Open Access Article
Open Access ArticleAlabOS: a Python-based reconfigurable workflow management framework for autonomous laboratories
Yuxing
Fei†
 ab,
Bernardus
Rendy†
ab,
Bernardus
Rendy†
 ab,
Rishi
Kumar†
c,
Olympia
Dartsi
c,
Hrushikesh P.
Sahasrabuddhe
ac,
Matthew J.
McDermott
ab,
Rishi
Kumar†
c,
Olympia
Dartsi
c,
Hrushikesh P.
Sahasrabuddhe
ac,
Matthew J.
McDermott
 b,
Zheren
Wang
b,
Zheren
Wang
 ab,
Nathan J.
Szymanski
ab,
Lauren N.
Walters
ab,
Nathan J.
Szymanski
ab,
Lauren N.
Walters
 b,
David
Milsted
b,
David
Milsted
 b,
Yan
Zeng
*bd,
Anubhav
Jain
*c and
Gerbrand
Ceder
b,
Yan
Zeng
*bd,
Anubhav
Jain
*c and
Gerbrand
Ceder
 *ab
*ab
aDepartment of Materials Science & Engineering, University of California, Berkeley, CA 94720, USA. E-mail: gceder@berkeley.edu
bMaterials Sciences Division, Lawrence Berkeley National Laboratory, Berkeley, CA 94720, USA. E-mail: yanzeng@lbl.gov
cEnergy Technologies Area, Lawrence Berkeley National Laboratory, Berkeley, CA 94720, USA. E-mail: ajain@lbl.gov
dDepartment of Chemistry & Biochemistry, Florida State University, Tallahassee, Florida 32306, USA. E-mail: zeng@chem.fsu.edu
First published on 3rd October 2024
Abstract
The recent advent of autonomous laboratories, coupled with algorithms for high-throughput screening and active learning, promises to accelerate materials discovery and innovation. As these autonomous systems grow in complexity, the demand for robust and efficient workflow management software becomes increasingly critical. In this paper, we introduce AlabOS, a general-purpose software framework for orchestrating experiments and managing resources, with an emphasis on automated laboratories for materials synthesis and characterization. AlabOS features a reconfigurable experiment workflow model and a resource reservation mechanism, enabling the simultaneous execution of varied workflows composed of modular tasks while eliminating conflicts between tasks. To showcase its capability, we demonstrate the implementation of AlabOS in a prototype autonomous materials laboratory, the A-Lab, with around 3500 samples synthesized over 1.5 years.
1 Introduction
Computational and data-driven approaches have shaped a new paradigm for materials discovery, leveraging recent advances in high-performance computing and machine learning.1–7 Despite these advances, the experimental realization of computationally predicted compounds remains a slow, yet essential step in materials discovery and development.8–10 Substantial progress has been made to increase experimental throughput while reducing human involvement, as demonstrated by several automated laboratories that utilize synthesis methods based on flow chemistry,11–14 thin-film deposition,15,16 solution-based synthesis,17 and solid-state synthesis.18,19 These laboratories have also been integrated with predictive computational tools such as high-throughput density functional theory (DFT) calculations,1,20–22 machine learning algorithms for automated interpretation of characterization data,23–27 and optimization algorithms that can perform decision making.28–32 The integration of computational modeling and artificial intelligence into autonomous laboratories gives rise to what is known as self-driving laboratories (SDLs), which can explore material spaces with minimal human intervention.33–35 The rising prominence of SDLs has elevated the significance of robotics and machine learning for materials research. To accommodate this changing landscape from the human-centric experimentation diagram, improved tools are needed to manage robotics and the data they rapidly produce.36–38In designing workflow management software, one can learn from the automated workflows developed for high-throughput DFT calculations.39–41 These workflows provide a high-level user interface from which materials and calculation types can be specified, abstracting the lower-level tasks required to perform such calculations.42,43 These computational management software need to effectively organize large datasets, a task similarly required for experimental management. However, orchestrating an autonomous laboratory creates additional challenges, in particular, requiring the seamless integration of tasks and data with physical hardware and experimental samples, which is crucial for maintaining uninterrupted operation.
Several workflow management programs have previously been developed and implemented in different autonomous laboratories with diverse applications. For example, ChemOS 2.0 (ref. 44) proposes an integral platform between DFT calculation, Bayesian optimization, and automated equipment communicated through the SiLA2 protocol.45 Helao46 and Helao-async47 present a highly modularized design by implementing each component as a web server, enabling workflow management across laboratories. Bluesky48 has been applied to synchrotron facilities, with a strong emphasis on controlling synchrotron characterization hardware, to improve data collection efficiency but not targeted to materials synthesis. Additional software packages have supported laboratories involving automated experiments, featured with expandability and reconfigurability across different experiment stations,49 standardized data specification and storage,50 and seamless integration with other science tools as a “science factory”.51 These software solutions have successfully orchestrated experiment workflows in autonomous laboratories within their domains.
On the other hand, recent trends toward increased throughput and greater complexity in autonomous laboratories necessitate workflow software to manage more diverse workflows with a higher volume of autonomous devices. For instance, in our use case as well as others aiming to boost overall throughput, a laboratory can feature multiple equivalent devices for a single task, such as several furnaces for heating or two X-ray diffractometers for characterization. Due to the lack of knowledge about the availability of devices during runtime, the workflow management system must dynamically allocate the devices to each task based on their needs, thereby maximizing the utilization rate of the autonomous laboratory. Moreover, as general-purpose research laboratories often run different workflows with varied task sequences simultaneously, a reconfigurable workflow model has become essential, with certain parts of the workflow being executed in parallel. For example, after a sample is synthesized, multiple characterizations can be performed concurrently while storage operation has to wait until all the characterizations are done. Depending on the needs of the experiments, different experiment procedures need to be executed in the same laboratory setting. Thus, an expressive workflow model is required to effectively encode such task order dependencies while preserving flexibility when composing the workflow. Efforts have been made to handle some of these challenges. For example, a scheduling strategy is developed to tackle the multi-device problem, which requires an accurate time model for each task to ensure efficiency.52 However, only by addressing all of these practical needs, a workflow management software can fit better into an autonomous laboratory with increased complexity and throughput and allow researchers greater freedom to push the boundaries of autonomous experimentation.
Herein, we present AlabOS, a versatile and accessible workflow management framework for autonomous laboratories. The system features a graph-based experimental workflow model with tasks being the nodes and task dependencies being the edge. A central resource manager is built into the system, to track the status of devices and allocate resources properly to each task, thus eliminating possible conflicts between tasks running at the same time in the laboratory. Furthermore, AlabOS proposes the concept of sample position, representing a position in the laboratory that can hold one sample. By tracking the sample's position, AlabOS makes it possible to track individual samples in real time. This Python-based software is platform-independent and designed to be user-friendly, requiring basic knowledge about database and parallel programming for community adoption. Considering the errors and maintenance demands in the daily operations of an autonomous laboratory, a status monitoring and notification system is built into the software along with a browser-based graphic user interface (GUI). AlabOS serves as a general framework for managing workflows, designed to simplify the programming required to establish an autonomous laboratory. At the same time, it maintains the flexibility to accommodate various workflows within the laboratory. Other features of AlabOS include:
• Provide a solution for autonomous laboratory workflow management, with common functionalities like resource management, and notification built in.
