Open Access Article
Open Access ArticleDiscovery and engineering of ribosomally synthesized and post-translationally modified peptide (RiPP) natural products
He
Li
a,
Wei
Ding
b and
Qi
Zhang
 *a
*a
aDepartment of Chemistry, Fudan University, Shanghai, 200433, China. E-mail: qizhang@sioc.ac.cn
bState Key Laboratory of Microbial Metabolism, School of Life Sciences & Biotechnology, Shanghai Jiao Tong University, Shanghai, 200240, China
First published on 21st November 2023
Abstract
Ribosomally synthesized and post-translationally modified peptides (RiPPs) represent a diverse superfamily of natural products with immense potential for drug development. This review provides a concise overview of the recent advances in the discovery of RiPP natural products, focusing on rational strategies such as bioactivity guided screening, enzyme or precursor-based genome mining, and biosynthetic engineering. The challenges associated with activating silent biosynthetic gene clusters and the development of elaborate catalytic systems are also discussed. The logical frameworks emerging from these research studies offer valuable insights into RiPP biosynthesis and engineering, paving the way for broader pharmaceutic applications of these peptide natural products.
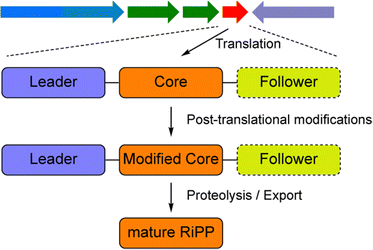
Global antimicrobial resistance is a critical public health concern today, and is primarily caused by the misuse and overuse of antimicrobial agents.1 As a result, there is a strong demand for the development of novel antimicrobial molecules. Natural products (NPs) have historically played a major role in drug discovery. In recent years, ribosomally synthesized and post-translationally modified peptides (RiPPs) hold immense promise in the discovery of new therapeutic bioactive molecules.2–5 RiPPs are derived from ribosomally synthesized precursor peptides, which, in most cases, consist of an N-terminal region called the leader peptide, and a C-terminal region called the core peptide (Fig. 1). A leader peptide is generally essential for the recognition by post-translationally modifying enzymes and will later be removed by proteolysis, whereas the core peptide is the site on which post-translational modifications occur and will be finally transformed to the mature product. Sometimes the precursor peptides also contain additional sequences at the C-termini (Fig. 1), which are called follower peptides and would be proteolytically removed for RiPP maturation. RiPPs have been found in all three kingdoms of life with remarkable structural diversity and wide-ranging biological functions, which arise from the considerable variability in precursor peptide sequences and the evolving catalytic reactions mediated by the modification enzymes.6–8
Compared to the overwhelming demand, the approaches for discovering new RiPPs are currently limited. Historically, the methods of bioassay-guided screening have played a major role in RiPP discovery, but recently this kind of strategy is continuously showing signs of reaching its limits.9,10 This is largely due to the fact that most of the RiPPs biosynthetic gene clusters (BGCs) in microbial genomes are inactive under laboratory culture conditions, and our knowledge about the majority of microbial communities remains limited. With significant advancements in genome sequencing and bioinformatics tools, genome mining is playing an increasingly important role in RiPP discovery.6,11,12 Despite its success in numerous cases, this approach also has its limits, as it often relies on specific modification enzymes or known types of leader peptides, and is hence less effective in discovering entirely new RiPP families.10,13 RiPPs engineering serves as a crucial technology for creating RiPP analogs that do not exist naturally.14 Techniques such as site-directed mutagenesis and the introduction of noncanonical amino acids are well-established methods employed in RiPPs engineering.15,16 Additionally, the flexibility and catalytic promiscuity of post-translational modification (PTM) enzymes allow for the production of RiPP variants and hybrids.17 Overall, each of these strategies has advantages and disadvantages, but they all play an important role in RiPP discovery.
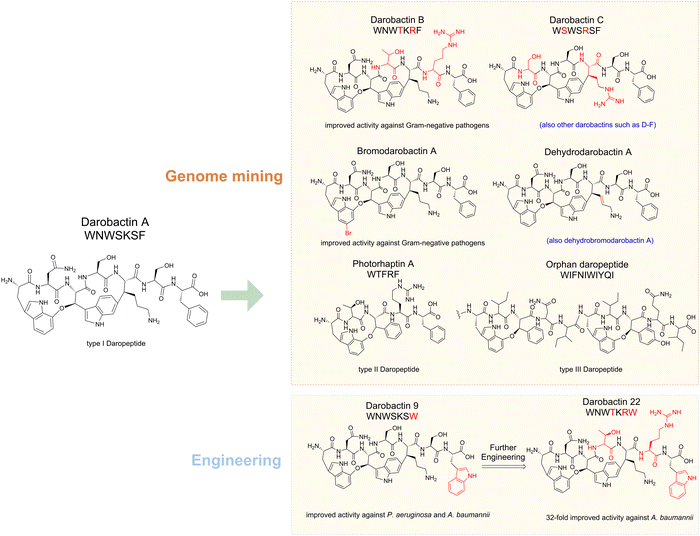
An excellent example in RiPP discovery is darobactin A and the darobactin-like peptide (daropeptide) family (Fig. 2). Darobactin A was initially identified through a bioactivity guided strategy,18 and many of its analogs and derivatives were later identified by genome mining and bioengineering approaches.19–21 Very recently, a combined genome mining strategy revealed that the daropeptide family can be grouped into three subfamilies, each with distinct structural features.22 Meanwhile unnatural darobactins were also created by engineering utilizing the substrate promiscuity of the biosynthetic enzymes (Fig. 2).23 These studies allowed discovery of new daropeptides with significantly improved activities, spurring various efforts in analog exploration, bioengineering, and mechanistic studies.19,20,23–26
Herein, we summarize the significant advancements made in the discovery and engineering of new RiPPs in recent years. Readers with an interest in the historical background of RiPPs and previous research can find extensive information in established reviews.11,27 Moreover, for those focusing on specific facets of RiPPs discovery, such as genome mining,28–30 engineering,15,17 or biological activity,14 consulting specialized reviews is recommended. A discernible trend in RiPPs research is the increasing adoption of more rationalized strategies, including delicate selection of microorganisms for screening, genome mining by comprehensive exploration of RiPP biosynthetic genes, and biosynthetic engineering and chemical modification. It is expected that future implementation of these strategies may hold great promise to usher in a new era in the study of RiPPs and facilitate practical and clinical applications of this remarkable superfamily of NPs.
1. Top-down approaches
Top-down approaches begin with the collection of biological samples from diverse environments, and it does not require genome sequencing or sophisticated genetic manipulation efforts. Top-down approaches involve direct screening of molecules from the biological samples and/or from artificially cultivated sources.31 Following the isolation and identification of bioactive molecules, the related BGCs are usually subjected to functional analysis to understand the biosynthetic pathways.32,33 Despite several drawbacks, the bioactivity guide strategy still remains a powerful and robust approach for discovering new RiPP families, which also offers valuable insights for genome mining and engineering efforts for discovering novel RiPPs.341.1 Bioactive molecules from various habitats
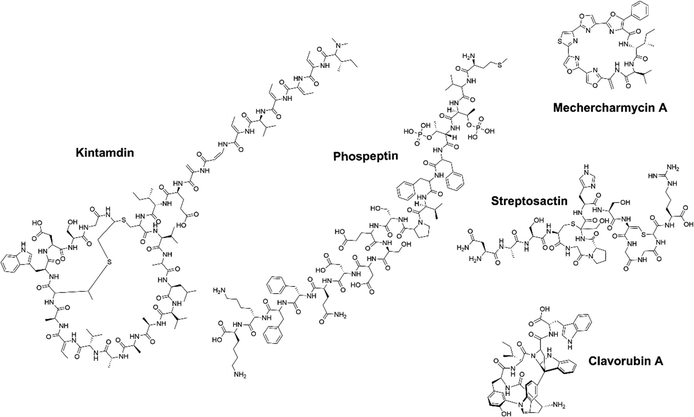
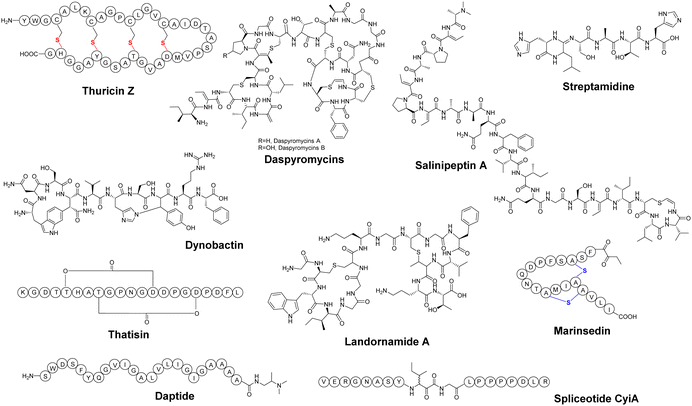
Microorganisms flourish globally due to their diverse metabolism, enabling them to thrive in highly competitive environments.35 This diverse metabolic capacity has led to the exploration of microbial natural products as a valuable resource for medicinal applications over the past decades.9 Among all microbial communities, soil microorganisms, marine microorganisms, and human-associated microorganisms have emerged as prominent sources in NP research. In recent years, a series of novel RiPPs exhibiting important biological activities have been isolated and identified from the metabolites of these microbial communities, expanding the growing knowledge in the field.Soil microbes represent a significant portion of the biodiversity on earth, and are tremendous producers of NPs, with a significant proportion being RiPPs.36 Mining these microbes can be fruitful for discovering novel RiPPs with unique structures and biological activities. For example, Streptomyces sp. CT34, isolated from a rhizosphere sample, showed the ability to produce a previously unknown high molecular-weight compound under acidic culture conditions.37 Mass spectrometry (MS) and nuclear magnetic resonance (NMR) analyses unveiled this compound, named legonaridin, which is similar to the canonical linaridin member cypemycin but features a unique carboxylic acid at the C-terminus. Legonaridin was later classified as a class B linaridin, which inspired the genome mining efforts for other class B linaridins.38–42 Similarly, kintamdin, a distinctive RiPPs compound characterized by a bis-thioether macrocyclic crosslink (MAbi) and a β-enamino acid residue (Fig. 3), was identified through metabolite profiling of Streptomyces sp. RK44 isolated from a soil sample.43 Through comprehensive analysis of kintamdin BGC, additional members of the previously uncharacterized β-bithionins family, sharing similarities with kintamdin, were identified through genomic analysis.43
Marine microbes represent a significant and diverse group of organisms.44,45 Within the marine environment, there is a rich microbial source of BGCs, where many novel PTMs were first identified in RiPPs.46 Over the past few years, a significant number of RiPPs have been successfully isolated and identified from marine microorganisms.47–49 For instance, Paoli et al. analyzed 1038 metagenomes from seawater samples, resulting in the discovery of 39![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 055 BGCs and 3861 previously unknown gene clusters.50 A substantial proportion (78%) of these gene cluster families encode terpenes, RiPPs and other NPs, with 47% originating from phyla with untapped biosynthetic potential. A closer examination of the genome of one of these phyla, Candidatus eudoremicrobium, led to the identification of two novel RiPPs natural products, namely phospeptin (Fig. 3) and pythonamide. The characterization of these two new RiPP pathways revealed unique bioactive compound structures and unprecedented enzymology.47,50 The highly cytotoxic natural product polytheonamide B, isolated from bacterial symbionts of the marine sponge Theonella swinhoei, was found to be a RiPP.51,52 Subsequent investigations into the polytheonamides biosynthetic pathway unveiled a series of bioactive molecules belonging to the polytheonamide family.53,54 The biosynthesis characterization of another marine RiPP, mechercharmycin A (Fig. 3), provided insights into the azol(in)e-containing RiPPs family and a new biosynthetic enzymology.55 Mechercharmycin A and its congeners demonstrated notable antitumor activities, opening up possibilities for further exploration in the development of anti-cancer drugs.
055 BGCs and 3861 previously unknown gene clusters.50 A substantial proportion (78%) of these gene cluster families encode terpenes, RiPPs and other NPs, with 47% originating from phyla with untapped biosynthetic potential. A closer examination of the genome of one of these phyla, Candidatus eudoremicrobium, led to the identification of two novel RiPPs natural products, namely phospeptin (Fig. 3) and pythonamide. The characterization of these two new RiPP pathways revealed unique bioactive compound structures and unprecedented enzymology.47,50 The highly cytotoxic natural product polytheonamide B, isolated from bacterial symbionts of the marine sponge Theonella swinhoei, was found to be a RiPP.51,52 Subsequent investigations into the polytheonamides biosynthetic pathway unveiled a series of bioactive molecules belonging to the polytheonamide family.53,54 The biosynthesis characterization of another marine RiPP, mechercharmycin A (Fig. 3), provided insights into the azol(in)e-containing RiPPs family and a new biosynthetic enzymology.55 Mechercharmycin A and its congeners demonstrated notable antitumor activities, opening up possibilities for further exploration in the development of anti-cancer drugs.
