Open Access Article
Open Access ArticleDNAsome with self-boosting ROS generation via tumour acidosis for enhanced and targeted chemodynamic cancer therapy†
Gowtham
Raj
,
Justin
Prasad
,
Tamraparni
Ghosh
,
Vasudev
D. S.
,
Athul
V. B.
,
Joyraj
Kalita
,
Devu B.
Kumar
and
Reji
Varghese
 *
*
School of Chemistry Indian Institute of Science Education and Research (IISER), Thiruvananthapuram, Trivandrum 695551, Kerala, India. E-mail: reji@iisertvm.ac.in
First published on 4th October 2024
Abstract
The anticancer efficacy of chemodynamic therapy (CDT) is significantly reduced owing to the mild acidic nature of the tumour microenvironment (TME). Typically, Fenton catalysts require a strong acidic microenvironment for effective radical generation at the tumour site. Hence the development of new strategies to achieve efficient Fenton reactions by increasing the acidity of the TME is highly demanded for the advancement of CDT-based cancer treatment. Herein, we demonstrate that the loading of the pH-regulator tamoxifen (TAM) into a CDT nanoagent (DNA1some) could significantly boost the efficiency of CDT action by increasing the acidity at the TME. The integration of nucleolin specific aptamer DNA (DNA2) onto the surface of DNA1some (DNA1some/TAM/DNA2) permitted the targeted internalization of the nanoformulation selectively into cancer cells, and consequently, a very efficient Fenton reaction was demonstrated inside the cancer cells selectively, which reduced the “off-target” toxicity of the nanoformulation to the surrounding normal cells. Enhanced cytotoxicity was observed for the TAM-loaded DNA1some compared to DNA1some and TAM alone, which was attributed to the very efficient Fenton reaction by DNA1some due to the increase in acidity caused by the release of TAM. Hence, the pH-regulator-loaded CDT-active DNAsome can potentially overcome the intrinsically insufficient acidity of the TME for enabling efficient Fenton reactions.
Introduction
Chemodynamic therapy (CDT) is an emerging non-invasive therapeutic approach for the treatment of cancer.1–6 It involves the use of a transition metal as a catalyst for the conversion of endogenous hydrogen peroxide (H2O2) present in the tumor microenvironment (TME) to yield highly oxidizing hydroxyl radicals (˙OH) via Fenton/Fenton-like reactions. The oxidative nature of ˙OH disrupts redox homeostasis and ultimately leads to cell death.7–9 Compared to other conventional therapeutic approaches, such as chemo, radiation, photothermal and photodynamic, the therapeutic action of CDT solely depends on the abundantly available transition metal as a catalyst and endogenous H2O2. Hence, CDT does not require any complicated instrumentation, which makes it a superior therapeutic choice over other conventional strategies for cancer treatment.10,11Ideally, the Fenton reaction requires an acidic pH (pH 3.0–5.0) for the efficient generation of ˙OH.12–14 However, because the pH range of the TME is only 6.5–6.9, the efficiency of the Fenton reaction has been found to be poor in the TME, which in turn reduces the therapeutic performance of CDT-based cancer therapy.15,16 Off-target toxicity to the surrounding normal tissues due to non-targeted accumulation of metal catalysts is another challenge associated with CDT-based cancer therapy. Hence the development of novel strategies to achieve efficient Fenton reactions by increasing the acidity of the TME in a targeted fashion is highly demanded for the advancement of CDT-based cancer treatment.
DNA-based nanostructures have recently emerged as potential nanocarriers for the delivery of metal ions (Fenton catalyst) to the TME because of their excellent biocompatibility, water solubility and, most importantly, their strong non-covalent interactions with metal ions through their negatively charged phosphate backbone and with nucleobases.17,18 Among the various metal ions, Cu2+ has received particular attention as a Fenton catalyst owing to its beneficial characteristics.19–22 For instance, Cu2+ exhibits good catalytic activity even in weakly acidic conditions.23–26 Moreover, Cu2+ reacts with endogenous glutathione (GSH) to form Cu+, which is the catalytically active form of copper for the Fenton reaction.27,28 Hence the active form of copper would be generated preferably inside cancer cells due to the high concentration of GSH inside cancer cells compared to the normal cells. This leads to the preferential activation of the Fenton reaction inside cancer cells, which would also be less likely to occur inside normal cells. This potentially reduces the off-target toxicity to the surrounding healthy cells. It should also be noted that Cu2+ not only produces catalytically active Cu+ but also downregulates GSH (antioxidant), with both reactions highly beneficial for efficient CDT. In addition, the surface of DNA nanostructures can be integrated with cell-targeting DNA aptamers via sequence-specific DNA hybridization, allowing the targeted delivery of the Fenton catalyst into the cancerous cells selectively.29–39
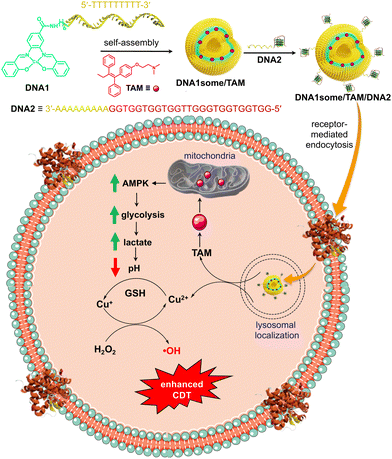
Very recently, we reported a class of DNA amphiphiles consisting of a Cu2+–salen complex as a hydrophobic Fenton reaction centre and ssDNA as the hydrophilic segment.40 Self-assembly of the amphiphile (DNA1) resulted in the formation of CDT-active vesicles (DNA1some) having a Cu2+-based Fenton reaction centre as the membrane of the vesicle and hydrophilic ssDNA as the shell. One of the challenges associated with CDT for cancer therapy is the poor yield of the Fenton reaction due to the nearly neutral pH range around the TME (pH 6.5–6.9).16 In this work, we wanted to improve the CDT efficacy of DNA1some by increasing the acidity of the TME. Tamoxifen (TAM) is a popular anti-oestrogen drug and is extensively used in treatment of oestrogen receptor-positive breast cancer.12,41 It has also been shown that TAM can inhibit the function of mitochondrial complex I and thereby increase the AMP to ATP ratio, which triggers the AMP-activated protein kinase signalling pathway.12 This in turn causes an enhancement in the rate of glycolysis reactions and lactate generation, which leads to a significant decrease in the pH in the TME. Hence, we envisioned that the delivery of the pH-regulator TAM along with the CDT-active DNA1some into cancer cells would be a promising approach to boost the efficiency of CDT by decreasing the pH of the TME by the action TAM.