• Built with MongoDB (NoSQL) backend, supporting a flexible schema, and allowing standardized continuous development of each task input, output, and data storage operations.
• A standard way to define devices and tasks with base classes when setting up a new lab using AlabOS. A simulation mode is also built in, allowing quick debugging of the devices and tasks.
• Submission and status monitoring APIs with JSON format. The users can set up scripts for job submission and queries on top of AlabOS. The input and output of the experiments can be validated by Pydantic53 before being stored in the database.
The AlabOS system is actively deployed in the A-Lab,18 an autonomous laboratory for inorganic materials synthesis housed at Lawrence Berkeley National Laboratory (LBNL). At the time of writing, it has synthesized and characterized over 3500 distinct samples under the control of AlabOS.
2 Key concepts and system design
2.1 Status representations
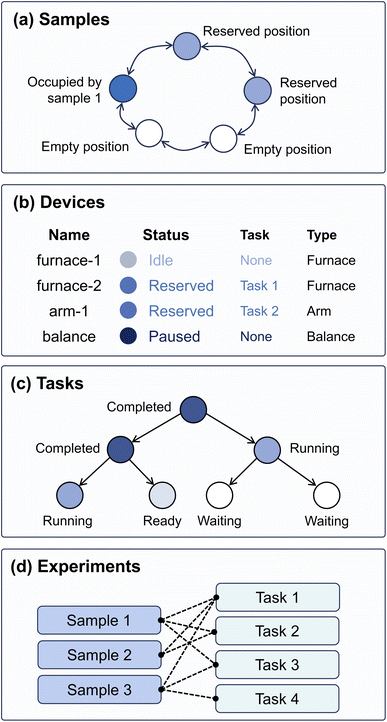
In AlabOS, a laboratory is represented as a combination of samples, devices, tasks, and experiments entities (Fig. 1). Each entity record is maintained in a separate collection in a MongoDB instance and updated during runtime to synchronize with the autonomous laboratory.A sample entity describes its name, position, and other metadata specified during submission (e.g., the composition of the sample and the project that the sample belongs to). A sample position is a space in the laboratory that can be occupied by one sample at a time. The sample positions are defined by operators before the system is up. When robots (or humans) move samples, the positions of the samples in the database are updated to track its physical location throughout the experiments. Every sample is assigned a human-readable name and a unique global ID for tracking purposes. Within autonomous laboratories, transferring a sample from one container to another is common. In this case, the old container will be disposed of in the physical laboratory while the sample itself will be updated to the position of the new container. When a sample is completely removed from the laboratory, its position will be set to null while the database entry describing its position history is kept for future reference.
A device refers to a piece of hardware to process or collect data from sample(s). Each device entity is linked to hardware in the lab that can send commands, perform physical operations, and collect data. Some examples of these operations in materials synthesis include sample dispensing, weighing, heating, and grinding. In AlabOS, a device is defined in Python code that provides the methods to communicate with the data transfer protocols like MODBUS,54 TCP/IP, and serial. Each device will have an entry in the database that indicates its status: whether it is occupied by a task, or whether it is paused for maintenance.
A task entity contains the procedure to execute a sequence of operations on a set of samples using the specified device. In a task, all relevant devices are orchestrated to achieve a high-level objective. For instance, to carry out a Heating task, the process begins by sending a request to the furnace to open its door. Following this, a robotic arm moves the designated samples into the now-open furnace. After the samples are loaded, a program is activated to heat the furnace to a temperature specified by the operator. Considering the parallel nature of an autonomous laboratory, where multiple samples may be processed simultaneously, a resource assignment mechanism is used to avoid conflicts between tasks in resource assignment. Before initiating control over one device or sample, each task has to request devices and sample positions. Once the resources are assigned, the task runs the procedures. After completion, the associated resources are released, allowing other tasks to request and use them. Apart from processing, a task could also be used for data generation and analysis for characterization or decision-making. In this case, the output of one task is directed as an input for another task by utilizing sample metadata as an information proxy.
An experiment is composed of one or multiple series of tasks to obtain conclusions. When submitting an experiment, the operator can specify a directed acyclic graph (DAG) of tasks to be performed on each sample. The execution sequence is guaranteed by the directed edge in DAG, where each node represents a task and each edge represents its order. A task can only be started when it does not have any unfinished parent tasks, which are defined to happen before this task. Relationships among tasks are stored in the task collection, where each task has a previous_tasks field and a next_tasks field. Whenever a task is completed, the system will initiate, request resources, and launch any descendent tasks when they are ready.
To maximize throughput, an autonomous laboratory often processes samples in batches. In AlabOS, an experiment can contain multiple samples and tasks. A task can accept one or more samples as input, depending on its predefined capacity. For example, if a furnace has a capacity for up to eight samples at a time, the Heating task will not accept more than eight samples.
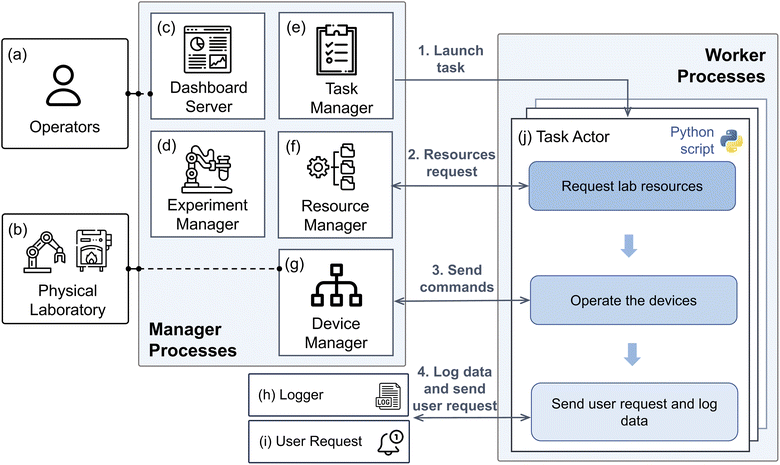
2.2 AlabOS architecture
AlabOS is designed to be a manager-worker architecture (Fig. 2). When a task is ready to run within this architecture, a worker object (named task actor) is instantiated in a new process by a task manager that executes the task logic. The managers monitor the status of the task actors and respond to each actor's requests for resources in the laboratory. Since the task actors only communicate with the managers, the interaction between ongoing laboratory tasks is eliminated. As a result, no conflicts between concurrent tasks (race conditions) need to be considered when defining the tasks' logic. The core services also run a dashboard server, which provides operators with a browser-based GUI and a set of APIs. The lab operator (Fig. 2a) can easily submit, view, and cancel experiments in the autonomous laboratory using these interfaces.There are four manager processes used to monitor and manipulate the status of the physical laboratory Fig. 2b at different levels: experiments, tasks, devices, and resources (the assignment of sample positions and devices). In addition, a dashboard server is working as the manager process to receive commands from the operators. All the managers communicate with one another through a database instance that is hosted on either a local computer or a cloud-based server. The experiment manager (Fig. 2d) receives experimental submissions from the dashboard server (Fig. 2c) and parses them into task graphs. The task manager (Fig. 2e) verifies and launches these tasks ready in the laboratory. It also monitors the status of each task as it is carried out. The task manager is alerted when a task is completed, or an error is encountered. It then marks all subsequent tasks as being ready or canceled accordingly. The resource manager (Fig. 2f) responds to the tasks' requests to occupy certain devices and sample positions in the lab. When a request is received, the manager checks the availability of all requested devices and sample positions. If the request can be met, the resource manager assigns the devices and sample positions to the task. The assignment will be valid until the signal is received that the task's resources are released. The device manager (Fig. 2g) is the intermediate layer between each task and the physical lab device it affects. When a task requests the device manager to send a command to some device, it first checks whether the device is occupied before sending the commands required to complete the task.
