Fast proton transport enables the magnetic relaxation response of graphene quantum dots for monitoring the oxidative environment in vivo†
Yongqiang
Li‡
 ab,
Hang
Wang‡
ab,
Hang
Wang‡
 ab,
Caichao
Ye
ab,
Caichao
Ye
 c,
Xuelian
Wang
d,
Peng
He
ab,
Siwei
Yang
c,
Xuelian
Wang
d,
Peng
He
ab,
Siwei
Yang
 *ab,
Hui
Dong
*ab,
Hui
Dong
 *ab and
Guqiao
Ding
*ab and
Guqiao
Ding
 *ab
*ab
aState Key Laboratory of Materials for Integrated Circuits, Shanghai Institute of Microsystem and Information Technology (SIMIT), Chinese Academy of Sciences (CAS), Shanghai 200050, People's Republic of China. E-mail: yangsiwei@mail.sim.ac.cn; donghui@mail.sim.ac.cn; gqding@mail.sim.ac.cn
bCenter of Materials Science and Optoelectronics Engineering, University of Chinese Academy of Sciences (UCAS), Beijing 100049, People's Republic of China
cAcademy for Advanced Interdisciplinary Studies & Department of Materials Science and Engineering, Guangdong Provincial Key Laboratory of Computational Science and Material Design, Southern University of Science and Technology, Shenzhen, Guangdong 518055, People's Republic of China
dDepartment of Cardiology, Ruijin Hospital, Shanghai Jiaotong University School of Medicine, Shanghai 200025, People's Republic of China
First published on 27th December 2023
Abstract
A magnetic relaxation switch (MRS) that targets small molecules such as H2O2 is difficult to realize because of the small size of the targets, which cannot gather enough MRS probes to form aggregates and generate a difference in magnetic relaxation times. Therefore, the development of small molecule-targeted MRS is strongly dependent on changes in the interfacial structure of the probe, which modulates the proton transport behavior near the probe. Herein, functionalized graphene quantum dots (GQDs) consisting of GQDs with disulfide bonds, polyethylene glycol (PEG), and paramagnetic Gd3+ were used as the MRS probe to sense H2O2. The structure of GQDs changed after reacting with H2O2. The PEG assembled a tube for transmitting changes in GQDs via proton transport and thus enabled the magnetic relaxation response of the probe towards H2O2. Pentaethylene glycol was experimentally and theoretically proven to have the strongest ability to transport protons. Such a probe can be applied in the differentiation of healthy and senescent cells/tissues using in vitro fluorescent imaging and in vivo magnetic resonance imaging. This work provides a reliable solution for building a proton transport route, which not only enables the response of the MRS probe towards the targets but also demonstrates the design of carbon nanostructures with proton transport behaviors.
Introduction
The use of a magnetic relaxation switch (MRS) belongs to a class of sensing techniques based on changes in magnetic relaxation times with and without target molecules.1–3 Compared to sensing techniques employing optical or electrical signals,4,5 MRS is advantageous in avoiding signal interfaces caused by the turbidity and background of the testing sample. In MRS, the technique used to record the signal is usually nuclear magnetic resonance (NMR) relaxometry, wherein the magnetic probe is taken as the basis to convert interactions among target molecules; moreover, it probes the changes in magnetic relaxation times.6,7 Therefore, the development of a target-specific, highly sensitive MRS probe is important for the future development of MRS technology.8–10According to the Solomon–Bloembergen–Morgan (SBM) theory that describes the magnetic relaxation of paramagnetic molecules,11 the magnetic relaxation of paramagnetic molecules is governed by the rotational dynamics of the nanoparticles (τR), the number of water molecules in the inner-coordination sphere (q), and the kinetics of water exchange (1/τm). Therefore, the sensing mechanism and the design of the MRS probe can be classified into two categories:
(I) Changing the state of aggregation of the MRS probe.8,12–15 The probe usually exhibits different aggregation states before and after interacting with the targets; the τR of the probe is different because of the size change of the probe, which leads to the change in magnetic relaxation time. Using this mechanism, the MRS probe can be designed to generate aggregation. Typically, antibodies are used to form the MRS probe, which specifically bind with the biomarkers and aggregate on the surface of target molecules. However, such a strategy is limited to designing MRS targets for small molecules (e.g., ions, anions, and H2O2) because of their poor aggregation capacity.