Herein, we report on the formulation of TAM-loaded DNA1some (DNA1some/TAM) and investigations into the therapeutic performance of the nanoformulation for targeted CDT-based cancer therapy using MDA-MB-231 as a representative cancer cell (Scheme 1). The self-assembly of DNA1 in the presence of TAM resulted in the formation of the TAM-loaded DNA1some (DNA1some/TAM), where the hydrophobic TAM most likely resides in the hydrophobic membrane of DNA1some. The DNA-based surface addressability of DNA1some was used for the integration of nucleolin-targeting DNA aptamer (DNA2) via sequence-specific DNA hybridization to form DNA1some/TAM/DNA2. Nucleolin is known to be overexpressed on the surfaces of several cancer cells, including MDA-MB-231.42 Following the nucleolin receptor-mediated endocytosis, DNA1some/TAM/DNA2 disassembles inside the lysosome and leads to the release of TAM and free Cu2+ at the cytoplasm. Free Cu2+ is then in situ reduced to Cu+ by the cellular GSH, and undergoes a Fenton reaction using endogenous H2O2 to produce ˙OH. Enhanced cytotoxicity was observed for DNA1some/TAM/DNA2 when compared to DNA1some alone. The enhanced cytotoxicity of DNA1some/TAM/DNA2 was attributed to the efficient Fenton reaction due to the more acidic TME caused by the action of TAM.
Results and discussion
Synthesis and characterization of the DNA1 amphiphile were reported in our recent report.40 Specifically, the amphiphilicity-driven self-assembly of DNA1 in aqueous medium (pH 7.0) led to the formation of CDT-active vesicular nanostructures (DNA1some), which were fully characterized using various spectroscopic, microscopic, and light scattering analyses, with the details of the analysis procedures also reported in our recent paper.40 The self-assembly of DNA1 (5 μM) in the presence of the pH-regulator TAM (10 μM) resulted in the formation of TAM-loaded DNA1some (DNA1some/TAM). The loading of TAM in the hydrophobic membrane of DNA1some was probed by UV-vis absorption spectroscopy by comparing the absorbance of a concentration-matched solution of free TAM with the TAM concentration of the filtrate of DNA1some/TAM (Fig. 1a).The absorption spectrum of free TAM (10 μM in MeOH) showed three major bands at 210, 250, and 290 nm. On the other hand, a significant reduction in the intensity of the absorption bands was observed for the filtrate obtained after the centrifugal filtration of DNA1some/TAM (DNA1some = 5 μM in H2O, TAM = 10 μM in MeOH) using a molecular weight cut-off filter. This reduction in intensity for the filtrate of DNA1some/TAM clearly indicated the efficient loading of TAM into the hydrophobic membrane of DNA1some. The loading efficiency of TAM was calculated to be ∼80%. Transmission electron microscopy (TEM) (Fig. 1b) and atomic force microscopy (AFM) (Fig. 1c) analyses were applied to reveal the vesicular morphology of DNA1some/TAM (DNA1some = 5 μM in H2O, TAM = 10 μM in MeOH), indicating that the vesicular morphology of DNA1some was retained even after loading TAM. The average diameter of the vesicles of DNA1some/TAM was found to be ∼200 nm, which was nearly the same as that of the diameter of DNA1some alone, suggesting that the loading of TAM did not affect the diameter of DNA1some. In support of the microscopic analyses, dynamic light scattering (DLS) analysis of DNA1some/TAM showed a unimodal distribution of spherical aggerates with an average diameter of ∼200 nm (Fig. 1d). These results collectively allowed concluding that the self-assembly of DNA1 in the presence of TAM resulted in the formation of TAM-loaded DNA1some, with TAM most likely residing in the hydrophobic membrane of DNA1some.
