A nicking enzyme-assisted allosteric strategy for self-resetting DNA switching circuits†
Haoliang
Wang‡
a,
Xiaokang
Zhang‡
 b,
Yuan
Liu
b and
Shihua
Zhou
b,
Yuan
Liu
b and
Shihua
Zhou
 *a
*a
aKey Laboratory of Advanced Design and Intelligent Computing, Ministry of Education, School of Software Engineering, Dalian University, Dalian 116622, China. E-mail: wanghaoliang715@gmail.com; zhoushihua@dlu.edu.cn
bSchool of Computer Science and Technology, Dalian University of Technology, Dalian 116024, China. E-mail: xiaokangzhangdl@gmail.com; liuyuan.dlut@gmail.com
First published on 24th November 2023
Abstract
The self-regulation of biochemical reaction networks is crucial for maintaining balance, stability, and adaptability within biological systems. DNA switching circuits, serving as basic units, play essential roles in regulating pathways, facilitating signal transduction, and processing biochemical reaction networks. However, the non-reusability of DNA switching circuits hinders its application in current complex information processing. Herein, we proposed a nicking enzyme-assisted allosteric strategy for constructing self-resetting DNA switching circuits to realize complex information processing. This strategy utilizes the unique cleavage ability of the nicking enzyme to achieve the automatic restoration of states. Based on this strategy, we implemented a self-resetting DNA switch. By leveraging the reusability of the DNA switch, we constructed a DNA switching circuit with selective activation characteristics and further extended its functionality to include fan-out and fan-in processes by expanding the number of functional modules and connection modes. Furthermore, we demonstrated the complex information processing capabilities of these switching circuits by integrating recognition, translation, and decision functional modules, which could analyze and transmit multiple input signals and realize parallel logic operations. This strategy simplifies the design of switching circuits and promotes the future development of biosensing, molecular computing, and nanomachines.
Introduction
In nature, biological systems can respond to various external environmental stimuli and process and transmit them through biochemical reaction networks to maintain the normal functioning of life.1–3 Inspired by nature, researchers are utilizing biomacromolecules to implement various artificial chemical reaction networks, aiming to emulate the complex information processing.4–7 Among them, DNA molecules as programmable materials have attracted widespread attention owing to their high stability,8,9 addressability,10,11 and modifiability.12,13 So far, various DNA chemical reaction networks have been constructed by combining dynamic DNA nanotechnologies, such as DNA strand displacement,14–16 enzyme cleavage,17,18 and self-assembly.19 For instance, DNA-based neural networks can memorize four single-stranded DNA patterns through simulated training and recall the most similar one while receiving an incomplete pattern.20 On this basis, winner-take-all neural networks further expand the number of DNA patterns that can be classified into and recognized as nine different patterns.21 Subsequently, G-quadruplex-based DNAzyme networks achieved the dynamic assembly and disassembly of structures through fuel strand activation and enzyme assistance.22 It can be seen that DNA chemical reaction networks have shown tremendous potential in dynamic control and logical computing and have been widely applied in intelligent biosensing,23,24 DNA storage,25,26 information processing,27 and other applications.Molecular circuits, the basis of complex information processing, can be used for molecular information recognition,28,29 processing,30,31 and decision-making.32,33 Therefore, they have been widely studied by researchers. In the study of molecular circuits, switching circuits not only transmit information in response to external stimuli but also represent information through various switching states, which is particularly important for realizing more complex information processing. Currently, substantial progress has been made in the research of switching circuits. For example, Qian et al.34 proposed an unbiased DNA switch using DNA strand displacement reactions to achieve circuit outputs with any desired probability. Taking advantage of the inherent stochasticity of molecular interactions, the DNA switch can be connected in parallel or series to perform more complex information processing. In addition, Fan et al.35 utilized the programmability of DNA sequences to propose a modular DNA molecular switch. They used this switch to design a fully functional switch canvas that was capable of realizing arbitrary logic functions by mapping input combinations from the truth table to the current paths on the canvas. Subsequently, a photochemically controlled DNA switching circuit was constructed by integrating photosensitive molecules with a CG-C + triplex structure-based DNA switch into one system, and this switching circuit can achieve multiple logic computation and probabilistic computation under UV irradiation.36 However, the processing of complex information requires the introduction of multiple DNA molecular switches with a uniform format,37 which not only increases the complexity of the circuit but also leads to signal crosstalk, thereby affecting the accuracy and stability of the computations.
These problems limit the development of more complex and powerful information processing systems. To simplify the complexity of system design, researchers have developed various DNA switching circuits with recyclable state transitions.38–46 However, in order to restore the switch to its initial state and achieve reusability, it requires the repeated manual addition of fuel. Therefore, how to achieve the dynamic self-restoration of switching circuits to simplify system design and realize complex information processing currently remains a major challenge.
In this study, we proposed a nicking enzyme-assisted allosteric strategy for constructing self-resetting DNA switching circuits to realize complex information processing. Firstly, we designed an allosteric strategy based on the cleavage specificity of the nicking enzyme, which could automatically be restored to the initial state with the assistance of the enzyme. Based on this strategy, we implemented a DNA switch with a self-resetting function, which realized the transmission of downstream signals and could automatically be reset. Secondly, by utilizing the programmable design and reusability of the DNA switch, a DNA switching circuit with selective activation characteristics was constructed, and this circuit could selectively activate the downstream reaction path according to the input orders of the signal. Thirdly, by extending the functional modules and connection mode, the self-resetting DNA switching circuits achieved fan-out and fan-in functions, which could easily transmit a DNA signal to multiple downstream modules (fan-out) and multiple signals to a single module (fan-in), significantly enhancing the logic computing capability of the circuit. Finally, we further designed the recognition, translation, and decision functional modules. The recognition module utilized reusability to perform parallel analysis of multiple input signals; the translation module took advantage of its fan-out feature to translate the upstream signals; the decision module relied on its logical processing capability to calculate the translated signals. By integrating these functional modules into the DNA switching circuit, the complex information processing was realized. The strategy increases the reusability of the switching circuit, enriches the signal transmission strategy, and promotes the development of molecular switching circuits.
