−1 for IgG 0.14 ng mL−1 for IgM | 45 min | 0.9928 | 217 | ||
| SARS-CoV-2 N-protein | 56 fg mL−1 | 1.5 min | 0.99 | 218 |
SERS is an optic method with high accuracy because it reveals the finger print information of the molecules. During the SERS experiments, the characteristic peaks of the bio-probes and the target analytes can be observed. Due to the bio-interaction, the SERS intensity of the characteristic peaks of the probe can change or shift. By analyzing the appearance of the new peaks caused by the analytes and the changes in the probe SERS peaks, the existence of the SARS-CoV-2 can be detected. The SERS substrates are the key to enhanced Raman scattering. The chemically synthesized metallic nanostructures are applied in SERS, but the introduced chemical bond in synthesis always shows a strong fluorescence background or even a strong Raman signal. This unexpected optic matter highly affected SERS performance. Moreover, the repeatability of the SERS substrate has also affected the application of the SERS.
The SPR biosensor is highly affected by the RI of the sensing surface. The wavelength-dependent and angle-dependent biosensors are easily fabricated, while the architecture of the phase-dependent biosensors with higher sensitivity is more complicated. The SPR biosensor can be applied to detect SARS-CoV-2 in real-time. The SPR biosensor is highly affected by the temperature and the absorption of non-target analytes. In the measurement of the SRR, the reference liquid solvent has to be the same as the solvent that is applied for dissolving the analytes. This property limits the application of the SRR biosensors in the real sample.
FET biosensors rely on the changes in mobilities of channel materials caused by the bio-interaction, and the SPR biosensors are based on the “mass changing” of the sensing surface. FET is a continuous measurement that can be utilized in detecting SARS-CoV-2 in real-time. The channel material is the most important factor, which affects the performance and sensitivity. For 2D materials such as MoS2 and WSe2, it is hard to synthesize large areas of continuous 2D film. The application of the 2D flake is possible, but it needs a photograph to fabricate the source and drain electrodes. The large-scale chemical vapor deposition (CVD) graphene can be directly transferred on top of the prefabricated electrode, which reduces the fabrication difficulty. Moreover, the large-scale graphene with a uniform property makes the fabrication of the FET much easier, and the FET has more stable performance. Some other materials, such as carbon nanotubes and reduced graphene oxide, are spin-coated on the substrate and form a thin film. The inhomogeneous spin-coated film highly affects the performance of the FET and its reproducibility.
Fluorescence biosensors can detect SAR-CoV-2 at low concentrations with good sensitivity. For fluorescence biosensors, either bio-probes or analytes need to be modified with fluorophores, which can be excited and emit strong fluorescence. Normally, the modification requires several steps, and it requires an experienced biological background. This disadvantage limits its application in detecting a large number of samples. Moreover, the unmodified fluorophores and the fluorescence background should be eliminated, which requires expensive equipment.
Electrochemical biosensors have properties of high selectivity and sensitivity, but they suffer from a lack of surface architectures, allowing the desired biochemical event. The electrode material, surface modification methods, or its dimensions heavily influence its detection ability. The proper architecture of the working electrodes can improve the specification and sensitivity of virus detection. The unexpected absorption of the organic or bio-compounds on the surface of working electrodes can result in the reduction of the working electrodes' mobilities and cause the failure of detection. As a surface sensing technique, it also is affected by the unexpected absorption of the non-target molecules. The detecting conditions, such as stirring, pH, and temperature, also affect the detecting reliabilities.
Until now, all of these nano-biosensors have been applied to detecting the real SARS-CoV-2 samples. Due to the applications of bio-probes, SARS-CoV-2 can be detected under good selectivity and sensitivity. In comparison with the pure analytes, the LOD is two or three orders of magnitudes larger. As surface sensing techniques, the bio events that occur on the surface are important and require some time. The “real-time” effects are always observed during the measurement in FET or SPR, but a cleaning step to remove the unexpectedly absorbed compounds is missing, which can confirm whether the signals are generated by the bio-interaction or not.
Until now, the re-utilization of the nano-biosensor has been rarely mentioned. For the SARS-CoV-2 nano-biosensor, the key scientific issue is to develop a reusable sensing surface such that the chemical or physically absorbed organic or biomolecules can be easily removed, and the sensitivity can be largely recovered. By conquering such a problem, the oropharyngeal swabs sampled from healthy people cannot affect the sensing surface, which enormously reduces the cost of detecting SARS-CoV-2.
12. Conclusion
COVID-19 has caused a huge disaster in the world. The pandemic of SARS-CoV-2 forced people to find a fast, efficient, easy, and cost-efficient detecting method. As a golden standard, the RT-PCR method meets the major bottleneck of lacking sufficient equipment, un-reusable reagents, and cost-consuming. Nowadays, there is a growing requirement for novel nano-biosensors with high selectivity and sensitivity. The principles of the SARS-CoV-2 nano-biosensor are an integration of physical/chemical principles with the bio-principle. The ACE2-S protein, ACE2-SARS-CoV-2 virus, S protein-antibody, antibody-IgG, antibody-IgM, and DNA probe-SARS-CoV-2 target oligonucleotide are the most used bio-pairs in the detection of SARS-CoV-2. By using the proper bio-probe, the nano-biosensors can selectively bind to the specific part of SARS-CoV-2, and the interactions between the probe and the target can be observed via optic or electrical signals. Moreover, some nano-biosensors possess the possibility of cyclic utilization, which reduces the cost of epidemiological investigation. Until now, the biosensor can reach a LOD of 1 copy per μL for the SARS-CoV-2 target oligonucleotide and 0.01 ag mL−1 for the SARS-CoV-2 spike antibody. The extremely low detection limits indicate a promising future for biosensors in detecting SARS-CoV-2.In conclusion, this review highlights the current trends in SARS-CoV-2 nano-biosensors combined with optic and electric methods, and the improvement of these nano-biosensors. To face the present COVID-19 pandemic or the effective virus epidemic, it is necessary to have efficient virus sensing techniques at hand to help aid in the detection and prevention of viral spread.
Author contributions
Conceptualization, Yansheng Liu; writing—original draft preparation, Yansheng Liu and Zhenle Qin; review and editing, Jin Zhou, Xiaobo Jia, Hongli Li, Xiaohong Wang, Yating Chen, Zijun Sun, Xiong He, and Hongda Li; funding acquisition, Yansheng Liu and Guofu Wang; supervision, Haixin Chang; all authors have read and agreed to the published version of the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work has been supported by the Guangxi Natural Science Foundation Project (No. 2021GXNSFBA196049), Guangxi Science and Technology Project (No. AD22035215), the National Key Research and Development Program of China (No. 2022YFE0134600 and No. 2021YFA0715404) and Guangxi Key Research and Development Program (No. 2021AB05083).References
- Y.-D. Li, W.-Y. Chi, J.-H. Su, L. Ferrall, C.-F. Hung and T. C. Wu, J. Biomed. Sci., 2020, 27, 104 CrossRef CAS PubMed.
- S. Lei, F. Jiang, W. Su, C. Chen, J. Chen, W. Mei, L.-Y. Zhan, Y. Jia, L. Zhang and D. Liu, EClinicalMedicine, 2020, 21, 100331 CrossRef PubMed.
- E. A. Meyerowitz, A. Richterman, R. T. Gandhi and P. E. Sax, Ann. Intern. Med., 2020, 174, 69–79 CrossRef PubMed.
- W. Trypsteen, J. Van Cleemput, W. V. Snippenberg, S. Gerlo and L. Vandekerckhove, PLoS Pathog., 2020, 16, e1009037 CrossRef CAS PubMed.