Human microbes are another fascinating reservoir of bioactive molecules because of their rich capacity in natural product biosynthesis, which could be essential for the microbial environment and directly impact human health. Recently, several active RiPPs molecules have been identified in commensal and pathogenic bacteria, primarily isolated from the human gut and skin.56 For example, Ruminococcus gnavus E1 strain, a Gram-positive bacterium isolated from human feces, was predicted to encode sactipeptides using in silico analysis.57 Previous research reported that the cecal contents of rats monoassociated with R. gnavus E1 displayed anti-Clostridium perfringens activity.58 Subsequently, the bioactive molecule named ruminococcin C1 (RumC1) was identified from feces samples of the monoassociated rats.59 Unlike most sactipeptides (e.g., subtilosin, thurincin H, thuricin CD, and thuricin Z) with a single hairpin-like structure, RumC1 possesses a unique double hairpin-like structure and demonstrates strong antimicrobial activity without toxicity toward eukaryotic cells, showing great promise as a candidate for drug development.60–63 In addition, a novel sactipeptide streptosactin was identified from the supernatants of a human-associated bacterium Streptococcus spp. (Fig. 3).64 Streptosactin possesses fratricidal activity against the producing host and the related strains. This finding not only identified streptosactin as the first sactipeptide from Streptococcus spp., but also suggests human streptococci may serve as a valuable source for the discovery of bioactive RiPPs.
1.2 New RiPPs obtained by activation of silent BGCs
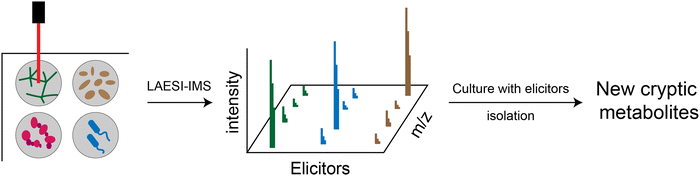
Silent or cryptic BGCs refer to those BGCs that are either not expressed or expressed at low levels under conventional laboratory conditions. It is estimated that the number of silent BGCs is significantly outnumbered by the number of active BGCs, indicating that a significant portion of the therapeutic potential of RiPPs remains untapped.54,65,66 There are several methods available to access the products of silent BGCs, including heterologous expression, promoter manipulation, reporter-guided mutant selection and many others.67,68 A particularly noteworthy application is the high-throughput elicitor screening (HiTES) strategy (Fig. 4).9 In the HiTES workflow, microbes are cultured in 96-well plates with hundreds of different elicitors, and the cultures are then analyzed using laser ablation coupled electrospray ionization imaging mass spectrometry (LAESI-IMS). By extracting signals from each well, the expression of specific metabolites in the presence of particular elicitors can be identified and compared to other conditions. With the recent advances in MS technologies, the LAESI-IMS inspection of a 96-well plate can now be completed in less than an hour. The simplicity of the HiTES approach has facilitated its broader application, including the study of rare actinomycetes that are traditionally difficult to genetically manipulate. Utilizing the HiTES strategy, dozens of cryptic bioactive metabolites were identified.69–74 For example, an unusual RiPP clavorubin A (Fig. 3) was identified through HiTES, which was stimulated by actinomycin.72 Moreover, the knowledge of the elicitors corresponding to these newly discovered metabolites provides insights into the mechanism of elicitation and aids in the further exploration of regulatory circuits in RiPP production (Table 1).| RiPP compound | RiPP class | Origin | Biological activity | Ref. |
|---|---|---|---|---|
| a Represent a new RiPPs class. | ||||
| Kintamdin | Kintamdina | Soil | Cytotoxic | 43 |
| Phospeptin | Phospeptina | Marine | Anti-neutrophil elastase | 50 |
| Mechercharmycin A | Polyazole cyclopeptides | Marine | Antitumor | 55 |
| Streptosactin | Sactipeptide | Human-related | Antimicrobial | 64 |
2. Genomics-based approaches
Genomics-based bottom up approaches utilize gene-based big data methods to discover specialized metabolites and study their corresponding biosynthetic pathways, which are in most cases referred to as genome mining.22 Genome mining offers a more rationalized and systematic alternative as opposed to the traditional bioactivity guided screening strategy, which is usually limited to the expressed molecules.13 The rationale behind genome mining lies in the understanding that the unique structural features of each RiPPs family are determined by the characteristic features of PTM enzymes and precursor peptides.Genome mining is usually performed by searching gene sequences that exhibit significant similarity to the characteristic genetic elements of known RiPPs, and BGCs associated with the same family RiPPs or similar RiPPs can be located.11,75 This approach enables access to metabolites that remain silent or minimally expressed under laboratory growth conditions, facilitating subsequent steps such as heterologous expression and large-scale production without relying solely on microbial cultivation.13 By leveraging the power of genome mining, scientists have made significant strides in uncovering novel molecules and unraveling the intricate enzyme mechanisms that underlie them.
The field of genome mining has witnessed significant advancements in tools and techniques over the past decade. Besides the early developed methods such as basic local alignment search (BLAST), the more recent approaches involve machine learning and neural network algorithms, allowing for the prediction of the new RiPPs with more unconventional BGCs.28,35,76,77 While genome mining tools have become increasingly versatile, limitations are also associated with these strategies. Because genome mining generally focuses on identifying gene similarity in conservative PTM enzymes and precursor peptides, its discovery is usually confined to the known classes of RiPPs. Also, as many PTM enzymes share significant homology with enzymes in other metabolic pathways, mining efforts can usually yield products that are unrelated to RiPPs.78
In recent years, strategies for exploring RiPPs using the PTM enzyme as the gene mining hallmark have been further developed. These methods have emerged to overcome the limitations of traditional approaches and discover new classes of RiPPs. Moreover, novel gene mining hallmarks associated with precursor peptides are increasingly being utilized. These new mining methods effectively address the aforementioned drawbacks and have the potential to uncover numerous latent RiPP BGCs that would have otherwise remained undiscovered (Table 2).
| RiPP compound | RiPP class | Genome mining hallmark | Biological activity | Ref. |
|---|---|---|---|---|
| a Represent a new RiPPs class. | ||||
| Thuricin Z/huazacin | Sactipeptide | rSAM | Antibacterial | 63 and 79 |
| Dynobactin | Dynobactina | rSAM | Anti-Gram-negative | 80 |
| Bromodarobactin A | Darobactin | rSAM | Anti-Gram-negative | 21 |
| Daspyromycins | Lanthipeptide | Cysteine decarboxylase | Antimicrobial | 81 |
| Landornamide A | Proteusin | NHLP | Anti-arenaviral | 54 |
| Daptides | Daptidea | RRE-short ORFs | Membrane targeting | 6 |
| Salinipeptins | Linaridin | Conserved motif | Antimicrobial/anticancer | 82 |
| Marinsedin | Lanthipeptide | BGC identification | Cytotoxic | 83 |
| Spliceotides | Spliceotidesa | rSAM-conserved motif | Protease inhibitory | 84 |
| Aeronamides | Polytheonamide | Precursor-epimerase-methyltransferase | Cytotoxic | 53 |
2.1 PTM enzyme-related genome mining
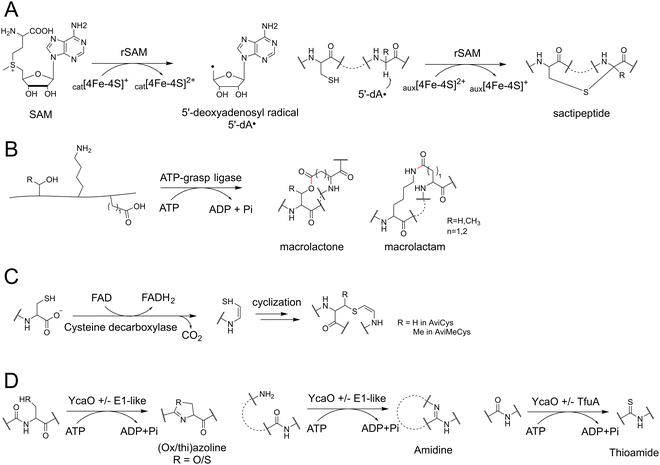
PTMs play a crucial role in determining the structural diversity of RiPPs, which is carried out by a range of specific PTM enzymes. Certain families of PTM enzymes are responsible for the formation of characteristic RiPP structures, which can serve as a hallmark to allow for the identification of specific RiPP families with distinct modifications.85 For example, [4+2]-cycloaddition enzymes have been employed to discover new thiopeptides, asparagine synthetase B-like proteins have been employed to explore lasso peptides, and α/β hydrolase fold proteins and horizontally transferred transmembrane helix (HTTH) proteins have facilitated the discovery of novel linaridin.38,86–89 The strategy of genome mining utilizing PTM enzymes has proven to be a well-established tool, yet it continues to yield surprising discoveries.Given the pivotal role of rSAM enzymes in RiPP biosynthesis, recent efforts have aimed to identify as many rSAM-based RiPPs as possible from the existing genome database.101 One study explored ∼15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 “RaS-RiPP” BGCs by examining the co-occurrence of rSAM enzymes and various transporters.102 Additionally, the SPECO (small peptide and enzyme co-occurrence analysis workflow) method was employed to identify 32
500 “RaS-RiPP” BGCs by examining the co-occurrence of rSAM enzymes and various transporters.102 Additionally, the SPECO (small peptide and enzyme co-occurrence analysis workflow) method was employed to identify 32![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 220 rSAM-based RiPP BGCs by detecting the co-occurrence of rSAM enzymes and hypothetical precursors encoded by small ORFs.103 The concept of utilizing the co-occurrence of multiple important genetic elements as a means of RiPP mining has proven to be a fruitful approach.
220 rSAM-based RiPP BGCs by detecting the co-occurrence of rSAM enzymes and hypothetical precursors encoded by small ORFs.103 The concept of utilizing the co-occurrence of multiple important genetic elements as a means of RiPP mining has proven to be a fruitful approach.
The discovery of darobactin A, the first antibiotic effective against Gram-negative bacteria since 1960, marked a significant milestone.18 Given its importance, there has been a strong focus on identifying darobactin A analogues. The key rSAM enzyme in darobactin biosynthesis, DarE, was utilized in the search for potential darobactin analog BGCs.24 Apart from the dominant darobactin A precursor sequence in all identified darobactin BGCs, a few darobactin analogs (darobactin B–F) were distinguished with one or two amino acid differences in the core sequence.20 Darobactin B (Fig. 2) has a different pharmacokinetic property and is slightly more effective on Acinetobacter baumannii isolates than darobactin A. Besides, three alternate groups of enzymes resembling DarE were identified.80 Subsequent screening efforts led to the isolation of a novel antibiotic against Gram-negative bacteria, dynobactin A, from the concentrated culture supernatant of Photorhabdus australis (Fig. 5). Dynobactin A demonstrated improved water solubility compared to darobactin and exhibited efficacy against systemic E. coli infection in mouse models. In a very recent search for BGCs containing DarE, some marine bacteria were found to possess additional genes beyond the darABCDE core, such as transporter gene darG, protease gene darF, and darH encoding a FAD-dependent oxidoreductase.21,23 Analysis of the culture broths of P. luteoviolacea strains (which are known to harbor the darH gene) resulted in identification of a series of darobactin derivatives, including bromodarobactin A, dehydrobromodarobactin A, and dehydrodarobactin A (Fig. 2).21 These compounds display distinct solubility and plasma protein binding profiles compared to darobactin A, resulting in greater effectiveness than darobactin A. DarH enzyme was identified as a new flavin-dependent halogenase responsible for halogenation of the darobactin core, whereas the enzyme responsible for the dehydrogenation remains unclear thus far.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 479. Thatisin features an isomeric behavior at the Asp14-Pro15 amide, which is rarely reported in known RiPPs. Recently, the graspeptides have been further extended to 174 families based on the signature motifs within the leader region of the graspeptides.109,110 The expanding graspetides RiPPs family, containing thousands of members with diverse topologies, provides valuable insights into the mechanism of macrocyclization in graspetide biosynthesis.
479. Thatisin features an isomeric behavior at the Asp14-Pro15 amide, which is rarely reported in known RiPPs. Recently, the graspeptides have been further extended to 174 families based on the signature motifs within the leader region of the graspeptides.109,110 The expanding graspetides RiPPs family, containing thousands of members with diverse topologies, provides valuable insights into the mechanism of macrocyclization in graspetide biosynthesis.
Furthermore, it has been observed that YcaO proteins can synergistically collaborate with the TfuA domain proteins to facilitate the formation of naturally rare thioamidated peptides.136 Extensive analyses employing the Rapid ORF Description & Evaluation Online (RODEO) and RiPP Precursor Peptide Enhanced Recognition (RiPPER) systems have demonstrated the wide distribution of YcaO and TfuA proteins in actinobacteria, leading to the discovery of a novel subfamily of thioamidated RiPPs known as thiovarsolins.136 Similarly, a global genome mining effort for thioamidated RiPPs by antiSMASH and RiPPER identified 613 YcaO-TfuA BGC families and 797 precursor families spanning across 14 different phyla.137,138 These findings highlight the immense potential for further investigations into the YcaO proteins and the discovery of YcaO-mediated RiPPs.
