Regulation of micropatterned curvature-dependent FA heterogeneity on cytoskeleton tension and nuclear DNA synthesis of malignant breast cancer cells†
Yongtao
Wang‡
 *a,
Nana
Wang‡
b,
Yazhou
Chen
c and
Yingjun
Yang
*a,
Nana
Wang‡
b,
Yazhou
Chen
c and
Yingjun
Yang
 *d
*d
aSchool of Medicine, Shanghai University, Shanghai, 200444, China. E-mail: yongtao_wang@shu.edu.cn
bDepartment of Pediatrics, Shanghai General Hospital, Shanghai Jiao Tong University, Shanghai, 200080, China
cMedical 3D Printing center, The First Affiliated Hospital of Zhengzhou University, Zhengzhou University, Zhengzhou, 450052, China
dMaterials Institute of Atomic and Molecular Science, Shaanxi University of Science and Technology, Xi’an, 710021, China. E-mail: yang.yingjun@sust.edu.cn
First published on 26th November 2022
Abstract
Breast cancer is considered as a worldwide disease due to its high incidence and malignant metastasis. Although numerous techniques have been developed well to conduct breast cancer therapy, the influence of micropattern-induced interfacial heterogeneity on the molecular mechanism and nuclear signalling transduction of carcinogenesis is rarely announced. In this study, PDMS stencil-assisted micropatterns were fabricated on tissue culture plates to manage cell clustering colony by adjusting initial cell seeding density and the size of microholes. The curvature of each microholes was controlled to construct the interfacial heterogeneity of MDA-MB231 cancer cells at the periphery of micropatterned colony. The distinguished focal adhesion (FA) and cytoskeleton distribution at the central and peripheral regions of the cell colony were regulated by heterogeneous properties. The interfacial heterogeneity of FA and cytoskeleton would induce the biased tension force to encourage more ezrin expression at the periphery and further promote DNA synthesis, therefore disclosing a stem-like phenotype in heterogeneous cells. This study will provide a value source of information for the development of micropattern-induced heterogeneity and the interpretation of metastatic mechanism in malignant breast cancer cells.
Introduction
The high morbidity and mortality of breast cancers have severely endangered female life during the last decade and has led to breast cancers becoming a worldwide malignant tumor.1–3 Excessive division and distant metastasis of cancerous cells are two predominant reasons that generate undesirable prognosis and even the malignant death of breast cancer patients.4,5 Although massive technologies of breast cancer treatment including surgery, postoperative radiotherapy/chemotherapy, photothermal/photodynamic therapy, and their combination have been developed well for efficient breast cancer ablation, exploring pathogenic inducement in breast cancer cells still remains a serious challenge due to the sophisticated multilevel stages of malignant metastasis.6–10 In the variant process, an abnormal modification of functional protein glycosylation occurs in breast cancer cells to regulate the conformation, positioning, and stability of glycoprotein.11,12 These revolutions can participate in governing receptor activation, cytoskeleton distribution, recognition, and response to stimulate intracellular genome and DNA activity of the nuclei.13–16 Therefore, it is urgently inspired to disclose the molecular mechanism and intracellular signalling transduction in metastatic carcinogenesis of breast cancer.The cells are enveloped in both neighbouring cells and extracellular matrix (ECM) in the surrounding microenvironment to sense numerous biochemical and biophysical cues.17,18 In addition to cell–cell interaction, the intracellular microenvironment can play a crucial role in affecting cell functions, such as cell adhesion and spreading, cell proliferation and differentiation, cell migration and apoptosis.19–23 To mimic microniches in vivo, various engineering strategies, including stencil-assisted patterning, photolithography, and microprinting, are exploited to reveal the influence of cell morphology and topography on cell behaviours in engineered micropatterns.24–26 In particular, it has been reported that the geometric performances of single cells including cell size, shape, elongation, and chirality enable the balancing of cell division, endocytosis internalization, and DNA synthesis.27–30 Compared with single-cell manipulation, cell clustering colony can accurately imitate multicellular complicated microenvironment to explore intracellular cytoskeleton tension distribution and nuclear molecule-level expression of genomes and DNA chains.31,32 Furthermore, the cells regularly display heterogeneity in the peripheral region of the micropatterned colony to communicate distinctive phenotypes.33–36 The spatial heterogeneity of interfacial geometry is recognized to determine morphogenesis and tumorigenesis in cancer stem cells.37
Although the heterogeneous architecture and tumorigenic evolution are extensively studied to ascertain the mechanism of recurrence and metastasis, the influence of morphologic cues of interfacial topography on cellular pluripotency and nuclear activity is obscure in micropatterned cancer cells. Therefore, in this study, PDMS stencil-assisted patterning method was applied to investigate the influence of cell-density-dependent FA heterogeneity on cytoskeleton tension distribution and nuclear DNA synthesis. PDMS stencils composed of various circular microholes with different diameters were prepared to precisely dominate the size of micropatterned colony. Breast cancer cells with different seeding densities were seeded on the stencils and cultured to construct the clustering colony with a particular patterning size and cell density. The heterogeneous FAs and cytoskeleton structures were formed at the peripheral region of the micropatterned colony to stimulate abundant ezrin expression on the breast cancer cell membrane. The interfacial heterogeneity could also transfer biased cytoskeleton tension into nuclei to induce DNA synthesis activity and a stem-like phenotype at the periphery. The results will inspire the establishment of micropattern-induced heterogeneity to understand unbalanced tension distribution and the tumorigenesis of breast cancer cells.