- A. S. Lauring, M. W. Tenforde, J. D. Chappell, M. Gaglani, A. A. Ginde, T. McNeal, S. Ghamande, D. J. Douin, H. K. Talbot, J. D. Casey, N. M. Mohr, A. Zepeski, N. I. Shapiro, K. W. Gibbs, D. C. Files, D. N. Hager, A. Shehu, M. E. Prekker, H. L. Erickson, M. C. Exline, M. N. Gong, A. Mohamed, N. J. Johnson, V. Srinivasan, J. S. Steingrub, I. D. Peltan, S. M. Brown, E. T. Martin, A. S. Monto, A. Khan, C. L. Hough, L. W. Busse, C. C. Ten Lohuis, A. Duggal, J. G. Wilson, A. J. Gordon, N. Qadir, S. Y. Chang, C. Mallow, C. Rivas, H. M. Babcock, J. H. Kwon, N. Halasa, C. G. Grijalva, T. W. Rice, W. B. Stubblefield, A. Baughman, K. N. Womack, J. P. Rhoads, C. J. Lindsell, K. W. Hart, Y. Zhu, K. Adams, S. J. Schrag, S. M. Olson, M. Kobayashi, J. R. Verani, M. M. Patel and W. H. Self, medRxiv: the preprint server for health sciences, 2022, DOI:10.1101/2022.02.06.22270558.
- T. P. Velavan and C. G. Meyer, Trop. Med. Int. Health, 2020, 25, 278–280 CrossRef CAS PubMed.
- J. Cui, F. Li and Z.-L. Shi, Nat. Rev. Microbiol., 2019, 17, 181–192 CrossRef CAS PubMed.
- M. Yuce, E. Filiztekin and K. G. Ozkaya, Biosens. Bioelectron., 2021, 172, 112752 CrossRef CAS PubMed.
- N. Zhu, D. Zhang, W. Wang, X. Li, B. Yang, J. Song, X. Zhao, B. Huang, W. Shi, R. Lu, P. Niu, F. Zhan, X. Ma, D. Wang, W. Xu, G. Wu, G. F. Gao and W. Tan, N. Engl. J. Med., 2020, 382, 727–733 CrossRef CAS PubMed.
- R. A. Khailany, M. Safdar and M. Ozaslan, Gene Rep., 2020, 19, 100682 CrossRef CAS PubMed.
- A. C. Walls, Y.-J. Park, M. A. Tortorici, A. Wall, A. T. McGuire and D. Veesler, Cell, 2020, 181, 281–292.e286 CrossRef CAS PubMed.
- S. Rosales-Mendoza, V. A. Márquez-Escobar, O. González-Ortega, R. Nieto-Gómez and J. I. Arévalo-Villalobos, Vaccines, 2020, 8, 183 CrossRef CAS PubMed.
- P. Yang and X. Wang, Cell. Mol. Immunol., 2020, 17, 555–557 CrossRef CAS PubMed.
- E. Ong, M. U. Wong, A. Huffman and Y. He, Front. Immunol., 2020, 11, 1581 CrossRef CAS PubMed.
- A. B. Ryzhikov, G. S. Onkhonova, I. R. Imatdinov, E. V. Gavrilova, R. A. Maksyutov, E. A. Gordeeva, G. V. Pazynina, I. M. Ryzhov, N. V. Shilova and N. V. Bovin, Biochemistry, 2021, 86, 243–247 CAS.
- L. Wang and Y. Xiang, Viruses, 2020, 12, 1289 CrossRef CAS PubMed.
- S. Xiaojie, L. Yu, Y. Lei, Y. Guang and Q. Min, Stem Cell Res., 2020, 50, 102125 CrossRef PubMed.
- A. C. Walls, Y. J. Park, M. A. Tortorici, A. Wall, A. T. McGuire and D. Veesler, Cell, 2020, 181, 281–292.e286 CrossRef CAS PubMed.
- Y. Huang, C. Yang, X.-f. Xu, W. Xu and S.-w. Liu, Acta Pharmacol. Sin., 2020, 41, 1141–1149 CrossRef PubMed.
- M. Hoffmann, H. Kleine-Weber, S. Schroeder, N. Krüger, T. Herrler, S. Erichsen, T. S. Schiergens, G. Herrler, N.-H. Wu, A. Nitsche, M. A. Müller, C. Drosten and S. Pöhlmann, Cell, 2020, 181, 271–280.e278 CrossRef CAS PubMed.
- S. Shahbaz, L. Xu, M. Osman, W. Sligl, J. Shields, M. Joyce, D. L. Tyrrell, O. Oyegbami and S. Elahi, Stem Cell Rep., 2021, 16, 1165–1181 CrossRef CAS PubMed.
- L. Shi, L. Wang, X. Ma, X. Fang, L. Xiang, Y. Yi, J. Li, Z. Luo and G. Li, Anal. Chem., 2021, 93, 16646–16654 CrossRef CAS PubMed.
- A. Tahamtan and A. Ardebili, Expert Rev. Mol. Diagn., 2020, 20, 453–454 CrossRef CAS PubMed.
- L. Lan, D. Xu, G. Ye, C. Xia, S. Wang, Y. Li and H. Xu, Jama, 2020, 323, 1502–1503 CrossRef CAS PubMed.
- A. Avula, K. Nalleballe, N. Narula, S. Sapozhnikov, V. Dandu, S. Toom, A. Glaser and D. Elsayegh, Brain, Behav., Immun., 2020, 87, 115–119 CrossRef CAS PubMed.
- A. Singanayagam, M. Patel, A. Charlett, J. L. Bernal, V. Saliba, J. Ellis, S. Ladhani, M. Zambon and R. Gopal, Eurosurveillance, 2020, 25, 2001483 CrossRef CAS PubMed.
- Y. Gong, K. Li, N. Copner, H. Liu, M. Zhao, B. Zhang, A. Pusch, D. L. Huffaker and S. S. Oh, Nanophotonics, 2021, 10, 1285–1293 CrossRef CAS.
- L. Guo, L. Ren, S. Yang, M. Xiao, D. Chang, F. Yang, C. S. Dela Cruz, Y. Wang, C. Wu, Y. Xiao, L. Zhang, L. Han, S. Dang, Y. Xu, Q.-W. Yang, S.-Y. Xu, H.-D. Zhu, Y.-C. Xu, Q. Jin, L. Sharma, L. Wang and J. Wang, Clin. Infect. Dis., 2020, 71, 778–785 CrossRef CAS PubMed.
- R. A. Marsh and J. S. Orange, Ann. Allergy, Asthma, Immunol., 2019, 123, 444–453 CrossRef CAS PubMed.
- M. Hofreiter, V. Jaenicke, D. Serre, A. v. Haeseler and S. Pääbo, Nucleic Acids Res., 2001, 29, 4793–4799 CrossRef CAS PubMed.
- J. Li, D. Wu, Y. Yu, T. Li, K. Li, M. M. Xiao, Y. Li, Z. Y. Zhang and G. J. Zhang, Biosens. Bioelectron., 2021, 183, 113206 CrossRef CAS PubMed.
- R. Yan, Y. Zhang, Y. Li, L. Xia, Y. Guo and Q. Zhou, Science, 2020, 367, 1444–1448 CrossRef CAS PubMed.
- F. Li, W. Li, M. Farzan and S. C. Harrison, Science, 2005, 309, 1864–1868 CrossRef CAS PubMed.
- J. F.-W. Chan, K.-H. Kok, Z. Zhu, H. Chu, K. K.-W. To, S. Yuan and K.-Y. Yuen, Emerging Microbes Infect., 2020, 9, 221–236 CrossRef CAS PubMed.
- J. Brand, Notes Rec. R. Soc. Lond., 1989, 43, 1–23 CrossRef.
- C. V. Raman, Indian J. Phys., 1928, 2, 387–398 CAS.
- R. Krishnan and R. Shankar, J. Raman Spectrosc., 1981, 10, 1–8 CrossRef CAS.
- W. E. Huang, M. Li, R. M. Jarvis, R. Goodacre and S. A. Banwart, in Advances in applied microbiology, Elsevier, 2010, vol. 70, pp. 153–186 Search PubMed.
- E. C. Le Ru and P. G. Etchegoin, Annu. Rev. Phys. Chem., 2012, 63, 65–87 CrossRef CAS PubMed.
- K. A. Willets, Chem. Soc. Rev., 2014, 43, 3854–3864 RSC.
- X. Ling, S. Huang, S. Deng, N. Mao, J. Kong, M. S. Dresselhaus and J. Zhang, Acc. Chem. Res., 2015, 48, 1862–1870 CrossRef CAS PubMed.
- Y. Liu and F. Luo, Nano Res., 2019, 12, 2788–2795 CrossRef CAS.