2.2 Precursor peptide related gene mining
RiPP precursor peptides typically consist of a leader peptide, a core peptide, and, in some cases, a follower peptide (Fig. 1).11 Upon extensive PTMs in the core peptide, the leader and follower peptides are proteolytically removed to release the modified cores. Despite a few successful cases, genome mining based on the core sequence usually poses challenges due to the relatively short length of the core.28,29 On the other hand, the leader not only plays a crucial role in enzyme recognition and ensuring the correct sequence of modifications, but can sometimes also serve as a valuable hallmark for gene mining and as a basis for the classification of RiPPs, as the leader could exhibit a significant level of similarity within a particular RiPP class.78,139,140 When combined with other characteristic hallmarks for gene mining, the genetic signatures associated with precursor peptides provide a straightforward approach to exploring RiPP families/subfamilies that were previously overlooked.6,141RRE-Finder, a bioinformatic tool designed for RRE domain prediction, demonstrates high sensitivity in RRE identification.141 By searching the UniProtKB protein database, RRE-Finder retrieved approximately 25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 RRE domains, some of which are associated with uncharacterized RiPP classes, suggesting the potential for the discovery of new RiPP subfamilies with novel structures. Recently, all predicted RRE domains from RRE-Finder were subjected to analysis using RODEO to determine their co-occurrence with short open reading frames (ORFs).6 Among them, one type of fusion protein combining a domain of unknown function (DUF) with an RRE was selected, leading to the identification of 1441 putative precursor peptides from 483 BGCs. The representative BGC encodes a unique product termed daptide (Fig. 5), which is characterized by a helical structure and net positive charged terminals. Daptide exhibit hemolytic activity and can aid in membrane targeting.
000 RRE domains, some of which are associated with uncharacterized RiPP classes, suggesting the potential for the discovery of new RiPP subfamilies with novel structures. Recently, all predicted RRE domains from RRE-Finder were subjected to analysis using RODEO to determine their co-occurrence with short open reading frames (ORFs).6 Among them, one type of fusion protein combining a domain of unknown function (DUF) with an RRE was selected, leading to the identification of 1441 putative precursor peptides from 483 BGCs. The representative BGC encodes a unique product termed daptide (Fig. 5), which is characterized by a helical structure and net positive charged terminals. Daptide exhibit hemolytic activity and can aid in membrane targeting.
Furthermore, the joint use of RRE-Finder and https://RadicalSAM.org has facilitated the compilation of all RRE-associated rSAM proteins, enabling the search for new RiPP families whose biosynthesis involves rSAM enzymes.146,147 As a result, ∼15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 RRE-associated rSAM proteins have been collected and categorized. Among them, six new sactipeptide groups that had eluded traditional genomic mining methods were identified. It is noteworthy that only 1/5 of the members of RRE-associated rSAM proteins overlap with the “RiPP-RaS” database, indicating the remaining proteins may belong to additional large and functionally diverse RiPPs families, which could potentially lead to significant research breakthroughs.12
000 RRE-associated rSAM proteins have been collected and categorized. Among them, six new sactipeptide groups that had eluded traditional genomic mining methods were identified. It is noteworthy that only 1/5 of the members of RRE-associated rSAM proteins overlap with the “RiPP-RaS” database, indicating the remaining proteins may belong to additional large and functionally diverse RiPPs families, which could potentially lead to significant research breakthroughs.12
2.3 Combined mining strategy
Genome mining using a single hallmark often yields a large amount of mixed mining results with reductant information. An approach to address this issue is to employ an evaluation system such as RODEO to score the excavated precursor peptides.40,106 Using this approach, highly reliable results can be selected to focus on the targeted RiPPs family. In addition, the combined mining strategy can also serve as a refining tool. As demonstrated in the search for RRE-associated rSAM proteins,146 combined gene mining can leverage a combination of PTM enzyme and precursor peptide as hallmarks, or multiple independent PTM enzymes as hallmarks, thereby ensuring the relevance of RiPPs in the mining efforts to explore new RiPP families.Spliceases are rSAM enzymes with a SPASM motif that introduces rare beta-amino acid residues into certain Nif11-type RiPPs. A known splicease, PlpX, for instance, works in collaboration with its auxiliary RRE protein, PlpY, to excise a tyramine equivalent from XYG sites and incorporate a truncated α-keto-β-amino acid moiety.157 In a study aimed at uncovering more RiPPs containing α-keto-β-amino acid moieties, a combined mining strategy was utilized.84 Initially, the result of a BLAST search for splicease using PlpX as an enquiry, was combined with all rSAM-SPASM proteins in the Uniprot RefProt database, resulting in 3418 candidates. Subsequently, the frequency of catalytic site YG motifs next to the SPASM domain was checked by RODEO v2.0.75 This analysis revealed the presence of YG-rich rSAM enzymes akin to PlpX in 189 different bacterial genera. Ultimately, 27 confirmed splicease-substrate pairs were designated as “spliceotides” (Fig. 5). Notably, all three identified spliceotides exhibited potent protease inhibitory activities, demonstrating significant potential in drug discovery.
Polytheonamides are a class of highly cytotoxic RiPPs initially isolated from a marine sponge.51,52 In early attempts to find molecules similar to polytheonamide, the epimerase PoyD was chosen as the genome mining signature. However, no polytheonamide-type compound was found.53 It was later noted that the stable β-helix structure characteristic of polytheonamides is formed through the methylation of Asn side-chains in repeated NX5N motifs by the N-methyltransferase PoyE. Based on these biosynthesis features, the BGCs containing clustered NX5N precursors, epimerases, and methyltransferases were considered as targeted candidates.53 As a result, aeronamides were identified from a deep-rock subsurface bacterium. Aeronamides were rich in D-amino acids and exhibited potent anti-HeLa cell activity, but they did not show any activity against bacteria or fungi.
Darobactins feature a unique ether cross-link between two aromatic residues. As a result, the Ω1–X2–Ω3 motifs (Ω denotes an aromatic residue while X denotes an non-aromatic residue) at the precursor's C-terminus could serve as a mining signature for daropeptides.22 By querying the DarE-like protein and Ω1–X2–Ω3 motifs simultaneously, a comprehensive dataset for daropeptides was revealed, containing 86 precursors across 11 bacterial genera. Daropeptides can be grouped into three subfamilies based on the sequence patterns. The canonical daropeptides (e.g. darobactin A) feature a Ω1–Ω3–X5 sequence for the formation of a bicyclic scaffold consisting of an ether crosslink and a C–C bond crosslink, whereas the other two groups only have an ether crosslink and lack the C–C crosslink. One of these daropeptides (photorhaptin A, Fig. 2) from Photorhabdus asymbiotica was recently identified through heterologous expression. Because the BGC (pas) also encodes a GNAT family acetytransferase (PasC) with thus far unknown function, it is very likely that photorhaptin A is not a final product but a biosynthetic intermediate. Intriguingly, the maturases of photorhaptin A possess intrinsic catalytic promiscuity that is controlled by the substrate sequence. The discovery of photorhaptin A has significantly expanded the darobopeptide family and our understanding of rSAM enzymology.
3. RiPPs engineering
RiPPs engineering has emerged as a rational approach for creating RiPP analogs with improved pharmaceutical properties, which is also an effective way to mass produce the desired RiPPs. Recent advances in molecular biology technologies, along with a better understanding of the substrate specificity and promiscuity of PTM enzymes, have significantly facilitated efforts in RiPPs engineering.34,1583.1 Hybrid RiPPs
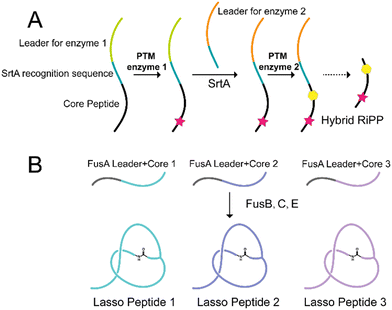
Hybrid RiPPs are unnatural molecules generated by the modification of precursor peptides using PTM enzymes from other pathways.46,159–161 To facilitate the generation of hybrid RiPPs, advanced strategies such as leader peptide exchange have been developed.162 For instance, transpeptidation mediated by sortase A with two PTM enzymes resulted in a doubly modified core peptide product, offering a cost-effective synthetic approach to accessing new-to-nature hybrid RiPPs (Fig. 7A).3.2 Engineered RiPPs formed by utilization of promiscuous PTM enzymes
Catalytic promiscuity refers to the ability of enzymes to catalyze reactions using mechanisms that differ from their native ones, while substrate promiscuity refers to the ability of enzymes to act on various similar substrates.163 In the context of RiPP, it has frequently been observed that RiPP biosynthetic enzymes are responsible for modifying different peptide substrates or different residues in the precursor peptide, indicating their inherent promiscuity.164 Exploiting the promiscuous nature of these enzymes allows the production of unnatural NPs by using them as modification tools to expand the chemical diversity of RiPPs, enabling the synthesis of novel compounds with potentially valuable properties.165,166 For example, the darobactin PTM enzymes darBCDE have been observed to have high substrate tolerance.23 Different darobactin precursor peptides with major amino acid substitutions can be tolerated in heterologous production with darBCDE or darE only.23 13 new unnatural darobactins were created in this way, and one of them, darobactin 9, carrying an unnatural C-terminal L-tryptophan, showed improved activity against Pseudomonas aeruginosa and Acinetobacter baumannii. Based on the cryo-EM structure of darobactin 9 and the antibiotic target Bam complex, darobactin 9 was further modified, and many of them exhibited stronger inhibition against Gram-negative pathogens, among which darobactin 22 exhibited 32-fold increased activity against Acinetobacter baumannii.167PapB is a substrate-tolerant rSAM RiPP maturase that catalyzes the formation of thioether crosslinks in Cys-X3-Asp/Glu motifs.168 PapB exhibits tolerance toward motif length variation and the incorporation of D-amino acids on either Cys or Asp residues.169 Leveraging these properties, Bandarian et al. have successfully utilized PapB to generate an analog of octreotide, an FDA-approved drug, with a thioether-cyclized Cys-Glu cross-link in place of the original disulfide. Although the bioactivity of this octreotide analogue requires further evaluation, this study represents an important milestone in utilizing rSAM enzymes for the modification of a therapeutic agent.
The substrate tolerance of several PTM enzymes of lasso peptides has been proven in the study investigating structural diversification of lasso peptides.170–175 Specifically, the PTM enzymes in the fusilassin BGC (FusB, C, E) have been found to tolerate chimeric substrates consisting of native leaders and unnatural cores.176 This remarkable substrate tolerance was also observed for FusC, which showed amazing substrate tolerance to accommodate diverse mutated substrates within the ring region (Fig. 7B).177 Additionally, microcin J25, another lasso peptide cyclase, exhibited tolerance toward variation in both the loop and tail regions.171 Leveraging these findings, a cell-free biosynthesis approach can generate millions of sequence-diverse lasso peptides by treating varied precursor peptides with these PTM enzymes in combination. The next phase of research will then focus on utilizing these substrate-tolerant PTM enzymes to generate lasso peptides with specific desired features.177
Type I sactipeptides are characterized by their nested hairpin structures formed by thioether cross-links between N-terminal cysteines and C-terminal acceptor amino acids.98 This structural feature confers heat and proteolysis resistance, making sactipeptides attractive scaffolds for various biotechnological applications.178 The sactipeptide subtilosin A features an exposed loop region that facilitates the insertion of target-binding peptide sequences. AlbA, the sacipeptie synthase responsible for the formation of thioether cross-links in subtilosin A, exhibits remarkable substrate promiscuity, allowing for generation of various subtilosin analogs by modification of precursor peptides with unnatural cores.179 Although successful engineering cases have been limited, which requires a deeper understanding of substrate recognition and catalytic mechanisms, these efforts have prepared the way for the directed evolution of studies aimed at optimizing sactipeptide properties.
3.3 Chemical editing of RiPPs
Following PTMs, certain residues, such as dehydroalanine (Dha), possess the potential for chemical editing. In an exciting development, several RiPPs from diverse families have been efficiently β-borylated using CuII-catalysis under mild conditions.180 This β-borylation of Dha residue significantly enhances the water solubility of thiostrepton, while retaining its antimicrobial activity. Moreover, the borylation group can serve as a useful handle for subsequent transformations and labeling, opening up possibilities for further functionalization of RiPPs.4. Conclusions and perspectives
RiPPs represent a highly diverse superfamily of natural products with remarkable diversity in structure and biological activity, showing great promise in the development of drug-leaders in pharmaceutical applications. Recent efforts in discovering novel RiPPs can be grouped into three major categories, the bioactivity guided strategies, genome mining, and biosynthetic engineering. Each of these approaches possesses distinct advantages and limitations, necessitating their optimization and utilization in various contexts.In the bioactivity guided strategy, researchers are increasingly turning their attention to microorganisms thriving in harsh environments characterized by limited resources and intense competition. The organisms from these environments usually represent a less tapped territory for the mining of novel natural products. However, a significant challenge arises from the likelihood of biosynthetic gene clusters being silenced under artificial culture conditions, resulting in a lack of antibacterial activity during screening. Consequently, there is a pressing need for efficient and reliable approaches to activating silent BGCs. Although the bioactivity guided strategy can be complex and time-consuming, it remains the most direct approach for discovering novel drugs when no initial clues are available. Incorporation of MS-based dereplication methods into the bioactivity guided process could be useful to ensure a more accurate and efficient discovery of novel RiPPs.181–186
The expanding wealth of knowledge regarding RiPP biosynthesis has propelled the genome mining strategy to an increasingly significant role in RiPP discovery. When specific details about a RiPP biosynthetic gene cluster are identified, the conserved and vital PTM enzymes within it can be utilized for comprehensive gene mining within the same RiPP family.187–189 The distinct structure and unique enzymology of each novel RiPP can also serve as valuable starting points for further gene mining and development. As showcased in this review, recent genome mining research appears to advance towards a more rational and multifaceted approach. To enhance the effectiveness of genome mining strategies, two or more gene mining hallmarks of RiPP BGCs are now being employed as indicators, which exponentially amplifies the probability of identifying structural analogs.