To interface with the manager processes in a user-friendly way, a dashboard server (Fig. 2c) with a GUI is provided. This allows the operator to control and monitor the progress of any experiments running in the laboratory, while also showing the current states of all devices in the lab. In addition to the GUI, this dashboard provides an API that can receive new experiment submissions in a JSON format.55 Each experiment submission is validated using Pydantic53 models to ensure the correctness of all formats and values.
In the worker processes, many task actors (Fig. 2j) run simultaneously to execute different tasks in the laboratory. In AlabOS, each task is configured in advance before the start of a new experiment. All tasks are defined as Python objects inherited from the BaseTask class available in the AlabOS package, providing universal methods for interacting with manager processes, the logger, and the user request module. To execute lab operations, the task actor must first request to occupy some devices and sample positions from the resource manager. When the request is approved, it continues to send commands to devices via the device manager while also updating each sample's position in the database. The data and device signal (e.g., the real-time temperature in a furnace) generated during the task will be logged to the database by the logger module (Fig. 2h). If the task requires any human intervention (for example, recovering a robot arm from error, or replacing consumables), it will generate a notification via the user request module, which the human operators can acknowledge once it is resolved.
3 Features and implementations
Several features are incorporated into AlabOS for resource management, human–machine interaction (HMI), lab device control, and data organization to ensure the autonomous laboratory runs efficiently and smoothly. These are detailed in the next few sections.3.1 Resource management
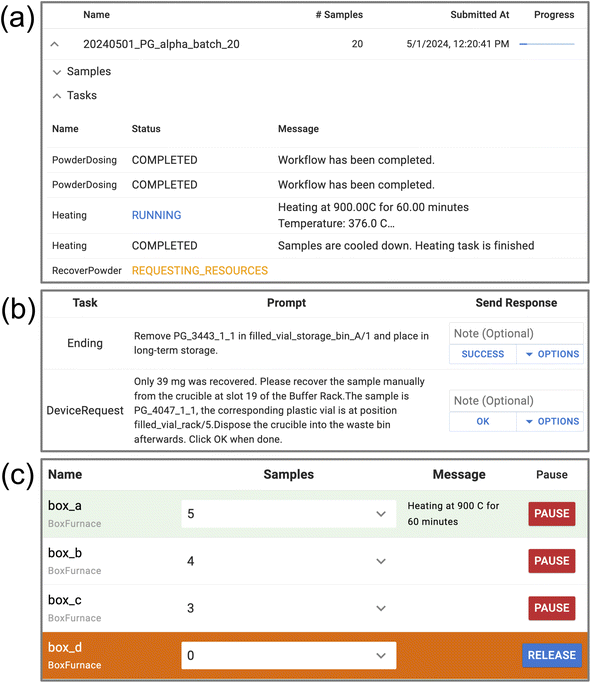
Unlike traditional materials research laboratories, where human experts perform various tasks (like sample preparation, heating, and characterization) serially, autonomous laboratories usually take a more distributed and parallelized approach. For example, a robot may dispense and weigh precursor powders while an automated furnace is heating a separate batch of samples that have already been weighed. There is typically a queue of pending tasks that the laboratory must plan and account for. To ensure the lab's continuous and successful operation without requiring human intervention, the system should avoid any possible conflicts between these tasks while also maximizing the overall throughput by organizing the distribution of tasks in a parallel fashion.In AlabOS, resource management is carried out using a cooperative multitasking schema.56 Before tasks perform any operations in the laboratory, it has to request the necessary resources from the AlabOS system. All the resource requests are handled by a module named resource manager, which will parse each request and check if there are any idle devices and sample positions that can fulfill the request. Once the request can be fulfilled, the resource manager will mark the assigned devices and sample positions as occupied and let the task know which devices and sample positions it can use. The task can then send commands to the assigned devices and move samples into the assigned sample positions. Once those operations are finished, the associated resources are released to be assigned to other tasks. To avoid the occurrence of “dead resources” where tasks are completed without releasing their resources properly, the system introduces a with-statement context for all requests, as shown in Fig. 3. Once resources are assigned to a task, they are recorded in a resource request context. All operations involving these resources need to be performed within this context to avoid a permission error. When the task is completed and exits the request context, it will automatically release all the requested resources, regardless of the task's success or failure.
A resource request can contain multiple devices and sample positions. In an autonomous laboratory, it is sometimes common to have multiple devices operating with the same functions (e.g., multiple furnaces for heating) to increase the lab's throughput. A task can operate on equivalent devices to obtain the desired results. In the task definition, a request can either specify the name of an exact device to use (e.g., Furnace A) or a type of device (e.g., muffle furnace). If a device type is specified, the resource manager will find any available device(s) under that type. The sample position request is then attached to the specific device. For example, one task may want to reserve the sample positions inside one furnace chamber. In this case, the sample positions request will be handled once the device request can be satisfied. When the resource manager finds an available combination of devices that can satisfy the request, it will advance to solve the available sample positions based on the proposed devices. Once both the devices and sample positions requests can be met, a request will be fulfilled and assigned to the corresponding tasks. With such a non-preemptive resource allocation strategy, the resource request serves in a “first-come-first-serve” strategy with the task priority taken into consideration. This does not need additional information like how long the task will occupy these resources. However, it also assumes that all the tasks do not occupy resources for an unreasonably long time which would lead to sample pileups in the lab. It is generally recommended for the laboratory developer to have a good knowledge of the bottlenecks and throughput of the laboratory and set up the right amount of tools to avoid this. On the other hand, it is always a good idea to break the resource request into many small pieces to avoid booking some resources for too long a time. Before the task definition is deployed in the physical laboratory, the lab developer should fully test the workflow definition in the simulation mode described in Section 3.3 to ensure the expected behavior of a task definition. More sophisticated scheduling of equipment reservation techniques52 could be integrated within AlabOS to further optimize the throughput.