- S. Xu, S. Jiang, J. Wang, J. Wei, W. Yue and Y. Ma, Sens. Actuators, B, 2016, 222, 1175–1183 CrossRef CAS.
- S. Nie and S. R. Emory, Science, 1997, 275, 1102–1106 CrossRef CAS PubMed.
- F. Schedin, E. Lidorikis, A. Lombardo, V. G. Kravets, A. K. Geim, A. N. Grigorenko, K. S. Novoselov and A. C. Ferrari, ACS Nano, 2010, 4, 5617–5626 CrossRef CAS PubMed.
- X. Huang, I. H. El-Sayed, W. Qian and M. A. El-Sayed, J. Am. Chem. Soc., 2006, 128, 2115–2120 CrossRef CAS PubMed.
- X. Ling, L. Xie, Y. Fang, H. Xu, H. Zhang, J. Kong, M. S. Dresselhaus, J. Zhang and Z. Liu, Nano Lett., 2010, 10, 553–561 CrossRef CAS PubMed.
- Y. Liu and F. Luo, Nano Res., 2020, 13, 138–144 CrossRef CAS.
- J. Langer, D. Jimenez de Aberasturi, J. Aizpurua, R. A. Alvarez-Puebla, B. Auguie, J. J. Baumberg, G. C. Bazan, S. E. J. Bell, A. Boisen, A. G. Brolo, J. Choo, D. Cialla-May, V. Deckert, L. Fabris, K. Faulds, F. J. Garcia de Abajo, R. Goodacre, D. Graham, A. J. Haes, C. L. Haynes, C. Huck, T. Itoh, M. Kall, J. Kneipp, N. A. Kotov, H. Kuang, E. C. Le Ru, H. K. Lee, J. F. Li, X. Y. Ling, S. A. Maier, T. Mayerhofer, M. Moskovits, K. Murakoshi, J. M. Nam, S. Nie, Y. Ozaki, I. Pastoriza-Santos, J. Perez-Juste, J. Popp, A. Pucci, S. Reich, B. Ren, G. C. Schatz, T. Shegai, S. Schlucker, L. L. Tay, K. G. Thomas, Z. Q. Tian, R. P. Van Duyne, T. Vo-Dinh, Y. Wang, K. A. Willets, C. Xu, H. Xu, Y. Xu, Y. S. Yamamoto, B. Zhao and L. M. Liz-Marzan, ACS Nano, 2020, 14, 28–117 CrossRef CAS PubMed.
- A. Otto, I. Mrozek, H. Grabhorn and W. Akemann, J. Phys.: Condens. Matter, 1992, 4, 1143 CrossRef CAS.
- S. Yang, X. Dai, B. B. Stogin and T.-S. Wong, Proc. Natl. Acad. Sci. U. S. A., 2016, 113, 268–273 CrossRef CAS PubMed.
- W. Fan, Y. H. Lee, S. Pedireddy, Q. Zhang, T. Liu and X. Y. Ling, Nanoscale, 2014, 6, 4843–4851 RSC.
- Y. Zhao, D. Yang, X. Li, Y. Liu, X. Hu, D. Zhou and Y. Lu, Nanoscale, 2017, 9, 1087–1096 RSC.
- A. A. Balandin, S. Ghosh, W. Bao, I. Calizo, D. Teweldebrhan, F. Miao and C. N. Lau, Nano Lett., 2008, 8, 902–907 CrossRef CAS PubMed.
- P. Reokrungruang, I. Chatnuntawech, T. Dharakul and S. Bamrungsap, Sens. Actuators, B, 2019, 285, 462–469 CrossRef CAS.
- Y. Liu, J. Deng, Z. Jin, T. Liu, J. Zhou, F. Luo and G. Wang, Nano Res., 2022, 15, 6062–6066 CrossRef CAS.
- Y. Wu, X. Wang, X. Wen, J. Zhu, X. Bai, T. Jia, H. Yang, L. Zhang and Y. Qi, Phys. Lett. A, 2020, 126544 CrossRef CAS.
- M. Vadai, D. K. Angell, F. Hayee, K. Sytwu and J. A. Dionne, Nat. Commun., 2018, 9, 1–8 CrossRef CAS PubMed.
- J.-A. Huang, M. Z. Mousavi, Y. Zhao, A. Hubarevich, F. Omeis, G. Giovannini, M. Schütte, D. Garoli and F. De Angelis, Nat. Commun., 2019, 10, 5321 CrossRef PubMed.
- Y. Liu and F. Luo, Nano Res., 2019, 1–8 Search PubMed.
- L. Zhang, H. Chang, A. Hirata, H. Wu, Q.-K. Xue and M. Chen, ACS Nano, 2013, 7, 4595–4600 CrossRef CAS PubMed.
- J. U. Lee, W. H. Kim, H. S. Lee, K. H. Park and S. J. Sim, Small, 2019, 15, 1804968 CrossRef PubMed.
- S. G. Park, X. Xiao, J. Min, C. Mun, H. S. Jung, V. Giannini, R. Weissleder, S. A. Maier, H. Im and D. H. Kim, Adv. Funct. Mater., 2019, 29, 1904257 CrossRef CAS.
- M. Q. He, S. Chen, K. Yao, K. Wang, Y. L. Yu and J. H. Wang, Small Methods, 2019, 3, 1900017 CrossRef.
- Q. Lv, H. Min, D. B. Duan, W. Fang, G. M. Pan, A. G. Shen, Q. Q. Wang, G. Nie and J. M. Hu, Adv. Healthcare Mater., 2019, 8, 1801257 Search PubMed.
- P. Wang, M. Xia, O. Liang, K. Sun, A. F. Cipriano, T. Schroeder, H. Liu and Y. H. Xie, Anal. Chem., 2015, 87, 10255–10261 CrossRef CAS PubMed.
- L. Zhang, X. Li, Y. Wang, Q. Fu, Y. Tan, H. Wang, H. Chen and J. Zhou, Sens. Actuators, B, 2019, 279, 503–508 CrossRef CAS.
- A. K. Nair, K. Bhavitha, S. Perumbilavil, P. Sankar, D. Rouxel, M. Kala, S. Thomas and N. Kalarikkal, Carbon, 2018, 132, 380–393 CrossRef CAS.
- C. Carrillo-Carrión, R. Martínez, M. F. Navarro Poupard, B. Pelaz, E. Polo, A. Arenas-Vivo, A. Olgiati, P. Taboada, M. G. Soliman and Ú. Catalán, Angew. Chem., Int. Ed., 2019, 58, 7078–7082 CrossRef PubMed.
- Q. Shi, D. E. Gómez, D. Dong, D. Sikdar, R. Fu, Y. Liu, Y. Zhao, D. M. Smilgies and W. Cheng, Adv. Mater., 2019, 31, 1900989 CrossRef PubMed.
- X. Xia, J. Zeng, B. McDearmon, Y. Zheng, Q. Li and Y. Xia, Angew. Chem., Int. Ed., 2011, 50, 12542–12546 CrossRef CAS PubMed.
- Z. Li, Y. Gao, L. Zhang, Y. Fang and P. Wang, Nanoscale, 2018, 10, 18720–18727 RSC.
- J. R. Wang, C. Xia, L. Yang, Y. F. Li, C. M. Li and C. Z. Huang, Anal. Chem., 2020, 92, 4046–4052 CrossRef CAS PubMed.
- S. Ganesh, K. Venkatakrishnan and B. Tan, Nat. Commun., 2020, 11, 1135 CrossRef CAS PubMed.
- J.-A. Huang, M. Z. Mousavi, Y. Zhao, A. Hubarevich, F. Omeis, G. Giovannini, M. Schütte, D. Garoli and F. De Angelis, Nat. Commun., 2019, 10, 5321 CrossRef PubMed.
- O. J. Achadu, N. Nwaji, D. Lee, J. Lee, E. M. Akinoglu, M. Giersig and E. Y. Park, Nanoscale Adv., 2022, 4, 871–883 RSC.
- Y. Peng, C. Lin, L. Long, T. Masaki, M. Tang, L. Yang, J. Liu, Z. Huang, Z. Li, X. Luo, J. R. Lombardi and Y. Yang, Nano-Micro Lett., 2021, 13, 52 CrossRef PubMed.