Materials and methods
Materials
All DNA samples in this paper were purchased from Sangon Bio-tech Co., Ltd (Shanghai, China). The DNA strands without modification were purified via polyacrylamide gel electrophoresis. The DNA strands modified with fluorophore and quencher were purified by high-performance liquid chromatography. The nicking enzyme Nb.BtsI was purchased from New England Biolabs. The sequences of all strands in the experiment are shown in ESI Table S1,† which were simulated using NUPACK to reduce unnecessary interference between sequences. All DNA strands were dissolved in 1× TAE/Mg2+ buffer (40 mM Tris, 20 mM acetic acid, 1 mM EDTA 2Na and 12.5 mM magnesium acetate, pH 8.0) as the stock solution. DNA strands were diluted using 1× CutSmart buffer (50 mM potassium acetate, 20 mM Tris-acetate, 10 mM magnesium acetate, 100 μg mL−1 BSA, pH 7.9). The DNA sequence concentration was determined using a Nanodrop 2000 instrument (Thermo Fisher Scientific Inc., Waltham, MA, USA) at an absorbance of λ = 260 nm.Annealing
All complementary DNA strands were dissolved in 1× CutSmart buffer. The final volume was 40 μL. The samples after mixing were annealed in a polymerase chain reaction (PCR) thermal cycler. The annealing assembly procedure was set at 90 °C for 5 minutes, held at 88 °C for 5 minutes and cooled to 24 °C at a rate of 0.8 °C per minute.Native PAGE
All reaction samples were run on 12% PAGE at a constant pressure of 85 V for 150 minutes in 1× TAE/Mg2+ buffer. After the polyacrylamide gel electrophoresis was completed, the gel was placed in Stains-All solution for 20 minutes. The gel was then allowed to fade in natural light, and imaged using a Canon scanner.Kinetic analysis of fluorescence
Fluorescence experiments were performed using a real-time PCR system (Bio-Rad, CFX96) equipped with a 96-well fluorescence plate reader. All samples were incubated in 1× CutSmart buffer at 37 °C. The FAM fluorescence signal was detected at 492 nm excitation and 518 nm emission. The ROX fluorescence signal was detected at 560 nm excitation and 606 nm emission. All fluorescence studies were performed in triplicate to ensure reproducibility. The fluorescence minima at five points of the initial scan of each reporter sample were measured as the baseline F1. Then, the input strand was added to the sample, and the fluorescence value obtained after scanning was denoted as F2. Consequently, the fluorescence increment value (ΔF) was calculated as F2 − F1. MaxΔF represents the maximum fluorescence increment. Therefore, ΔF/MaxΔF represents the result of the fluorescence normalization.Results and discussion
Nicking enzyme-assisted allosteric strategy
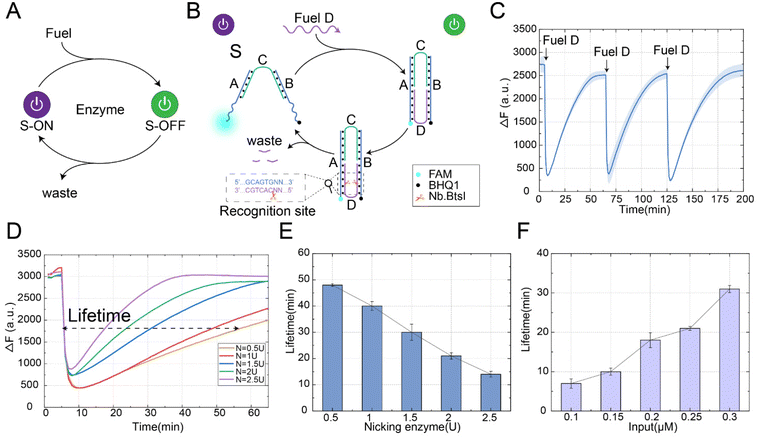
The automatic reset of the conformation is crucial for the reusability of the structure and the stability of the system. As shown in Fig. 1A, we proposed a nicking enzyme-assisted allosteric strategy. In this strategy, our goal was to introduce the enzyme as a consuming unit in the design of the structure, where the enzyme was able to recognize a specific site and perform the cleavage. Through the design of the enzyme recognition domain location, the conformation could be restored to the initial state under the action of the enzyme. As shown in Fig. 1B, this strategy included structure S, fuel D and nicking enzyme Nb.BtsI. Structure S was constructed by hybridization of three DNA strands: A, B, and C, where strand C had complementary pairs of 16 bases each with strands A and B, and the middle part contained 4 bases, which was the key part of the state transition. The allosteric process of this strategy proceeded as follows: firstly, the fuel strand D was combined with the two arm ends of structure S to form a stable double strand, which shortened the distance between the two arm ends, resulting in the transition of the conformational from the “S-ON” state to the “S-OFF” state. However, in the absence of the enzyme Nb.BtsI, the “S-OFF” state remained stable and could not return to its initial “S-ON” state. Secondly, since the recognition site of the enzyme Nb.BtsI was fully exposed in the “S-OFF” state, the strand D was specifically recognized and cleaved to generate waste in the presence of the nicking enzyme. Finally, due to the unstable combination of waste and structure S, the conformation gradually returned to its initial state.We performed a fluorescence kinetic analysis of the reaction process to demonstrate the feasibility of the nicking enzyme-assisted allosteric strategy, as shown in Fig. 1C. Initially, the structure S was in the “S-ON” state, and the distance between the fluorescence and the quenching group was far, resulting in a high fluorescence value. When the fuel strand D was introduced, it closed the distance between the ends of the arms of structure S, leading to a rapid decrease in fluorescence, indicating the conformational change from the “S-ON” to “S-OFF” state. At this time, the enzyme Nb.BtsI cleaved the strand D into small waste fragments, making the conformation automatically return to its initial state. With the reset of the conformation, the fluorescence signal gradually increased. To further verify the reusability of the allosteric strategy, we continued to input fuel strand D when the conformation was restored to the initial state and observed its fluorescence change. After three repeated inputs of fuel strand D, we observed that the conformation could consistently and effectively return to its initial state. This demonstrates the stability and reusability of structure S even after multiple experiments. The allosteric strategy was further verified by polyacrylamide gel electrophoresis experiments (Fig. S1†). In order to ensure that strand D could be automatically detached from structure S after being cleaved and the structure could be restored to the initial state, we investigated the number of bases in strand D after being cleaved by the enzyme Nb.BtsI. Through experiments, we found that when the number of bases of strand D was 6nt after being cleaved, the binding between strand D and structure S was weakest (Fig. S2†), so that the conformation could be restored to its initial state.