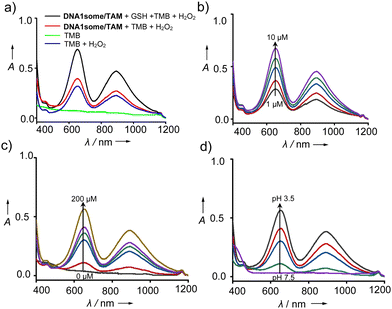
After the characterization of DNA1some/TAM, we evaluated the peroxidase-like (POD) activity of DNA1some/TAM, and found it was due to the presence of Cu2+ in the salen complex. According to our design, Cu2+ is in situ reduced to Cu+ by GSH at acidic pH, and then reacts with H2O2 to form ˙OH.40 Tetramethylbenzidine (TMB) was chosen as the substrate to probe the POD activity of DNA1some/TAM. For this, DNA1some/TAM ([DNA1some] = 20 μM, [TAM] = 10 μM) was treated with TMB (500 μM) in the presence of H2O2 (200 μM) and GSH (20 μM) at pH 5.0, and the emergence of the peak corresponding to oxidized TMB (ox-TMB) at 650 nm was monitored. As shown in Fig. 2a, the most intense peak of ox-TMB was observed for DNA1some/TAM and H2O2 in the presence of GSH compared to DNA1some/TAM and H2O2 in the absence of GSH, revealing the excellent POD activity of DNA1some/TAM in the presence of GSH. Moreover, DNA1some/TAM exhibited good activity even at a low concentration (1 μM) of DNA1some (Fig. 2b). As expected, the catalytic activity was found to increase with the increase in H2O2 concentration (Fig. 2c).
Next, we evaluated the effect of pH on the catalytic activity of DNA1some/TAM. Typically, the Fenton reaction requires acidic pH 3.0–5.0 to facilitate the catalytic action. When DNA1some/TAM was incubated in sodium acetate buffer with different pH values, the absorption peak of ox-TMB at 650 nm was found to increase with the decrease in pH. The maximum POD-like activity was observed at pH 3.5, while the lowest activity was observed at pH 7.5. These results strongly suggest that DNA1some/TAM exhibits excellent POD-like activity in an acidic pH environment (Fig. 2d).
After demonstrating the encapsulation of TAM inside DNA1some and the POD-mimicking activity of DNA1some/TAM, the cellular internalization of DNA1some/TAM was studied using MDA-MB-231 as a representative cancer cell. Towards this, a G-quadruplex-based cell-targeting aptamer DNA2 (5′-GGTGGTGGTGGTTGGGTGGTGGTGGAAAAAAAAA-3′) was integrated onto the surface of DNA1some/TAMvia sequence-specific DNA hybridization to yield DNA1some/TAM/DNA2. The DNA2 integrated on the surface of DNA1some/TAM can then act as a targeting ligand, targeting the membrane-overexpressed nucleolin proteins on the surface of MDA-MB-231 cells and allowing the selective internalization of DNA1some/TAM into MDA-MB-231 cells compared to healthy cells. In order to monitor the cellular internalization, DNA2 was fluorescently labelled with FAM at the 5′-end (DNA3) and the green fluorescence of DNA3 was used to investigate the internalization of the nanoformulation. For this, DNA1some/TAM/DNA3 ([DNA1] = 20 μM, [TAM] = 20 μM, [DNA3] = 1 μM) was incubated with MDA-MB-231 cells for 12 h and the internalization of the nanoformulation was studied by confocal laser scanning microscopy (CLSM). The CLSM images showed a strong overlay of the green fluorescence of DNA1some/TAM/DNA3 with the red fluorescence of lysotracker from the lysosome of the cells. This clearly suggests the lysosomal entrapment of DNA1some/TAM/DNA3 immediately after nucleolin-targeted receptor-mediated endocytosis (Fig. 3a). In support of this, the corresponding line analyses revealed the excellent colocalization of DNA1some/TAM/DNA3 with lysosome, with a high Pearson's correlation coefficient of 0.724 (Fig. S4, ESI†).
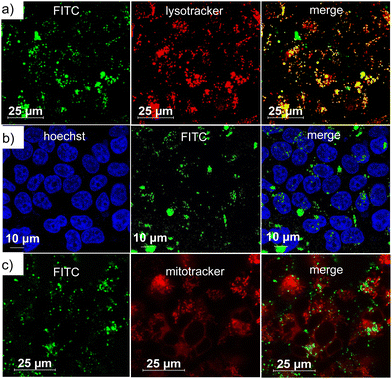 | ||
| Fig. 3 CLSM images of DNA1some/TAM/DNA3-treated MDA-MB-231 cells: (a) lysosomal, (b) nuclear and (c) mitochondrial colocalization of DNA1some/TAM/DNA3. | ||
In order to understand whether the nanoformulation undergoes localization in any other cell organelles, such as mitochondria and nucleus, colocalization experiments were performed after staining the nucleus and the mitochondria. The nucleus of MDA-MB-231 cells was stained with Hoechst (blue fluorescence) and mitochondria with MitoTracker deep red (red fluorescence). The corresponding CLSM images clearly revealed no colocalization of the green fluorescence of DNA1some/TAM/DNA3 with the blue fluorescence of Hoechst (Fig. 3b) and the red fluorescence of MitoTracker deep red (Fig. 3c). In accordance with these, very low Pearson's correlation coefficients were observed for the colocalization of the mitochondria (0.149) and nucleus (0.012) (Fig. S5 and S6, ESI†). These results verified the specific lysosomal entrapment of DNA1some/TAM/DNA3 and its subsequent degradation at the lysosome to release free Cu2+ and TAM. The selective internalization of DNA1some/TAM/DNA3 into nucleolin overexpressed cancer cells was then studied by comparing the cellular internalization efficiency of the nanoformulation between MDA-MB-231 (nucleolin overexpressed cancer cells) and HEK-293T (healthy cells) cell lines.42 The CLSM analyses disclosed an intense green fluorescence for the DNA1some/TAM/DNA3-treated MDA-MB-231 cells, whereas only a negligible green fluorescence was associated with the DNA1some/TAM/DNA3-treated HEK-293T cells (Fig. 4a). This clearly indicates the selective internalization of the nanoformulation into nucleolin overexpressed cancer cells.