- D. Zhang, X. Zhang, R. Ma, S. Deng, X. Wang, X. Wang, X. Zhang, X. Huang, Y. Liu, G. Li, J. Qu, Y. Zhu and J. Li, Water Res., 2021, 200, 117243 CrossRef CAS PubMed.
- M. Zhang, X. Li, J. Pan, Y. Zhang, L. Zhang, C. Wang, X. Yan, X. Liu and G. Lu, Biosens. Bioelectron., 2021, 190, 113421 CrossRef CAS PubMed.
- L. Tong, H. Xu and M. Käll, MRS Bull., 2014, 39, 163–168 CrossRef CAS.
- C. Zhang, C. Li, J. Yu, S. Jiang, S. Xu, C. Yang, Y. J. Liu, X. Gao, A. Liu and B. Man, Sens. Actuators, B, 2018, 258, 163–171 CrossRef CAS.
- L. Ma, Y.-L. Chen, D.-J. Yang, S.-J. Ding, L. Xiong, P.-L. Qin and X.-B. Chen, J. Phys. Chem. C, 2020, 124, 25473–25479 CrossRef CAS.
- U. Dinish, G. Balasundaram, Y.-T. Chang and M. Olivo, Sci. Rep., 2014, 4, 1–7 Search PubMed.
- H. Liu, E. Dai, R. Xiao, Z. Zhou, M. Zhang, Z. Bai, Y. Shao, K. Qi, J. Tu, C. Wang and S. Wang, Sens. Actuators, B, 2021, 329, 129196 CrossRef CAS PubMed.
- S. Chen, L. Meng, L. Wang, X. Huang, S. Ali, X. Chen, M. Yu, M. Yi, L. Li, X. Chen, L. Yuan, W. Shi and G. Huang, Sens. Actuators, B, 2021, 348, 130706 CrossRef CAS PubMed.
- W. Liu, L. Liu, G. Kou, Y. Zheng, Y. Ding, W. Ni, Q. Wang, L. Tan, W. Wu and S. Tang, J. Clin. Microbiol., 2020, 58(6) DOI:10.1128/jcm.00461-20.
- R. Banerjee and A. Jaiswal, Analyst, 2018, 143, 1970–1996 RSC.
- L. Perico, S. Tomasoni, T. Peracchi, A. Perna, A. Pezzotta, G. Remuzzi and A. Benigni, EBioMedicine, 2020, 61, 103069 CrossRef PubMed.
- T. Nicol, C. Lefeuvre, O. Serri, A. Pivert, F. Joubaud, V. Dubée, A. Kouatchet, A. Ducancelle, F. Lunel-Fabiani and H. Le Guillou-Guillemette, J. Clin. Virol., 2020, 129, 104511 CrossRef CAS PubMed.
- S. Srivastav, A. Dankov, M. Adanalic, R. Grzeschik, V. Tran, S. Pagel-Wieder, F. Gessler, I. Spreitzer, T. Scholz, B. Schnierle, O. E. Anastasiou, U. Dittmer and S. Schlücker, Anal. Chem., 2021, 93, 12391–12399 CrossRef CAS PubMed.
- S. Yadav, M. A. Sadique, P. Ranjan, N. Kumar, A. Singhal, A. K. Srivastava and R. Khan, ACS Appl. Bio Mater., 2021, 4, 2974–2995 CrossRef CAS PubMed.
- S. Sloan-Dennison, E. O’Connor, J. W. Dear, D. Graham and K. Faulds, Anal. Bioanal. Chem., 2022, 414, 4541–4549 CrossRef CAS PubMed.
- R. Chen, C. Ren, M. Liu, X. Ge, M. Qu, X. Zhou, M. Liang, Y. Liu and F. Li, ACS Nano, 2021, 15, 8996–9004 CrossRef CAS PubMed.
- K. Kim, L. Kashefi-Kheyrabadi, Y. Joung, K. Kim, H. Dang, S. G. Chavan, M.-H. Lee and J. Choo, Sens. Actuators, B, 2021, 329, 129214 CrossRef CAS PubMed.
- J. Huang, J. Wen, M. Zhou, S. Ni, W. Le, G. Chen, L. Wei, Y. Zeng, D. Qi, M. Pan, J. Xu, Y. Wu, Z. Li, Y. Feng, Z. Zhao, Z. He, B. Li, S. Zhao, B. Zhang, P. Xue, S. He, K. Fang, Y. Zhao and K. Du, Anal. Chem., 2021, 93, 9174–9182 CrossRef CAS PubMed.
- F. Lussier, V. Thibault, B. Charron, G. Q. Wallace and J.-F. Masson, TrAC, Trends Anal. Chem., 2020, 124, 115796 CrossRef CAS.
- X. Lin, D. Lin, Y. Chen, J. Lin, S. Weng, J. Song and S. Feng, Adv. Funct. Mater., 2021, 31, 2103382 CrossRef CAS.
- H. Shin, S. Oh, S. Hong, M. Kang, D. Kang, Y.-g. Ji, B. H. Choi, K.-W. Kang, H. Jeong and Y. Park, ACS Nano, 2020, 14, 5435–5444 CrossRef CAS PubMed.
- W. J. Thrift, S. Ronaghi, M. Samad, H. Wei, D. G. Nguyen, A. S. Cabuslay, C. E. Groome, P. J. Santiago, P. Baldi and A. I. Hochbaum, ACS Nano, 2020, 14, 15336–15348 CrossRef CAS PubMed.
- Z. Fang, W. Wang, A. Lu, Y. Wu, Y. Liu, C. Yan and C. Han, Rapid classification of honey varieties by surface enhanced Raman scattering combining with deep learning, 2018 Cross Strait Quad-Regional Radio Science and Wireless Technology Conference (CSQRWC), IEEE, 2018, pp. 1–3 Search PubMed.
- S. Weng, W. Zhu, X. Zhang, H. Yuan, L. Zheng, J. Zhao, L. Huang and P. Han, Artif. Intell. Agric., 2019, 3, 1–10 Search PubMed.
- C. Carlomagno, D. Bertazioli, A. Gualerzi, S. Picciolini, P. I. Banfi, A. Lax, E. Messina, J. Navarro, L. Bianchi, A. Caronni, F. Marenco, S. Monteleone, C. Arienti and M. Bedoni, Sci. Rep., 2021, 11, 4943 CrossRef CAS PubMed.
- S. X. Leong, Y. X. Leong, E. X. Tan, H. Y. F. Sim, C. S. L. Koh, Y. H. Lee, C. Chong, L. S. Ng, J. R. T. Chen, D. W. C. Pang, L. B. T. Nguyen, S. K. Boong, X. Han, Y. C. Kao, Y. H. Chua, G. C. Phan-Quang, I. Y. Phang, H. K. Lee, M. Y. Abdad, N. S. Tan and X. Y. Ling, ACS Nano, 2022, 16, 2629–2639 CrossRef CAS PubMed.
- N. Queralto, A. N. Berliner, B. Goldsmith, R. Martino, P. Rhodes and S. H. Lim, J. Breath Res., 2014, 8, 027112 CrossRef CAS PubMed.
- S. Grassin-Delyle, C. Roquencourt, P. Moine, G. Saffroy, S. Carn, N. Heming, J. Fleuriet, H. Salvator, E. Naline and L.-J. Couderc, EBioMedicine, 2021, 63, 103154 CrossRef CAS PubMed.
- D. M. Ruszkiewicz, D. Sanders, R. O'Brien, F. Hempel, M. J. Reed, A. C. Riepe, K. Bailie, E. Brodrick, K. Darnley and R. Ellerkmann, EClinicalMedicine, 2020, 29, 100609 CrossRef PubMed.
- A. Amann, M. Corradi, P. Mazzone and A. Mutti, Expert Rev. Mol. Diagn., 2011, 11, 207–217 CrossRef CAS PubMed.
- K. Badjagbo, Clin. Chem. Lab. Med., 2012, 50, 1893–1902 CrossRef CAS PubMed.
- B. Liedberg, C. Nylander and I. Lundström, Biosens. Bioelectron., 1995, 10, i–ix CrossRef CAS PubMed.