Materials and methods
Fabrication and characterization of PDMS stencils
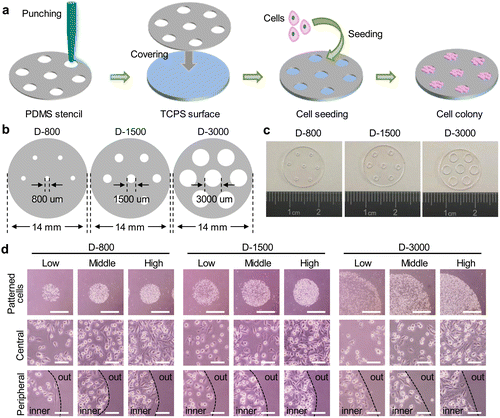
PDMS stencils were fabricated by the straightforward punching method.38 In brief, the PDMS substrates were a commercial film with the thickness of 100 μm (Hangzhou Bald Advanced Materials Technology Co., Ltd, China). First, the PDMS films were cut into a certain circle with a diameter of 1.4 cm using a scissor. The circle-shaped films were immersed in 75% ethanol for 30 min to remove the residual components of PDMS and washed with deionized water. After that, the differential punchers with inner diameters of 800, 1500, and 3000 μm were separately used to drill the through-film microholes on PDMS stencils. Three-type microholes were defined as D-800, D-1500, and D-3000, respectively. Before cell seeding, the perforated PDMS stencils were sterilized in 75% ethanol for 30 min and followed by with 3 PBS washes. Finally, the PDMS stencils were replaced into a 24-well cell culture plate for cell experiment.Cell culture
Human breast cancer cell line (MDA-MB231 cells) was purchased from Chinese Shanghai Aiyan Biology Science and Technology Co., Ltd. The cells were fast thawed in 37 °C water bath and subcultured in DMEM medium (Xi’an Mishu Biotechnology Co., Ltd, China). DMEM medium was supplemented with 10% FBS (Biological Industries Israel Beit Haemek Ltd, Israel) and 1% penicillin-streptomycin (Xi’an Mishu Biotechnology Co., Ltd, China). After cell confluence in a 75 cm2 flask, MDA-MB231 cells were detached with EDTA/trypsin to harvest the cell pellet through centrifugation. The obtained cell pellet was re-suspended in DMEM medium to form the uniform cell solution with 5 × 104, 10 × 104, and 20 × 104 cells mL−1 cell density. 1 mL cell suspension solution was respectively added onto each PDMS stencil for cell seeding to control the cell density. Three-type cell seeding densities were defined as low, middle, and high cell density. After 6 h cell incubation, the medium was replaced with fresh DMEM medium. The cells were further cultured for 18 h to maintain different cell densities and morphologies.Nucleus staining for cell density regulation
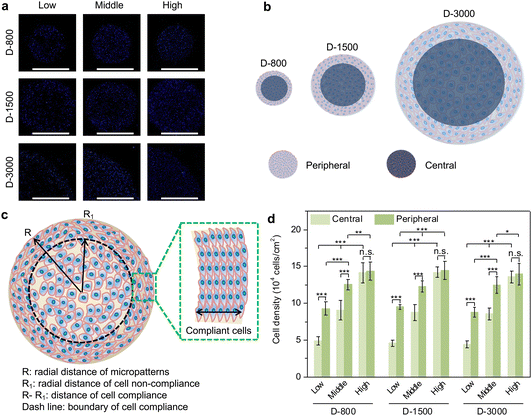
After the cells were cultured in the microholes of stencils for 24 h, the PDMS stencils were removed by direct peeling to show the cell colony. Then, the samples were followed with 3 PBS washes and fixed with 4% cold paraformaldehyde (Shanghai Aladdin Biochemical Technology Co., Ltd, China) for 10 min. The fixed cells were treated with 1% Triton X-100 for 10 min and the nucleus was stained with 1% DAPI (Shandong Sparkjade Scientific Instruments Co., Ltd, China) for 10 min. The DAPI fluorescence of the stained cells was observed by an MF52-N fluorescence microscope (Guangzhou Micro-shot Technology Co., Ltd, China). The fluorescence images were used to regulate the cell density of each micropattern. According to microscopic observation, the cell density was not only calculated due to different seeding densities but also differentiated between the central region and peripheral region of each colony. Considering that the central region was set to the same area with the peripheral region, the width of the peripheral ring was calculated to be 117 μm for 800 μm holes, 219 μm for 1500 μm holes and 439 μm for 3000 μm holes. The cell number at the peripheral and central regions was counted to analyze the cell density by particle analysis using the ImageJ software. Five independent assays were carried out to calculate the mean and standard deviation (SD).Immunofluorescent staining of vinculin