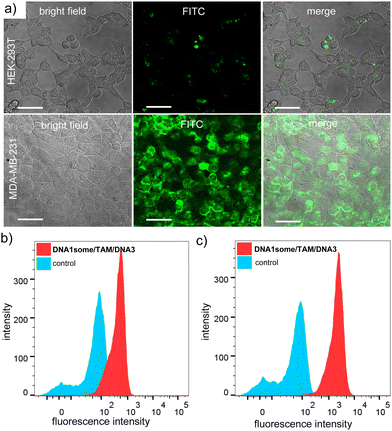 | ||
| Fig. 4 (a) CLSM images of DNA1some/TAM/DNA3-treated HEK-293T and MDA-MB-231 cells and (b) and (c) corresponding FACS analyses for HEK-293T and MDA-MB-231 cells. | ||
In support of this, the corresponding fluorescence-activated cell sorting (FACS) analyses revealed the high mean fluorescence intensity (MFI) shift for the DNA1some/TAM/DNA3-treated MDA-MB-231 cells (2153) when compared to the corresponding HEK-293T cells (436) (Fig. 4b). Furthermore, the cellular internalization of DNA1some/TAM/DNA3 was found to be directly proportional to the relative expression level of nucleolin on the cell membrane. In order to validate this, the efficiency of the cellular internalization of DNA1some/TAM/DNA3 was studied using three different cell lines having different nucleolin expression levels, namely MCF-7 (high nucleolin expression), HeLa (medium nucleolin expression), and HEK-293T (low nucleolin expression) cells.43 Accordingly, the CLSM analyses revealed the highest fluorescence intensity for MCF-7 cells, moderate intensity for HeLa cells, and the lowest intensity for HEK-293T cells; clearly revealing that the cellular internalization of DNA1some/TAM/DNA3 was directly proportional to the expression level of nucleolin on the membrane of the cells (Fig. S7, ESI†).
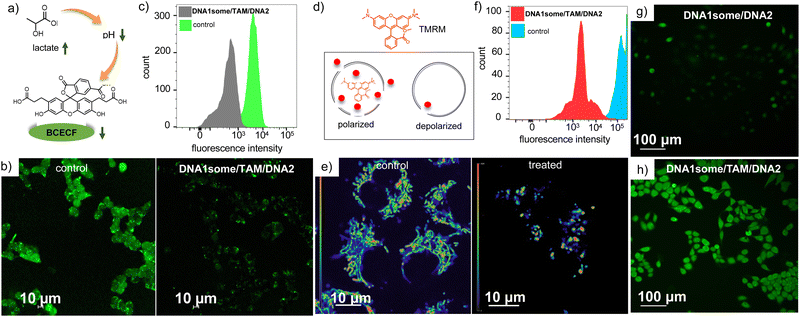
The pH-regulator TAM was incorporated into the nanoformulation to decrease the pH of the TME, and thereby enhance the efficacy of CDT action. To investigate the ability of DNA1some/TAM/DNA2 to decrease intracellular pH, MDA-MB-231 cells were incubated with DNA1some/TAM/DNA2 ([DNA1] = 20 μM, [TAM] = 20 μM, [DNA2] = 1 μM) and the decrease in intracellular pH was probed using a fluorescent pH indicator, namely 2′,7′-bis-(2-carboxyethyl)-5-(and-6)-carboxyfluorescein acetoxy-methyl ester (BCECF); whereby a decrease in the fluorescence intensity of BCECF-AM would be indicative of the decrease in intracellular pH. In accordance with our design, the CLSM images of DNA1some/TAM/DNA2-treated MDA-MB-231 cells showed a lower green fluorescence intensity compared to the untreated control cells (Fig. 5b). This result indicates that TAM released from DNA1some/TAM/DNA2 acts as an inhibitor of mitochondrial complex I, which enhances glycolysis and the lactate content and increases the acidity of TME. This was subsequently confirmed and quantified by FACS analyses, which displayed a lower MFI shift for DNA1some/TAM/DNA2-treated cells (502) compared to the untreated control cells (3889) (Fig. 5c).