When initiating a resource request, a priority can also be attached to determine the urgency of the request. The resource request's priority is kept the same as the priority of the task, which is specified by the user during submission as a task parameter (default to “NORMAL” priority). The resource requests' priority can also be overridden by supplying a customized value when creating a resource request inside the task. It is sometimes useful to force a higher priority to the post-processing operations in a task. For example, the sample must be unloaded from the X-ray diffractometer before the next sample can be loaded. In this case, it is necessary to prioritize the unload operation to avoid gridlock in the laboratory. The priority is encoded as an ordinal number ranging from 1 to 100. The priority is used to establish a ranking between resource requests, without any implication of proportional difference between the values. The default priority of all the tasks is 20, which indicates normal priority. When the resource manager polls the pending requests, it will first rank them by their priority and then, if they have the same priority number, by their submission time. The pending request that has a higher priority and was submitted earlier will generally be handled first.
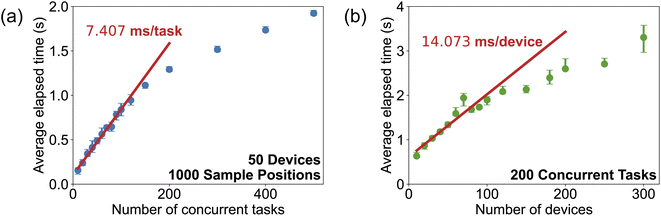
In an autonomous laboratory, many tasks can request resources at the same time. It is necessary to handle the large amount of possible resources on time so that the task can be executed faster. To understand the performance of the resource manager, a virtual lab with M devices, each of which has 20 associated sample positions, is defined. At each step, there are N tasks that request one device with one to twenty sample positions. Such configuration simulates the situation when the laboratory has a heavy workload. At each step, the resource manager will try to assign resources to each task. The CPU wall time is collected at different conditions. As shown in Fig. 4a, when the number of devices is fixed, the processing time for all the tasks' requests scales linearly with the task number at small task numbers (≤120). The average processing time for one task is 7.407 ms obtained from the slope of the fitted line. When the number of tasks continues to increase, the processing time deviates down from the fitted line. This may be attributed to a saturation of the resources. When there are way more tasks requesting resources compared to the available devices, most tasks' requests cannot be fulfilled in one step, leading to a processing time shorter than the time predicted by the linear relationship. The performance of the resource manager on different numbers of devices is shown in Fig. 4b. Similar to the performance under varied task numbers, with the number of devices increasing, the processing time first follows a linear relationship with a slope of 14.073 ms/device and then becomes flat due to the saturation of the tasks. When the devices are much more than the tasks, most devices remain idle and do not cost any time for resource assignment.
Compared to the time required for completing a normal operation in the laboratory for solid-state synthesis, which usually takes several minutes to hours, the time for assigning resources is nearly negligible, thus ensuring a higher turnover rate in the laboratory. In some cases, multiple operations can be short and complete in a few seconds, while requiring different resources to complete. If that is the case, it is generally recommended to combine these short operations and request the resources all at once to minimize the waiting time.
3.2 User interaction
To better facilitate daily maintenance and monitoring of experimental progress, AlabOS interacts with the operators through a web-based dashboard server (Fig. 5). In the dashboard, the operators can check the real-time status of each sample, device, and task running or queued in the lab. It also provides a variety of interfaces for human–machine interaction. These are detailed below.In addition to monitoring the real-time status of the lab, the dashboard also provides a cancellation button for each ongoing experiment and task, which can force the unfinished experiments/tasks to stop running. When the task receives a cancellation signal but has not started, it will be canceled directly. If the task is running, an error will be raised in the task process, where it is automatically decided how to halt the process without causing a sudden interruption of the lab's robotic operation.
In AlabOS, the notification can be initiated by either the system (manager instances) or the task processes. It can be used whenever a human action is needed. Each notification contains a prompt field and a list of possible options for the operators to select. The notification requires a receipt of acknowledgment from the operators with one option selected. Once that option is selected and the operator has completed the necessary tasks, the program will be carried out according to the selected actions. With clear notification messages designed when defining the tasks, the notification can be handled by the users without much knowledge about the autonomous laboratory. Thus, the maintenance jobs can be distributed among the lab developers and lab users.
Apart from the dashboard messages, notifications can also be configured to send over Slack bot or email. Further extensions can be made to other notification platforms, such as IFTTT,57 with minor programming efforts.
3.3 Device control
The AlabOS system uses a centralized manager to monitor and control all devices in a laboratory. The task actors do not communicate directly with the physical equipment to execute commands and read data. Instead, they send all commands to the device manager through a remote procedure call (RPC).58 In each task, an identical RPC proxy class will be created as an in-place replacement for the task to send commands. In this way, the RPC communication will not change the way driver methods are called in the task definition, as opposed to directly calling the devices' methods.Instead of creating the device driver objects in the task process, the indirect method call ensures the global singleton of each device. This helps to avoid the conflicts that can arise from multiple commands. Furthermore, the device manager checks the ownership of the requested devices and prevents them from being operated by tasks that do not reserve them. This minimizes the chance of conflicts and accidents in the laboratory.
To facilitate the rapid integration of new workflows in the autonomous laboratories, each device can be switched to simulation mode to allow tasks to run without connecting to the actual equipment. In the simulation mode, the operators can select to skip some methods (usually related to communication with the equipment) by applying a “mock” decorator to the method in the device definition. A mocked object with certain attributes will be specified as the return value in the “mock” decorator. In this way, the task can still proceed with the return values to test its functionalities, without requiring to talk with the hardware and their corresponding device drivers.
3.4 Data storage
The system provides a logger object for tracking and storing data. Each piece of data is saved as a document in MongoDB, with some metadata that includes information such as the data source and its time of logging. The logged data is classified as device signal, sample amount, characterization result, system log, and “other” which includes all the data that do not fall into the previous four catalogs. The classification of logged data improves the organization of the database. Apart from the classification, each logged data also has a level to indicate its importance, ranging from 10 to 50. In this scheme, level 10 is intended for debugging, while level 50 is intended for fatal errors. In each task actor process, a logger object will be created at the beginning of the task. During runtime, it can be called conveniently to log the data. For example, it can be used to record the real-time temperature of the oven at a regular interval during the Heating task, which can be read afterward from the database for debugging and post-analysis.Despite the logger's ease of use, logged data can still become unstructured and scattered in the database. For example, each point of the real-time temperature of the furnace can be a single log document in the database. To make the data more accessible to researchers, we also provide a “result” field for each task. At the end of each task, the task can gather data generated during the run and return them as a Python dictionary. AlabOS stores all returned data in the result field. For example, one can return the temperature–time curve at the end of the Heating task, which is especially useful when troubleshooting failed syntheses. If a large file needs to be stored in the database, AlabOS also provides a “LargeResult” object to handle the large file with MongoDB GridFS backend. The large file will be stored as chunks, with a reference ID generated and linked to the result collection.