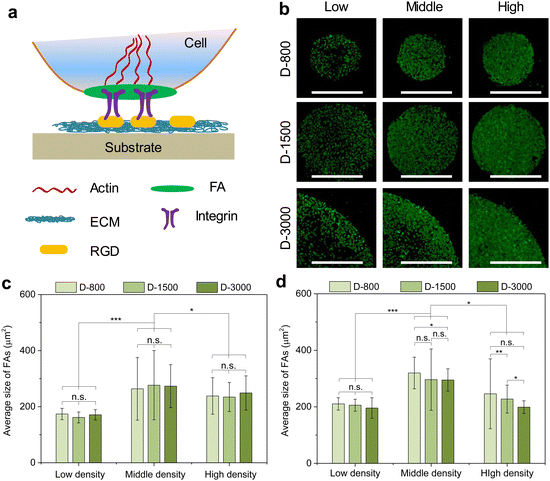
Vinculin was stained to evaluate the formation of focal adhesion at the peripheral and central regions of cell colonies. MDA-MB231 cells were cultured in the microholes of stencils for 24 h, and the stencils were directly peeled off to expose the cell colony and followed with 3 PBS washes. The samples were fixed with 4% cold paraformaldehyde solution for 10 min, treated with Triton X-100 for 10 min, and Tween-20 for 30 min. After that, the treated samples were blocked with 2% BSA solution for 30 min. 1% mouse antivinculin primary antibody (Merck KGaA, Germany) was used to stain the samples at 4 °C overnight. After 3 PBS washes, the cells were stained with 1% Alexa Fluor 488-labeled donkey antimouse IgG secondary antibody (Sigma-Aldrich Co. LLC., USA) at a 37 °C incubator for 1 h. Then, the nucleus was stained with 1% DAPI at room temperature in the dark for 10 min. A fluorescent microscope was used to observe the vinculin of stained cells. The total area and average area of focal adhesion were calculated based on the vinculin images though the step-by-step process using the ImageJ software, as previously reported.39 In brief, the images were converted to 16-bit type and the background was subtracted to enhance the local contrast. The images were treated by changing the bright and contrast processes. After Mexican hat filter, the images were adjusted by the threshold to analyze the total area and average size of vinculin. Five independent assays were carried out to calculate the mean and SD.Fluorescent staining of actin bundles and immunofluorescent staining of actinin and myosin
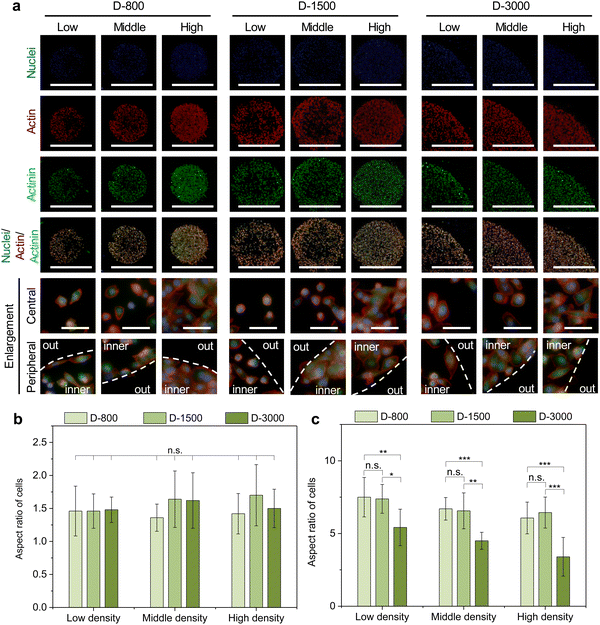
After 24 h cell culture, the MDA-MB231 cells were washed with PBS and fixed with 4% cold paraformaldehyde solution for 10 min. Subsequently, the fixed cells were treated with Triton X-100 for 10 min and blocked with 2% BSA solution for 30 min. Actin bundles and nuclei were stained with 2% Alexa Fluor-594 phalloidin for 20 min and 1% DAPI at room temperature in the dark for 10 min, respectively. The fluorescent images of stained cells were captured by a fluorescence microscope. The aspect ratio of breast cancer cells was analyzed according to the distribution of actin bundles in the ImageJ software. Five independent assays were used to calculate the mean and SD.For immunofluorescent staining of actinin and myosin, the cells were incubated with their primary antibody, 1% mouse antiactinin antibody (Abcam, USA) for actinin, and 1% rabbit antimyosin IIA antibody (Sigma-Aldrich Co., LLC.) for myosin at 4 °C overnight. After 3 PBS washes, the samples were stained with 1% Alexa Fluor 488-labeled donkey antimouse IgG antibody for actinin and 1% Alexa Fluor 488-labeled donkey antirabbit IgG antibody (Sigma-Aldrich Co. LLC., USA) for myosin in the dark for 1 h. The fluorescent images of stained cells were captured by a fluorescence microscope. Further, the heatmap of myosin staining was overlapped by more than 15 images to analyze the distribution of myosin intensity at peripheral and central regions using the ImageJ software.