We have previously reported that DNA1some alone could not induce any mitochondrial damage.40 In order to understand whether the in situ-released TAM can cause any mitochondrial damage as a result of the inhibition of mitochondrial complex I, a tetramethylrhodamine methyl ester perchlorate (TMRM) assay was performed on DNA1some/TAM/DNA2 ([DNA1some] = 20 μM, [TAM] = 20 μM, [DNA2] = 1 μM)-treated MDA-MB-231 cells. TMRM binds strongly to healthy and polarized mitochondria, whereas it binds weakly to unhealthy and depolarized mitochondria and hence a reduction in the florescence intensity of TMRM is an indication of damaged mitochondria. A significant decrease in TMRM fluorescence was observed for DNA1some/TAM/DNA2-treated cells when compared to the corresponding untreated control cells, revealing mitochondrial damage due to the release of TAM (Fig. 5e). This was further supported through FACS analyses, which exhibited a significant reduction in the MFI shift value for the DNA1some/TAM/DNA2-treated cells (3780) compared to the corresponding untreated controlled cells (129676) (Fig. 5f).
Having achieved a TAM-induced acidic TME inside the MDA-MB-231 cells, we evaluated the CDT action of DNA1some/TAM/DNA2 ([DNA1some] = 20 μM, [TAM] = 20 μM, [DNA2] = 1 μM) using 2,7-dichlorofluorescein-diacetate (DCFH-DA) as a fluorescent probe for the detection of ROS. This was achieved by probing the formation of green fluorescent 2,7-dichlorofluorescein (DCF) upon the reaction of ROS with DCFH-DA. As shown in Fig. 5g and h, the DNA1some/TAM/DNA2-treated MDA-MB-231 cells showed stronger DCF green fluorescence than the DNA1some/DNA2-treated cells. FACS analyses also revealed a high MFI shift for the DNA1some/TAM/DNA2-treated cells (620) compared to the DNA2/DNA1some-treated cells (99). These results indicate the enhanced ROS generation for the DNA1some/TAM/DNA2-treated cells due to the increased acidity induced by the release of TAM.
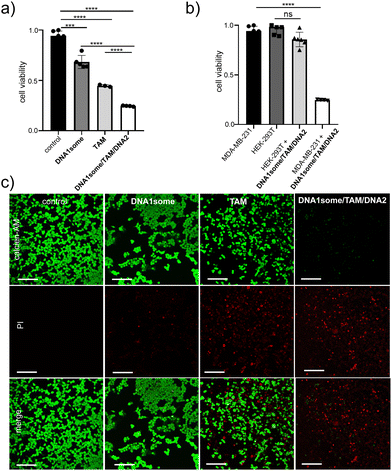
Subsequently, we tested the cytotoxicity of DNA1some/TAM/DNA2 ([DNA1some] = 20 μM, [TAM] = 20 μM, [DNA2] = 1 μM) via a methyl thiazolyl tetrazolium (MTT) assay. As expected, enhanced cytotoxicity was observed for DNA1some/TAM/DNA2 compared to DNA1some and TAM, revealing the synergetic combination of DNA1some and TAM (Fig. 6a). Cell deaths of 28% and 54% were observed for the DNA1some- and TAM-treated cells, respectively. On the other hand, a higher cell death of 74% was observed for the DNA1some/TAM/DNA2-treated sample. Very interestingly, only negligible cytotoxicity (13%) was observed for the DNA1some/TAM/DNA2-treated HEK-293T cells, indicating the high selectivity of the nanoformulation due to the presence of the aptamer DNA (DNA2) on the surface of the nanoformulation (Fig. 6b). The cytotoxicity was further visualized by calcein-AM/propidium iodide (PI) co-staining assay. Calcein-AM stains and imparts green fluorescence for viable cells, whereas PI stains the dead cells and imparts red fluorescence.44–46 In accordance with the MTT assay, the DNA1some/TAM/DNA2 ([DNA1some] = 20 μM, [TAM] = 20 μM, [DNA2] = 1 μM)-treated MDA-MB-231 cells exhibited mostly red fluorescent cells compared to the DNA1some- and TAM-treated cells, further confirming the excellent cytotoxicity of DNA1some/TAM/DNA2 (Fig. 6c).
Subsequently, an annexin V-FITC (AV)/PI assay was performed to understand the mechanism of cell death. Annexin V-FITC binds to the cell membrane during the early stages of apoptosis and gives rise to green fluorescence around the cell membrane. In contrast, PI binds to the nucleus at the late stage of apoptosis and gives rise to red fluorescence. For this, MDA-MB-231 cells were treated with DNA1some/TAM/DNA2 ([DNA1some] = 20 μM, [TAM] = 20 μM, [DNA2] = 1 μM) for 12 h and analysed by CLSM and FACS. The CLSM images clearly showed green and red fluorescence at the membrane and the nucleus of the cells, respectively, implying the apoptotic pathway of cell death (Fig. 7a). In accordance with this, the FACS studies showed the shift of the cell population to the quadrant corresponding to the apoptotic pathway (Fig. 7b). These results allowed concluding that DNA1some/TAM/DNA2 induces cell death via the apoptotic pathway.