- I. Alves, C. Park and V. Hruby, Curr. Protein Pept. Sci., 2005, 6, 293–312 CrossRef CAS PubMed.
- P. Singh, Sens. Actuators, B, 2016, 229, 110–130 CrossRef CAS.
- J. H. Qu, K. Leirs, W. Maes, M. Imbrechts, N. Callewaert, K. Lagrou, N. Geukens, J. Lammertyn and D. Spasic, ACS Sens., 2022, 7, 477–487 CrossRef CAS PubMed.
- J. N. Dash and R. Jha, IEEE Photonics Technol. Lett., 2014, 26, 595–598 CAS.
- K. Matsubara, S. Kawata and S. Minami, Appl. Opt., 1988, 27, 1160–1163 CrossRef CAS PubMed.
- L. D. Maria and M. Martinelli, Sens. Actuators, A, 1992, 32, 710–712 CrossRef.
- X. Zhao, Z. Wang, F. Liang, Y. Cao and H. Xu, Chinese J. Anal. Chem., 1998, 26, 1320–1323 CAS.
- Y.-F. Chang, W.-H. Wang, Y.-W. Hong, R.-Y. Yuan and K.-H. Chen, Anal. Chem., 2018, 90, 1861–1869 CrossRef CAS PubMed.
- Y.-F. Chang, W.-H. Wang, Y.-W. Hong, R.-Y. Yuan, K.-H. Chen, Y.-W. Huang, P.-L. Lu, Y.-H. Chen, Y.-M. A. Chen, L.-C. Su and S.-F. Wang, Anal. Chem., 2018, 90, 1861–1869 CrossRef CAS PubMed.
- K. E. B. Thornton, D. Uttamchandani and B. Culshaw, Electron. Lett., 1986, 22, 1232–1234 CrossRef.
- J. Homola, J. Dostálek, S. Chen, A. Rasooly, S. Jiang and S. S. Yee, Int. J. Food Microbiol., 2002, 75, 61–69 CrossRef CAS PubMed.
- X. Liu, Anal. Biochem., 2003, 314, 301–309 CrossRef CAS PubMed.
- Y. Tian, W. Zhang, D. Song, Y. Cao, X. Liu, H. Zhang and X. U. Han, Chem. Res. Chin. Univ., 2004, 25, 441–444 CAS.
- H. Yan, G. Lai-xu, L. Song-quan, S. Wen-ling and Y. Hong-an, Research on Surface Plasmon Resonance Sensor Based on Wavelength Modulation by Using Theoretical Simulation Resonance Sensor Based on Wavelength Modulation by Using Theoretical Simulation, 2011 Third International Conference on Measuring Technology and Mechatronics Automation, Shanghai, China, 2011, pp. 83–86, DOI:10.1109/ICMTMA.2011.592.
- S. P. Ng, C. M. L. Wu, S. Y. Wu and H. P. Ho, Opt. Express, 2011, 19, 4521–4527 CrossRef CAS PubMed.
- P. Hlubina, J. Luňáček, D. Ciprian and R. Chlebus, Opt. Commun., 2008, 281, 2349–2354 CrossRef CAS.
- Q. Kemao, Opt. Lasers Eng., 2007, 45, 304–317 CrossRef.
- G. Qiu, Z. Gai, Y. Tao, J. Schmitt, G. A. Kullak-Ublick and J. Wang, ACS Nano, 2020, 14, 5268–5277 CrossRef CAS PubMed.
- W. K. Yeung, S. C. Lo, S. H. Wang, P. K. Wei and J. Y. Cheng, Analyst, 2021, 146, 5584–5591 RSC.
- I. Plikusiene, V. Maciulis, A. Ramanaviciene, Z. Balevicius, E. Buzavaite-Verteliene, E. Ciplys, R. Slibinskas, M. Simanavicius, A. Zvirbliene and A. Ramanavicius, J. Colloid Interface Sci., 2021, 594, 195–203 CrossRef CAS PubMed.
- C. M. Das, Y. Guo, G. Yang, L. Kang, G. Xu, H. P. Ho and K. T. Yong, Adv. Theory Simul., 2020, 3, 2000185 CrossRef CAS PubMed.
- M. Couture, Y. Liang, H.-P. P. Richard, R. Faid, W. Peng and J.-F. Masson, Nanoscale, 2013, 5, 12399–12408 RSC.
- J. Song, P. Huang, H. Duan and X. Chen, J. Acc. Chem. Res., 2015, 48, 2506–2515 CrossRef CAS PubMed.
- A. Samavati, Z. Samavati, M. Velashjerdi, A. Fauzi Ismail, M. H. D. Othman, B. G. Eisaabadi, M. Sohaimi Abdullah, M. Bolurian and M. Bolurian, Chem. Eng. J., 2021, 420, 127655 CrossRef CAS PubMed.
- E. Cesewski and B. N. Johnson, Biosens. Bioelectron., 2020, 159, 112214 CrossRef CAS PubMed.
- Y. Liang, M. Xiao, D. Wu, Y. Lin, L. Liu, J. He, G. Zhang, L.-M. Peng and Z. Zhang, ACS Nano, 2020, 14, 8866–8874 CrossRef CAS PubMed.
- L. Syedmoradi, A. Ahmadi, M. L. Norton and K. Omidfar, Microchim. Acta, 2019, 186, 739 CrossRef CAS PubMed.
- Y. Song, Y. Luo, C. Zhu, H. Li, D. Du and Y. Lin, Biosens. Bioelectron., 2016, 76, 195–212 CrossRef CAS PubMed.
- S. Mao, J. Chang, H. Pu, G. Lu, Q. He, H. Zhang and J. Chen, Chem. Soc. Rev., 2017, 46, 6872–6904 RSC.
- G. Seo, G. Lee, M. J. Kim, S. H. Baek, M. Choi, K. B. Ku, C. S. Lee, S. Jun, D. Park, H. G. Kim, S. J. Kim, J. O. Lee, B. T. Kim, E. C. Park and S. I. Kim, ACS Nano, 2020, 14, 5135–5142 CrossRef CAS PubMed.
- P. Blake, E. Hill, A. Castro Neto, K. Novoselov, D. Jiang, R. Yang, T. Booth and A. Geim, Appl. Phys. Lett., 2007, 91, 063124 CrossRef.
- C. N. R. Rao, A. K. Sood, K. S. Subrahmanyam and A. Govindaraj, Angew. Chem., Int. Ed., 2009, 48, 7752–7777 CrossRef CAS PubMed.
- K. S. Novoselov, A. K. Geim, S. V. Morozov, D. Jiang, Y. Zhang, S. V. Dubonos, I. V. Grigorieva and A. A. Firsov, Science, 2004, 306, 666–669 CrossRef CAS PubMed.
- K. S. Novoselov, A. K. Geim, S. V. Morozov, D. Jiang, M. I. Katsnelson, I. V. Grigorieva, S. V. Dubonos and A. A. Firsov, Nature, 2005, 438, 197–200 CrossRef CAS PubMed.
- Y. Zhang, Y.-W. Tan, H. L. Stormer and P. Kim, Nature, 2005, 438, 201–204 CrossRef CAS PubMed.
- K. S. Novoselov, Z. Jiang, Y. Zhang, S. Morozov, H. L. Stormer, U. Zeitler, J. Maan, G. Boebinger, P. Kim and A. K. Geim, Science, 2007, 315, 1379 CrossRef CAS PubMed.
- M. Y. Han, B. Özyilmaz, Y. Zhang and P. Kim, Phys. Rev. Lett., 2007, 98, 206805 CrossRef PubMed.
- M. C. Lemme, T. J. Echtermeyer, M. Baus and H. Kurz, IEEE Electron Device Lett., 2007, 28, 282–284 CAS.
- Y. Zhang, J. P. Small, W. V. Pontius and P. Kim, Appl. Phys. Lett., 2005, 86, 073104 CrossRef.
- A. C. Ferrari, J. Meyer, V. Scardaci, C. Casiraghi, M. Lazzeri, F. Mauri, S. Piscanec, D. Jiang, K. Novoselov and S. Roth, Phys. Rev. Lett., 2006, 97, 187401 CrossRef CAS PubMed.