Regulation of DNA synthesis by BrdU staining
The DNA synthesis activity of MDA-MB231 cells was regulated by 5-bromo-2′-deoxyuridine (BrdU, Thermo Fisher Scientific, USA) staining. Before cell seeding in the microholes of stencils, the cells were cultured in FBS-free DMEM medium for 24 h to maintain the cells in the G0 state. After that, the cells were harvested from the culture flask, seeded in the stencil microholes, and cultured for 6 h. Then, DMEM medium containing 1% BrdU labelling reagent (Thermo Fisher Scientific) was applied to change the FBS-free DMEM medium. The cells were further cultured for another 18 h. Then, the cells were rinsed with 3 PBS washes and fixed with 75% ethanol solution for 30 min, denatured with 2 M HCl aqueous solution for 30 min, treated with Triton X-100 for 10 min, and blocked with 2% BSA for 30 min. For the immunofluorescent staining of BrdU, 1% mouse antiBrdU primary antibody (Abcam, USA) was applied to incubate the cells at 4 °C overnight. Then, the samples were stained with 1% Alexa Fluor 488-labeled antimouse IgG secondary antibody in the dark for 1 h. In addition, the nuclei were stained with 1% DAPI at room temperature for 10 min. The fluorescent images were captured by a fluorescence microscope. The percentage of BrdU-positive cells was calculated by counting the number of BrdU-positive cells in all the checked cells. The percentage of BrdU-positive cells was defined as the DNA synthesis activity. Five independent assays were used to calculate the mean and SD.Immunofluorescent staining of ezrin
The membrane-protein expression was studied by ezrin staining. After the cells were cultured in the stencil holes for 24 h, the cells were fixed with 4% cold paraformaldehyde solution for 10 min and blocked with 2% BSA solution for 30 min. Rabbit ezrin primary antibody (Cell Signalling Technology, USA) was used to incubate the samples at 4 °C overnight. The samples were followed with PBS washing and stained with 1% Alexa Fluor 488-labeled antirabbit IgG antibody at room temperature in the dark for 1 h. A fluorescence microscope was used to capture the fluorescent images of stained ezrin. The fluorescent images were analyzed by the ImageJ software. More than five images were converted into 16-bit type and multiple images were overlapped into one image to present the intensity of ezrin protein.Statistical analysis
Statistical analysis was used to evaluate the quantitative and fluorescent results. In this study, the data presented the mean ± SD. Significant differences were regulated by the one-way analysis of variance (ANOVA) method. Significant difference was considered when p < 0.05.Results and discussion
Characteristics of stencils and cell culture
The stencil technique has been widely applied to control the cell morphology.40 In this study, the PDMS stencils were fabricated by a straightforward punching method for cell culture (Fig. 1(a)). Specifically, 100 μm-thickness PDMS films were punched to prepare the through-film microholes on PDMS stencils using the differential punchers. The diameters of microholes were respectively designed to be 800 μm, 1500 μm, and 3000 μm on the circular PDMS films (Fig. 1(b)). The formation of microholes on PDMS stencils was confirmed by the optical images, and the circular microholes were established on the stencils (Fig. 1(c)). To further affirm the favorable performance for the control of the hole size, the diameter of PDMS holes was measured to be 811.7 ± 12.0 μm, 1554.0 ± 42.0 μm, and 3238.5 ± 125.5 μm and the circularity of the holes was calculated to be 0.99 ± 0.03, 0.97 ± 0.02, and 1.10 ± 0.25, respectively. The results showed that the through-film microholes could be well-controlled on the PDMS stencils.Before cell seeding, the prepared stencils were firstly sterilized in 75% ethanol solution. The sterilized PDMS stencils were tightly covered on the bottom of 24-well plates for cell culture. The cell suspension solution was homogeneously added on the stencils, and the cell colonies were formed within PDMS microholes. The morphology of the colonies was observed by an optical microscope (Fig. 1(d)). The circular cell colonies were constructed on the tissue culture surface. The magnified images showed that the cells were irregularly arranged at the central region, while peripheral cells presented strong compliance along the periphery of the microholes. In addition, the diameter of the cell colony was also measured to be 816 ± 10 μm, 1612 ± 44 μm, and 3207 ± 111 μm, and the circularity was 0.98 ± 0.02, 1.09 ± 0.03, and 0.99 ± 0.04, respectively. The cell colony indicated the same characteristics as the microholes of PDMS stencils.