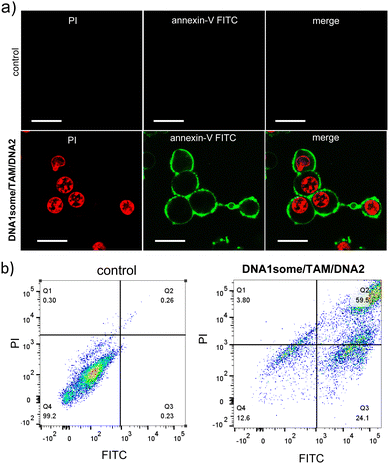 | ||
| Fig. 7 (a) Annexin V-FITC/PI assay of DNA1some/TAM/DNA2-treated MDA-MB-231 cells and (b) the corresponding FACS analyses. Scale bars correspond to 25 μm. | ||
After demonstrating the in vitro performance of DNA1some/TAM/DNA2, we studied the efficiency of the CDT agent in a multicellular tumour spheroid model using a 3D cell culture. The 3D-multicellular tumour spheroid mimics the in vivo tumour, and it undergoes proliferation in all directions similar to malignant tumours. In 3D cell culture, the cells clump together to form a stable 3D spheroid-like structure, wherein the communication and signalling between the cells are maximized, leading to efficient proliferation and invasion over time.47–49 In our study of triple-negative human breast cancer cells, MDA-MB-231 was used to prepare tumour 3D spheroids using the hanging drop method.50,51 The invasion potential of the spheroids was then studied in the presence of DNA1some/TAM/DNA2 to understand the effect on the spheroid invasion. For this, 3D spheroids were incubated with DNA1some/TAM/DNA2 ([DNA1some] = 20 μM, [TAM] = 20 μM, [DNA2] = 1 μM) at 37 °C for 24 h. Untreated spheroids were considered as the controls and showed the maximum invasion potential as they exhibited migration in all directions. On the other hand, the invasion was significantly reduced for the DNA1some/TAM/DNA2-treated sample due to the combined therapeutic actions. A nearly nil invasion index was observed for the DNA1some/TAM/DNA2-treated sample compared to the untreated sample, which showed an invasion index of 1.92 ± 0.5 (Fig. 8).
 | ||
| Fig. 8 CLSM images of the DNA2/TAM/DNA1some-treated MDA-MB-231 3D spheroids (left) and the corresponding untreated control spheroids (right). | ||
Conclusions
In summary, we report a CDT nanoagent loaded with a pH regulator for the improvement of CDT action by decreasing the pH at the TME for targeted cancer therapy. Self-assembly of a DNA amphiphile containing a Fenton reaction centre as the hydrophobic segment resulted in the formation of CDT-active DNAsome. One of the unique features of DNAsome is the dense surface decoration of ssDNA with a defined sequence, which permitted the integration of the DNA aptamer for nucleolin onto the surface of the DNAsome via sequence-specific DNA hybridization. The aptamer-decorated DNAsome exhibited excellent targeted internalization towards nucleolin overexpressed cancer cells, whereas negligible internalization was observed for normal cell lines, indicating the significantly reduced “off-target” toxicity to the surrounding normal cells. The cytotoxicity of the CDT agent was found to be significantly increased by the loading of the pH regulator and excellent targeted cytotoxicity was shown by the nanoformulation when compared to the CDT agent alone. This was attributed to the efficient Fenton reaction due to the acidic TME caused by the action of the pH regulator. We presented a simple and promising strategy to increase the efficiency of CDT action of CDT-active DNAsome by the non-covalent incorporation of a pH regulator into the hydrophobic membrane of the DNAsome. We strongly feel that this strategy would definitely help in the advancement of CDT-based targeted cancer therapy and will also encourage other researchers in this area to explore this approach for the advancement of cancer therapy in general.Author contributions
The manuscript was written through the contributions of all authors. All authors have given approval to the final version of the manuscript.Data availability
The data supporting this article have been included as part of the ESI.†Conflicts of interest
There are no conflicts to declare.Acknowledgements
Financial support from SERB (CRG/2022/002612) is gratefully acknowledged. The help of Sarika Mohan S. is acknowledged for the FACS analyses.References
- Z. Tang, Y. Liu, M. He and W. Bu, Angew. Chem., Int. Ed., 2019, 58, 946–956 CrossRef CAS PubMed.
- K. Wei, Y. Wu, X. Zheng, L. Ouyang, G. Ma, C. Ji and M. Yin, Angew. Chem., Int. Ed., 2024, 63, e202404395 CrossRef CAS PubMed.
- C. Zheng, Z. Wang, H. Xu, H. Huang, X. Tao, Y. Hu, Y. He, Z. Zhang and X. Huang, Small Methods, 2024, 8, 2301099 CrossRef CAS PubMed.
- B. Zhao, Z. Ma, S. Ding, Y. Cao, J. Du, L. Zeng, Y. Hu, J. Zhou, X. Zhang, X. Bian and G. Tian, Adv. Funct. Mater., 2023, 33, 2306328 CrossRef CAS.
- L. Zhang, C.-X. Li, S.-S. Wan and X.-Z. Zhang, Adv. Healthcare Mater., 2022, 11, 2101971 CrossRef CAS PubMed.
- A. Silswal and A. L. Koner, Chem. Commun., 2023, 59, 1769–1772 RSC.
- M. Peng, E. Ju, Y. Xu, Y. Wang, S. Lv, D. Shao, H. Wang, Y. Tao, Y. Zheng and M. Li, NPG Asia Mater., 2022, 14, 95 CrossRef CAS.
- H. Lin, Y. Chen and J. Shi, Chem. Soc. Rev., 2018, 47, 1938–1958 RSC.