- J. S. Bunch, A. M. Van Der Zande, S. S. Verbridge, I. W. Frank, D. M. Tanenbaum, J. M. Parpia, H. G. Craighead and P. L. McEuen, Science, 2007, 315, 490–493 CrossRef CAS PubMed.
- P. Blake, P. D. Brimicombe, R. R. Nair, T. J. Booth, D. Jiang, F. Schedin, L. A. Ponomarenko, S. V. Morozov, H. F. Gleeson and E. W. Hill, Nano Lett., 2008, 8, 1704–1708 CrossRef PubMed.
- X. Huang, X. Qi, F. Boey and H. Zhang, Chem. Soc. Rev., 2012, 41, 666–686 RSC.
- X. Du, I. Skachko, A. Barker and E. Y. Andrei, Nat. Nanotechnol., 2008, 3, 491 CrossRef CAS PubMed.
- X. Jin, H. Zhang, Y. T. Li, M. M. Xiao, Z. L. Zhang, D. W. Pang, G. Wong, Z. Y. Zhang and G. J. Zhang, Microchim. Acta, 2019, 186, 223 CrossRef PubMed.
- L. Wu, J. Guo, Q. Wang, S. Lu, X. Dai, Y. Xiang and D. Fan, Sens. Actuators, B, 2017, 249, 542–548 CrossRef CAS.
- S. Islam, S. Shukla, V. K. Bajpai, Y.-K. Han, Y. S. Huh, A. Kumar, A. Ghosh and S. Gandhi, Biosens. Bioelectron., 2019, 126, 792–799 CrossRef CAS PubMed.
- D. Wu, Y. Yu, D. Jin, M.-M. Xiao, Z.-Y. Zhang and G.-J. Zhang, Anal. Chem., 2020, 92, 4006–4015 CrossRef CAS PubMed.
- Y. Yu, Y. T. Li, D. Jin, F. Yang, D. Wu, M. M. Xiao, H. Zhang, Z. Y. Zhang and G. J. Zhang, Anal. Chem., 2019, 91, 10679–10686 CrossRef CAS PubMed.
- M. T. Hwang, I. Park, M. Heiranian, A. Taqieddin, S. You, V. Faramarzi, A. A. Pak, A. M. van der Zande, N. R. Aluru and R. Bashir, Adv. Mater. Technol., 2021, 6, 2100712 CrossRef CAS PubMed.
- E. Piccinini, G. E. Fenoy, A. L. Cantillo, J. A. Allegretto, J. Scotto, J. M. Piccinini, W. A. Marmisollé and O. Azzaroni, Adv. Mater. Interfaces, 2022, 9, 2102526 CrossRef CAS PubMed.
- M. A. Zamzami, G. Rabbani, A. Ahmad, A. A. Basalah, W. H. Al-Sabban, S. Nate Ahn and H. Choudhry, Bioelectrochemistry, 2022, 143, 107982 CrossRef CAS PubMed.
- W. Shao, M. R. Shurin, S. E. Wheeler, X. He and A. Star, ACS Appl. Mater. Interfaces, 2021, 13, 10321–10327 CrossRef CAS PubMed.
- J. Kong, R. Franklin Nathan, C. Zhou, G. Chapline Michael, S. Peng, K. Cho and H. Dai, Science, 2000, 287, 622–625 CrossRef CAS PubMed.
- M. S. Dresselhaus, G. Dresselhaus and P. C. Eklund, Science of fullerenes and carbon nanotubes: their properties and applications, Elsevier, 1996 Search PubMed.
- K. Balasubramanian and K. Kern, Adv. Mater., 2014, 26, 1154–1175 CrossRef CAS PubMed.
- M. Thanihaichelvan, S. N. Surendran, T. Kumanan, U. Sutharsini, P. Ravirajan, R. Valluvan and T. Tharsika, Mater. Today: Proc., 2022, 49, 2546–2549 CAS.
- J. Wei, Z. Zhao, F. Luo, K. Lan, R. Chen and G. Qin, 2D Mater., 2021, 9, 015030 CrossRef.
- P. Fathi-Hafshejani, N. Azam, L. Wang, M. A. Kuroda, M. C. Hamilton, S. Hasim and M. Mahjouri-Samani, ACS Nano, 2021, 15, 11461–11469 CrossRef CAS PubMed.
- N. Masurkar, N. K. Thangavel, S. Yurgelevic, S. Varma, G. W. Auner and L. M. Reddy Arava, Biosens. Bioelectron., 2021, 172, 112724 CrossRef CAS PubMed.
- K. Kalantar-zadeh and J. Z. Ou, ACS Sens., 2016, 1, 5–16 CrossRef CAS.
- K. S. Novoselov, A. K. Geim, S. V. Morozov, D. Jiang, M. I. Katsnelson, I. Grigorieva and S. Dubonos, Firsov and AA, Nature, 2005, 438, 197–200 CrossRef CAS PubMed.
- A. K. Geim and K. S. Novoselov, in Nanoscience and Technology: A Collection of Reviews from Nature Journals, World Scientific, 2010, pp. 11–19 Search PubMed.
- X. Ling, W. Fang, Y. H. Lee, P. T. Araujo, X. Zhang, J. F. Rodriguez-Nieva, Y. Lin, J. Zhang, J. Kong and M. S. Dresselhaus, Nano Lett., 2014, 14, 3033–3040 CrossRef CAS PubMed.
- T. Shimada, K. Hamaguchi, A. Koma and F. Ohuchi, Appl. Phys. Lett., 1998, 72, 1869–1871 CrossRef CAS.
- H. Jiang, J. Phys. Chem. C, 2012, 116, 7664–7671 CrossRef CAS.
- W. Liu, J. Kang, D. Sarkar, Y. Khatami, D. Jena and K. Banerjee, Nano Lett., 2013, 13, 1983–1990 CrossRef CAS PubMed.
- B. Doppagne, T. Neuman, R. Soria-Martinez, L. E. P. López, H. Bulou, M. Romeo, S. Berciaud, F. Scheurer, J. Aizpurua and G. Schull, Nat. Nanotechnol., 2020, 15, 207–211 CrossRef CAS PubMed.
- R. Weissleder and V. Ntziachristos, Nat. Med., 2003, 9, 123–128 CrossRef CAS PubMed.
- X. Chen, K. Xu, J. Li, M. Yang, X. Li, Q. Chen, C. Lu and H. Yang, Biosens. Bioelectron., 2020, 155, 112104 CrossRef CAS PubMed.
- E. Benito-Pena, M. G. Valdes, B. Glahn-Martinez and M. C. Moreno-Bondi, Anal. Chim. Acta, 2016, 943, 17–40 CrossRef CAS PubMed.
- M. Bauch, K. Toma, M. Toma, Q. Zhang and J. Dostalek, Plasmonics, 2014, 9, 781–799 CrossRef CAS PubMed.
- M. Wang, Y. Lin, J. Lu, Z. Sun, Y. Deng, L. Wang, Y. Yi, J. Li, J. Yang and G. Li, Chem. Eng. J., 2022, 429, 132332 CrossRef CAS PubMed.
- S. K. Krishnan, E. Singh, P. Singh, M. Meyyappan and H. S. Nalwa, RSC Adv., 2019, 9, 8778–8881 RSC.
- N. C. Cady, N. Tokranova, A. Minor, N. Nikvand, K. Strle, W. T. Lee, W. Page, E. Guignon, A. Pilar and G. N. Gibson, Biosens. Bioelectron., 2021, 171, 112679 CrossRef CAS PubMed.
- B. Kang, Y. Lee, J. Lim, D. Yong, Y. Ki Choi, S. Woo Yoon, S. Seo, S. Jang, S. Uk Son, T. Kang, J. Jung, K.-S. Lee, M. H. Kim and E.-K. Lim, Chem. Eng. J., 2022, 442, 136143 CrossRef CAS PubMed.
- G. R. Bardajee, M. Zamani, M. Sharifi, H. Rezanejad and M. Motallebi, J. Fluoresc., 2022, 32, 1959–1967 CrossRef CAS PubMed.
- G. R. Bardajee, M. Zamani, H. Mahmoodian, H. Elmizadeh, H. Yari, L. Jouyandeh, R. Shirkavand and M. Sharifi, Spectrochim. Acta, Part Ac, 2022, 269, 120702 CrossRef CAS PubMed.