When an experiment is completed, all data involving sample information, task results, and metadata can be copied to a backup database. This can be configured as a remote database on a cloud server. After being copied, no change will be made to these data. The operators can later query the data needed from this backup database. In this way, AlabOS ensures data safety and alleviates concerns about any potential file loss in the local database.
3.5 Experiment submission
In AlabOS, all the experiments are submitted through the dashboard server, where an API is exposed to receive all the experiment submissions. The format of the experiment is first validated to ensure the correct processing afterward. The validated experiment is then dumped into an experiment collection, where the experiment manager instance keeps polling and builds samples and task graphs according to the experiment specifications. The raw input format is in JSON format, which includes the sample and task information. Although it is easy for the system to parse the JSON format, it is usually not straightforward for the users to compose the input file directly. An experiment builder class is introduced to help users to define experiments in a Python script. When an experiment builder is created, the name and metadata of the experiment should be provided. With an “add_sample” method, the users can create and add samples to the experiments. Then the tasks are created and attached to the samples. The experiment builder will record the sequence of the tasks added to each sample and build the task graph with the dependency specified in the sequence. Finally, the experiment can be converted to a JSON format string and submitted to the submission API with a “submit” method. The users can also query the experiment status and results from the experiment information API.The submission API and the experiment builder serve as a high-level abstraction to the autonomous laboratory. It resembles the queue management system in high-performance computing (HPC) clusters. When submitting jobs to HPC, the users do not need to pay attention to how and where their jobs are run. Similarly, when an experiment is submitted to AlabOS, the users can focus on designing experiments with proper parameters without knowing the details of how the experiment will be run.
The submission infrastructure of AlabOS opens up the possibility of conducting a close-loop experiment campaign inside an autonomous laboratory, where AI agents analyze the results and make plans for future experiments. With a structured input format that is sent to an API endpoint, there is no distinction between experiments submitted by humans and by AI. When designing a close-loop experiment campaign, the user can wrap the submission and result query script into an objective function with the inputs and outputs to be explored. The objective function can be directly used in various experiment planning software for self-driving laboratories like Chimera59 and Altas.60
4 AlabOS in practice: orchestrating an autonomous laboratory for solid-state synthesis
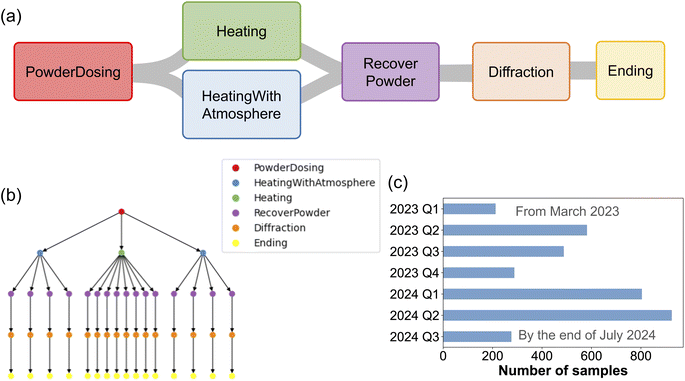
AlabOS is actively used to orchestrate the A-Lab,18 an autonomous synthesis platform for inorganic material synthesis. The A-Lab focuses specifically on preparing samples in powder form using solid-state synthesis and characterizes them using X-ray diffraction. A typical workflow of the A-Lab's operations is shown in Fig. 6a. The workflow is divided into several tasks, which are the building blocks of each experiment in the A-Lab. A PowderDosing task weighs certain amounts of precursor powders and mixes them using ball-milling. A Heating task sends the samples into a box furnace, which is heated according to a pre-defined temperature profile. Similarly, a HeatingWithAtmosphere task uses tube furnaces to heat the samples under a controlled gas flow atmosphere. After the samples are heated, they are sent to the RecoverPowder task to be ground into fine powders. A small portion of the sample powder is extracted and sent to an X-ray diffractometer to measure the powder X-ray diffraction (XRD) spectrum in the Diffraction task. At the end of one experimental cycle, the Ending task notifies the operators to store each completed sample in a designated position, and this position is updated in the database accordingly.To set up the A-Lab workflows using AlabOS, a project folder is first created. A toml format configuration file is made to store all the connection information to the database, the notification service, and the message queue service. The devices' driver codes are stored in a devices folder. Each type of device is defined in a Python object class inherited from BaseDevice to include some basic functions like connecting and status checking. Apart from these, one can define as many methods as needed to operate the device. We also define the associated sample position names in the device. They are read by AlabOS and recorded in the database when launching. To build an automated lab for solid-state synthesis and powder XRD characterization, there are in total 16 types of devices with 28 device instances and 289 device-related sample positions. The full list of defined devices, their communication protocols, the number of associated sample positions, and numbers of handled exceptions are listed in Table 1. Most devices communicate with the control PC through Ethernet, with various protocols including HTTP, MODBUS, XML-RPC, etc. Each device (and control PC) is assigned to a unique IP within an intranet. The control PC can send commands to the devices by specifying the IP addresses of the devices. The box furnaces, as an exception, are connected directly to the control PC through its serial interfaces, with a COM port assigned to each furnace.
| Device type | Physical device | Communication protocol | Number of devices | Number of associated sample positions per device |
|---|---|---|---|---|
| BallDispenser | Arduino | HTTP | 1 | 1 |
| BoxFurnace | Thermo Scientific F4805560 | MODBUS over serial | 4 | 8 |
| CapDispenser | Arduino | HTTP | 1 | 3 |
| CappingGripper | Arduino | HTTP | 1 | 1 |
| Diffractometer | Malvern Panalytical Aeris Mineral Edition | Socket | 1 | 1 |
| LabmanQuadrant | Labman powder dosing system | HTTP | 4 | 48 |
| ManualFurnace | N/A | N/A | 4 | 8 |
| RobotArmCharacterization | Universal robot 5e | Socket & SSH | 1 | 1 |
| RobotArmFurnaces | Universal robot 5e | Socket & SSH | 1 | 1 |
| Scale | OHAUS Scout | HTTP | 1 | 1 |
| Shaker | Arduino | HTTP | 1 | 1 |
| TransferRack | N/A | N/A | 1 | 6 |
| TubeFurnace | MTI OTF1200X5ASD | XML-RPC | 4 | 4 |
| VialDispenserRack | N/A | N/A | 1 | 0 |
| VialLabeler | Reiner jetStamp 1025 | Serial | 1 | 1 |
| XRDDispenserRack | N/A | N/A | 1 | 0 |
After the devices are set, another folder named tasks is created to store all the task procedures. Each task inherits from BaseTask in AlabOS, which implements methods to report progress, reserve resources, and get samples' information. In each task, the device and sample position resources are reserved before any actions are taken in the A-Lab. A device handle is returned after the resource is assigned in AlabOS. The task can call any method in the device class to perform operations. Finally, the system can be launched via a terminal's command line interface (CLI). If any changes are made to the task definition, the system will need to be restarted to take the changes into effect.