In order to further investigate the factors influencing the time required for the conformation to return to its initial state, we defined the lifetime of the transient state as the full width at half maximum of the fluorescence curve. The lifetime for the conformation to return to its initial state could be controlled by the fuel consumption rate (controlled by the enzyme concentration), the fuel concentration, and temperature. First, we explored the effect of enzyme concentration on the lifetime of the conformation to return to its initial state, as shown in Fig. 1D. Under the fixed concentration of the fuel strand (0.2 μM), we observed that the lifetime for the conformation to return to its initial state decreased from 48 minutes to 14 minutes when the concentration of enzyme Nb.BtsI increased from 0.5 U to 2.5 U, as shown in Fig. 1E. With the increase of enzyme concentration, the time for the conformation to return to the initial state was gradually shortened. This is because a higher enzyme concentration results in a faster cleavage rate of the fuel strand. It is worth noting that at high concentrations of the enzyme Nb.BtsI, the energy was dissipated so rapidly that it was impossible to completely transition all structures from the “S-ON” to the “S-OFF” state. We also explored the effect of fuel concentration on the time required for the conformation to return to its initial state. Similarly, under a fixed enzyme concentration (2 U), the lifetime for the conformation to return to its initial state increased from 7 minutes to 31 minutes when the fuel strand concentration increased from 0.1 μM to 0.3 μM, as shown in Fig. 1F. This was because the enzyme requires more time to remove the fuel molecules. Finally, the time for the conformation to return to its initial state could also be adjusted by changing the temperature. As the temperature increased, the stability of the binding between the waste strand and structure S becomes weaker, and the lifetime for the conformational to return to its initial state gradually shortened. To observe the reaction rates in detail, the real-time fluorescence traces of different factors influencing the lifetime for the conformational to return to its initial state are shown in Fig. S3.† The above experimental results prove that the nicking enzyme-assisted allosteric strategy can be adjusted by fuel strand concentration, enzyme concentration, and temperature, and make it a programmable allosteric process in a time-controllable manner.
Design of the DNA switch with self-resetting function
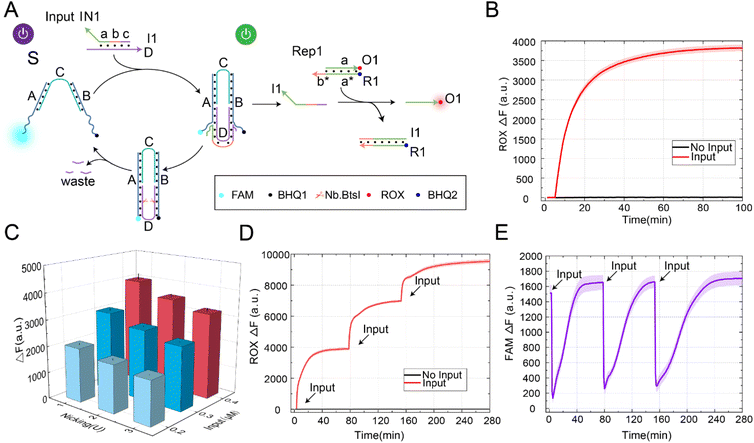
To verify the scalability of the allosteric strategies, we constructed a DNA switch with a self-resetting function, as shown in Fig. 2A. In this design, our core goal was to utilize the nicking enzyme-assisted allosteric strategy, allowing the switch to return to its initial state after signal transmission. This switch achieved the self-resetting function through structure S, enzyme Nb.BtsI, and the input signal, which consisted of a double strand formed by strand D and strand I1. According to the structure of the self-resetting DNA switch, the process from switch startup to self-resetting can be divided into two steps. Firstly, the input signal IN1 and structure S generated downstream signal strand I1 through the strand displacement reaction, and the switch state changed from “S-ON” to “S-OFF”. By programmable design of domain b in signal IN1 (Fig. S4†), the generated signal strand I1 could further react with Rep1 through toehold domain b to release the strand O1. Secondly, under the action of the enzyme Nb.BtsI, strand D was specifically cleaved to form waste, and the DNA switch gradually returned to its initial state.In order to demonstrate the feasibility of the DNA switch with a self-resetting function, we conducted a fluorescence kinetic analysis of the reaction process, as shown in Fig. 2B. Without the input signal IN1, the fluorescence did not increase (black curve). In the presence of the input signal IN1, IN1 reacted with the DNA switch to produce the downstream signal strand I1. Furthermore, strand I1 reacted with Rep1 through strand displacement to generate the fluorescence strand O1, and the fluorescence gradually rose (red curve). At the same time, we verified the self-resetting function of the DNA switch through the PAGE experiment (Fig. S5†). Through the analysis of the above experimental results, we successfully constructed a switch with a self-resetting function and realized activation of the downstream signaling pathway. This indicates that the strategy is feasible and provides a practical approach for constructing a DNA switch with self-resetting capability.