- N. H. T. Tran, K. T. L. Trinh, J. H. Lee, W. J. Yoon and H. Ju, Small, 2018, 14, 1801385 CrossRef PubMed.
- Z. Zhu, P. Yuan, S. Li, M. Garai, M. Hong and Q.-H. Xu, ACS Appl. Bio Mater., 2018, 1, 118–124 CrossRef CAS.
- K. E. Sapsford, L. Berti and I. L. Medintz, Angew. Chem., Int. Ed. Engl., 2006, 45, 4562–4589 CrossRef CAS PubMed.
- J. Han, H. Y. Zou, M. X. Gao and C. Z. Huang, Talanta, 2016, 148, 279–284 CrossRef CAS PubMed.
- F. Tian, J. Lyu, J. Shi and M. Yang, Biosens. Bioelectron., 2017, 89, 123–135 CrossRef CAS PubMed.
- E. Ploetz, E. Lerner, F. Husada, M. Roelfs, S. Chung, J. Hohlbein, S. Weiss and T. Cordes, Sci. Rep., 2016, 6, 33257 CrossRef CAS PubMed.
- R. Roy, S. Hohng and T. Ha, Nat. Methods, 2008, 5, 507 CrossRef CAS PubMed.
- E. A. Jares-Erijman and T. M. Jovin, Nat. Biotechnol., 2003, 21, 1387 CrossRef CAS PubMed.
- K. Girigoswami and N. Akhtar, Int. J. Nano Dimens., 2019, 10, 1–17 CAS.
- H. Ueda and J. Dong, Biochim. Biophys. Acta, Proteins Proteomics, 2014, 1844, 1951–1959 CrossRef CAS PubMed.
- X. Li, Z.-Q. Dong, P. Yu, L.-P. Wang, X.-D. Niu, H. Yamaguchi and D.-C. Li, Phys. Fluids, 2021, 33, 042004 CrossRef CAS PubMed.
- G. Balkourani, A. Brouzgou, M. Archonti, N. Papandrianos, S. Song and P. Tsiakaras, J. Electroanal. Chem., 2021, 893, 115289 CrossRef CAS PubMed.
- F. Zhao, Y. Bai, L. Cao, G. Han, C. Fang, S. Wei and Z. Chen, J. Electroanal. Chem., 2020, 867, 114184 CrossRef CAS.
- H. Ilkhani and S. Farhad, Anal. Biochem., 2018, 557, 151–155 CrossRef CAS PubMed.
- Q. Gong, H. Han, H. Yang, M. Zhang, X. Sun, Y. Liang, Z. Liu, W. Zhang and J. Qiao, J. Materiomics, 2019, 5, 313–319 CrossRef.
- G. Martins, J. L. Gogola, F. R. Caetano, C. Kalinke, T. R. Jorge, C. N. D. Santos, M. F. Bergamini and L. H. Marcolino-Junior, Talanta, 2019, 204, 163–171 CrossRef CAS PubMed.
- J. Huang, J. Meng, S. Chen, S. Zhang, T. Liu, C. Li and F. Wang, Biosens. Bioelectron., 2020, 164, 112310 CrossRef CAS PubMed.
- J. Wang and J. W. Schultze, Angew. Chem., Int. Ed. Engl., 1996, 35, 1998 CrossRef.
- L. Liv, Microchem. J., 2021, 168, 106445 CrossRef CAS PubMed.
- H. Zhao, F. Liu, W. Xie, T.-C. Zhou, J. OuYang, L. Jin, H. Li, C.-Y. Zhao, L. Zhang, J. Wei, Y.-P. Zhang and C.-P. Li, Sens. Actuators, B, 2021, 327, 128899 CrossRef CAS PubMed.
- L. Farzin, S. Sadjadi, A. Sheini and E. Mohagheghpour, Mikrochim. Acta, 2021, 188, 121 CrossRef CAS PubMed.
- N. J. Ronkainen, H. B. Halsall and W. R. Heineman, Chem. Soc. Rev., 2010, 39, 1747–1763 RSC.
- V. K. Gupta, R. Jain, K. Radhapyari, N. Jadon and S. Agarwal, Anal. Biochem., 2011, 408, 179 CrossRef CAS PubMed.
- F. Patolsky, Y. Weizmann and I. Willner, J. Am. Chem. Soc., 2002, 124, 770–772 CrossRef CAS PubMed.
- A. J. Blasco, M. C. González and A. Escarpa, Anal. Chim. Acta, 2004, 511, 71–81 CrossRef CAS.
- B. J. Venton and D. J. DiScenza, in Electrochemistry for Bioanalysis, ed. B. Patel, Elsevier, 2020, pp. 27–50, DOI:10.1016/B978-0-12-821203-5.00004-X.
- P. Westbroek, in Analytical Electrochemistry in Textiles, ed. P. Westbroek, G. Priniotakis and P. Kiekens, Woodhead Publishing, 2005, pp. 37–69, DOI:10.1533/9781845690878.1.37.
- J. C. Abrego-Martinez, M. Jafari, S. Chergui, C. Pavel, D. Che and M. Siaj, Biosens. Bioelectron., 2022, 195, 113595 CrossRef CAS PubMed.
- J. Zeng, P. A. Duarte, Y. Ma, O. Savchenko, L. Shoute, Y. Khaniani, S. Babiuk, R. Zhuo, G. N. Abdelrasoul, C. Charlton, J. N. Kanji, L. Babiuk, C. Edward and J. Chen, Biosens. Bioelectron., 2022, 213, 114476 CrossRef CAS PubMed.
- A. Yakoh, U. Pimpitak, S. Rengpipat, N. Hirankarn, O. Chailapakul and S. Chaiyo, Biosens. Bioelectron., 2021, 176, 112912 CrossRef CAS PubMed.
- R. Salahandish, P. Jalali, H. O. Tabrizi, J. E. Hyun, F. Haghayegh, M. Khalghollah, A. Zare, B. M. Berenger, Y. D. Niu, E. Ghafar-Zadeh and A. Sanati-Nezhad, Biosens. Bioelectron., 2022, 213, 114459 CrossRef CAS PubMed.
- S. D. Bukkitgar, N. P. Shetti, R. M. Kulkarni, K. R. Reddy, S. S. Shukla, V. S. Saji and T. M. Aminabhavi, J. Electrochem. Soc., 2019, 166, B3072–B3078 CrossRef CAS.
- N. P. Shetti, S. J. Malode, D. S. Nayak, G. B. Bagihalli, S. S. Kalanur, R. S. Malladi, C. V. Reddy, T. M. Aminabhavi and K. R. Reddy, Appl. Surf. Sci., 2019, 496, 143656 CrossRef CAS.
- S. D. Bukkitgar, N. P. Shetti and T. M. Aminabhavi, Chem. Eng. J., 2021, 420 CrossRef CAS PubMed.
- Y. Peng, Y. Pan, Z. Sun, J. Li, Y. Yi, J. Yang and G. Li, Biosens. Bioelectron., 2021, 186, 113309 CrossRef CAS PubMed.
- Y. Deng, T. Zhou, Y. Peng, M. Wang, L. Xiang, Y. Zhang, J. Li, J. Yang and G. Li, Anal. Chem., 2023, 95, 3358–3362 CrossRef CAS PubMed.
- Y. Deng, Y. Peng, L. Wang, M. Wang, T. Zhou, L. Xiang, J. Li, J. Yang and G. Li, Anal. Chim. Acta, 2022, 1208, 339846 CrossRef CAS PubMed.
- Q. Liang, Y. Huang, M. Wang, D. Kuang, J. Yang, Y. Yi, H. Shi, J. Li, J. Yang and G. Li, Chem. Eng. J., 2023, 452, 139646 CrossRef CAS PubMed.
- L. Yin, S. Man, S. Ye, G. Liu and L. Ma, Biosens. Bioelectron., 2021, 193, 113541 CrossRef CAS PubMed.
- J. S. Park, K. Hsieh, L. Chen, A. Kaushik, A. Y. Trick and T. H. Wang, Adv. Sci., 2021, 8, 2003564 CrossRef CAS PubMed.