(II) Changing the interfacial properties of the MRS probe.16,17 The probe undergoes changes in the interfacial structure before and after contact with the targets, resulting in different proton transport behaviors near the probe. The proton transport behavior can be ascribed to q and 1/τm, both of which affect the magnetic relaxation time. This approach can be thus utilized to sense the small molecules that bring changes to the interfacial structures of the probe. Once the change in the interfacial properties of the MRS probe is no longer adequate to generate a strong change in the magnetic relaxation time, MRS will exhibit poor sensitivity. Therefore, enlarging the difference in proton transport of the MRS probe and the magnetic relaxation time are important to enhancing the sensitivity of MRS, which can be developed by modulating the structure of the MRS probe.
In this work, an antibody-free probe comprising graphene quantum dots (GQDs) and polyethylene glycol (PEG) was developed for MRS use based on the modulation of proton transport near paramagnetic nanoparticles. In the probe, GQDs were designed to change the structure specifically when sensing the oxidative environment. PEG was chosen to transmit the change in GQDs to the paramagnetic center (Gd3+) and then regulate the proton transport near Gd3+. Such a hybrid structure can achieve both fluorescence and magnetic relaxation responses when reacting with oxidative species (H2O2). The disulfide bonds in GQDs can be oxidized to form sulfoxide/sulfone structures, resulting in the suppression of fluorescent intensity. Moreover, PEG transmits the structural change in GQDs to the microenvironment of Gd3+ and provides a proton-transport tube, which leads to a decrease in the magnetic relaxation time. Based on differences between healthy and senescent cells/tissues, in vitro fluorescence experiments that differentiated healthy and senescent cells and in vivo MRI scanning that brought improved contrast were conducted.
Results and discussion
Design, characterization, and optimization of SGQDs
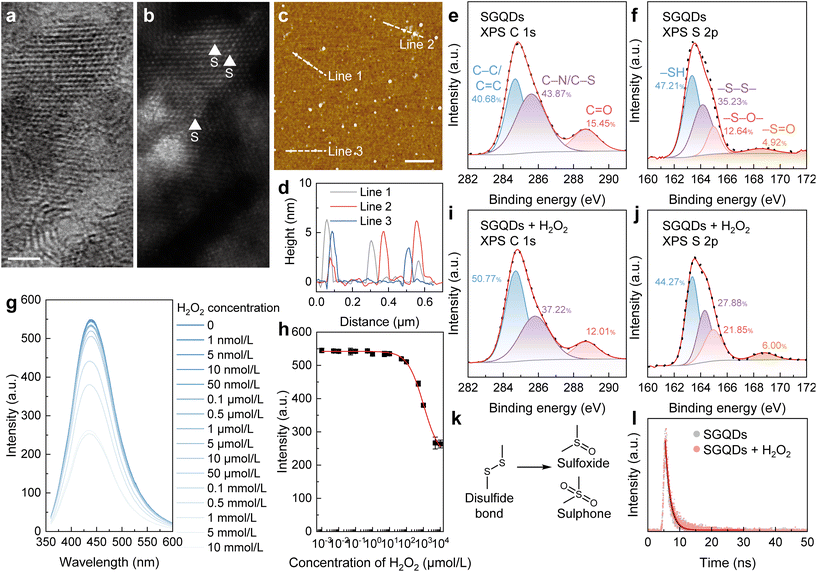
GQDs are zero-dimensional carbon materials at the nanometre scale.18 In the “bottom-up” synthesis of GQDs, their structures can be easily modulated by selecting factors such as the precursors, synthesis route, and after treatment.19 Disulfide bonds (–S–S–) widely exist in proteins, which are sensitive to the oxidative environment.20 With the tunable structures of GQDs and oxidation-sensitive disulfide bonds, GQDs with disulfide bonds (SGQDs) were synthesized using L-cysteine as the precursor (Fig. S1a†).Fig. S1b† shows the morphological image of SGQDs captured using transmission electron microscopy (TEM) with an average diameter of 8.89 nm (Fig. S1c†). Fig. S1d† reveals that the lattice spacing of SGQDs is 0.21 nm, corresponding to the [1120] lattice fringes of graphene. In the high-resolution TEM and the corresponding spherical aberration-corrected high-angle annular dark-field (HAADF) scanning TEM (STEM) images of SGQDs (Fig. 1a and b), the typical honeycomb structure and the presence of S atoms can be observed. Moreover, the height of the SGQDs, as evinced by atomic force microscopy (AFM), is 2 to 6 nm, corresponding to 6–18 graphene layers (Fig. 1c and d). The X-ray photoelectron spectroscopy (XPS) spectrum of SGQDs (Fig. S2†) indicates the elements in SGQDs are C, N, O, and S. Fig. 1e and f show the XPS C 1s and S 2p spectra of SGQDs, respectively. Peaks located at 284.69, 285.67, and 288.70 eV can be assigned to C–C/C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C, C–N/C–S, and C