Leveraging the gene-encoded nature of RiPP biosynthesis, RiPP engineering offers a potent approach for generating novel analogs through straightforward manipulation of precursor peptide sequences. By harnessing the promiscuity of PTM enzymes, either within or outside BGCs, more advanced catalytic systems can be developed for RiPP engineering. However, achieving future success in RiPP engineering requires a deeper comprehension of structure–activity relationships, the substrate and catalytic promiscuity of enzymes, and the creation of a chemical toolbox for precise structural modifications.
In conclusion, the recent advances in RiPP discovery have not only deepened our understanding of RiPP biosynthesis but also sparked the development of more comprehensive approaches for designing novel RiPPs. The insights gained from these studies will serve as guiding principles for future endeavors in RiPP research, bringing us closer to the ambitious objective of widespread and extensive discovery of novel RiPP natural products. These efforts hold immense potential for exploring the applications of RiPPs in medicine and human health, opening up new avenues for therapeutic development and positively impacting our well-being.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
We thank Prof. Liujie Huo (Shandong University) for the helpful discussion on the bacteria types harboring significant potential for RiPPs discovery. This work is supported by grants from the National Key Research and Development Program (2018Y F A0900402, 2021YFA0910501), from the National Natural Science Foundation of China (31870050, 32270070, U22A20451), from the funding of Innovative research team of high-level local universities in Shanghai and a key laboratory program of the Education Commission of Shanghai Municipality (ZDSYS14005), and from the West Light Foundation of The Chinese Academy of Sciences (xbzg-zdsys-202105).References
- F. A. E. de Brito, A. P. P. de Freitas and M. S. Nascimento, Multidrug-Resistant Biofilms (MDR): Main Mechanisms of Tolerance and Resistance in the Food Supply Chain, Pathogens, 2022, 11(12), 1416 CrossRef CAS PubMed
.
- A. G. Atanasov, S. B. Zotchev, V. M. Dirsch, T. International Natural Product Sciences and C. T. Supuran, Natural products in drug discovery: advances and opportunities, Nat. Rev. Drug Discovery, 2021, 20(3), 200–216 CrossRef CAS PubMed
.
- D. J. Newman and G. M. Cragg, Natural Products as Sources of New Drugs from 1981 to 2014, J. Nat. Prod., 2016, 79(3), 629–661 CrossRef CAS PubMed
.
- C. Ongpipattanakul, E. K. Desormeaux, A. DiCaprio, W. A. van der Donk, D. A. Mitchell and S. K. Nair, Mechanism of Action of Ribosomally Synthesized and Post-Translationally Modified Peptides, Chem. Rev., 2022, 122(18), 14722–14814 CrossRef CAS PubMed
.
- L. Cao, T. Do and A. J. Link, Mechanisms of action of ribosomally synthesized and posttranslationally modified peptides (RiPPs), J. Ind. Microbiol. Biotechnol., 2021, 48(3–4), kuab00 Search PubMed
.
- H. Ren, S. R. Dommaraju, C. Huang, H. Cui, Y. Pan, M. Nesic, L. Zhu, D. Sarlah, D. A. Mitchell and H. Zhao, Genome mining unveils a class of ribosomal peptides with two amino termini, Nat. Commun., 2023, 14(1), 1624 CrossRef CAS PubMed
.
- S. M. Rowe and D. R. Spring, The role of chemical synthesis in developing RiPP antibiotics, Chem. Soc. Rev., 2021, 50(7), 4245–4258 RSC
.
- H. Lee and W. A. van der Donk, Macrocyclization and Backbone Modification in RiPP Biosynthesis, Annu. Rev. Biochem., 2022, 91, 269–294 CrossRef CAS PubMed
.
- F. Xu, Y. Wu, C. Zhang, K. M. Davis, K. Moon, L. B. Bushin and M. R. Seyedsayamdost, A genetics-free method for high-throughput discovery of cryptic microbial metabolites, Nat. Chem. Biol., 2019, 15(2), 161–168 CrossRef CAS PubMed
.
- K. D. Bauman, K. S. Butler, B. S. Moore and J. R. Chekan, Genome mining methods to discover bioactive natural products, Nat. Prod. Rep., 2021, 38(11), 2100–2129 RSC
.
- M. Montalban-Lopez, T. A. Scott, S. Ramesh, I. R. Rahman, A. J. van Heel, J. H. Viel, V. Bandarian, E. Dittmann, O. Genilloud, Y. Goto, M. J. Grande Burgos, C. Hill, S. Kim, J. Koehnke, J. A. Latham, A. J. Link, B. Martinez, S. K. Nair, Y. Nicolet, S. Rebuffat, H. G. Sahl, D. Sareen, E. W. Schmidt, L. Schmitt, K. Severinov, R. D. Sussmuth, A. W. Truman, H. Wang, J. K. Weng, G. P. van Wezel, Q. Zhang, J. Zhong, J. Piel, D. A. Mitchell, O. P. Kuipers and W. A. van der Donk, New developments in RiPP discovery, enzymology and engineering, Nat. Prod. Rep., 2021, 38(1), 130–239 RSC
.
- K. A. Clark, L. B. Bushin and M. R. Seyedsayamdost, RaS-RiPPs in Streptococci and the Human Microbiome, ACS Bio Med Chem Au, 2022, 2(4), 328–339 CrossRef CAS PubMed
.
- M. H. Medema, T. de Rond and B. S. Moore, Mining genomes to illuminate the specialized chemistry of life, Nat. Rev. Genet., 2021, 22(9), 553–571 CrossRef CAS PubMed
.
- G. Zhong, Z.-J. Wang, F. Yan, Y. Zhang and L. Huo, Recent Advances in Discovery, Bioengineering, and Bioactivity-Evaluation of Ribosomally Synthesized and Post-translationally Modified Peptides, ACS Bio Med Chem Au, 2022, 3(1), 1–31 Search PubMed
.
- T. Do and A. J. Link, Protein Engineering in Ribosomally Synthesized and Post-translationally Modified Peptides (RiPPs), Biochemistry, 2023, 62(2), 201–209 CrossRef CAS PubMed
.
- T. Liu, X. Ma, J. Yu, W. Yang, G. Wang, Z. Wang, Y. Ge, J. Song, H. Han, W. Zhang, D. Yang, X. Liu and M. Ma, Rational generation of lasso peptides based on biosynthetic gene mutations and site-selective chemical modifications, Chem. Sci., 2021, 12(37), 12353–12364 RSC
.
- C. Wu and W. A. van der Donk, Engineering of new-to-nature ribosomally synthesized and post-translationally modified peptide natural products, Curr. Opin. Biotechnol., 2021, 69, 221–231 CrossRef CAS PubMed
.
- Y. Imai, K. J. Meyer, A. Iinishi, Q. Favre-Godal, R. Green, S. Manuse, M. Caboni, M. Mori, S. Niles, M. Ghiglieri, C. Honrao, X. Ma, J. J. Guo, A. Makriyannis, L. Linares-Otoya, N. Bohringer, Z. G. Wuisan, H. Kaur, R. Wu, A. Mateus, A. Typas, M. M. Savitski, J. L. Espinoza, A. O'Rourke, K. E. Nelson, S. Hiller, N. Noinaj, T. F. Schaberle, A. D'Onofrio and K. Lewis, A new antibiotic selectively kills Gram-negative pathogens, Nature, 2019, 576(7787), 459–464 CrossRef CAS PubMed
.
- H. Kaur, R. P. Jakob, J. K. Marzinek, R. Green, Y. Imai, J. R. Bolla, E. Agustoni, C. V. Robinson, P. J. Bond, K. Lewis, T. Maier and S. Hiller, The antibiotic darobactin mimics a beta-strand to inhibit outer membrane insertase, Nature, 2021, 593(7857), 125–129 CrossRef CAS PubMed
.
- N. Böhringer, R. Green, Y. Liu, U. Mettal, M. Marner, M. Modaresi Seyed, P. Jakob Roman, G. Wuisan Zerlina, T. Maier, A. Iinishi, S. Hiller, K. Lewis and F. Schäberle Till, Mutasynthetic Production and Antimicrobial Characterization of Darobactin Analogs, Microbiol. Spectrum, 2021, 9(3), e01535 Search PubMed
.
- N. Bohringer, J. C. Kramer, E. de la Mora, L. Padva, Z. G. Wuisan, Y. Liu, M. Kurz, M. Marner, H. Nguyen, P. Amara, K. Yokoyama, Y. Nicolet, U. Mettal and T. F. Schaberle, Genome- and metabolome-guided discovery of marine BamA inhibitors revealed a dedicated darobactin halogenase, Cell Chem. Biol., 2023, 30(8), 943–952 e7 CrossRef PubMed
.
- S. Ma, W. Xi, S. Wang, H. Chen, S. Guo, T. Mo, W. Chen, Z. Deng, F. Chen, W. Ding and Q. Zhang, Substrate-Controlled Catalysis in the Ether Cross-Link-Forming Radical SAM Enzymes, J. Am. Chem. Soc., 2023, 145(42), 22945–22953 CrossRef CAS PubMed
.
- S. Gross, F. Panter, D. Pogorevc, C. E. Seyfert, S. Deckarm, C. D. Bader, J. Herrmann and R. Muller, Improved broad-spectrum antibiotics against Gram-negative pathogens via darobactin biosynthetic pathway engineering, Chem. Sci., 2021, 12(35), 11882–11893 RSC
.
- S. Guo, S. Wang, S. Ma, Z. Deng, W. Ding and Q. Zhang, Radical SAM-dependent ether crosslink in daropeptide biosynthesis, Nat. Commun., 2022, 13(1), 2361 CrossRef CAS PubMed
.
- H. Nguyen, I. D. Made Kresna, N. Bohringer, J. Ruel, E. Mora, J. C. Kramer, K. Lewis, Y. Nicolet, T. F. Schaberle and K. Yokoyama, Characterization of a Radical SAM Oxygenase for the Ether Crosslinking in Darobactin Biosynthesis, J. Am. Chem. Soc., 2022, 144(41), 18876–18886 CrossRef CAS PubMed
.
- N. Ritzmann, S. Manioglu, S. Hiller and D. J. Muller, Monitoring the antibiotic darobactin modulating the beta-barrel assembly factor BamA, Structure, 2022, 30(3), 350–359 CrossRef CAS PubMed
e3.
- P. G. Arnison, M. J. Bibb, G. Bierbaum, A. A. Bowers, T. S. Bugni, G. Bulaj, J. A. Camarero, D. J. Campopiano, G. L. Challis, J. Clardy, P. D. Cotter, D. J. Craik, M. Dawson, E. Dittmann, S. Donadio, P. C. Dorrestein, K. D. Entian, M. A. Fischbach, J. S. Garavelli, U. Goransson, C. W. Gruber, D. H. Haft, T. K. Hemscheidt, C. Hertweck, C. Hill, A. R. Horswill, M. Jaspars, W. L. Kelly, J. P. Klinman, O. P. Kuipers, A. J. Link, W. Liu, M. A. Marahiel, D. A. Mitchell, G. N. Moll, B. S. Moore, R. Muller, S. K. Nair, I. F. Nes, G. E. Norris, B. M. Olivera, H. Onaka, M. L. Patchett, J. Piel, M. J. Reaney, S. Rebuffat, R. P. Ross, H. G. Sahl, E. W. Schmidt, M. E. Selsted, K. Severinov, B. Shen, K. Sivonen, L. Smith, T. Stein, R. D. Sussmuth, J. R. Tagg, G. L. Tang, A. W. Truman, J. C. Vederas, C. T. Walsh, J. D. Walton, S. C. Wenzel, J. M. Willey and W. A. van der Donk, Ribosomally synthesized and post-translationally modified peptide natural products: overview and recommendations for a universal nomenclature, Nat. Prod. Rep., 2013, 30(1), 108–160 RSC
.
- Z. Zhong, B. He, J. Li and Y. X. Li, Challenges and advances in genome mining of ribosomally synthesized and post-translationally modified peptides (RiPPs), Synth. Syst. Biotechnol., 2020, 5(3), 155–172 CrossRef PubMed
.
- A. H. Russell and A. W. Truman, Genome mining strategies for ribosomally synthesised and post-translationally modified peptides, Comput. Struct. Biotechnol. J., 2020, 18, 1838–1851 CrossRef CAS PubMed
.
- K. J. Hetrick and W. A. van der Donk, Ribosomally synthesized and post-translationally modified peptide natural product discovery in the genomic era, Curr. Opin. Chem. Biol., 2017, 38, 36–44 CrossRef CAS PubMed
.