To optimize the throughput, several tasks in the A-Lab can handle multiple samples, with the capacity of each task and the number of successfully completed tasks shown in Table 2. For example, samples are loaded into a ceramic rack with eight slots in the Heating task. A robot arm then inserts this entire ceramic rack into a box furnace. Such batch processing approach gives the Heating task a maximum capacity of eight. When submitting the experiment, one node in the task graph can take multiple samples, which indicates that they are processed in the same batch. For example, an experiment with sixteen samples has the task graph shown in Fig. 6b. All of the sixteen samples are processed in one PowderDosing task. The samples are then divided into three heating batches with four, four, and eight samples, respectively. These are heated in different furnaces according to the specified heating profiles. Samples with different heating profiles cannot share the same furnace.
| Task name | Capacity | Number of tasks | Exception handling routines in the definition | % With exceptions | % With unrecoverable exceptions |
|---|---|---|---|---|---|
| PowderDosing | 16 | 333 | 1 | 1.20 | 1.20 |
| Heating | 8 | 403 | 2 | 4.96 | 4.22 |
| ManualHeating | 8 | 75 | 0 | 0 | 0 |
| HeatingWithAtmosphere | 4 | 27 | 0 | 22.22 | 7.41 |
| RecoverPowder | 1 | 2818 | 15 | 4.19 | 0.99 |
| Diffraction | 1 | 2658 | 11 | 4.74 | 1.20 |
| Ending | 1 | 2574 | 3 | 0.12 | 0.04 |
While a common workflow of tasks is shown in Fig. 6a, many other workflows are possible in the A-Lab. Several of these are shown in Table 3. For example, when the operators wish to heat samples manually while still using the rest of the automated processes in the A-Lab, a ManualHeating task can inform the operators (via the notification system) of the position of each sample after the PowderDosing task is completed. It will also notify the operators where to place the heated samples so that any downstream tasks can process them. As another example, if the operators wish to obtain the XRD pattern of the precursors used in the A-Lab, they can skip the Heating tasks and move the sample (precursor powders) directly to the RecoverPowder and Diffraction tasks. Also, if the operators would like to process a set of samples that are heated outside of the A-Lab, they can use a Starting task to create the record of the sample in the database and specify the position of the sample so that tasks can find the sample.
| Workflow | Note | |
|---|---|---|
| 1 | PowderDosing – ManualHeating – RecoverPowder – Diffraction – Ending | Heat the samples in the external furnaces for higher throughput |
| 2 | PowderDosing – RecoverPowder –Diffraction – Ending | Skip the heating to characterize the precursors |
| 3 | Starting – RecoverPowder – Diffraction – Ending | Process samples that are made outside of the A-Lab |
By the time of writing, AlabOS has been driving the A-Lab to synthesize and characterize over 3500 distinct samples over a period of approximately one and half years, as shown in Fig. 6c. The maximum number of samples submitted in one single day was 149 samples on Feb 9, 2024. The system is designed to be able to handle a large amount of sample submissions at a time. By dividing the experiments composed of many samples into modular tasks, AlabOS is able to schedule and manage experiments at a finer granularity, thus ensuring the high efficiency of completing the complicated workflows.
To maintain smooth operation, the A-Lab task definitions incorporate handling routines for common exceptions. The number of tasks with exceptions with each of the A-Lab task name is listed in Table 2. Depending on the complexity of the task, different exception-handling codes are inserted into the task definitions. Among them, RecoverPowder and Diffraction require the most exception handling as many steps are involved in the operation. The number of exception-handling routines also aligns with the percentage of recovered tasks in all the exceptions-raised ones. RecoverPowder and Diffraction demonstrate a higher exception recovery rate of 76% and 75%, respectively. On the contrary, Heating and PowderDosing have a lower recovery rate of 15% and 0% due to limited exception handling. The lack of routines to address exceptions in these tasks can be attributed to the fact that some are due to hardware that is worn out, or to newly emerged software communication exceptions, which were not detected in the soak test. Over time, these tasks will suffer less from unrecoverable errors in the future with more exception-handling routines included.
5 Conclusion and outlook
We have outlined the development and application of AlabOS as an orchestration software for managing workflows in autonomous laboratories. This package is designed to meet the fast-changing requirements of a typical materials research program. With the graph-based experiment input format, independent task actor design, and the resource occupation mechanism, human operators can define and submit workflows in AlabOS without concerning themselves with possible conflicts between concurrent tasks. The dashboard and notification system provide a general solution for human–machine interaction in the laboratory, which becomes especially useful when performing maintenance and error recovery jobs. With the public availability of AlabOS, we hope that researchers will no longer need to write complex workflow management codes but instead focus their time on developing high-level logic followed by autonomous laboratories for accelerated materials discovery and optimization. Only in this way, the autonomous laboratory can be more accessible to general researchers and become a more powerful tool for accelerating material discovery.Data availability
The code for AlabOS is available at https://github.com/CederGroupHub/alabos. The version of the AlabOS code employed for this study is v1.0.1. The online document is hosted at https://cedergrouphub.github.io/alabos/. The implementation of device communication driver is available at https://github.com/CederGroupHub/alab_control. The A-Lab-specific device and task definitions are available at https://github.com/CederGroupHub/alabos/tree/main/examples/alab_example.Author contributions
Y. F. and B. R.: conceptualization, software, writing – original draft, writing – review and editing. R. K.: conceptualization, software, writing – review and editing. O. D., H. P. S., and M. J. M: software, writing – review and editing. Z. W., N. J. S., L. N. W., and D. M.: investigation, writing – review and editing. Y. Z.: methodology, supervision, writing – review and editing. A. J.: methodology, supervision, resources, writing – review and editing. G. C.: resources, supervision, methodology, project administration, writing – review and editing.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was primarily financed by the U.S. Department of Energy, Office of Science, Office of Basic Energy Sciences, Materials Sciences and Engineering Division under contract no. DE-AC02-05-CH11231 (D2S2 programme, KCD2S2), the Laboratory Directed Research and Development Program of Lawrence Berkeley National Laboratory, and Materials Project. Work done at UC Berkeley was supported by Umicore Specialty Oxides and Chemicals.References
- A. Jain, S. P. Ong, G. Hautier, W. Chen, W. D. Richards, S. Dacek, S. Cholia, D. Gunter, D. Skinner, G. Ceder and K. A. Persson, APL Mater., 2013, 1, 011002 CrossRef.
- J. Schmidt, M. R. G. Marques, S. Botti and M. A. L. Marques, npj Comput. Mater., 2019, 5, 83 CrossRef.