C, C–N/C–S, and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O bonds, respectively, whereas peaks at 163.34, 164.14, 167.98, and 168.50 eV can assigned to the –SH (47.21%), –S–S– (35.23%), –S–O– (12.64%), and –S
O bonds, respectively, whereas peaks at 163.34, 164.14, 167.98, and 168.50 eV can assigned to the –SH (47.21%), –S–S– (35.23%), –S–O– (12.64%), and –S![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O (4.92%) bonds, respectively.
O (4.92%) bonds, respectively.
The SGQDs were synthesized by a hydrothermal treatment. The time of hydrothermal treatment affects the structure and thus the properties of SGQDs.21 As shown in Fig. S3a and b,† the fluorescence intensity of SGQDs increases with the increase of hydrothermal treatment time within 24 h and then remains constant with the additional reaction time. To study the changes in the fluorescence of SGQDs in an oxidative environment, H2O2 was used. SGQDs with different hydrothermal treatment times exhibited differences when reacting with H2O2. As can be seen in Fig. S3c–j,† SGQDs showed a decreased fluorescence intensity after the reaction with H2O2 but with varying degrees of reduction. Among the SGQDs, those synthesized over 12 h exhibited the greatest reduction in fluorescence intensity after reacting with H2O2. Therefore, 12 h of hydrothermal treatment was used to synthesize SGQDs. Moreover, the reaction time between SGQDs and H2O2 influences the degree of the reaction and therefore the fluorescence intensity of SGQDs (Fig. S4†). Next, H2O2 titration experiments were conducted to reveal the relationship between the fluorescence intensity and H2O2 concentration. As can be seen in Fig. 1g, when the H2O2 concentration increases, the fluorescence intensity decreases. The quantum yield (φ) of SGQDs with an H2O2 concentration of 0 was 0.42. To quantify the aforementioned relationship, the Hill coefficient (n = 0.92 ± 0.05, adjusted R2 = 0.99493) was determined (Fig. 1h). The Hill coefficient indicates negative cooperativity in the binding of one molecular component, suppressing the reaction of subsequent molecules on the other sites of SGQDs.22
Fluorescence sensing mechanism of SGQDs towards an oxidative environment
Structural differences in SGQDs before and after the reaction with H2O2 were revealed using XPS. As can be seen in Fig. 1i and j, the XPS C 1s spectrum of SGQDs + H2O2 shows no obvious change compared to that of SGQDs (Fig. 1e); however, the XPS S 2p spectrum shows a decrease in –S–S– bonding (27.88%) and an increase of –S–O– (21.85%) and –S![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O (6.00%) bonding, indicating the oxidation of –S–S– bond in SGQDs. The increased proportions of –S–O– and –S