- M. M. Zhang, Y. Qiao, E. L. Ang and H. Zhao, Using natural products for drug discovery: the impact of the genomics era, Expert Opin. Drug Discovery, 2017, 12(5), 475–487 CrossRef CAS PubMed
.
- M. Aftab Uddin, S. Akter, M. Ferdous, B. Haidar, A. Amin, A. H. M. Shofiul Islam Molla, H. Khan and M. R. Islam, A plant endophyte Staphylococcus hominis strain MBL_AB63 produces a novel lantibiotic, homicorcin and a position one variant, Sci. Rep., 2021, 11(1), 11211 CrossRef CAS PubMed
.
- F. J. Ortiz-Lopez, D. Carretero-Molina, M. Sanchez-Hidalgo, J. Martin, I. Gonzalez, F. Roman-Hurtado, M. de la Cruz, S. Garcia-Fernandez, F. Reyes, J. P. Deisinger, A. Muller, T. Schneider and O. Genilloud, Cacaoidin, First Member of the New Lanthidin RiPP Family, Angew. Chem., Int. Ed., 2020, 59(31), 12654–12658 CrossRef CAS PubMed
.
- Y. Fu, A. H. Jaarsma and O. P. Kuipers, Antiviral activities and applications
of ribosomally synthesized and post-translationally modified peptides (RiPPs), Cell. Mol. Life Sci., 2021, 78(8), 3921–3940 CrossRef CAS PubMed
.
- F. Hemmerling and J. Piel, Strategies to access biosynthetic novelty in bacterial genomes for drug discovery, Nat. Rev. Drug Discovery, 2022, 21(5), 359–378 CrossRef CAS PubMed
.
- N. W. Sokol, E. Slessarev, G. L. Marschmann, A. Nicolas, S. J. Blazewicz, E. L. Brodie, M. K. Firestone, M. M. Foley, R. Hestrin, B. A. Hungate, B. J. Koch, B. W. Stone, M. B. Sullivan, O. Zablocki, L. S. M. Consortium and J. Pett-Ridge, Life and death in the soil microbiome: how ecological processes influence biogeochemistry, Nat. Rev. Microbiol., 2022, 20(7), 415–430 CrossRef CAS PubMed
.
- M. E. Rateb, Y. Zhai, E. Ehrner, C. M. Rath, X. Wang, J. Tabudravu, R. Ebel, M. Bibb, K. Kyeremeh, P. C. Dorrestein, K. Hong, M. Jaspars and H. Deng, Legonaridin, a new member of linaridin RiPP from a Ghanaian Streptomyces isolate, Org. Biomol. Chem., 2015, 13(37), 9585–9592 RSC
.
- T. Mo, W. Q. Liu, W. Ji, J. Zhao, T. Chen, W. Ding, S. Yu and Q. Zhang, Biosynthetic Insights into Linaridin Natural Products from Genome Mining and Precursor Peptide Mutagenesis, ACS Chem. Biol., 2017, 12(6), 1484–1488 CrossRef CAS PubMed
.
- F. Wang, W. Wei, J. Zhao, T. Mo, X. Wang, X. Huang, S. Ma, S. Wang, Z. Deng, W. Ding, Y. Liang and Q. Zhang, Genome Mining and Biosynthesis Study of a Type B Linaridin Reveals a Highly Versatile α-N-Methyltransferase, CCS Chem., 2021, 3(3), 1049–1057 CrossRef CAS
.
- M. A. Georgiou, S. R. Dommaraju, X. Guo, D. H. Mast and D. A. Mitchell, Bioinformatic and Reactivity-Based Discovery of Linaridins, ACS Chem. Biol., 2020, 15(11), 2976–2985 CrossRef CAS PubMed
.
- E. Pashou, S. J. Reich, A. Reiter, D. Weixler, B. J. Eikmanns, M. Oldiges, C. U. Riedel and O. Goldbeck, Identification and Characterization of Corynaridin, a Novel Linaridin from Corynebacterium lactis, Microbiol. Spectrum, 2023, 11(1), e0175622 CrossRef PubMed
.
- M. X. Guo, M. M. Zhang, K. Sun, J. J. Cui, Y. C. Liu, K. Gao, S. H. Dong and S. Luo, Genome Mining of Linaridins Provides Insights into the Widely Distributed LinC Oxidoreductases, J. Nat. Prod., 2023, 86(10), 2333–2341 CrossRef CAS PubMed
.
- S. Wang, S. Lin, Q. Fang, R. Gyampoh, Z. Lu, Y. Gao, D. J. Clarke, K. Wu, L. Trembleau, Y. Yu, K. Kyeremeh, B. F. Milne, J. Tabudravu and H. Deng, A ribosomally synthesised and post-translationally modified peptide containing a beta-enamino acid and a macrocyclic motif, Nat. Commun., 2022, 13(1), 5044 CrossRef CAS PubMed
.
- S. Sunagawa, L. P. Coelho, S. Chaffron, J. R. Kultima, K. Labadie, G. Salazar, B. Djahanschiri, G. Zeller, D. R. Mende, A. Alberti, F. M. Cornejo-Castillo, P. I. Costea, C. Cruaud, F. d'Ovidio, S. Engelen, I. Ferrera, J. M. Gasol, L. Guidi, F. Hildebrand, F. Kokoszka, C. Lepoivre, G. Lima-Mendez, J. Poulain, B. T. Poulos, M. Royo-Llonch, H. Sarmento, S. Vieira-Silva, C. Dimier, M. Picheral, S. Searson, S. Kandels-Lewis, N. Null, C. Bowler, C. de Vargas, G. Gorsky, N. Grimsley, P. Hingamp, D. Iudicone, O. Jaillon, F. Not, H. Ogata, S. Pesant, S. Speich, L. Stemmann, M. B. Sullivan, J. Weissenbach, P. Wincker, E. Karsenti, J. Raes, S. G. Acinas, P. Bork, E. Boss, C. Bowler, M. Follows, L. Karp-Boss, U. Krzic, E. G. Reynaud, C. Sardet, M. Sieracki and D. Velayoudon, Structure and function of the global ocean microbiome, Science, 2015, 348(6237), 1261359 CrossRef PubMed
.
- Y. Gao, X. Du, H. Li and Y. Wang, Genome sequence of Aspergillus flavus A7, a marine-derived fungus with antibacterial activity, Genome, 2021, 64(7), 719–733 CrossRef CAS PubMed
.
- D. Sardar and E. W. Schmidt, Combinatorial biosynthesis of RiPPs: docking with marine life, Curr. Opin. Chem. Biol., 2016, 31, 15–21 CrossRef CAS PubMed
.
- L. Sukmarini, Marine Bacterial Ribosomal Peptides: Recent Genomics- and Synthetic Biology-Based Discoveries and Biosynthetic Studies, Mar. Drugs, 2022, 20(9), 544 CrossRef CAS PubMed
.
- P. A. Jordan and B. S. Moore, Biosynthetic Pathway Connects Cryptic Ribosomally Synthesized Posttranslationally Modified Peptide Genes with Pyrroloquinoline Alkaloids, Cell Chem. Biol., 2016, 23(12), 1504–1514 CrossRef CAS PubMed
.
- T. E. Smith, C. D. Pond, E. Pierce, Z. P. Harmer, J. Kwan, M. M. Zachariah, M. K. Harper, T. P. Wyche, T. K. Matainaho, T. S. Bugni, L. R. Barrows, C. M. Ireland and E. W. Schmidt, Accessing chemical diversity from the uncultivated symbionts of small marine animals, Nat. Chem. Biol., 2018, 14(2), 179–185 CrossRef CAS PubMed
.
- L. Paoli, H. J. Ruscheweyh, C. C. Forneris, F. Hubrich, S. Kautsar, A. Bhushan, A. Lotti, Q. Clayssen, G. Salazar, A. Milanese, C. I. Carlstrom, C. Papadopoulou, D. Gehrig, M. Karasikov, H. Mustafa, M. Larralde, L. M. Carroll, P. Sanchez, A. A. Zayed, D. R. Cronin, S. G. Acinas, P. Bork, C. Bowler, T. O. Delmont, J. M. Gasol, A. D. Gossert, A. Kahles, M. B. Sullivan, P. Wincker, G. Zeller, S. L. Robinson, J. Piel and S. Sunagawa, Biosynthetic potential of the global ocean microbiome, Nature, 2022, 607(7917), 111–118 CrossRef CAS PubMed
.
- M. F. Freeman, C. Gurgui, M. J. Helf, B. I. Morinaka, A. R. Uria, N. J. Oldham, H.-G. Sahl, S. Matsunaga and J. Piel, Metagenome Mining Reveals Polytheonamides as posttranslationally Modified Ribosomal Peptides, Science, 2012, 338, 6105 CrossRef PubMed
.
- T. Hamada, S. Matsunaga, M. Fujiwara, K. Fujita, H. Hirota, R. Schmucki, P. Güntert and N. Fusetani, Solution Structure of Polytheonamide B, a Highly Cytotoxic Nonribosomal Polypeptide from Marine Sponge, J. Am. Chem. Soc., 2010, 132(37), 12941–12945 CrossRef CAS PubMed
.
- A. Bhushan, P. J. Egli, E. E. Peters, M. F. Freeman and J. Piel, Genome mining- and synthetic biology-enabled production of hypermodified peptides, Nat. Chem., 2019, 11(10), 931–939 CrossRef CAS PubMed
.
- N. M. Bosch, M. Borsa, U. Greczmiel, B. I. Morinaka, M. Gugger, A. Oxenius, A. L. Vagstad and J. Piel, Landornamides: Antiviral Ornithine-Containing Ribosomal Peptides Discovered through Genome Mining, Angew. Chem., Int. Ed., 2020, 59(29), 11763–11768 CrossRef PubMed
.
- Z. F. Pei, M. J. Yang, K. Zhang, X. H. Jian and G. L. Tang, Heterologous characterization of mechercharmycin A biosynthesis reveals alternative insights into post-translational modifications for RiPPs, Cell Chem. Biol., 2022, 29(4), 650–659 CrossRef CAS PubMed
e5.
- M. S. Donia and M. A. Fischbach, HUMAN MICROBIOTA. Small molecules from the human microbiota, Science, 2015, 349(6246), 1254766 CrossRef PubMed
.
- M. G. Lamarche, B. L. Wanner, S. Crepin and J. Harel, The phosphate regulon and bacterial virulence: a regulatory network connecting phosphate homeostasis and pathogenesis, FEMS Microbiol. Rev., 2008, 32(3), 461–473 CrossRef CAS PubMed
.
- A. Pujol, E. H. Crost, G. Simon, V. Barbe, D. Vallenet, A. Gomez and M. Fons, Characterization and distribution of the gene cluster encoding RumC, an anti-Clostridium perfringens bacteriocin produced in the gut, FEMS Microbiol. Ecol., 2011, 78(2), 405–415 CrossRef CAS PubMed
.
- S. Chiumento, C. Roblin, S. Kieffer-Jaquinod, S. Tachon, C. Leprètre, C. Basset, D. Aditiyarini, H. Olleik, C. Nicoletti, O. Bornet, O. Iranzo, M. Maresca, R. Hardré, M. Fons, T. Giardina, E. Devillard, F. Guerlesquin, Y. Couté, M. Atta and J. Perrier,
et al., A promising antibiotic produced by a human gut symbiont, Sci. Adv., 2019, 5(9), eaaw9969 CrossRef CAS PubMed
.
- K. E. Kawulka, T. Sprules, C. M. Diaper, R. M. Whittal, R. T. McKay, P. Mercier, P. Zuber and J. C. Vederas, Structure of Subtilosin A, a Cyclic Antimicrobial Peptide from Bacillus subtilis with Unusual Sulfur to α-Carbon Cross-Links: Formation and Reduction of α-Thio-α-Amino Acid Derivatives, Biochemistry, 2004, 43(12), 3385–3395 CrossRef CAS PubMed
.
- M. C. Rea, C. S. Sit, E. Clayton, P. M. O'Connor, R. M. Whittal, J. Zheng, J. C. Vederas, R. P. Ross, C. Hill and C. D. Thuricin, a posttranslationally modified bacteriocin with a narrow spectrum of activity against Clostridium difficile, Proc. Natl. Acad. Sci. U. S. A., 2010, 107(20), 9352–9357 CrossRef CAS PubMed
.
- H. Lee, J. J. Churey and R. W. Worobo, Biosynthesis and transcriptional analysis of thurincin H, a tandem repeated bacteriocin genetic locus, produced by Bacillus thuringiensis SF361, FEMS Microbiol. Lett., 2009, 299(2), 205–213 CrossRef CAS PubMed
.
- T. Mo, X. Ji, W. Yuan, D. Mandalapu, F. Wang, Y. Zhong, F. Li, Q. Chen, W. Ding, Z. Deng, S. Yu, Q. Zhang, Z. Thuricin and A. Narrow-Spectrum, Sactibiotic that Targets the Cell Membrane, Angew. Chem., Int. Ed., 2019, 58(52), 18793–18797 CrossRef CAS PubMed
.
- L. B. Bushin, B. C. Covington, B. E. Rued, M. J. Federle and M. R. Seyedsayamdost, Discovery and Biosynthesis of Streptosactin, a Sactipeptide with an Alternative Topology Encoded by Commensal Bacteria in the Human Microbiome, J. Am. Chem. Soc., 2020, 142(38), 16265–16275 CrossRef CAS PubMed
.