- A. Ramachandran, D. A. Huyke, E. Sharma, M. K. Sahoo, C. Huang, N. Banaei, B. A. Pinsky and J. G. Santiago, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 29518–29525 CrossRef CAS PubMed.
- A. Santiago-Frangos, A. Nemudryi, A. Nemudraia, T. Wiegand, J. E. Nichols, P. Krishna, A. M. Scherffius, T. R. Zahl, R. A. Wilkinson and B. Wiedenheft, Methods, 2022, 205, 1–10 CrossRef CAS PubMed.
- L. Wang, R. He, B. Lv, X. Yu, Y. Liu, J. Yang, W. Li, Y. Wang, H. Zhang, G. Yan, W. Mao, L. Liu, F. Wang and L. Ma, Talanta, 2021, 227, 122154 CrossRef CAS PubMed.
- X. Ye, H. Zhou, X. Guo, D. Liu, Z. Li, J. Sun, J. Huang, T. Liu, P. Zhao, H. Xu, K. Li, H. Wang, J. Wang, L. Wang, W. Zhao, Q. Liu, S. Xu and Y. Feng, Biosens. Bioelectron., 2022, 207, 114169 CrossRef CAS PubMed.
- Y. Li, J. Qiao, X. Han, Z. Zhao, J. Kou, W. Zhang, S. Man and L. Ma, Viruses, 2022, 15, 120 CrossRef PubMed.
- F. E. Chen, P. W. Lee, A. Y. Trick, J. S. Park, L. Chen, K. Shah, H. Mostafa, K. C. Carroll, K. Hsieh and T. H. Wang, Biosens. Bioelectron., 2021, 190, 113390 CrossRef CAS PubMed.
- A. Santiago-Frangos, L. N. Hall, A. Nemudraia, A. Nemudryi, P. Krishna, T. Wiegand, R. A. Wilkinson, D. T. Snyder, J. F. Hedges, C. Cicha, H. H. Lee, A. Graham, M. A. Jutila, M. P. Taylor and B. Wiedenheft, Cell Rep. Med., 2021, 2, 100319 CrossRef CAS PubMed.
- L. Ma, W. Zhang, L. Yin, Y. Li, J. Zhuang, L. Shen and S. Man, J. Hazard. Mater., 2023, 452, 131195 CrossRef CAS PubMed.
- F. Wang, J. Yang, R. He, X. Yu, S. Chen, Y. Liu, L. Wang, A. Li, L. Liu, C. Zhai and L. Ma, Biosens. Bioelectron., 2021, 177, 112932 CrossRef CAS PubMed.
- J. Li, L. Tang, T. Li, K. Li, Y. Zhang, W. Ni, M. M. Xiao, Y. Zhao, Z. Y. Zhang and G. J. Zhang, ACS Sens., 2022, 7, 2680–2690 CrossRef CAS PubMed.
- Q. H. Nguyen and M. I. Kim, Trends Anal. Chem., 2020, 132, 116038 CrossRef CAS PubMed.
- L. Tessaro, A. Aquino, P. Panzenhagen, N. Joshi and C. A. Conte-Junior, J. Pharm. Biomed. Anal., 2023, 222, 115087 CrossRef CAS PubMed.
- Z. Ali, R. Aman, A. Mahas, G. S. Rao, M. Tehseen, T. Marsic, R. Salunke, A. K. Subudhi, S. M. Hala, S. M. Hamdan, A. Pain, F. S. Alofi, A. Alsomali, A. M. Hashem, A. Khogeer, N. A. M. Almontashiri, M. Abedalthagafi, N. Hassan and M. M. Mahfouz, Virus Res., 2020, 288, 198129 CrossRef CAS PubMed.
- E. Karakus, E. Erdemir, N. Demirbilek and L. Liv, Anal. Chim. Acta, 2021, 1182, 338939 CrossRef CAS PubMed.
- M. Alafeef, P. Moitra, K. Dighe and D. Pan, Nat. Protoc., 2021, 16, 3141–3162 CrossRef CAS PubMed.
- W. Ren and J. Irudayaraj, Biosensors, 2021, 11, 488 CrossRef CAS PubMed.
- K. Behrouzi and L. Lin, Biosens. Bioelectron., 2022, 195, 113669 CrossRef CAS PubMed.
- A. Vaquer, A. Alba-Patino, C. Adrover-Jaume, S. M. Russell, M. Aranda, M. Borges, J. Mena, A. Del Castillo, A. Socias, L. Martin, M. M. Arellano, M. Agudo, M. Gonzalez-Freire, M. Besalduch, A. Clemente, E. Baron and R. de la Rica, Sens. Actuators, B, 2021, 345, 130347 CrossRef CAS PubMed.
- A. L. Ferreira, L. F. de Lima, M. T. Torres, W. R. de Araujo and C. de la Fuente-Nunez, ACS Nano, 2021, 15, 17453–17462 CrossRef CAS PubMed.
- S. Cavalera, B. Colitti, S. Rosati, G. Ferrara, L. Bertolotti, C. Nogarol, C. Guiotto, C. Cagnazzo, M. Denina, F. Fagioli, F. Di Nardo, M. Chiarello, C. Baggiani and L. Anfossi, Talanta, 2021, 223, 121737 CrossRef CAS PubMed.
- A. Bhattacharjee, R. M. Sabino, J. Gangwish, V. K. Manivasagam, S. James, K. C. Popat, M. Reynolds and Y. V. Li, In Vitro Models, 2022, 1, 241–247 CrossRef.
- L. Wu, A. Garrido-Maestu, J. R. L. Guerreiro, S. Carvalho, S. Abalde-Cela, M. Prado and L. Diéguez, Nanoscale, 2019, 11, 7781–7789 RSC.
- J.-A. Huang, M. Z. Mousavi, Y. Zhao, A. Hubarevich, F. Omeis, G. Giovannini, M. Schütte, D. Garoli and F. De Angelis, Nat. Commun., 2019, 10, 1–10 CrossRef PubMed.
- Z. He, Z. Han, M. Kizer, R. J. Linhardt, X. Wang, A. M. Sinyukov, J. Wang, V. Deckert, A. V. Sokolov, J. Hu and M. O. Scully, J. Am. Chem. Soc., 2019, 141, 753–757 CrossRef CAS PubMed.
- T. Zhang, R. Deng, Y. Wang, C. Wu, K. Zhang, C. Wang, N. Gong, R. Ledesma-Amaro, X. Teng and C. Yang, Nat. Biomed. Eng., 2022, 1–11 Search PubMed.
- K. M. Cheung, J. M. Abendroth, N. Nakatsuka, B. Zhu, Y. Yang, A. M. Andrews and P. S. Weiss, Nano Lett., 2020, 20, 5982–5990 CrossRef CAS PubMed.
- S. Tripathy, R. Gangwar, P. Supraja, A. N. Rao, S. R. K. Vanjari and S. G. Singh, Electroanalysis, 2018, 30, 2110–2120 CrossRef CAS.
- R. Zhang, X. Zhang, H. Wang, Y. Zhang, S. Jiang, C. Hu, Y. Zhang, Y. Luo and Z. Dong, Angew. Chem., Int. Ed., 2017, 56, 5561–5564 CrossRef CAS PubMed.
- F. Pashaee, M. Tabatabaei, F. A. Caetano, S. S. Ferguson and F. Lagugne-Labarthet, Analyst, 2016, 141, 3251–3258 RSC.
- K. H. Ooi, M. M. Liu, J. W. D. Tay, S. Y. Teo, P. Kaewsapsak, S. Jin, C. K. Lee, J. Hou, S. Maurer-Stroh and W. Lin, Nat. Commun., 2021, 12, 1–23 CrossRef PubMed.
- T. A. Yano, T. Kajisa, M. Ono, Y. Miyasaka, Y. Hasegawa, A. Saito, K. Otsuka, A. Sakane, T. Sasaki, K. Yasutomo, R. Hamajima, Y. Kanai, T. Kobayashi, Y. Matsuura, M. Itonaga and T. Yasui, Sci. Rep., 2022, 12, 1060 CrossRef CAS PubMed.
- D. Song, J. Liu, W. Xu, X. Han, H. Wang, Y. Cheng, Y. Zhuo and F. Long, Talanta, 2021, 235, 122800 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2023 |