The yield of signal strand I1 is crucial for further programmable control of downstream reactions. To generate a sufficient amount of signal strand I1, we investigated the factors that influence its yield, including enzyme concentration and input signal concentration, as shown in Fig. 2C. When the substrate concentration was fixed at 0.2 μM, the fluorescence yield gradually increased with the increase in input signal concentration. When the input-to-substrate concentration ratio reached 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, the fluorescence yield reached its maximum and no longer increased with further increases in input concentration, as shown in Fig. S6.† At the same time, the concentration of the enzyme also affected the yield of the output signal. When the input-to-substrate concentration ratio reached 2
1, the fluorescence yield reached its maximum and no longer increased with further increases in input concentration, as shown in Fig. S6.† At the same time, the concentration of the enzyme also affected the yield of the output signal. When the input-to-substrate concentration ratio reached 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, the fluorescence yield decreased with an increase in enzyme concentration. This is because the increase in enzyme concentration enhances the cleavage rate of strand D, leading to the formation of waste that can rebind with strand I1, thereby inhibiting the generation of output signals. To observe reaction rates in detail, the real-time fluorescence traces of different factors influencing the yield of strand I1 are shown in Fig. S7.† The above experimental results show that the output signal was optimal when the enzyme concentration was 1 U, and the input-to-substrate concentration ratio was 2
1, the fluorescence yield decreased with an increase in enzyme concentration. This is because the increase in enzyme concentration enhances the cleavage rate of strand D, leading to the formation of waste that can rebind with strand I1, thereby inhibiting the generation of output signals. To observe reaction rates in detail, the real-time fluorescence traces of different factors influencing the yield of strand I1 are shown in Fig. S7.† The above experimental results show that the output signal was optimal when the enzyme concentration was 1 U, and the input-to-substrate concentration ratio was 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. Therefore, in subsequent experiments, we selected an enzyme concentration of 1 U and an input-to-substrate concentration ratio of 2
1. Therefore, in subsequent experiments, we selected an enzyme concentration of 1 U and an input-to-substrate concentration ratio of 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 for further study.
1 for further study.
In building a large complex circuit, it was inevitable to increase the number of DNA switch components to achieve complex logic calculation functions, which limited the scalability of the circuit. Increasing the reusability of the DNA switch components became particularly important at this point. Therefore, we further validated the reusability of the DNA switch by continuously inputting the signal IN1, as shown in Fig. 2D. As a result, we observed that the fluorescence increased gradually (red curve). In the absence of the input signal IN1, no leakage was observed even with the increase in time (black curve). Labeling fluorescence and quenching groups at the end of the DNA switch, the state change of the DNA switch in the process of self-resetting could be observed, as shown in Fig. 2E. The DNA switch automatically returns to its initial state after three rounds of signal input. The above experimental results prove that it is feasible to construct a DNA switch with a self-resetting function. The self-resetting function and multiple reusabilities of DNA switches provide a new scalable method for constructing large molecular circuits to realize complex molecular computation.
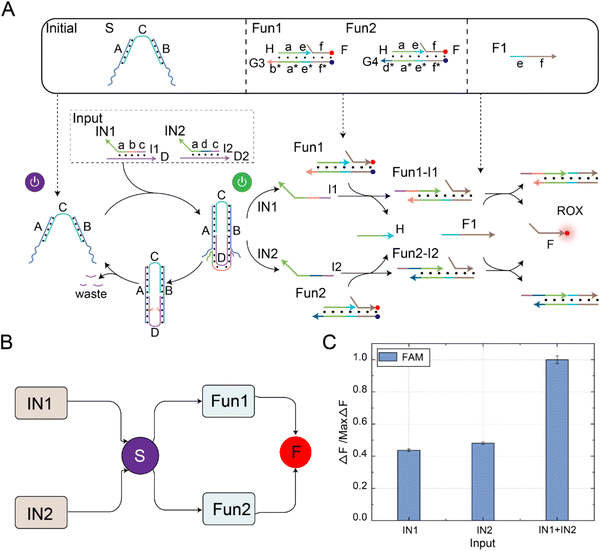
A self-resetting DNA switching circuit with selective activation characteristics
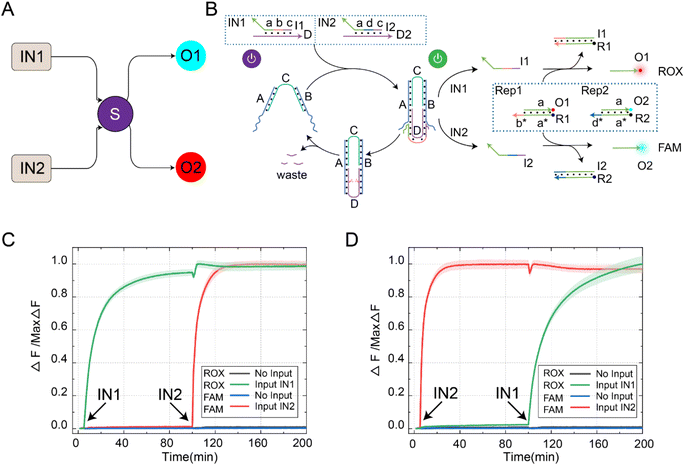
In order to further extend the reusability of this function, we programmatically designed the DNA switch to recognize different input signals, enabling the selective activation of downstream signal pathways, as shown in Fig. 3A. The reaction principle of the switching circuit with selective activation features is shown in Fig. 3B. In this experiment, we set up four reactants, including inputs (IN1 and IN2), DNA switch elements (S), reporters (ROX-labeled Rep1 and FAM-labeled Rep2), and enzyme Nb.BtsI. Programmatically designing the domains b and d of input IN1 and IN2 can be used for cascading downstream reaction pathways. The reusability of the DNA switch self-reset function enabled selective activation of downstream reaction pathways. When the signal IN1 was input, the two-arm ends of switch S bind with strand D in IN1, releasing strand I1 through a strand displacement reaction. Once strand I1 was generated, it could consume Rep1 and release strand O1. At the same time, the state of the switch changed from “S-ON” to “S-OFF” and returned to “S-ON” under the action of Nb.BtsI. We continued to input the signal IN2, and the strand I2 generated by the switch reacted with Rep2 and released O2. By controlling the input sequence of IN1 and IN2, we could achieve selective activation of the downstream reaction path.Further fluorescence experiments were performed to determine the selective activation characteristics of the switching circuit, as shown in Fig. 3C. We first input IN1, which activates the DNA switch to produce output strand I1. Then, strand I1 and Rep1 generated strand O1 through a strand displacement reaction, resulting in a rising ROX fluorescence curve (green curve). Due to the self-resetting function of the switch, the DNA switch automatically returned to its initial state after the reaction was completed. When the signal IN2 was input, the switch was reactivated, producing output I2. Then, strand I2 and Rep2 generated strand O2 through a strand displacement reaction, leading to an increase in the FAM fluorescence curve (red curve). Similarly, changing the input order of IN1 and IN2 resulted in the signal activation shown in Fig. 3D. The feasibility of the selective activation feature of the DNA switch circuit was verified through real-time kinetic fluorescence curves with different input sequences, providing possibilities for achieving complex logic computing functions.