Cell-density-dependent distribution
After the micropatterned cells were cultured in the microholes of the stencils for 24 h, the nuclei were stained blue to analyze the cell-density-dependent distribution (Fig. 2(a)). MDA-MB231 cells preferred attaching to the peripheral regions of microholes when cell initial seeding density was low and middle density, while the cells were homogenously adhered on central and peripheral regions in high cell seeding density. In addition, cellular attachment behavior presented similar results in PDMS microholes with different diameters. The clustered cells assembled their morphology to adapt to the microenvironment, especially at the peripheral region (Fig. 2(b)). As observed above, the cells were elongated at the peripheral region, while they were distributed freely at the central region of the microholes. Therefore, in this study, the PDMS holes were separated into two parts of central and peripheral regions (Fig. 2(c)), assuming that the area of central region was the same as the peripheral region to quantitatively analyze the cell density. Further, the cell density was systematically compared to investigate the relationship between the hole size, cell seeding density or cell position, and cell density in micropatterns (Fig. 2(d)). MDA-MB231 cells had different cell densities at the central and peripheral regions in the micropatterns, independent of the diameter of the PDMS microholes. The cell density increased from the central regions to the peripheral regions in low and middle cell initial seeding density, but showed little significant difference in high cell seeding density. Both the cell densities at the central and peripheral regions were enhanced with increasing cell initial seeding density from low to high. The results showed that heterogeneous cell density could be affected by cell initial seeding density and cell position, but not affected by the hole size.Formation of heterogeneous focal adhesion in micropatterns
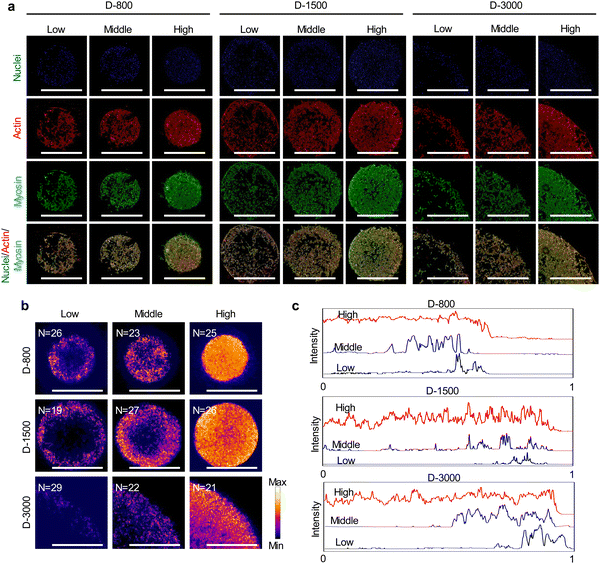
The formation and arrangement of cellular adhesion proteins can be regulated by cell density-dependent cell–cell interactions and cellular microenvironment.41–43 Focal adhesion (FA) serves as the most important adhesion proteins of cells to affect the cell behaviors.44 Herein, the cells cultured in various microholes reorganized their FAs to deliver biophysical signals from the extracellular microenvironment into the cells; thus, FA acted as the mechanosensor between the extracellular matrix (ECM) and the cytoskeleton to affect the nuclear functions (Fig. 3(a)). Vinculin was stained to study the influence of cell colony on the formation of FA in microholes and co-stained with nuclei (Fig. S1, ESI†). Immunofluorescent staining showed that FA could be formed in all the micropatterned cells (Fig. 3(b)). To further investigate the interaction of cellular colony and FA formation, the total area and average size of FAs were calculated by the ImageJ software. The total area of the FAs in the whole cell colony increased with increasing initial cell seeding density and hole size, while the average size of FA showed no significant difference. The colony was further separated into central and peripheral regions to disclose their respective FA formation. At the central regions, the average size of FA was enhanced with increasing cell seeding density independently of the hole size (Fig. 3(c)). On the other hand, the average size of FA could be affected by both cell seeding density and hole size at the peripheral regions (Fig. 3(d)). Interestingly, the peripheral average size presented a decreasing tendency with increasing hole size, which was associated with the reduced curvature of the microholes. Therefore, the formation of heterogeneous FA could be controlled by initial cell seeding density, hole size, and cell position.Curvature-dependent cytoskeletal heterogeneity in micropatterns
As mentioned above, extracellular cues are induced into cells by FAs to guide the cytoskeletal structures in micropatterned cells.45 Cytoskeletal structures including actin, actinin, and myosin not only serve as the basal components to support the cellular framework but also play a crucial role in regulating the cell functions.46–48 In this study, the cytoskeleton was stained to investigate the influence of cell density, hole size, and cell position on cytoskeletal heterogeneity. Actin bundles were arranged along with the periphery of micropatterns and actin bonding protein of actinin also showed similar results with actin bundles in the enlarged images of the central and peripheral regions (Fig. 4(a)). Based on the heterogeneous difference of the cytoskeleton at the central and peripheral regions, the aspect ratio of cells was further investigated to disclose the alignment of micropatterned cells. At the central regions, the aspect ratio of cells presented no significant difference (Fig. 4(b)). The aspect ratio decreased with increasing cell density and hole size at the peripheral regions (Fig. 4(c)). In addition, the peripheral aspect ratio was much longer than the central aspect ratio. The cytoskeleton results revealed that the cell density, hole size, and cell position could change the cellular aspect ratio and tension formation to affect the cell functions.Myosin, as the motor protein, could affect signal transduction in micropatterned cells. Myosin was also reorganized at the peripheral regions of micropatterns, showing similar results to actin filaments (Fig. 5(a)). The heatmap of myosin demonstrated that micropatterned cells preferred forming myosin structures at peripheral regions rather than the central regions (Fig. 5(b)). Further, the intensity curves of the myosin heatmap were evaluated from the center to the periphery of the heatmap images (Fig. S2, ESI†). The quantitative results disclosed that the peripheral intensity of myosin was much stronger than that of the central intensity (Fig. 5(c)). The high cell density at the heterogenous periphery could form more myosin structures, while the hole size did not affect myosin formation. These staining results together with vinculin staining suggested that heterogeneous FA and cytoskeleton could be controlled by the PDMS stencils to develop the peripheral heterogeneity in the micropatterns.