- D. Mao, B. K. Okada, Y. Wu, F. Xu and M. R. Seyedsayamdost, Recent advances in activating silent biosynthetic gene clusters in bacteria, Curr. Opin. Microbiol., 2018, 45, 156–163 CrossRef CAS PubMed
.
- G. A. Hudson, A. R. Hooper, A. J. DiCaprio, D. Sarlah and D. A. Mitchell, Structure Prediction and Synthesis of Pyridine-Based
Macrocyclic Peptide Natural Products, Org. Lett., 2021, 23(2), 253–256 CrossRef CAS PubMed
.
- R. E. Cobb, Y. Wang and H. Zhao, High-efficiency multiplex genome editing of Streptomyces species using an engineered CRISPR/Cas system, ACS Synth. Biol., 2015, 4(6), 723–728 CrossRef CAS PubMed
.
- F. Guo, S. Xiang, L. Li, B. Wang, J. Rajasarkka, K. Grondahl-Yli-Hannuksela, G. Ai, M. Metsa-Ketela and K. Yang, Targeted activation of silent natural product biosynthesis pathways by reporter-guided mutant selection, Metab. Eng., 2015, 28, 134–142 CrossRef CAS PubMed
.
- K. Moon, F. Xu, C. Zhang and M. R. Seyedsayamdost, Bioactivity-HiTES Unveils Cryptic Antibiotics Encoded in Actinomycete Bacteria, ACS Chem. Biol., 2019, 14(4), 767–774 CrossRef CAS PubMed
.
- K. Moon, F. Xu and M. R. Seyedsayamdost, Cebulantin, a Cryptic Lanthipeptide Antibiotic Uncovered Using Bioactivity-Coupled HiTES, Angew. Chem., Int. Ed., 2019, 58(18), 5973–5977 CrossRef CAS PubMed
.
- B. K. Okada, A. Li and M. R. Seyedsayamdost, Identification of the Hypertension Drug Guanfacine as an Antivirulence Agent in Pseudomonas aeruginosa, ChemBioChem, 2019, 20(15), 2005–2011 CrossRef CAS PubMed
.
- E. J. Han, S. R. Lee, S. Hoshino and M. R. Seyedsayamdost, Targeted Discovery of Cryptic Metabolites with Antiproliferative Activity, ACS Chem. Biol., 2022, 17(11), 3121–3130 CrossRef CAS PubMed
.
- C. Zhang and M. R. Seyedsayamdost, Discovery of a Cryptic Depsipeptide from Streptomyces ghanaensis via MALDI-MS-Guided High-Throughput Elicitor Screening, Angew. Chem., Int. Ed., 2020, 59(51), 23005–23009 CrossRef CAS PubMed
.
- Y. Li, S. R. Lee, E. J. Han and M. R. Seyedsayamdost, Momomycin, an Antiproliferative Cryptic Metabolite from the Oxytetracycline Producer Streptomyces rimosus, Angew. Chem., Int. Ed., 2022, 61(39), e202208573 CrossRef CAS PubMed
.
- J. I. Tietz, C. J. Schwalen, P. S. Patel, T. Maxson, P. M. Blair, H. C. Tai, U. I. Zakai and D. A. Mitchell, A new genome-mining tool redefines the lasso peptide biosynthetic landscape, Nat. Chem. Biol., 2017, 13(5), 470–478 CrossRef CAS PubMed
.
- F. Biermann, S. L. Wenski and E. J. N. Helfrich, Navigating and expanding the roadmap of natural product genome mining tools, Beilstein J. Org. Chem., 2022, 18, 1656–1671 CrossRef CAS PubMed
.
- E. L. C. de Los Santos, NeuRiPP: Neural network identification of RiPP precursor peptides, Sci. Rep., 2019, 9(1), 13406 CrossRef PubMed
.
- K. E. Shelton and D. A. Mitchell, Bioinformatic prediction and experimental validation of RiPP recognition elements, Methods Enzymol., 2023, 679, 191–233 CAS
.
- G. A. Hudson, B. J. Burkhart, A. J. DiCaprio, C. J. Schwalen, B. Kille, T. V. Pogorelov and D. A. Mitchell, Bioinformatic Mapping of Radical S-Adenosylmethionine-Dependent Ribosomally Synthesized and Post-Translationally Modified Peptides Identifies New Calpha, Cbeta, and Cgamma-Linked Thioether-Containing Peptides, J. Am. Chem. Soc., 2019, 141(20), 8228–8238 CrossRef CAS PubMed
.
- R. D. Miller, A. Iinishi, S. M. Modaresi, B. K. Yoo, T. D. Curtis, P. J. Lariviere, L. Liang, S. Son, S. Nicolau, R. Bargabos, M. Morrissette, M. F. Gates, N. Pitt, R. P. Jakob, P. Rath, T. Maier, A. G. Malyutin, J. T. Kaiser, S. Niles, B. Karavas, M. Ghiglieri, S. E. J. Bowman, D. C. Rees, S. Hiller and K. Lewis, Computational identification of a systemic antibiotic for Gram-negative bacteria, Nat. Microbiol., 2022, 7(10), 1661–1672 CrossRef CAS PubMed
.
- J. Shi, J.-Q. Ma, Y.-C. Wang, Z.-F. Xu, B. Zhang, R.-H. Jiao, R.-X. Tan and H.-M. Ge, Discovery of daspyromycins A and B, 2-aminovinyl-cysteine containing lanthipeptides, through a genomics-based approach, Chin. Chem. Lett., 2022, 33(1), 511–515 CrossRef CAS
.
- Z. Shang, J. M. Winter, C. A. Kauffman, I. Yang and W. Fenical, Salinipeptins: Integrated Genomic and Chemical Approaches Reveal Unusual d-Amino Acid-Containing Ribosomally Synthesized and Post-Translationally Modified Peptides (RiPPs) from a Great Salt Lake Streptomyces sp, ACS Chem. Biol., 2019, 14(3), 415–425 CrossRef CAS PubMed
.
- Y. Han, X. Wang, Y. Zhang and L. Huo, Discovery and Characterization of Marinsedin, a New Class II Lanthipeptide Derived from Marine Bacterium Marinicella sediminis F2(T), ACS Chem. Biol., 2022, 17(4), 785–790 CrossRef CAS PubMed
.
- T. A. Scott, M. Verest, J. Farnung, C. C. Forneris, S. L. Robinson, X. Ji, F. Hubrich, C. Chepkirui, D. U. Richter, S. Huber, P. Rust, A. B. Streiff, Q. Zhang, J. W. Bode and J. Piel, Widespread microbial utilization of ribosomal β-amino acid-containing peptides and proteins, Chem, 2022, 8(10), 2659–2677 CAS
.
- M. C. Walker, S. M. Eslami, K. J. Hetrick, S. E. Ackenhusen, D. A. Mitchell and W. A. van der Donk, Precursor peptide-targeted mining of more than one hundred thousand genomes expands the lanthipeptide natural product family, BMC Genomics, 2020, 21(1), 387 CrossRef CAS PubMed
.
- C. J. Schwalen, G. A. Hudson, B. Kille and D. A. Mitchell, Bioinformatic Expansion and Discovery of Thiopeptide Antibiotics, J. Am. Chem. Soc., 2018, 140(30), 9494–9501 CrossRef CAS PubMed
.
- T. A. Knappe, U. Linne, S. Zirah, S. Rebuffat, X. Xie and M. A. Marahiel, Isolation and Structural Characterization of Capistruin, a Lasso Peptide Predicted from the Genome Sequence of Burkholderia thailandensis E264, J. Am. Chem. Soc., 2008, 130(34), 11446–11454 CrossRef CAS PubMed
.
- M. O. Maksimov, I. Pelczer and A. J. Link, Precursor-centric genome-mining approach for lasso peptide discovery, Proc. Natl. Acad. Sci. U. S. A., 2012, 109(38), 15223–15228 CrossRef CAS PubMed
.
- S. S. Elsayed, F. Trusch, H. Deng, A. Raab, I. Prokes, K. Busarakam, J. A. Asenjo, B. A. Andrews, P. van West, A. T. Bull, M. Goodfellow, Y. Yi, R. Ebel, M. Jaspars and M. E. Rateb, Chaxapeptin, a Lasso Peptide from Extremotolerant Streptomyces leeuwenhoekii Strain C58 from the Hyperarid Atacama Desert, J. Org. Chem., 2015, 80(20), 10252–10260 CrossRef CAS PubMed
.
- A. Caruso, L. B. Bushin, K. A. Clark, R. J. Martinie and M. R. Seyedsayamdost, Radical Approach to Enzymatic beta-Thioether Bond Formation, J. Am. Chem. Soc., 2019, 141(2), 990–997 CrossRef CAS PubMed
.
- K. R. Schramma, L. B. Bushin and M. R. Seyedsayamdost, Structure and biosynthesis of a macrocyclic peptide containing an unprecedented lysine-to-tryptophan crosslink, Nat. Chem., 2015, 7(5), 431–437 CrossRef CAS PubMed
.
- T. Q. N. Nguyen, Y. W. Tooh, R. Sugiyama, T. P. D. Nguyen, M. Purushothaman, L. C. Leow, K. Hanif, R. H. S. Yong, I. Agatha, F. R. Winnerdy, M. Gugger, A. T. Phan and B. I. Morinaka, Post-translational formation of strained cyclophanes in bacteria, Nat. Chem., 2020, 12(11), 1042–1053 CrossRef CAS PubMed
.
- A. Benjdia, C. Balty and O. Berteau, Radical SAM Enzymes in the Biosynthesis of Ribosomally Synthesized and Post-translationally Modified Peptides (RiPPs), Front. Chem., 2017, 5, 87 CrossRef PubMed
.
- N. Mahanta, G. A. Hudson and D. A. Mitchell, Radical S-Adenosylmethionine Enzymes Involved in RiPP Biosynthesis, Biochemistry, 2017, 56(40), 5229–5244 CrossRef CAS PubMed
.
- L. Fluhe, T. A. Knappe, M. J. Gattner, A. Schafer, O. Burghaus, U. Linne and M. A. Marahiel, The radical SAM enzyme AlbA catalyzes thioether bond formation in subtilosin A, Nat. Chem. Biol., 2012, 8(4), 350–357 CrossRef PubMed
.
- K. Murphy, O. O'Sullivan, M. C. Rea, P. D. Cotter, R. P. Ross and C. Hill, Genome mining for radical SAM protein determinants reveals multiple sactibiotic-like gene clusters, PLoS One, 2011, 6(7), e20852 CrossRef CAS PubMed
.
- L. Fluhe and M. A. Marahiel, Radical S-adenosylmethionine enzyme catalyzed thioether bond formation in sactipeptide biosynthesis, Curr. Opin. Chem. Biol., 2013, 17(4), 605–612 CrossRef PubMed
.
- Y. Chen, J. Wang, G. Li, Y. Yang and W. Ding, Current Advancements in Sactipeptide Natural Products, Front. Chem., 2021, 9, 595991 CrossRef CAS PubMed
.
- W. Ding, Y. Li and Q. Zhang, Substrate-Controlled Stereochemistry in Natural Product Biosynthesis, ACS Chem. Biol., 2015, 10(7), 1590–1598 CrossRef CAS PubMed
.
- P. Alvarez-Sieiro, M. Montalban-Lopez, D. Mu and O. P. Kuipers, Bacteriocins of lactic acid bacteria: extending the family, Appl. Microbiol. Biotechnol., 2016, 100(7), 2939–2951 CrossRef CAS PubMed
.
- K. A. Clark, B. C. Covington and M. R. Seyedsayamdost, Biosynthesis-guided discovery reveals enteropeptins as alternative sactipeptides containing N-methylornithine, Nat. Chem., 2022, 14(12), 1390–1398 CrossRef CAS PubMed
.
- K. A. Clark and M. R. Seyedsayamdost, Bioinformatic Atlas of Radical SAM Enzyme-Modified RiPP Natural Products Reveals an Isoleucine-Tryptophan Crosslink, J. Am. Chem. Soc., 2022, 144(39), 17876–17888 CrossRef CAS PubMed
.
- B. B. He, Z. Cheng, Z. Zhong, Y. Gao, H. Liu and Y. X. Li, Expanded Sequence Space of Radical S-Adenosylmethionine-Dependent Enzymes Involved in Post-translational Macrocyclization, Angew. Chem., Int. Ed., 2022, 61(48), e202212447 CrossRef CAS PubMed
.
- I. Song, Y. Kim, J. Yu, S. Y. Go, H. G. Lee, W. J. Song and S. Kim, Molecular mechanism underlying substrate recognition of the peptide macrocyclase PsnB, Nat. Chem. Biol., 2021, 17(11), 1123–1131 CrossRef CAS PubMed
.
- H. E. Elashal, J. D. Koos, W. L. Cheung-Lee, B. Choi, L. Cao, M. A. Richardson, H. L. White and A. J. Link, Biosynthesis and characterization of fuscimiditide, an aspartimidylated graspetide, Nat. Chem., 2022, 14(11), 1325–1334 CrossRef CAS PubMed
.