Fan-out and fan-in functions of self-resetting DNA switching circuits
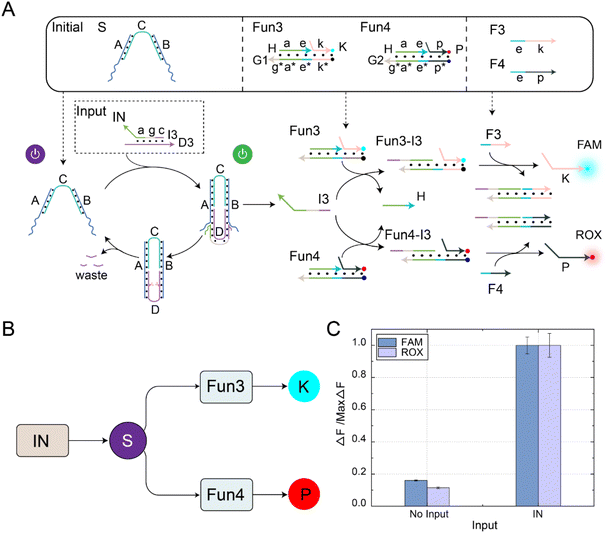
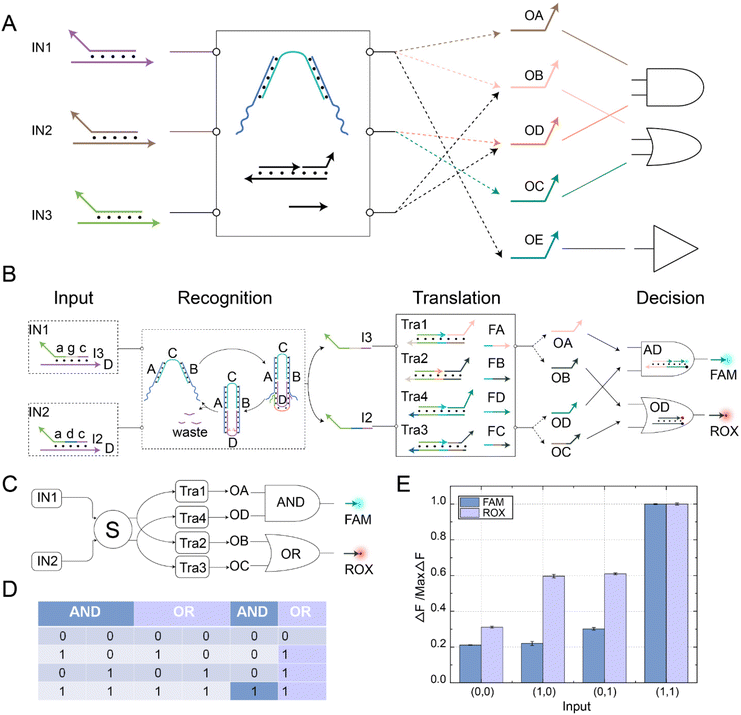
The fan-out mechanism allows for the construction of complex logic and functional structures in DNA circuits. As shown in Fig. 4A, we designed and implemented a fan-out function using self-resetting DNA switches to increase the flexibility and functional expandability of the DNA circuits. The core elements in the circuit were self-resetting DNA switches S, functional substrates Fun3 and Fun4, and fuels F3 and F4. Among them, there was the same toehold domain g* in the substrates Fun3 and Fun4. When the signal IN was input, the same domain g* in Fun3 and Fun4 can simultaneously receive the output signal I3 of the DNA switch, thus performing a strand displacement reaction and exposing the toehold domain e*. At this time, fuel strands F3 and F4 underwent strand displacement reactions with the products Fun3-I3 and Fun4-I3 through the toehold domain e*, releasing signal strands K and P. An abstract diagram of the self-resetting DNA switch with fan-out function is shown in Fig. 4B. Using the programmable design of functional substrates Fun3 and Fun4, the self-resetting DNA switching circuit realizes the one-to-many signal transmission mode.The fluorescence histogram analysis of the fan-out circuit is shown in Fig. 4C. When IN was not introduced, no significant fluorescent signal was observed. Upon introduction of IN, the two downstream signal paths were activated under the self-reset function of the DNA switch, and the output of the FAM and ROX signals increased. The PAGE experiment results and real-time fluorescence kinetic analysis of the fan-out circuit are shown in Fig. S8.† The above experiments show that the output signal generated by the input signal activation switch further interacts with the downstream substrates Fun3 and Fun4 to realize the activation of the two downstream signal paths.