Regulation of DNA synthesis and ezrin expression in heterogeneous micropatterns
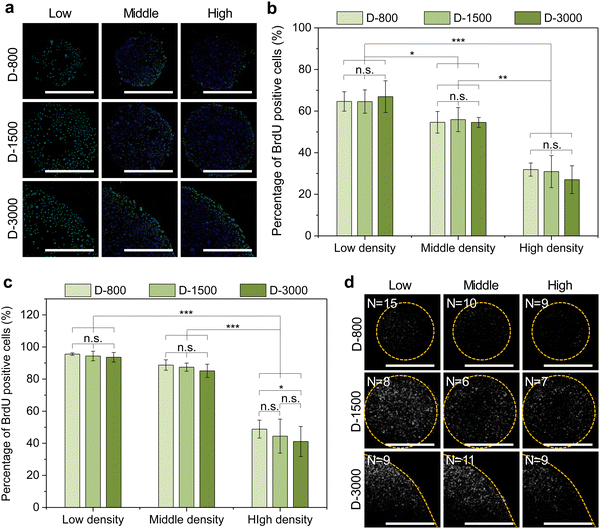
Stencil-induced cell density and curvature could induce heterogeneous FA formation and cytoskeletal structures to affect cellular behaviors, especially the cell nuclear activity. In this study, the nuclear activity was evaluated by BrdU staining to analyze DNA synthesis activity of the micropatterned cells. The BrdU staining results showed that the cells presented green BrdU-positive nuclei with low cell density, especially at the periphery of micropatterns (Fig. 6(a)). To further investigate the influence of cell density on the nuclear activity of cells, DNA synthesis activity was quantitatively analyzed by calculating the percentage of BrdU-positive cells in all the checked cells (Fig. 6(b) and (c)). DNA synthesis activity presented a decreased tendency with increasing cell density in stencil-induced micropatterns, whether at the central regions (Fig. 6(b)) or the peripheral regions (Fig. 6(c)). Nevertheless, the cells at the periphery showed higher percentage of BrdU-positive nuclei than that of the central regions to present a stem-like phenotype of the breast cancer cells. More interestingly, the contribution of interfacial curvature to the proliferation of the breast cancer cells may affect the cell density. Unlike stem cells and somatic cells, breast cancer cells are immortal and show infinite division ability. Thus, the results showed that cell density and curvature play a crucial role in regulating DNA synthesis activity.As previously reported, cytoskeleton tension force and cytoplasm membrane are predominately generated at the boundary region of micropatterned colonies or cells to induce interfacial tension force and biased tension force.49–51 In this study, membrane-protein ezrin served as the biolinker between the cell membrane and actin filaments to sense the extracellular stimuli and transmit these biased tension signals into the nuclei by cytoskeleton-induced mechanotransduction to affect the DNA synthesis activity of micropatterned cells. The staining results demonstrated that the cells with low cell density and at the periphery had stronger intensity of ezrin (Fig. 6(d)). The stained ezrin showed similar results with BrdU staining. Therefore, DNA synthesis could be affected by heterogeneous FA formation, cytoskeleton structures, and ezrin expression in stencil-assisted micropatterns.
Conclusion
The micropatterns produced by PDMS stencil-assisted method were applied to precisely control the cell density ranging from low to high cell density at the central and peripheral regions of micropatterns. The cell colony presented the same parameters with the prepared micropatterns on the PDMS stencils. The interfacial heterogeneity at the periphery of the cell colony was formed to manage FA formation and cytoskeleton structures. The biased tension force induced by FAs and cytoskeleton was stimulated to enhance ezrin expression at the peripheral region of the micropatterns. Micropattern-induced heterogeneity could also deliver the interfacial tension force through the polar cytoskeleton structures into the cells to guide nuclear DNA synthesis activity. The interfacial cells had higher percentage of proliferation and showed a stem-like phenotype. The results will pave the way to the construction of cellular heterogeneity and the understating of the molecular mechanism in breast cancer cells.Author contributions
Conceptualization, Y. W., N. W. and Y. Y.; formal analysis, Y. W. and N. W.; funding acquisition, Y. W. and Y. Y.; investigation, Y. W. and N. W.; methodology, all co-authors; project administration, Y. W. and Y. Y.; resources, Y. W.; data curation, all co-authors; supervision, Y. W. and Y. Y.; visualization, Y. W. and N. W.; writing-original draft preparation, Y. W. and N. W.; and writing-review and editing, all co-authors.Conflicts of interest
The authors declare no conflicts of interest.Acknowledgements
This work was supported by the National Natural Science Foundation of China (Grant no. 82102219), the Foundation of Shaanxi University of Science and Technology (Grant no. 126021993), the Natural Science Foundation of Shaanxi Province (Grant no. 2021JQ-545) and the Young Researchers’ Program of Zhengzhou University (Grant no. 32212777).References
- M. Hu, J. Yao, L. Cai, K. E. Bachman, F. Brûle, V. Velculescu and K. Polyak, Nat. Genet., 2005, 37, 899–905 CrossRef CAS PubMed.