O (6.00%) bonding, indicating the oxidation of –S–S– bond in SGQDs. The increased proportions of –S–O– and –S![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O bonds revealed that the –S–S– bond is oxidized to form sulfoxide/sulphone structures (Fig. 1k). Moreover, the fluorescent lifetime of SGQDs before and after reacting with H2O2 shows no difference (Fig. 1l, from 1.54 to 1.60 ns), indicating the suppression of the fluorescence intensity of SGQDs is static quenching.23
O bonds revealed that the –S–S– bond is oxidized to form sulfoxide/sulphone structures (Fig. 1k). Moreover, the fluorescent lifetime of SGQDs before and after reacting with H2O2 shows no difference (Fig. 1l, from 1.54 to 1.60 ns), indicating the suppression of the fluorescence intensity of SGQDs is static quenching.23
Design of SGQDs with a magnetic relaxation response for oxidative environment sensing
Based on the changes in the fluorescence intensity of SGQDs in an oxidative environment and the development of GQD-based paramagnetic nanoparticles, the SGQDs were designed to bind paramagnetic Gd3+ to sense H2O2 using a magnetic relaxation signal as the readout. A homemade ultra-low-field (ULF) NMR relaxometry was used to measure the longitudinal relaxation time (T1) of samples.24,25 Initially, Gd3+ was directly bound to the SGQDs using hydrothermal treatment (Fig. S5†). Unfortunately, the obtained SGQDs-Gd showed no significant change in T1 after reacting with H2O2 at a high concentration (Fig. S6a–c†) but exhibited a reduced fluorescence intensity (Fig. S6d†). This phenomenon is primarily attributed to the lack of a proton transport route from SGQDs to Gd3+.The pathways for transporting protons can be built by assembling carbon nanomaterials, including GQDs with other proton-conductive materials.26–29 To enable the change in T1 when SGQDs react with H2O2, PEG is introduced to connect SGQDs and Gd3+ because the oxygen-containing chain structure of PEG can build a bridge for proton transport.30,31 To synthesize SGQDs-PEG-Gd, hydrothermal treatment was undertaken (Fig. S7a†).24 The morphological and chemical characterizations of SGQD-pentaethylene glycol (PEG5) and SGQDs-PEG5-Gd are shown in Fig. S7b–e, S8, S9,† and Fig. 2a. Data in matrix-assisted laser desorption ionization time-of-flight mass (MALDI-TOF-MASS) spectra show that the Δm/z values between the peaks of SGQDs and SGQDs-PEG5 are 716.02, 950.83, 1186.52, 1426.59, 1663.95, 1913.10, 2147.55, and 2385.70 m/z, indicating that 3 to 10 PEG5 chains are connected to SGQDs. Moreover, the mass distribution pattern from the SGQDs-PEG5-Gd shifts to even higher masses by amounts equal to between one and three times the molecular weight of Gd3+, indicating that each SGQDs-PEG5 binds 1–3 Gd3+. By considering the characterization of SGQDs-PEG5-Gd, an illustration of SGQDs-PEG5-Gd is displayed in Fig. 2b. PEG5 chains are attached to the surface of SGQDs, and Gd3+ is bound to PEG5.
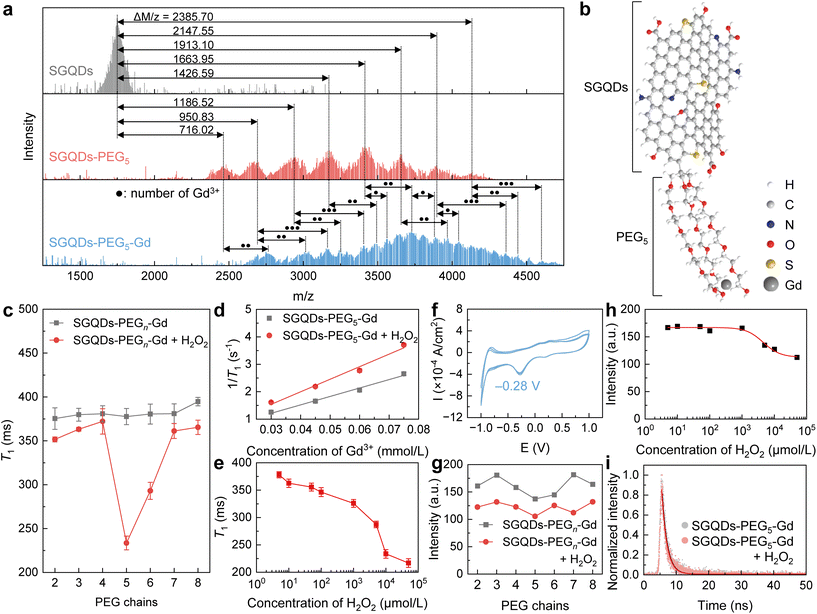 | ||