The specific recognition ability of molecular circuits makes them a powerful tool for control and operation at the molecular level, contributing to the development of biosensors, drug delivery, and nanomachines. In order to verify the specificity of the circuit, we modified the base sequences of the input signal IN (named IN1, IN2, and IN3), and conducted fluorescence kinetic analysis. The experimental results show that the circuit has a specific recognition ability for the input signal IN. Only when the correct input signal IN was input, the fan-out circuit could produce two output signals, FAM and ROX. In addition, no output signal was generated when other signals were input. Therefore, the circuit we designed demonstrates excellent specificity, as shown in Fig. S9.†
The fan-in mechanism allows multiple input signals to be passed to the same downstream reaction or output part. By fan-in function, the number of DNA switches that need to be designed and controlled can be reduced, thus simplifying the circuit design, optimizing the experimental verification process, and reducing the complexity and difficulty of the system. As shown in Fig. 5A, to combine, process, or judge multiple inputs and achieve more complex computational and logical operations, we designed and implemented a fan-in function using a self-resetting DNA switch circuit. The critical components in the circuit were self-resetting DNA switches, functional substrates Fun1 and Fun2, and fuel F1. Firstly, because the sequences of the domain b and domain d in the input signal strand were independent of the sequences in the DNA switch, we programmatically designed the domain b and domain d of the input signal strand. Using the reusable self-resetting function of the DNA switch, the input strands D from signals IN1 and IN2 could bind with the DNA switch, resulting in the generation of output strands I1 and I2, respectively. The generated output signals could react with Fun1 and Fun2 in the downstream signal pathway, exposing the toehold domain e*. Secondly, fuel strand F1 reacted with the products Fun1-I1 and Fun1-I2 using the toehold domain e*, releasing the common signal strand F. An abstract diagram of the self-resetting DNA switch with fan-in function is shown in Fig. 5B. Using the programmable design of functional substrates Fun1 and Fun2, the self-resetting DNA switching circuit realized the many-to-one signal transmission mode.
The fluorescence histogram analysis of the fan-in circuit is shown in Fig. 5C. When only IN1 or IN2 was input, the fluorescence yield in the presence of only one input signal was about half of that when both IN1 and IN2 were simultaneously input. This circuit exhibited the same functionality as an OR logic gate. The PAGE experiment results and real-time fluorescence kinetic analysis of the fan-in circuit implemented using the self-resetting DNA switch are shown in Fig. S10.† Using the reusability of the self-resetting DNA switch, we implemented the fan-in and fan-out functions and verified the scalability of the switching circuit. The experimental results show that the switching circuit performs well in expansibility, and provides feasibility verification for constructing large logic circuits and realizing complex logic operations.
A parallel DNA switching circuit for complex information processing
In order to demonstrate the complex information processing capabilities of this DNA switching circuit, we further constructed a parallel DNA switching circuit, as shown in Fig. 6A. Different logical operations could be implemented by changing the input combination. For instance, inputting IN1 and IN2 could realize the OR logic operation, while inputting IN1 and IN3 could achieve the AND logic operation. The switching circuit also realized parallel logic operations using its reusability and fan-in and fan-out characteristics. Among them, the parallel AND OR logic operations controlled by a self-resetting DNA switching circuit is shown in Fig. 6B. The switching circuit integrated three functional modules: recognition, translation, and decision. The self-resetting DNA switch received the upstream signals IN1 and IN2 in the recognition module. Subsequently, the recognition module generated downstream output strands I2 and I3 while also returning to the initial state, achieving signal perception. In the translation module, the output signals I2 and I3 reacted with downstream substrates Tra1, Tra2, Tra3, and Tra4, releasing the toehold domains. FA, FB, FC, and FD fuel strands underwent strand displacement reactions with the substrates through the toehold domain to generate signal strands OA, OB, OC, and OD, facilitating the transmission of signals. In the decision module, OA and OD signal strands reacted with the AND gate substrate AD, producing the FAM fluorescence signal. OB and OC signal strands reacted with the OR gate substrate OD, producing the ROX fluorescence signal. Complex information processing was achieved through parallel logic operations. The abstract diagram of the self-resetting DNA switching circuits implementing parallel AND OR logic operations is shown in Fig. 6C. Using the fan-out feature of the self-resetting DNA switch, IN1 could fan out to OA/OB and IN2 to OC/OD simultaneously. Then, OA and OD performed the AND logic operation, while OB and OC perform the OR logic operation. The fluorescent FAM and ROX were the output signals for the AND OR gates, respectively. The truth table for the AND OR parallel logic operations is shown in Fig. 6D. At this point, the reaction process of the parallel AND OR logic operations were completed, and the specific reaction processes of each module are shown in Fig. S11 to S13.†To visually observe the implementation of parallel logic operations in the switch circuit, we performed fluorescence analysis, as shown in Fig. 6E. When there was no input, no significant fluorescent signal was observed. A significant increase in ROX fluorescence could be observed if only IN1 or IN2 was present. However, when IN1 and IN2 were input simultaneously, significant FAM and ROX fluorescence signals were observed, and the experimental results were consistent with the operation logic of the AND and OR gates. The fluorescence histogram shows that both gates have strong robustness and negligible crosstalk, which indicates that the switching circuit could achieve parallel information processing by flexibly combining two circuit units. The real-time fluorescence kinetic analysis of the parallel logic operations of the AND and OR gates is shown in Fig. S14.† To verify the potential of the self-resetting switching circuits for complex information processing, we implemented different logic operations by changing the input signal. The AND logic operation was performed when the signal strands IN1 and IN3 were input. The OR logic operation was achieved by inputting IN2 and IN3. The YES logic operation was performed by inputting IN1 alone (Fig. S15†). The above experimental results demonstrate the potential of self-resetting switching circuits to realize complex operations.