- S. Pece, D. Tosoni, S. Confalonieri, G. Mazzarol, M. Vecchi, S. Ronzoni, L. Bernard, G. Viale, P. G. Pelicci and P. P. Fiore, Cell, 2010, 140, 62–73 CrossRef CAS PubMed.
- F. Bertucci, C. K. Y. Ng, A. Patsouris, N. Droin, S. Piscuoglio, N. Carbuccia, J. C. Soria, A. T. Dien, Y. Adnani, M. Kamal, S. Garnier, G. Meurice, M. Jimenez, S. Dogan, B. Verret, M. Chaffanet, T. Bachelot, M. Campone, C. Lefeuvre, H. Bonnefoi, F. Dalenc, A. Jacquet, M. R. De Filippo, N. Babbar, D. Birnbaum, T. Filleron, C. Le Tourneau and F. André, Nature, 2019, 569, 560–564 CrossRef CAS.
- T. Iwamoto, N. Niikura, R. Ogiya, H. Yasojima, K. Watanabe, C. Kanbayashi, M. Tsuneizumi, A. Matsui, T. Fujisawa, T. Iwasa, T. Shien, S. Saji, N. Masuda and H. Iwata, Sci. Rep., 2019, 9, 13343 CrossRef.
- X. Deng, Z. Shao and Y. Zhao, Sci. Adv., 2021, 8, 2002504 CrossRef CAS.
- S. Goel, C. A. Ferreira, F. Chen, P. A. Ellison, C. M. Siamof, T. E. Barnhart and W. Cai, Adv. Mater., 2018, 30, 1704367 CrossRef PubMed.
- Z. Yuan, B. Tao, Y. He, C. Mu, G. Liu, J. Zhang, Q. Liao, P. Liu and K. Cai, Biomaterials, 2019, 223, 119479 CrossRef CAS.
- R. L. Sousa, B. L. Melo, C. G. Alves, A. F. Moreira, A. G. Mendonça, I. J. Correia and D. M. Diogo, Adv. Funct. Mater., 2021, 31, 2010777 CrossRef.
- N. Pasha and N. C. Turner, Nat. Cancer, 2021, 2, 680–692 CrossRef CAS.
- F. Wang, J. Zhu, Y. Wang and J. Li, Nanomaterials, 2022, 12, 1656 CrossRef CAS.
- S. Esmail and M. F. Manolson, Eur. J. Cell Biol., 2021, 100, 151186 CrossRef CAS.
- M. M. Ali, S. Zhu, F. R. Amin, D. Hussain, Z. Du and L. Hu, Theranostics, 2022, 12, 2406–2426 CrossRef CAS.
- T. M. J. Evers, L. J. Holt, S. Alberti and A. Mashaghi, Nat. Metab., 2021, 3, 456–468 CrossRef.
- P. Abdollahiyan, F. Oroojalian, B. Baradaran, M. Guardia and A. Mokhtarzadeh, J. Controlled Release, 2021, 335, 596–618 CrossRef CAS PubMed.
- V. Sharma, O. Letson and S. Furuta, Sci. Signaling, 2022, 15, 724 CrossRef PubMed.
- F. Weiss, D. Lauffenburger and P. Friedl, Nat. Rev. Cancer, 2022, 22, 157–173 CrossRef CAS.
- X. Wang, W. Song, N. Kawazoe and G. Chen, J. Biomed. Mater. Res., Part A, 2013, 102, 3388 CrossRef.
- S. Helvert, C. Storm and P. Friedl, Nat. Cell Biol., 2018, 20, 8–20 CrossRef PubMed.
- X. Wang, X. Hu, N. Kawazoe, Y. Yang and G. Chen, Adv. Funct. Mater., 2016, 26, 7634–7643 CrossRef CAS.
- M. Uroz, S. Wistorf, X. S. Picamal, V. Conte, M. S. Pardo, P. R. Cusachs, R. Guimerà and X. Trepat, Nat. Cell Biol., 2018, 20, 646–654 CrossRef CAS.
- Y. Yang, X. Wang, T. C. Huang, X. Hu, N. Kawazoe, W. B. Tsai, Y. Yang and G. Chen, J. Mater. Chem. B, 2018, 6, 5424–5434 RSC.
- K. M. Yamada and M. Sixt, Nat. Rev. Mol. Cell Biol., 2019, 20, 738–752 CrossRef CAS PubMed.
- J. Winkler, A. A. Ogunniyan, K. J. Metcalf and Z. Werb, Nat. Commun., 2020, 11, 5120 CrossRef CAS PubMed.
- K. Lee, Y. Chen, X. Li, Y. Wang, N. Kawazoe, Y. Yang and G. Chen, J. Mater. Chem. B, 2019, 7, 7713–7722 RSC.
- S. N. Esfahani, A. M. R. Irizarry, X. Xue, S. B. Lee, Y. Shao and J. Fu, Nano Today, 2021, 41, 101310 CrossRef CAS PubMed.
- Y. Wang, Y. Yang, X. Wang, N. Kawazoe, Y. Yang and G. Chen, Acta Biomater., 2021, 125, 100–111 CrossRef CAS PubMed.
- Y. Yang, X. Wang, X. Hu, N. Kawazoe, Y. Yang and G. Chen, ACS Appl. Mater. Interfaces, 2018, 11, 1932–1943 CrossRef.