| Fig. 2 Characterization, magnetic, and optical responses of SGQDs-PEGn-Gd (n = 2–8). (a) MALDI-TOF-MASS spectra of SGQDs, SGQDs-PEG5, and SGQDs-PEG5-Gd. The solid black dots marked in the MALDI-TOF-MASS spectrum of SGQDs-PEG5-Gd represent the numbers of Gd3+. (b) Illustration of SGQDs-PEG5-Gd. (c) Comparison of the T1 values of SGQDs-PEGn-Gd (n = 2–8) before and after reacting with H2O2 (10 mmol L−1) for 12 h. Error bars indicate the fitting error of the T1 fitting. (d) The r1 fittings of SGQDs-PEG5-Gd before and after reacting with H2O2 (10 mmol L−1) for 12 h. Fittings of T1 are shown in Fig. S10.† (e) Change in T1 of SGQDs-PEG5-Gd after reacting with H2O2 at different concentrations. Error bars indicate the fitting error of T1 fitting. (f) The oxidation–reduction potential of SGQDs-PEG5-Gd. (g) Comparison of the fluorescence intensities of SGQDs-PEGn-Gd (n = 2–8) before and after reacting with H2O2 (10 mmol L−1) for 12 h. (h) Change in fluorescence intensity of SGQDs-PEG5-Gd after 12 h reacting at H2O2 with different concentrations. A Hill fit was used to fit the data. (i) Fluorescence lifetimes of SGQDs-PEG5-Gd and SGQDs-PEG5-Gd + H2O2. | ||
Next, SGQDs-PEGn-Gd (where PEGn represents HO(CH2CH2O)nH, n = 2–8) with different PEG chains were synthesized and their T1 values before and after reacting with H2O2 were obtained using ULF NMR relaxometry. As can be seen in Fig. 2c, H2O2 reduces the T1 of SGQDs-PEGn-Gd (n = 2–8). More importantly, SGQDs-PEG5-Gd showed the largest change in T1 (from 385.03 to 233.50 ms) after the reaction with H2O2. To quantify the change in T1, the longitudinal relaxivity (r1) of SGQDs-PEG5-Gd is shown in Fig. 2d. The r1 of SGQDs-PEG5-Gd before the reaction with H2O2 is 30.72 ± 2.22 L mmol−1 s−1 (adjusted R2 = 0.98446), which increased to 45.75 ± 4.09 L mmol−1 s−1 (adjusted R2 = 0.97637) after the reaction. Note that the Gd3+ aqueous solution shows no significant change in r1 before and after the reaction with H2O2 (Fig. S11†). Next, H2O2 with different concentrations was used to show the ability of SGQDs-PEG5-Gd to decrease T1 to different extents (Fig. 2e). The T1 of SGQDs-PEG5-Gd decreases (from 378.03 to 216.82 ms) after reacting with H2O2 at an increasing concentration (from 5 μmol L−1 to 50 mmol L−1). The above results indicate that the structural change in SGQDs can be reflected as the T1 change. The negative oxidation–reduction potential (−0.28 V, Fig. 2f) also proves that SGQDs-PEG5-Gd can be oxidized. After connecting PEG and Gd3+, the obtained SGQDs-PEGn-Gd (n = 2–8) still exhibit a fluorescent response. As can be seen in Fig. 2g and S12a–g,† all SGQDs-PEGn-Gd (n = 2–8) nanostructures undergo a reduction in fluorescence after reacting with H2O2. Among them, the fluorescence intensity of SGQDs-PEG5-Gd tended to decrease with an increasing concentration of H2O2 (Fig. 2h and S12h†). When the H2O2 concentration is 0, the value of φ of SGQDs-PEG5-Gd is 0.44, which is close to that of SGQDs, indicating that the structure of SGQDs remains unchanged after connecting PEG5 and Gd3+. The Hill coefficient (n = 1.65 ± 0.61, adjusted R2 = 0.96389) indicates an increased binding ability of SGQDs-PEG5-Gd to H2O2 compared with that of SGQDs (Fig. 1h). Moreover, the fluorescence lifetimes of SGQDs-PEG5-Gd before and after reacting with H2O2 were 1.60 and 1.59 ns, respectively. This is the same as SGQDs that possess a static quenching process.