Conclusions
In this paper, we proposed a nicking enzyme-assisted allosteric strategy. In this strategy, we utilized the specific cleavage ability of nicking enzymes to cleavage double-stranded DNA, resulting in a conformational change of the structure. By precisely controlling the enzyme concentration and fuel strand concentration, we could enable the structure to revert to its initial state in a time-controllable manner. Building upon this foundation, we applied it to a DNA switching circuit with a self-resetting capability. Different input signals could be identified by taking advantage of the resetting ability and programmable design of the DNA switch, and selective activation of downstream pathways can be achieved. In addition, we extended self-resetting DNA switching circuits to fan-out and fan-in functions with many-to-one and one-to-many signaling modes. Finally, we demonstrated the powerful information processing capabilities of the switching circuits by constructing logic units capable of performing different operations and implementing their parallel computation.Compared to the previous studies on switching circuits,36,37 our switching circuits design introduced a self-resetting allosteric strategy. This allowed the DNA switching circuits to automatically return to its initial state after use, increasing the reusability of the switching circuits. This improvement made the circuit design simpler and more modular, facilitating the construction of more complex information processing systems. In theory, our proposed self-resetting DNA switching circuits were scalable and sustainable. However, as the number of input cycles increases, the efficiency of the enzyme cleavage decreases, which can affect the information processing capability of the switching circuits. By optimizing the cutting efficiency of the enzyme, the stability of the switching circuits can be effectively improved, and the information processing capacity can be increased. In the future design, we will continue our research based on the reusability and parallel information processing capability of self-resetting DNA switching circuits to explore multi-signal detection technologies in actual biological samples. In addition, we will further combine the selective function of self-resetting DNA switches with self-assembled structures47,48 and macromolecular proteins such as antibodies49,50 to provide programmable tools for building intelligent nanomachines, highly specific drug delivery systems, and multi-signal detection and reusable biosensors.
Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
This work is supported by 111 Project (No. D23006), the National Natural Science Foundation of China (No. 62272079), Natural Science Foundation of Liaoning Province (No. 2022-KF-12-14), the Postgraduate Education Reform Project of Liaoning Province (No. LNYJG2022493), the Dalian Outstanding Young Science and Technology Talent Support Program (No. 2022RJ08), the Fundamental Research Funds for the Central Universities under grant (No. DUT23YG122),the Science and Technology Project of Liaoning Province under grant (2021JH1/10400009).References
- B. Kholodenko, M. B. Yaffe and W. Kolch, Sci. Signaling, 2012, 5, 14 CrossRef.
- B. D. Manning and A. Toker, Cell, 2017, 169, 381–405 CrossRef CAS.
- Y. J. Chen, N. Dalchau, N. Srinivas, A. Phillips, L. Cardelli, D. Soloveichik and G. Seelig, Nat. Nanotechnol., 2013, 8, 755–762 CrossRef CAS.
- S. Y. S. Wang and A. D. Ellington, Chem. Rev., 2019, 119, 6370–6383 CrossRef CAS PubMed.
- L. He, F. M. Chen, D. L. Zhang, S. T. Xie, S. J. Xu, Z. M. Wang, L. L. Zhang, C. Cui, Y. L. Liu and W. H. Tan, J. Am. Chem. Soc., 2020, 142, 14234–14239 CrossRef CAS PubMed.
- N. Li, Y. Zhao, Y. Liu, Z. Yin, R. Liu, L. H. Zhang, L. Ma, X. C. Dai, D. S. Zhou and X. Su, Nano Today, 2021, 41, 10 Search PubMed.
- S. W. Schaffter and R. Schulman, Nat. Chem., 2019, 11, 829–838 CrossRef CAS.
- D. Mariottini, A. Idili, G. Ercolani and F. Ricci, ACS Nano, 2023, 17, 1998–2006 CrossRef CAS PubMed.
- L. J. Sun, B. Cao, Y. Liu, P. J. Shi, Y. F. Zheng, B. Wang and Q. Zhang, J. Phys. Chem. B, 2022, 12, DOI:10.1021/acs.jpcb.2c05611.
- L. N. Green, H. K. K. Subramanian, V. Mardanlou, J. Kim, R. F. Hariadi and E. Franco, Nat. Chem., 2019, 11, 510–520 CrossRef CAS.
- Y. A. Zhang, X. Y. Yin, C. J. Cui, K. He, F. Wang, J. Chao, T. Li, X. L. Zuo, A. L. Li, L. H. Wang, N. Wang, X. C. Bo and C. H. Fan, Sci. Adv., 2023, 9, 12 Search PubMed.
- L. Zhou, M. X. Gao, W. L. Fu, Y. X. Wang, D. Luo, K. Chang and M. Chen, Sci. Adv., 2020, 6, 8 Search PubMed.
- A. P. Diaz, S. Bracaglia, S. Ranallo, T. Patino, A. Porchetta and F. Ricci, J. Am. Chem. Soc., 2022, 144, 5820–5826 CrossRef PubMed.
- D. Y. Zhang, A. J. Turberfield, B. Yurke and E. Winfree, Science, 2007, 318, 1121–1125 CrossRef CAS PubMed.
- A. P. Lapteva, N. Sarraf and L. L. Qian, J. Am. Chem. Soc., 2022, 144, 12443–12449 CrossRef CAS.
- J. Cabello-Garcia, W. Bae, G. B. V. Stan and T. E. Ouldridge, ACS Nano, 2021, 15, 3272–3283 CrossRef CAS PubMed.
- J. Yang, R. F. Wu, Y. F. Li, Z. Y. Wang, L. Q. Pan, Q. Zhang, Z. H. Lu and C. Zhang, Nucleic Acids Res., 2018, 46, 8532–8541 CrossRef CAS PubMed.
- J. B. Wang, Z. Z. Li and I. Willner, Nat. Commun., 2022, 13, 10 CrossRef.
- S. Ranallo, D. Sorrentino and F. Ricci, Nat. Commun., 2019, 10, 9 CrossRef PubMed.
- L. Qian, E. Winfree and J. Bruck, Nature, 2011, 475, 368–372 CrossRef CAS.
- K. M. Cherry and L. L. Qian, Nature, 2018, 559, 370–376 CrossRef CAS.