- S. Ramesh, X. Guo, A. J. DiCaprio, A. M. De Lio, L. A. Harris, B. L. Kille, T. V. Pogorelov and D. A. Mitchell, Bioinformatics-Guided Expansion and Discovery of Graspetides, ACS Chem. Biol., 2021, 16(12), 2787–2797 CrossRef CAS PubMed
.
- H. Lee, M. Choi, J. U. Park, H. Roh and S. Kim, Genome Mining Reveals High Topological Diversity of omega-Ester-Containing Peptides and Divergent Evolution of ATP-Grasp Macrocyclases, J. Am. Chem. Soc., 2020, 142(6), 3013–3023 CrossRef CAS PubMed
.
- M. V. Fawaz, M. E. Topper and S. M. Firestine, The ATP-grasp enzymes, Bioorg. Chem., 2011, 39(5), 185–191 CrossRef CAS PubMed
.
- K. S. Makarova, B. Blackburne, Y. I. Wolf, A. Nikolskaya, S. Karamycheva, M. Espinoza, C. E. Barry III, C. A. Bewley and E. V. Koonin, Phylogenomic analysis of the diversity of graspetides and proteins involved in their biosynthesis, Biol. Direct, 2022, 17(1), 7 CrossRef CAS PubMed
.
- S. Scholz, S. Kerestetzopoulou, V. Wiebach, R. Schnegotzki, B. Schmid, E. Reyna-Gonzalez, L. Ding, R. D. Sussmuth, E. Dittmann and M. Baunach, One-Pot Chemoenzymatic Synthesis of Microviridin
Analogs Containing Functional Tags, ChemBioChem, 2022, 23(20), e202200345 CrossRef CAS PubMed
.
- E. S. Grant-Mackie, E. T. Williams, P. W. R. Harris and M. A. Brimble, Aminovinyl Cysteine Containing Peptides: A Unique Motif That Imparts Key Biological Activity, JACS Au, 2021, 1(10), 1527–1540 CrossRef CAS PubMed
.
- C. S. Sit, S. Yoganathan and J. C. Vederas, Biosynthesis of Aminovinyl-Cysteine-Containing Peptides and Its Application in the Production of Potential Drug Candidates, Acc. Chem. Res., 2011, 44(4), 261–268 CrossRef CAS PubMed
.
- T. Mo, H. Yuan, F. Wang, S. Ma, J. Wang, T. Li, G. Liu, S. Yu, X. Tan, W. Ding and Q. Zhang, Convergent evolution of the Cys decarboxylases involved in aminovinyl-cysteine (AviCys) biosynthesis, FEBS Lett., 2019, 593(6), 573–580 CrossRef CAS PubMed
.
- J. Lu, J. Li, Y. Wu, X. Fang, J. Zhu and H. Wang, Characterization of the FMN-Dependent Cysteine Decarboxylase from Thioviridamide Biosynthesis, Org. Lett., 2019, 21(12), 4676–4679 CrossRef CAS PubMed
.
- M. A. Ortega, D. P. Cogan, S. Mukherjee, N. Garg, B. Li, G. N. Thibodeaux, S. I. Maffioli, S. Donadio, M. Sosio, J. Escano, L. Smith, S. K. Nair and W. A. van der Donk, Two Flavoenzymes Catalyze the Post-Translational Generation of 5-Chlorotryptophan and 2-Aminovinyl-Cysteine during NAI-107 Biosynthesis, ACS Chem. Biol., 2017, 12(2), 548–557 CrossRef CAS PubMed
.
- L. M. Repka, J. R. Chekan, S. K. Nair and W. A. van der Donk, Mechanistic Understanding of Lanthipeptide Biosynthetic Enzymes, Chem. Rev., 2017, 117(8), 5457–5520 CrossRef CAS PubMed
.
- Q. Zhang, Y. Yu, J. E. Velasquez and W. A. van der Donk, Evolution of lanthipeptide synthetases, Proc. Natl. Acad. Sci. U. S. A., 2012, 109(45), 18361–18366 CrossRef CAS PubMed
.
- W. A. van der Donk and S. K. Nair, Structure and mechanism of lanthipeptide biosynthetic enzymes, Curr. Opin. Struct. Biol., 2014, 29, 58–66 CrossRef CAS PubMed
.
- J. D. Hegemann and R. D. Sussmuth, Matters of class: coming of age of class III and IV lanthipeptides, RSC Chem. Biol., 2020, 1(3), 110–127 RSC
.
- A. D. P. van Staden, W. F. van Zyl, M. Trindade, L. M. T. Dicks and C. Smith, Therapeutic Application of Lantibiotics and Other Lanthipeptides: Old and New Findings, Appl. Environ. Microbiol., 2021, 87(14), e0018621 CrossRef PubMed
.
- J. Barbosa, T. Caetano and S. Mendo, Class I and Class II Lanthipeptides Produced by Bacillus spp, J. Nat. Prod., 2015, 78(11), 2850–2866 CrossRef CAS PubMed
.
- E. L. Ongey and P. Neubauer, Lanthipeptides: chemical synthesis versus in vivo biosynthesis as tools for pharmaceutical production, Microb. Cell Fact., 2016, 15, 97 CrossRef PubMed
.
- M. Lagedroste, J. Reiners, C. V. Knospe, S. H. J. Smits and L. Schmitt, A Structural View on the Maturation of Lanthipeptides, Front. Microbiol., 2020, 11, 1183 CrossRef PubMed
.
- C. Li, K. Alam, Y. Zhao, J. Hao, Q. Yang, Y. Zhang, R. Li and A. Li, Mining and Biosynthesis of Bioactive Lanthipeptides From Microorganisms, Front. Bioeng. Biotechnol., 2021, 9, 692466 CrossRef PubMed
.
- E. L. Ongey, H. Yassi, S. Pflugmacher and P. Neubauer, Pharmacological and pharmacokinetic properties of lanthipeptides undergoing clinical studies, Biotechnol. Lett., 2017, 39(4), 473–482 CrossRef PubMed
.
- H. R. Karbalaei-Heidari and N. Budisa, Combating Antimicrobial Resistance With New-To-Nature Lanthipeptides Created by Genetic Code Expansion, Front. Microbiol., 2020, 11, 590522 CrossRef PubMed
.
- Y. Fu, Y. Xu, F. Ruijne and O.
P. Kuipers, Engineering lanthipeptides by introducing a large variety of RiPP modifications to obtain new-to-nature bioactive peptides, FEMS Microbiol. Rev., 2023, 47(3), fuad017 CrossRef CAS PubMed
.
- J. Z. Acedo, I. R. Bothwell, L. An, A. Trouth, C. Frazier and W. A. van der Donk, O-Methyltransferase-Mediated Incorporation of a beta-Amino Acid in Lanthipeptides, J. Am. Chem. Soc., 2019, 141(42), 16790–16801 CrossRef CAS PubMed
.
- I. R. Bothwell, T. Caetano, R. Sarksian, S. Mendo and W. A. van der Donk, Structural Analysis of Class I Lanthipeptides from Pedobacter lusitanus NL19 Reveals an Unusual Ring Pattern, ACS Chem. Biol., 2021, 16(6), 1019–1029 CrossRef CAS PubMed
.
- Y. Zhang, Z. Hong, L. Zhou, Z. Zhang, T. Tang, E. Guo, J. Zheng, C. Wang, L. Dai, T. Si and H. Wang, Biosynthesis of Gut-Microbiota-Derived Lantibiotics Reveals a Subgroup of S8 Family Proteases for Class III Leader Removal, Angew. Chem., Int. Ed., 2022, 61(6), e202114414 CrossRef CAS PubMed
.
- B. J. Burkhart, C. J. Schwalen, G. Mann, J. H. Naismith and D. A. Mitchell, YcaO-Dependent Posttranslational Amide Activation: Biosynthesis, Structure, and Function, Chem. Rev., 2017, 117(8), 5389–5456 CrossRef CAS PubMed
.
- K. L. Dunbar, J. R. Chekan, C. L. Cox, B. J. Burkhart, S. K. Nair and D. A. Mitchell, Discovery of a new ATP-binding motif involved in peptidic azoline biosynthesis, Nat. Chem. Biol., 2014, 10(10), 823–829 CrossRef CAS PubMed
.
- Y. Du, Y. Qiu, X. Meng, J. Feng, J. Tao, W. Liu and A. Heterotrimeric, Dehydrogenase Complex Functions with 2 Distinct YcaO Proteins to Install 5 Azole Heterocycles into 35-Membered Sulfomycin Thiopeptides, J. Am. Chem. Soc., 2020, 142(18), 8454–8463 CrossRef CAS PubMed
.
- Y. Zheng and S. K. Nair, YcaO-mediated ATP-dependent peptidase activity in ribosomal peptide biosynthesis, Nat. Chem. Biol., 2023, 19(1), 111–119 CrossRef CAS PubMed
.
- A. H. Russell, N. M. Vior, E. S. Hems, R. Lacret and A. W. Truman, Discovery and characterisation of an amidine-containing ribosomally-synthesised peptide that is widely distributed in nature, Chem. Sci., 2021, 12(35), 11769–11778 RSC
.
- J. Santos-Aberturas, G. Chandra, L. Frattaruolo, R. Lacret, T. H. Pham, N. M. Vior, T. H. Eyles and A. W. Truman, Uncovering the unexplored diversity of thioamidated ribosomal peptides in Actinobacteria using the RiPPER genome mining tool, Nucleic Acids Res., 2019, 47(9), 4624–4637 CrossRef CAS PubMed
.
- J. J. L. Malit, C. Wu, L. L. Liu and P. Y. Qian, Global Genome Mining Reveals the Distribution of Diverse Thioamidated RiPP Biosynthesis Gene Clusters, Front. Microbiol., 2021, 12, 635389 CrossRef PubMed
.
- K. Blin, S. Shaw, K. Steinke, R. Villebro, N. Ziemert, S. Y. Lee, M. H. Medema and T. Weber, antiSMASH 5.0: updates to the secondary metabolite genome mining pipeline, Nucleic Acids Res., 2019, 47(W1), W81–W87 CrossRef CAS PubMed
.
- D. H. Haft, M. K. Basu and D. A. Mitchell, Research article Expansion of ribosomally produced natural products: a nitrile hydratase-and Nif11-related precursor family, BMC Biol., 2010, 8, 70 CrossRef PubMed
.
- X. Yang and W. A. van der Donk, Ribosomally synthesized and post-translationally modified peptide natural products: new insights into the role of leader and core peptides during biosynthesis, Chemistry, 2013, 19(24), 7662–7677 CrossRef CAS PubMed
.
- M. Kloosterman Alexander, E. Shelton Kyle, P. van Wezel Gilles, H. Medema Marnix and A. Mitchell Douglas, RRE-Finder: a Genome-Mining Tool for Class-Independent RiPP Discovery, mSystems, 2020, 5(5), e00267 Search PubMed
.
- A.-C. Letzel, S. J. Pidot and C. Hsrtweck, Genome mining for ribosomally synthesized and post-translationally modified peptides (RiPPs) in anaerobic bacteria, BMC Genomics, 2014, 15, 983 CrossRef PubMed
.
- F. Hubrich, N. M. Bosch, C. Chepkirui, B. I. Morinaka, M. Rust, M. Gugger, S. L. Robinson, A. L. Vagstad and J. Piel, Ribosomally derived lipopeptides containing distinct fatty acyl moieties, Proc. Natl. Acad. Sci. U. S. A., 2022, 119(3), e2113120119 CrossRef CAS PubMed
.
- B. J. Burkhart, G. A. Hudson, K. L. Dunbar and D. A. Mitchell, A prevalent peptide-binding domain guides ribosomal natural product biosynthesis, Nat. Chem. Biol., 2015, 11(8), 564–570 CrossRef CAS PubMed
.
- C. Zhang and M. R. Seyedsayamdost, CanE, an Iron/2-Oxoglutarate-Dependent Lasso Peptide Hydroxylase from Streptomyces canus, ACS Chem. Biol., 2020, 15(4), 890–894 CrossRef CAS PubMed
.
- T. W. Precord, S. Ramesh, S. R. Dommaraju, L. A. Harris, B. L. Kille and D. A. Mitchell, Catalytic Site Proximity Profiling for Functional Unification of Sequence-Diverse Radical S-Adenosylmethionine Enzymes, ACS Bio Med Chem Au, 2023, 3(3), 240–251 CrossRef CAS PubMed
.
- N. Oberg, T. W. Precord, D. A. Mitchell and J. A. Gerlt, RadicalSAM.org: A Resource to Interpret Sequence-Function Space and Discover New Radical SAM Enzyme Chemistry, ACS Bio Med Chem Au, 2022, 2(1), 22–35 CrossRef CAS PubMed
.
- K. Komiyama, K. Otoguro., T. Segawa, K. Shiomi, H. Yang, Y. Takahashi, M. Hayashi, T. Oxani and S. Omura, A new antibiotic, cypemycin. Taxonomy, fermentation,isolation and biological charactetistics, J. Antibiot., 1993, 46(11), 1666–1671 CrossRef CAS PubMed
.
- Y. Minami, K.-i Yoshida, R. Azuma, A. Urakawa, T. Kawauchi, T. Otani, K. Komiyama and S. Ōmura, Structure of cypemycin, a new peptide antibiotic, Tetrahedron Lett., 1994, 35(43), 8001–8004 CrossRef
.