Proton transport in a PEG tube
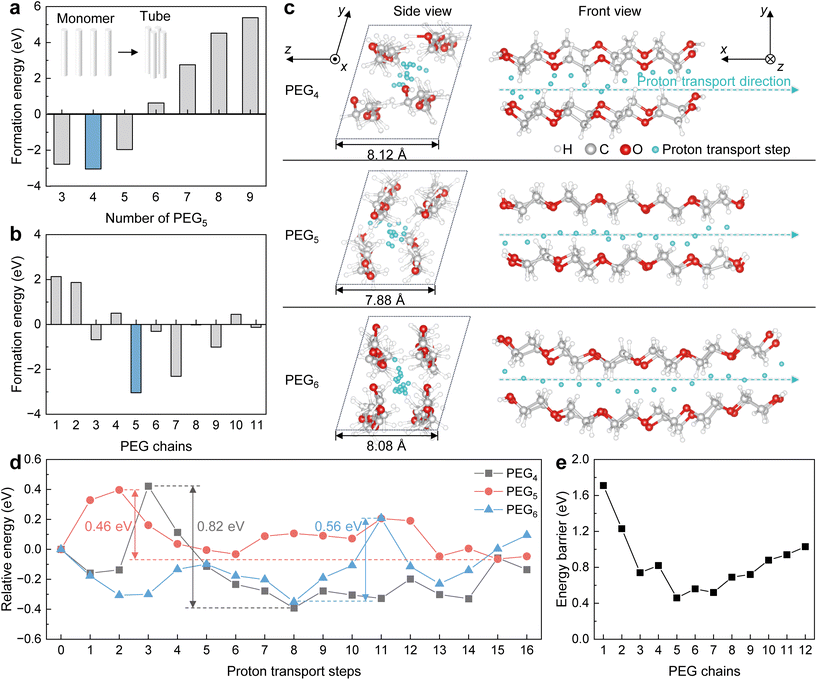
Thermodynamic and dynamic calculations based on density functional theory (DFT) were performed to demonstrate proton transport in a PEG tube. Before simulating the proton transport in a PEG tube, the assembly of PEG chains was optimized. In the thermodynamics simulation, PEG chains were initially assembled as a tube via the hydrogen bond and van der Waals force (inset illustration in Fig. 3a). To determine the optimal quantity of PEG that forms the tube, different quantities of PEG5 were used to simulate the formation energy. As can be seen in Fig. 3a, four PEG5 chains have the lowest formation energy to form the tube. PEGs with different chain lengths (1–11) were then adopted to simulate the formation energy during tube formation. The results show that PEG5 has the lowest formation energy when forming the tube (Fig. 4b).Under optimized conditions, proton transport within the PEG tube was simulated. As can be seen in Fig. 3c, protons can be transported in the tubular structure formed by PEG. In the side view of the tubular structures of PEGn (n = 4–6) tubes, the sizes of the tubular structures differ and the PEG5 tube is the smallest. Such a small tube leaves little room within (e.g. in a PEG5 tube), giving rise to rapid proton transport. In the front view of the proton transport processes of PEGn (n = 4–6) tubes, each cyan particle represents a step of proton transport in the tube. In the first step of the proton transport, the energy is set to 0, and the energy associated with each of the following steps is shown as the relative energy (Fig. 3d). In each step, the relative energy of the proton transport fluctuates, the proton transport in the PEG5 tube shows the smallest energy fluctuation compared to that in the PEG4 and PEG5 tubes. Such an energy fluctuation can be described using a maximum energy barrier. The maximum energy barrier for proton transport in the PEG tubes is calculated as the maximum relative energy minus the minimum relative energy during the proton transport. The PEG4, PEG5, and PEG6 tubes show maximum energy barriers of 0.82, 0.46, and 0.56 eV, respectively. Moreover, the tube formed using PEG5 also showed the lowest energy barrier in proton transport compared to PEG1 to PEG12 (Fig. 3e).