- K. Choudhary, B. DeCost, C. Chen, A. Jain, F. Tavazza, R. Cohn, C. W. Park, A. Choudhary, A. Agrawal, S. J. L. Billinge, E. Holm, S. P. Ong and C. Wolverton, npj Comput. Mater., 2022, 8, 59 CrossRef.
- K. T. Butler, D. W. Davies, H. Cartwright, O. Isayev and A. Walsh, Nature, 2018, 559, 547–555 CrossRef CAS PubMed.
- G. R. Schleder, A. C. Padilha, C. M. Acosta, M. Costa and A. Fazzio, J. Phys.: Mater., 2019, 2, 032001 CAS.
- D. Morgan and R. Jacobs, Annu. Rev. Mater. Res., 2020, 50, 71–103 CrossRef CAS.
- S. Kirklin, J. E. Saal, B. Meredig, A. Thompson, J. W. Doak, M. Aykol, S. Rühl and C. Wolverton, npj Comput. Mater., 2015, 1, 1–15 Search PubMed.
- B. G. Sumpter, R. K. Vasudevan, T. Potok and S. V. Kalinin, npj Comput. Mater., 2015, 1, 15008 CrossRef CAS.
- J. R. Chamorro and T. M. McQueen, Acc. Chem. Res., 2018, 51, 2918–2925 CrossRef CAS PubMed.
- Z. Wang, Y. Sun, K. Cruse, Y. Zeng, Y. Fei, Z. Liu, J. Shangguan, Y.-W. Byeon, K. Jun, T. He, W. Sun and G. Ceder, Nat. Synth., 2024, 3, 527–536 CrossRef.
- S. Steiner, J. Wolf, S. Glatzel, A. Andreou, J. M. Granda, G. Keenan, T. Hinkley, G. Aragon-Camarasa, P. J. Kitson, D. Angelone and L. Cronin, Science, 2019, 363, eaav2211 CrossRef CAS PubMed.
- N. Hartrampf, A. Saebi, M. Poskus, Z. P. Gates, A. J. Callahan, A. E. Cowfer, S. Hanna, S. Antilla, C. K. Schissel, A. J. Quartararo, X. Ye, A. J. Mijalis, M. D. Simon, A. Loas, S. Liu, C. Jessen, T. E. Nielsen and B. L. Pentelute, Science, 2020, 368, 980–987 CrossRef CAS PubMed.
- J. S. Manzano, W. Hou, S. S. Zalesskiy, P. Frei, H. Wang, P. J. Kitson and L. Cronin, Nat. Chem., 2022, 14, 1311–1318 CrossRef CAS PubMed.
- J. Bennett, N. Orouji, M. Khan, S. Sadeghi, J. Rodgers and M. Abolhasani, Nat. Chem. Eng., 2024, 1–11 Search PubMed.
- B. P. MacLeod, F. G. Parlane, T. D. Morrissey, F. Häse, L. M. Roch, K. E. Dettelbach, R. Moreira, L. P. Yunker, M. B. Rooney and J. R. Deeth, et al. , Chem, 2021, 7, 2541–2545 Search PubMed.
- A. G. Kusne, H. Yu, C. Wu, H. Zhang, J. Hattrick-Simpers, B. DeCost, S. Sarker, C. Oses, C. Toher, S. Curtarolo, A. V. Davydov, R. Agarwal, L. A. Bendersky, M. Li, A. Mehta and I. Takeuchi, Nat. Commun., 2020, 11, 5966 CrossRef CAS PubMed.
- B. Burger, P. M. Maffettone, V. V. Gusev, C. M. Aitchison, Y. Bai, X. Wang, X. Li, B. M. Alston, B. Li and R. Clowes, et al. , Nature, 2020, 583, 237–241 CrossRef CAS PubMed.
- N. J. Szymanski, B. Rendy, Y. Fei, R. E. Kumar, T. He, D. Milsted, M. J. McDermott, M. Gallant, E. D. Cubuk, A. Merchant, H. Kim, A. Jain, C. J. Bartel, K. Persson, Y. Zeng and G. Ceder, Nature, 2023, 624, 86–91 CrossRef CAS PubMed.
- J. Chen, S. R. Cross, L. J. Miara, J.-J. Cho, Y. Wang and W. Sun, Nat. Synth., 2024, 1–9 CrossRef PubMed.
- J.-P. Correa-Baena, K. Hippalgaonkar, J. van Duren, S. Jaffer, V. R. Chandrasekhar, V. Stevanovic, C. Wadia, S. Guha and T. Buonassisi, Joule, 2018, 2, 1410–1420 CrossRef CAS.
- J. E. Saal, S. Kirklin, M. Aykol, B. Meredig and C. Wolverton, Jom, 2013, 65, 1501–1509 CrossRef CAS.
- B. Liu, J. Zhao, Y. Liu, J. Xi, Q. Li, H. Xiang and Y. Zhou, J. Mater. Sci. Technol., 2021, 88, 143–157 CrossRef CAS.
- N. J. Szymanski, C. J. Bartel, Y. Zeng, Q. Tu and G. Ceder, Chem. Mater., 2021, 33, 4204–4215 CrossRef CAS.
- V. Stanev, V. V. Vesselinov, A. G. Kusne, G. Antoszewski, I. Takeuchi and B. S. Alexandrov, npj Comput. Mater., 2018, 4, 43 CrossRef.
- D. Chen, Y. Bai, S. Ament, W. Zhao, D. Guevarra, L. Zhou, B. Selman, R. B. van Dover, J. M. Gregoire and C. P. Gomes, Nat. Mach. Intell., 2021, 3, 812–822 CrossRef.
- J. Liu, M. Osadchy, L. Ashton, M. Foster, C. J. Solomon and S. J. Gibson, Analyst, 2017, 142, 4067–4074 RSC.
- F. Oviedo, Z. Ren, S. Sun, C. Settens, Z. Liu, N. T. P. Hartono, S. Ramasamy, B. L. DeCost, S. I. Tian and G. Romano, et al. , npj Comput. Mater., 2019, 5, 60 CrossRef.
- N. J. Szymanski, P. Nevatia, C. J. Bartel, Y. Zeng and G. Ceder, Nat. Commun., 2023, 14, 6956 CrossRef CAS PubMed.
- M. Aykol, J. H. Montoya and J. S. Hummelshøj, J. Am. Chem. Soc., 2021, 143, 9244–9259 CrossRef CAS PubMed.
- F. Strieth-Kalthoff, H. Hao, V. Rathore, J. Derasp, T. Gaudin, N. H. Angello, M. Seifrid, E. Trushina, M. Guy, J. Liu, X. Tang, M. Mamada, W. Wang, T. Tsagaantsooj, C. Lavigne, R. Pollice, T. C. Wu, K. Hotta, L. Bodo, S. Li, M. Haddadnia, A. Wolos, R. Roszak, C.-T. Ser, C. Bozal-Ginesta, R. J. Hickman, J. Vestfrid, A. Aguilar-Gránda, E. L. Klimareva, R. C. Sigerson, W. Hou, D. Gahler, S. Lach, A. Warzybok, O. Borodin, S. Rohrbach, B. Sanchez-Lengeling, C. Adachi, B. A. Grzybowski, L. Cronin, J. E. Hein, M. D. Burke and A. Aspuru-Guzik, Science, 2024, 384, eadk9227 CrossRef CAS PubMed.