- C. Jia, Y. Guo and F.-G. Wu, Small, 2022, 18, 2103868 CrossRef CAS PubMed.
- C. Cao, X. Wang, N. Yang, X. Song and X. Dong, Chem. Sci., 2022, 13, 863–889 RSC.
- L.-S. Lin, J. Song, L. Song, K. Ke, Y. Liu, Z. Zhou, Z. Shen, J. Li, Z. Yang, W. Tang, G. Niu, H.-H. Yang and X. Chen, Angew. Chem., Int. Ed., 2018, 57, 4902–4906 CrossRef CAS PubMed.
- L. Shi, Y. Wang, C. Zhang, Y. Zhao, C. Lu, B. Yin, Y. Yang, X. Gong, L. Teng, Y. Liu, X. Zhang and G. Song, Angew. Chem., Int. Ed., 2021, 60, 9562–9572 CrossRef CAS PubMed.
- S. Fu, R. Yang, L. Zhang, W. Liu, G. Du, Y. Cao, Z. Xu, H. Cui, Y. Kang and P. Xue, Biomaterials, 2020, 257, 120279 CrossRef CAS PubMed.
- Y. Liu, J. Wu, Y. Jin, W. Zhen, Y. Wang, J. Liu, L. Jin, S. Zhang, Y. Zhao, S. Song, Y. Yang and H. Zhang, Adv. Funct. Mater., 2019, 29, 1904678 CrossRef CAS.
- B. Lin, H. Chen, D. Liang, W. Lin, X. Qi, H. Liu and X. Deng, ACS Appl. Mater. Interfaces, 2019, 11, 11157–11166 CrossRef CAS PubMed.
- B. Ma, S. Wang, F. Liu, S. Zhang, J. Duan, Z. Li, Y. Kong, Y. Sang, H. Liu, W. Bu and L. Li, J. Am. Chem. Soc., 2019, 141, 849–857 CrossRef CAS PubMed.
- L. Lin, J. Yu, H. Lu, Z. Wei, Z. Chao, Z. Wang, W. Wu, H. Jiang and L. Tian, Chem. Commun., 2021, 57, 1734–1737 RSC.
- M. Li, C. Wang, Z. Di, H. Li, J. Zhang, W. Xue, M. Zhao, K. Zhang, Y. Zhao and L. Li, Angew. Chem., Int. Ed., 2019, 58, 1350–1354 CrossRef CAS PubMed.
- C. Liu, Y. Chen, J. Zhao, Y. Wang, Y. Shao, Z. Gu, L. Li and Y. Zhao, Angew. Chem., Int. Ed., 2021, 60, 14324–14328 CrossRef CAS PubMed.
- C. Liu, S. Jia, L. Tu, P. Yang, Y. Wang, S. Ke, W. Shi and S. Ye, ACS Biomater. Sci. Eng., 2022, 8, 1942–1955 CrossRef CAS PubMed.
- C.-K. Sun, Y.-H. Wang, Y.-L. Chen, T.-Y. Lu, H.-Y. Chen, S.-C. Pan, P.-C. Chen, M.-Y. Liao and J. Yu, Sci. Rep., 2022, 12, 18729 CrossRef CAS PubMed.
- W.-X. Zhang, Y.-N. Hao, Y.-R. Gao, Y. Shu and J.-H. Wang, ACS Appl. Mater. Interfaces, 2021, 13, 38127–38137 CrossRef CAS PubMed.
- M. Chen, S. Zhao, J. Zhu, E. Feng, F. Lv, W. Chen, S. Lv, Y. Wu, X. Peng and F. Song, ACS Appl. Mater. Interfaces, 2022, 14, 20682–20692 CrossRef CAS PubMed.
- Q. Li, J. Yu, L. Lin, Y. Zhu, Z. Wei, F. Wan, X. Zhang, F. He and L. Tian, ACS Appl. Mater. Interfaces, 2023, 15, 16482–16491 CrossRef CAS PubMed.
- W. B. Dirersa, T.-C. Kan, J. Chang, G. Getachew, S. Ochirbat, S. Kizhepat, A. Wibrianto, A. Rasal, H.-A. Chen, A. V. Ghule, T.-H. Chou and J.-Y. Chang, ACS Appl. Mater. Interfaces, 2024, 16, 24172–24190 CrossRef CAS PubMed.
- Q. Yu, J. Zhou, Y. Liu, X. Q. Li, S. Li, H. Zhou, B. Kang, H.-Y. Chen and J.-J. Xu, Adv. Healthcare. Mater., 2023, 12, 2301429 CrossRef CAS PubMed.
- Y.-N. Hao, W.-X. Zhang, Y.-R. Gao, Y.-N. Wei, Y. Shu and J.-H. Wang, J. Mater. Chem. B, 2021, 9, 250–266 RSC.
- Y. You, H. Liu, J. Zhu, Y. Wang, F. Pu, J. Ren and X. Qu, Chem. Sci., 2022, 13, 7829–7836 RSC.
- Q. Li, F. Wang, L. Shi, Q. Tang, B. Li, X. Wang and Y. Jin, ACS Appl. Mater. Interfaces, 2022, 14, 37280–37290 CrossRef CAS PubMed.