- Y. Yang, X. Wang, Y. Wang, X. Hu, N. Kawazoe, Y. Yang and G. Chen, Sci. Rep., 2019, 9, 6891 CrossRef.
- Y. Wang, Y. Yang, X. Wang, T. Yoshitomi, N. Kawazoe, Y. Yang and G. Chen, Biomaterials, 2021, 271, 120751 CrossRef CAS.
- Y. Wang, Y. Yang, T. Yoshitomi, N. Kawazoe, Y. Yang and G. Chen, J. Mater. Chem. B, 2021, 9, 4329–4339 RSC.
- D. Park, E. Wershof, S. Boeing, A. Labernadie, R. P. Jenkins, S. George, X. Trepat, P. A. Bates and E. Sahai, Nat. Mater., 2020, 19, 227–238 CrossRef CAS.
- Y. Wang, T. Yoshitomi, N. Kawazoe, Y. Yang and G. Chen, Adv. Mater. Interfaces, 2022, 9, 2101978 CrossRef.
- I. Y. Wong, S. Javaid, E. A. Wong, S. Perk, D. A. Haber, M. Toner and D. Irimia, Nat. Mater., 2014, 13, 1063–1071 CrossRef CAS PubMed.
- J. Lee, A. A. Abdeen, K. L. Wycislo, T. M. Fan and K. A. Kilian, Nat. Mater., 2016, 15, 856–862 CrossRef CAS.
- F. Pelon, B. Bourachot, Y. Kieffer, I. Magagna, F. M. Meillon, I. Bonnet, A. Costa, A. M. Givel, Y. Attieh, J. Barbazan, C. Bonneau, L. Fuhrmann, S. Descroix, D. Vignjevic, P. Silberzan, M. C. Parrini, A. V. Salomon and F. M. Grigoriou, Nat. Commun., 2020, 11, 404 CrossRef CAS PubMed.
- H. W. Jackson, J. R. Fischer, V. R. T. Zanotelli, H. R. Ali, R. Mechera, S. D. Soysal, H. Moch, S. Muenst, Z. Varga, W. P. Weber and B. Bodenmiller, Nature, 2020, 578, 615–620 CrossRef CAS.
- J. E. Visvader and G. J. Lindeman, Nat. Rev. Cancer, 2008, 8, 755–768 CrossRef CAS PubMed.
- B. A. T. Quang, R. Peters, D. A. D. Cassani, P. Chugh, A. G. Clark, M. Agnew, G. Charras and E. K. Paluch, Nat. Commun., 2021, 12, 6511 CrossRef.
- U. Horzum, B. Ozdil and D. P. Okvur, MethodsX, 2014, 1, 56–59 CrossRef PubMed.
- S. Huang, Q. Su, X. Hou, K. Han, S. Ma, B. Xu and Y. Yang, Front. Bioeng. Biotechnol., 2022, 10, 922159 CrossRef.
- J. Zheng, Y. Wang, N. Kawazoe, Y. Yang and G. Chen, J. Mater. Chem. B, 2022, 10, 3989–4001 RSC.
- H. Che, M. Selig and B. Rolauffs, Adv. Drug Delivery Rev., 2022, 184, 114169 CrossRef CAS PubMed.
- J. Lou, R. Stowers, S. Nam, Y. Xia and O. Chaudhuri, Biomaterials, 2018, 154, 213–222 CrossRef CAS PubMed.
- S. Wang, E. Englund, P. Kjellman, Z. Li, J. K. Ahnlide, C. R. Cupello, M. Saggioro, R. Kanzaki, K. Pietras, D. Lindgren, H. Axelson, C. N. Prinz, V. Swaminathan and C. D. Madsen, Nat. Cell Biol., 2021, 23, 758–770 CrossRef CAS PubMed.
- M. F. M. Rooij, Y. J. Thus, N. Swier, R. L. Beijersbergen, S. T. Pals and M. Spaargaren, Nat. Commun., 2022, 13, 2136 CrossRef PubMed.
- M. L. Cagigas, N. S. Bryce, N. Ariotti, S. Brayford, P. W. Gunning and E. C. Hardeman, Nat. Mater., 2022, 21, 120–128 CrossRef CAS.
- C. S. Hansel, S. W. Crowder, S. Cooper, S. Gopal, M. J. P. Cruz, L. O. Martins, D. Keller, S. Rothery, M. Becce, A. E. G. Cass, C. Bakal, C. Chiappini and M. M. Stevens, ACS Nano, 2019, 13, 2913–2926 CrossRef CAS.
- S. D. Cio, T. Iskratsch, J. T. Connelly and J. E. Gautrot, Biomaterials, 2020, 232, 119683 CrossRef.
- M. Thery, J. Cell Sci., 2010, 123, 4201–4213 CrossRef CAS.
- C. Loebel, R. L. Mauck and J. A. Burdick, Nat. Mater., 2019, 18, 883–891 CrossRef CAS PubMed.
- S. Huang, Z. Chen, X. Hou, K. Han, B. Xu, M. Zhang, S. Ding, Y. Wang and Y. Yang, Int. J. Mol. Sci., 2022, 23, 11884 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2tb01774a |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2023 |