These results reveal that the PEG5 can form a tube to transport protons and provide the lowest energy barrier, which positively accelerates the proton transport process and accelerates the kinetics of water exchange.11 Such processes are beneficial to enable the magnetic relaxation responses of SGQDs-PEG5-Gd when reacting with H2O2, thereby generating a large difference in T1 before and after reacting with H2O2.
Monitoring the oxidative environment in vivo using SGQDs-PEG5-Gd
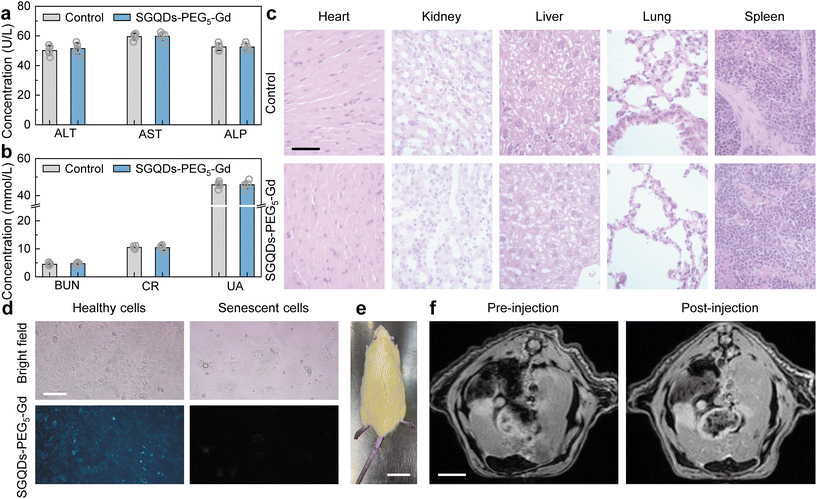
Before executing the bio-application of SGQDs-PEG5-Gd, the biotoxicity of SGQDs-PEG5-Gd should be assessed both in vitro and in vivo. The cell viability of various tumor and healthy cells demonstrated no significant changes in the cell survival rate (Fig. S13†). The effects of SGQDs-PEG5-Gd on liver and kidney functions were then studied by analyzing the blood from nude mice. Compared with the control group, there was no significant change in the hepatotoxicity index (Fig. 4a, serum alanine aminotransferase, ALT; aspartate aminotransferase, AST; alkaline phosphatase, ALP), and nephrotoxicity index (Fig. 4b, blood urea nitrogen, BUN; creatinine, CR; urate, UA) of the experimental group (Fig. 4a and b). Moreover, there was no obvious cell necrosis, cell apoptosis, or tissue inflammation in various tissue sections (Fig. 4c, liver, lung, spleen, kidney, and heart) of nude mice. Therefore, SGQDs-PEG5-Gd have low biological toxicity.Aging is a major risk factor for cognitive decline, cancer, cardiovascular disease, metabolic disease, sarcopenia, and frailty.32,33 At the cellular level, senescence is a permanent proliferative arrest with multiple phenotypic changes.34 Senescent cells possess a complex, multi-component senescent-associated secretory phenotype (SASP). SASP acts on the cell non-autonomously to alter the behavior of neighboring cells and the tissue microenvironment.35 The metabolic drivers of senescence mainly include mitochondrial dysfunction, oxygen, disrupted autophagy, hyperglycemia, loss of nicotinamide adenine dinucleotide (NAD+), and accumulation of metals.34 Among them, oxygen that oxidizes products of cell metabolism or known oxidative agents (e.g., H2O2) can cause senescence.36 Although oxidizing agents exert their effect partly through DNA damage, other cellular components and processes are also affected.37
Because the fluorescence of SGQDs-PEG5-Gd can be suppressed in an oxidative environment, it will display a dark region showing senescent cells or tissues. Fortunately, the T1 of SGQDs-PEG5-Gd reduces in the oxidative environment, resulting in a bright region to show the senescent parts in the magnetic resonance image (MRI) after injection. As shown in Fig. 4d, healthy and senescent human umbilical vein endothelial cells (HUVECs) display different cellular morphologies. The senescent cells possess a larger volume than healthy cells. After incubation with SGQDs-PEG5-Gd (100 μg mL−1) for 48 h, the healthy cells emitted blue fluorescence. However, the high expression of oxidative species in senescent cells suppresses the fluorescence of SGQDs-PEG5-Gd. Although the healthy and senescent cells show different fluorescent intensities after incubation with SGQDs-PEG5-Gd, the suppression of fluorescence provides limited information for determining the senescent cells. The in vivo MRI of a naturally aging rat (1-year-old, body weight: 550 g, Fig. 4e) is shown in Fig. 4f. After 30 min of injecting SGQDs-PEG5-Gd, the image of the aging rat displayed more information and the boundary of the heart became clear. The above results in vitro and in vivo exhibit the application prospect of the H2O2-sensitive SGQDs-PEG5-Gd, suggesting potential for use in both MRI and precision medicine.