- L. Zhang, R. Abdullah, X. Hu, H. Bai, H. Fan, L. He, H. Liang, J. Zou, Y. Liu, Y. Sun, X. Zhang and W. Tan, J. Am. Chem. Soc., 2019, 141, 4282–4290 CrossRef CAS PubMed.
- C. Ji, H. Li, L. Zhang, P. Wang, Y. Lv, Z. Sun, J. Tan, Q. Yuan and W. Tan, Angew. Chem., Int. Ed., 2022, 61, e202200237 CrossRef CAS PubMed.
- J. Tan, H. Li, X. Hu, R. Abdullah, S. Xie, L. Zhang, M. Zhao, Q. Luo, Y. Li, Z. Sun, Q. Yuan and W. Tan, Chem, 2019, 5, 1775–1792 CAS.
- Y. Ouyang, M. Fadeev, P. Zhang, R. Carmieli, J. Li, Y. S. Sohn, O. Karmi, R. Nechushtai, E. Pikarsky, C. Fan and I. Willner, ACS Nano, 2022, 16, 18232–18243 CrossRef CAS PubMed.
- W. Song, P. Song, Y. Sun, Z. Zhang, H. Zhou, X. Zhang and P. He, ACS Biomater. Sci. Eng., 2021, 7, 5165–5174 CrossRef CAS PubMed.
- W. Xuan, Y. Xia, T. Li, L. Wang, Y. Liu and W. Tan, J. Am. Chem. Soc., 2020, 142, 937–944 CrossRef CAS PubMed.
- C. Yao, H. Qi, X. Jia, Y. Xu, Z. Tong, Z. Gu and D. Yang, Angew. Chem., Int. Ed., 2022, 61, e202113619 CrossRef CAS PubMed.
- Q. Tang, Q. Li, L. Shi, W. Liu, B. Li and Y. Jin, Nanoscale Horiz., 2023, 8, 1106–1112 RSC.
- W. Tang, L. Han, X. Lu, Z. Wang, F. Liu, Y. Li, S. Liu, S. Liu, R. Tian, J. Liu and B. Ding, ACS Appl. Mater. Interfaces, 2021, 13, 20974–20981 CrossRef CAS PubMed.
- J. Y. Lee, Q. Yang, X. Chang, H. Wisniewski, T. R. Olivera, M. Saji, S. Kim, D. Perumal and F. Zhang, J. Mater. Chem. B, 2022, 10, 7460–7472 RSC.
- G. Raj, A. P. Vasantha, V. D. Sreekumar, A. V. Beena, V. K. K. Dommeti, H. Perozhy, A. T. Jose, S. Khurana and R. Varghese, Adv. Healthcare Mater., 2024, 2400256 CrossRef CAS PubMed.
- V. C. Jordan, Nat. Rev. Drug Discovery, 2003, 2, 205–213 CrossRef CAS PubMed.
- J. He, T. Peng, Y. Peng, L. Ai, Z. Deng, X.-Q. Wang and W. Tan, J. Am. Chem. Soc., 2020, 142, 2699–2703 CrossRef CAS PubMed.
- S. Sun, Y. Yang, Z. Gao, H. Jiang, L. Ye, Y. Lai, Z. Shen and Z.-S. Wu, ACS Appl. Mater. Interfaces, 2022, 14, 45201–45216 CrossRef CAS PubMed.
- A. Konieva, V. Deineka, K. Diedkova, D. Aguilar-Ferrer, M. Lyndin, G. Wennemuth, V. Korniienko, S. Kyrylenko, A. Lihachev, V. Zahorodna, I. Baginskiy, E. Coy, O. Gogotsi, A. Blacha-Grzechnik, W. Simka, I. Kube-Golovin, I. Iatsunskyi and M. Pogorielov, ACS Appl. Mater. Interfaces, 2024, 16, 43302–43316 CrossRef CAS PubMed.
- A. Joe, P. Manivasagan, J. K. Park, H.-W. Han, S.-H. Seo, T. Thambi, V. H. Giang Phan, S. A. Kang, J. Conde and E.-S. Jang, ACS Nano, 2024, 18, 19581–19596 CAS.
- T. Xia, Z. Xia, P. Tang, J. Fan and X. Peng, J. Am. Chem. Soc., 2024, 146, 12941–12949 CrossRef CAS PubMed.
- A. Aung, S. K. Davey, J. Theprungsirikul, V. Kumar and S. Varghese, Adv. Healthcare Mater., 2023, 12, 2201842 CrossRef CAS PubMed.
- L. Yu, Y. Xu, M. Al-Amin, S. Jiang, M. Sample, A. Prasad, N. Stephanopoulos, P. Šulc and H. Yan, J. Am. Chem. Soc., 2023, 145, 27336–27347 CrossRef CAS PubMed.
- M. L. Janssen, T. Liu, M. Özel, M. Bril, H. V. Prasad Thelu and R. E. Kieltyka, Angew. Chem., Int. Ed., 2024, 63, e202314738 CrossRef CAS PubMed.
- H. NaveenaA and D. D. Bhatia, ChemBioChem, 2023, 24, e202300506 CrossRef PubMed.
- A. Rajwar, S. R. Shetty, P. Vaswani, V. Morya, A. Barai, S. Sen, M. Sonawane and D. Bhatia, ACS Nano, 2022, 16, 10496–10508 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4ma00822g |
| This journal is © The Royal Society of Chemistry 2024 |