- J. T. Dong, Y. Ouyang, J. B. Wang, M. P. O'Hagan and I. Willner, ACS Nano, 2022, 16, 6153–6164 CrossRef CAS.
- F. Chen, Q. J. Lu, L. N. Huang, B. W. Liu, M. L. Liu, Y. Y. Zhang and J. W. Liu, Angew. Chem., Int. Ed., 2021, 60, 5453–5458 CrossRef CAS PubMed.
- C. Zhang, T. T. Zheng, Q. Ma, L. L. Yang, M. Z. Zhang, J. Y. Wang, X. Y. Teng, Y. Y. Miao, H. C. Lin, Y. Yang and D. Han, Angew. Chem., Int. Ed., 2022, 61, 9 Search PubMed.
- Y. F. Zheng, B. Cao, J. Q. Wu, B. Wang and Q. Zhang, IEEE/ACM Trans. Comput. Biol. Bioinf., 2023, 20, 2992–3000 Search PubMed.
- B. Cao, B. Wang and Q. Zhang, iScience, 2023, 26, 106231 CrossRef CAS.
- D. Huang, H. Y. Han, C. Guo, X. Lin, D. Chen, S. Yang, Q. F. Yang and F. Li, Nanoscale, 2021, 13, 5706–5713 RSC.
- X. Y. Liu, X. M. Zhou, X. Y. Xia and H. Xiang, Anal. Chim. Acta, 2020, 1096, 159–165 CrossRef CAS.
- L. N. Zou, Q. Wu, Y. J. Zhou, X. Gong, X. Q. Liu and F. Wang, Chem. Commun., 2019, 55, 6519–6522 RSC.
- D. N. Taylor, S. R. Davidson and L. L. Qian, J. Am. Chem. Soc., 2021, 143, 15567–15571 CrossRef CAS PubMed.
- B. M. G. Janssen, M. van Rosmalen, L. van Beek and M. Merkx, Angew. Chem., Int. Ed., 2015, 54, 2530–2533 CrossRef CAS PubMed.
- T. C. Xie, Y. H. Deng, J. R. Zhang, Z. Zhang, Z. Hu and T. B. Wu, Nucleic Acids Res., 2022, 50, 8431–8440 CrossRef CAS PubMed.
- N. L. Xie, M. Q. Li, Y. Wang, H. Lv, J. Y. Shi, J. Li, Q. Li, F. Wang and C. H. Fan, J. Am. Chem. Soc., 2022, 144, 9479–9488 CrossRef CAS.
- D. Wilhelm, J. Bruck and L. L. Qian, Proc. Natl. Acad. Sci. U. S. A., 2018, 115, 903–908 CrossRef CAS PubMed.
- F. Wang, H. Lv, Q. Li, J. Li, X. L. Zhang, J. Y. Shi, L. H. Wang and C. H. Fan, Nat. Commun., 2020, 11, 8 CrossRef.
- X. W. Xiong, M. S. Xiao, W. Lai, L. Li, C. H. Fan and H. Pei, Angew. Chem., Int. Ed., 2021, 60, 3397–3401 CrossRef CAS.
- X. Liu, X. Zhang, Y. Yao, P. J. Shi, C. Y. Zeng and Q. Zhang, Nanoscale, 2023, 15, 7755–7764 RSC.
- R. M. Liu, J. Yang, J. Yao, Z. Zhao, W. He, N. Su, Z. Y. Zhang, C. X. Zhang, Z. Zhang, H. B. Cai, L. Y. Zhu, Y. Z. Zhao, S. Quan, X. J. Chen and Y. Yang, Nat. Biotechnol., 2022, 40, 779–786 CrossRef CAS.
- J. Wang, Q. T. Song, X. G. Guo, X. Cui, L. X. Tan and L. C. Dong, Anal. Chem., 2019, 91, 14530–14537 CrossRef CAS PubMed.
- I. A. P. Thompson, L. W. Zheng, M. Eisenstein and H. T. Soh, Nat. Commun., 2020, 11, 7 CrossRef PubMed.
- T. Patino, A. Porchetta, A. Jannasch, A. Llado, T. Stumpp, E. Schaffer, F. Ricci and S. Sanchez, Nano Lett., 2019, 19, 3440–3447 CrossRef CAS.
- X. X. Chen, T. S. Chen, L. J. Ren, G. F. Chen, X. H. Gao, G. X. Li and X. L. Zhu, ACS Nano, 2019, 13, 7333–7344 CrossRef CAS.
- Y. Xiang and Y. Lu, Inorg. Chem., 2014, 53, 1925–1942 CrossRef CAS.
- X. Q. Liu, A. Niazov-Elkan, F. A. Wang and I. Willner, Nano Lett., 2013, 13, 219–225 CrossRef CAS.
- Z. W. Xiong, Q. Wang, J. F. Zhang, W. Yun, X. M. Wang, X. Ha and L. Z. Yang, Spectrochim. Acta A Mol. Biomol. Spectrosc., 2020, 229, 5 CrossRef PubMed.
- Y. Zhao, Y. Q. Zhang and G. F. Jie, Sens. Actuators, B, 2021, 326, 8 Search PubMed.
- X. D. Cui, Y. Liu and Q. Zhang, Analyst, 2022, 147, 2223–2230 RSC.
- K. F. Wagenbauer, C. Sigl and H. Dietz, Nature, 2017, 552, 78–83 CrossRef CAS.
- W. Engelen, L. H. H. Meijer, B. Somers, T. F. A. de Greef and M. Merkx, Nat. Commun., 2017, 8, 8 CrossRef PubMed.
- S. Ranallo, D. Sorrentino, E. Delibato, G. Ercolani, K. W. Plaxco and F. Ricci, Angew. Chem., Int. Ed., 2022, 61, 5 Search PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3an01677c |
| ‡ These authors contribute equally to this work. |
| This journal is © The Royal Society of Chemistry 2024 |