- J. Claesen and M. Bibb, Genome mining and genetic analysis of cypemycin biosynthesis reveal an unusual class of posttranslationally modified peptides, Proc. Natl. Acad. Sci. U. S. A., 2010, 107(37), 16297–16302 CrossRef CAS PubMed
.
- Q. Zhang and W. A. van der Donk, Catalytic promiscuity of a bacterial alpha-N-methyltransferase, FEBS Lett., 2012, 586(19), 3391–3397 CrossRef CAS PubMed
.
- S. Ma and Q. Zhang, Linaridin natural products, Nat. Prod. Rep., 2020, 37(9), 1152–1163 RSC
.
- Y. Xue, X. Wang and W. Liu, Reconstitution of the Linaridin Pathway Provides Access to the Family-Determining Activity of Two Membrane-Associated Proteins in the Formation of Structurally Underestimated Cypemycin, J. Am. Chem. Soc., 2023, 145(12), 7040–7047 CrossRef CAS PubMed
.
- L. Chu, J. Cheng, C. Zhou, T. Mo, X. Ji, T. Zhu, J. Chen, S. Ma, J. Gao and Q. Zhang, Hijacking a Linaridin Biosynthetic Intermediate for Lanthipeptide Production, ACS Chem. Biol., 2022, 17(11), 3198–3206 CrossRef CAS PubMed
.
- W. Xiao, Y. Satoh, Y. Ogasawara and T. Dairi, Biosynthetic Gene Cluster of Linaridin Peptides Contains Epimerase Gene, ChemBioChem, 2022, 23(12), e202100705 CrossRef CAS PubMed
.
- J. Claesen and M. J. Bibb, Biosynthesis and regulation of grisemycin, a new member of the linaridin family of ribosomally synthesized peptides produced by Streptomyces griseus IFO 13350, J. Bacteriol., 2011, 193(10), 2510–2516 CrossRef CAS PubMed
.
- B. I. Morinaka, E. Lakis, M. Verest, M. J. Helf, T. Scalvenzi, A. L. Vagstad, J. Sims, S. Sunagawa, M. Gugger and J. Piel, Natural noncanonical protein splicing yields products with diverse β-amino acid residues, Science, 2018, 359(6377), 779–782 CrossRef CAS PubMed
.
- A. A. Vinogradov, Y. Zhang, K. Hamada, J. S. Chang, C. Okada, H. Nishimura, N. Terasaka, Y. Goto, K. Ogata, T. Sengoku, H. Onaka and H. Suga, De Novo Discovery of Thiopeptide Pseudo-natural Products Acting as Potent and Selective TNIK Kinase Inhibitors, J. Am. Chem. Soc., 2022, 144(44), 20332–20341 CrossRef CAS PubMed
.
- S. R. Fleming, T. E. Bartges, A. A. Vinogradov, C. L. Kirkpatrick, Y. Goto, H. Suga, L. M. Hicks and A. A. Bowers, Flexizyme-Enabled Benchtop Biosynthesis of Thiopeptides, J. Am. Chem. Soc., 2019, 141(2), 758–762 CrossRef CAS PubMed
.
- B. J. Burkhart, N. Kakkar, G. A. Hudson, W. A. van der Donk and D. A. Mitchell, Chimeric Leader Peptides for the Generation of Non-Natural Hybrid RiPP Products, ACS Cent. Sci., 2017, 3(6), 629–638 CrossRef CAS PubMed
.
- J. A. Walker, N. Hamlish, A. Tytla, D. D. Brauer, M. B. Francis and A. Schepartz, Redirecting RiPP Biosynthetic Enzymes to Proteins and Backbone-Modified Substrates, ACS Cent. Sci., 2022, 8(4), 473–482 CrossRef CAS PubMed
.
- L. Franz and J. Koehnke, Leader peptide exchange to produce hybrid, new-to-nature ribosomal natural products, Chem. Commun., 2021, 57(52), 6372–6375 RSC
.
- E. Janzen, C. Blanco, H. Peng, J. Kenchel and I. A. Chen, Promiscuous Ribozymes and Their Proposed Role in Prebiotic Evolution, Chem. Rev., 2020, 120(11), 4879–4897 CrossRef CAS PubMed
.
- B. Li, D. Sher, L. Kelly, Y. Shi, K. Huang, P. J. Knerr, I. Joewono, D. Rusch, S. W. Chisholm and W. A. van der Donk, Catalytic promiscuity in the biosynthesis of cyclic peptide secondary metabolites in planktonic marine cyanobacteria, Proc. Natl. Acad. Sci. U. S. A., 2010, 107(23), 10430–10435 CrossRef CAS PubMed
.
- N. A. Nguyen, Y. Cong, R. C. Hurrell, N. Arias, N. Garg, A. W. Puri, E. W. Schmidt and V. Agarwal, A Silent Biosynthetic Gene Cluster from a Methanotrophic Bacterium Potentiates Discovery of a Substrate Promiscuous Proteusin Cyclodehydratase, ACS Chem. Biol., 2022, 17(6), 1577–1585 CrossRef CAS PubMed
.
- S. Ma, H. Chen, H. Li, X. Ji, Z. Deng, W. Ding and Q. Zhang, Post-Translational Formation of Aminomalonate by a Promiscuous Peptide-Modifying Radical SAM Enzyme, Angew. Chem., Int. Ed., 2021, 60(36), 19957–19964 CrossRef CAS PubMed
.
- C. E. Seyfert, C. Porten, B. Yuan, S. Deckarm, F. Panter, C. D. Bader, J. Coetzee, F. Deschner, K. Tehrani, P. G. Higgins, H. Seifert, T. C. Marlovits, J. Herrmann and R. Muller, Darobactins Exhibiting Superior Antibiotic Activity by Cryo-EM Structure Guided Biosynthetic Engineering, Angew. Chem., Int. Ed., 2023, 62(2), e202214094 CrossRef CAS PubMed
.
- T. W. Precord, N. Mahanta and D. A. Mitchell, Reconstitution and Substrate Specificity of the Thioether-Forming Radical S-Adenosylmethionine Enzyme in Freyrasin Biosynthesis, ACS Chem. Biol., 2019, 14(9), 1981–1989 CrossRef CAS PubMed
.
- K. A. S. Eastman, W. M. Kincannon and V. Bandarian, Leveraging Substrate Promiscuity of a Radical S-Adenosyl-L-methionine RiPP Maturase toward Intramolecular Peptide Cross-Linking Applications, ACS Cent. Sci., 2022, 8(8), 1209–1217 CrossRef CAS PubMed
.
- O. Pavlova, J. Mukhopadhyay, E. Sineva, R. H. Ebright and K. Severinov, Systematic structure-activity analysis of microcin J25, J. Biol. Chem., 2008, 283(37), 25589–25595 CrossRef CAS PubMed
.
- S. J. Pan and A. J. Link, Sequence diversity in the lasso peptide framework: discovery of functional microcin J25 variants with multiple amino acid substitutions, J. Am. Chem. Soc., 2011, 133(13), 5016–5023 CrossRef CAS PubMed
.
- R. Ducasse, K. P. Yan, C. Goulard, A. Blond, Y. Li, E. Lescop, E. Guittet, S. Rebuffat and S. Zirah, Sequence determinants governing the topology and biological activity of a lasso peptide, microcin J25, ChemBioChem, 2012, 13(3), 371–380 CrossRef CAS PubMed
.
- J. D. Hegemann, M. De Simone, M. Zimmermann, T. A. Knappe, X. Xie, F. S. Di Leva, L. Marinelli, E. Novellino, S. Zahler, H. Kessler and M. A. Marahiel, Rational improvement of the affinity and selectivity of integrin binding of grafted lasso peptides, J. Med. Chem., 2014, 57(13), 5829–5834 CrossRef CAS PubMed
.
- F. J. Piscotta, J. M. Tharp, W. R. Liu and A. J. Link, Expanding the chemical diversity of lasso peptide MccJ25 with genetically encoded noncanonical amino acids, Chem. Commun., 2015, 51(2), 409–412 RSC
.
- R. S. Al Toma, A. Kuthning, M. P. Exner, A. Denisiuk, J. Ziegler, N. Budisa and R. D. Sussmuth, Site-directed and global incorporation of orthogonal and isostructural noncanonical amino acids into the ribosomal lasso peptide capistruin, ChemBioChem, 2015, 16(3), 503–509 CrossRef CAS PubMed
.
- A. J. DiCaprio, A. Firouzbakht, G. A. Hudson and D. A. Mitchell, Enzymatic Reconstitution and Biosynthetic Investigation of the Lasso Peptide Fusilassin, J. Am. Chem. Soc., 2019, 141(1), 290–297 CrossRef CAS PubMed
.
- Y. Si, A. M. Kretsch, L. M. Daigh, M. J. Burk and D. A. Mitchell, Cell-Free Biosynthesis to Evaluate Lasso Peptide Formation and Enzyme-Substrate Tolerance, J. Am. Chem. Soc., 2021, 143(15), 5917–5927 CrossRef CAS PubMed
.
- P. M. Himes, S. E. Allen, S. Hwang and A. A. Bowers, Production of Sactipeptides in Escherichia coli: Probing the Substrate Promiscuity of Subtilosin A Biosynthesis, ACS Chem. Biol., 2016, 11(6), 1737–1744 CrossRef CAS PubMed
.
- A. Ali, D. Happel, J. Habermann, K. Schoenfeld, A. Macarron Palacios, S. Bitsch, S. Englert, H. Schneider, O. Avrutina, S. Fabritz and H. Kolmar, Sactipeptide Engineering by Probing the Substrate Tolerance of a Thioether-Bond-Forming Sactisynthase, Angew. Chem., Int. Ed., 2022, 61(45), e202210883 CrossRef CAS PubMed
.
- R. H. de Vries, J. H. Viel, O. P. Kuipers and G. Roelfes, Rapid and Selective Chemical Editing of Ribosomally Synthesized and Post-Translationally Modified Peptides (RiPPs) via Cu(II) -Catalyzed beta-Borylation of Dehydroamino Acids, Angew. Chem., Int. Ed., 2021, 60(8), 3946–3950 CrossRef CAS PubMed
.
- S. P. Gaudêncio and F. Pereira, Dereplication: racing to speed up the natural products discovery process, Nat. Prod. Rep., 2015, 32(6), 779–810 RSC
.
- G. F. Qin, X. Zhang, F. Zhu, Z. Q. Huo, Q. Q. Yao, Q. Feng, Z. Liu, G. M. Zhang, J. C. Yao and H. B. Liang, MS/MS-Based Molecular Networking: An Efficient Approach for Natural Products Dereplication, Molecules, 2023, 28(1), 157 CrossRef CAS PubMed
.
- S. R. Lee, F. Schalk, J. W. Schwitalla, H. Guo, J. S. Yu, M. Song, W. H. Jung, Z. W. de Beer, C. Beemelmanns and K. H. Kim, GNPS-Guided Discovery of Madurastatin Siderophores from the Termite-Associated Actinomadura sp. RB99, Chemistry, 2022, 28(36), e202200612 CrossRef CAS PubMed
.
- P. M. Allard, T. Peresse, J. Bisson, K. Gindro, L. Marcourt, V. C. Pham, F. Roussi, M. Litaudon and J. L. Wolfender, Integration of Molecular Networking and In-Silico MS/MS Fragmentation for Natural Products Dereplication, Anal. Chem., 2016, 88(6), 3317–3323 CrossRef CAS PubMed
.
- T. El-Elimat, M. Figueroa, B. M. Ehrmann, N. B. Cech, C. J. Pearce and N. H. Oberlies, High-resolution MS, MS/MS, and UV database of fungal secondary metabolites as a dereplication protocol for bioactive natural products, J. Nat. Prod., 2013, 76(9), 1709–1716 CrossRef CAS PubMed
.
- J. L. Lopez-Perez, R. Theron, E. del Olmo and D. Diaz, NAPROC-13: a database for the dereplication of natural product mixtures in bioassay-guided protocols, Bioinformatics, 2007, 23(23), 3256–3257 CrossRef CAS PubMed
.
- W. L. Cheung-Lee, M. E. Parry, A. Jaramillo Cartagena, S. A. Darst and A. J. Link, Discovery and structure of the antimicrobial lasso peptide citrocin, J. Biol. Chem., 2019, 294(17), 6822–6830 CrossRef CAS PubMed
.
- L. M. Stariha and D. G. McCafferty, Discovery of the Class I Antimicrobial Lasso Peptide Arcumycin, ChemBioChem, 2021, 22(16), 2632–2640 CrossRef CAS PubMed
.
- D. Y. Travin, Z. L. Watson, M. Metelev, F. R. Ward, I. A. Osterman, I. M. Khven, N. F. Khabibullina, M. Serebryakova, P. Mergaert, Y. S. Polikanov, J. H. D. Cate and K. Severinov, Structure of ribosome-bound azole-modified peptide phazolicin rationalizes its species-specific mode of bacterial translation inhibition, Nat. Commun., 2019, 10(1), 4563 CrossRef PubMed
.
| This journal is © The Royal Society of Chemistry 2024 |