- J. M. Granda, L. Donina, V. Dragone, D.-L. Long and L. Cronin, Nature, 2018, 559, 377–381 CrossRef CAS PubMed.
- Y. Xie, K. Sattari, C. Zhang and J. Lin, Prog. Mater. Sci., 2023, 132, 101043 CrossRef.
- M. Abolhasani and E. Kumacheva, Nat. Synth., 2023, 2, 483–492 CrossRef.
- H. G. Martin, T. Radivojevic, J. Zucker, K. Bouchard, J. Sustarich, S. Peisert, D. Arnold, N. Hillson, G. Babnigg and J. M. Marti, et al. , Curr. Opin. Biotechnol., 2023, 79, 102881 CrossRef CAS PubMed.
- Y. Xie, K. Sattari, C. Zhang and J. Lin, Prog. Mater. Sci., 2023, 132, 101043 CrossRef.
- P. Raccuglia, K. C. Elbert, P. D. Adler, C. Falk, M. B. Wenny, A. Mollo, M. Zeller, S. A. Friedler, J. Schrier and A. J. Norquist, Nature, 2016, 533, 73–76 CrossRef CAS PubMed.
- M. D. Zimmerman, M. Grabowski, M. J. Domagalski, E. M. MacLean, M. Chruszcz and W. Minor, Structural Genomics and Drug Discovery: Methods and Protocols, 2014, 1–25 Search PubMed.
- L. Talirz, S. Kumbhar, E. Passaro, A. V. Yakutovich, V. Granata, F. Gargiulo, M. Borelli, M. Uhrin, S. P. Huber and S. Zoupanos, et al. , Sci. Data, 2020, 7, 299 CrossRef PubMed.
- A. Jain, S. P. Ong, W. Chen, B. Medasani, X. Qu, M. Kocher, M. Brafman, G. Petretto, G.-M. Rignanese and G. Hautier, et al. , Concurr. Comp.-Pract. E, 2015, 27, 5037–5059 CrossRef.
- L. Himanen, A. Geurts, A. S. Foster and P. Rinke, Adv. Sci., 2019, 6, 1900808 CrossRef PubMed.
- S. Curtarolo, W. Setyawan, G. L. Hart, M. Jahnatek, R. V. Chepulskii, R. H. Taylor, S. Wang, J. Xue, K. Yang and O. Levy, et al. , Comput. Mater. Sci., 2012, 58, 218–226 CrossRef CAS.
- K. Mathew, J. H. Montoya, A. Faghaninia, S. Dwarakanath, M. Aykol, H. Tang, I.-h. Chu, T. Smidt, B. Bocklund and M. Horton, et al. , Comput. Mater. Sci., 2017, 139, 140–152 CrossRef.
- S. P. Huber, E. Bosoni, M. Bercx, J. Bröder, A. Degomme, V. Dikan, K. Eimre, E. Flage-Larsen, A. Garcia and L. Genovese, et al. , npj Comput. Mater., 2021, 7, 136 CrossRef.
- M. Sim, M. G. Vakili, F. Strieth-Kalthoff, H. Hao, R. Hickman, S. Miret, S. Pablo-García and A. Aspuru-Guzik, Matter, 2024, 7, 2959–2977 CrossRef CAS.
- D. Juchli, Adv. Biochem. Eng./Biotechnol., 2022, 182, 147–174 CrossRef CAS PubMed.
- F. Rahmanian, J. Flowers, D. Guevarra, M. Richter, M. Fichtner, P. Donnely, J. M. Gregoire and H. S. Stein, Adv. Mater. Interfaces, 2022, 9, 2101987 CrossRef.
- D. Guevarra, K. Kan, Y. Lai, R. J. Jones, L. Zhou, P. Donnelly, M. Richter, H. S. Stein and J. M. Gregoire, Digital Discovery, 2023, 2, 1806–1812 RSC.
- D. Allan, T. Caswell, S. Campbell and M. Rakitin, Synchrotron Radiat. News, 2019, 32, 19–22 CrossRef.
- H. Fakhruldeen, G. Pizzuto, J. Glowacki and A. I. Cooper, 2022 International Conference on Robotics and Automation (ICRA), 2022, pp. 6013–6019 Search PubMed.
- I. M. Pendleton, G. Cattabriga, Z. Li, M. A. Najeeb, S. A. Friedler, A. J. Norquist, E. M. Chan and J. Schrier, MRS Commun., 2019, 9, 846–859 CrossRef CAS.
- R. Vescovi, T. Ginsburg, K. Hippe, D. Ozgulbas, C. Stone, A. Stroka, R. Butler, B. Blaiszik, T. Brettin and K. Chard, et al. , Digital Discovery, 2023, 2, 1980–1998 RSC.
- J. Zhou, M. Luo, L. Chen, Q. Zhu, S. Jiang, F. Zhang, W. Shang and J. Jiang, A multi-robot-multi-task scheduling system for autonomous chemistry laboratories, ChemRxiv, 2024, preprint, DOI:10.26434/chemrxiv-2024-0g4sz.
- The Pydantic development team, Pydantic, https://github.com/pydantic/pydantic, 2024, accessed: 2024-02-05 Search PubMed.
- I. C. Bertolotti and T. Hu, Embedded Software Development, 2017, 29, 343–374 Search PubMed.
- F. Pezoa, J. L. Reutter, F. Suarez, M. Ugarte and D. Vrgoč, Proceedings of the 25th International Conference on World Wide Web, 2016, pp. 263–273 Search PubMed.
- K. Jeffay, D. F. Stanat and C. U. Martel, IEEE real-time systems symposium, 1991, pp. 129–139 Search PubMed.
- X. Mi, F. Qian, Y. Zhang and X. Wang, Proceedings of the 2017 Internet Measurement Conference, 2017, pp. 398–404 Search PubMed.
- R. Srinivasan, RFC1831: RPC: Remote Procedure Call Protocol Specification Version 2, 1995 Search PubMed.
- F. Häse, L. M. Roch and A. Aspuru-Guzik, Chem. Sci., 2018, 9, 7642–7655 RSC.
- R. Hickman, M. Sim, S. Pablo-García, I. Woolhouse, H. Hao, Z. Bao, P. Bannigan, C. Allen, M. Aldeghi and A. Aspuru-Guzik, A Brain for Self-driving Laboratories, ChemRxiv, 2023, preprint, DOI:10.26434/chemrxiv-2023-8nrxx.
Footnote |
| † Equal contribution. |
| This journal is © The Royal Society of Chemistry 2024 |