Experimental section
Synthesis and purification of SGQDs
SGQDs were synthesized by dissolving 100 mg of L-cysteine hydrochloride in 100 mL of deionized water and heating at 150 °C for 12 h. After cooling the resulting mixture, the product was sequentially filtered through a filter paper and an inorganic alumina membrane (220 nm). The filtrate was then dialyzed in a 3500 Da dialysis bag against deionized water to remove any remaining impurities. The SGQD solution was then concentrated and lyophilized into a powder.T 1 measurement by ULF NMR relaxometry
ULF NMR relaxometry24,25 works at a static magnetic field (B0) of 118 μT (corresponding to the proton Larmor frequency of 5030 Hz). During the measurement, 10 mL of the sample was placed in the centre of the system. A pre-polarization magnetic field (87 mT) orthogonal to B0 was first applied to the sample for 500 ms. Thereafter, the sample magnetization freely relaxed at B0 for a certain time (ΔTdelay1). Then, π/2 and π pulses were applied to excite the spin-echo signals. The pulse sequence ran ten times with different ΔTdelay1 values allowing the acquisition of ten signal intensities. A single exponential fit was used to derive the value of T1 of the sample.Computational method
All calculations were performed using DFT with the Perdew–Burke–Ernzerhof form of the generalized gradient approximation functional.38 The Vienna ab initio simulation package (VASP) was employed.39,40 The energy cutoff for plane wave expansions was set to 500 eV, and the energy (converged to 10−5 eV per atom) and force (converged to 0.01 eV Å−1) were set as the convergence criteria for geometry optimization. A 2 × 2 × 2 periodic model was constructed to simulate PEG4, PEG5, and PEG6. The Brillouin zones were sampled with the gamma-centered Monkhorst-Pack41 (2 × 2 × 1) k-point meshes for model optimization and (1 × 1 × 1) k-point for microcanonical ensemble (NVE) molecular dynamic simulation at 300 K for 2 ps. The DFT-D3 method42 considering the van der Waals interaction was adopted for the adsorption system.The climbing image nudged elastic band (CI-NEB) method implemented in the VASP transition state tools was used to determine the proton transport pathways and the corresponding energy barriers.43 In this step, the algorithm to relax the ions into their energy minimization transition state is required to coincide with the previous calculation of the initial and final states.
Cell culture
Human umbilical vein endothelial cells (HUVECs) were obtained from the American Type Culture Collection (ATCC) and cultured in 5% CO2 at 37 °C in high-glucose Dulbecco's modified Eagle's medium (Hyclone, USA) supplemented with 10% fetal bovine serum (Gibco, USA).To induce cell senescence, HUVECs were treated with 20 μmol L−1 etoposide for 24 h.44 After etoposide removal, cells were incubated with SGQDs-PEG5-Gd (1 μg mL−1) for 24 h before being collected for fluorescence intensity assay.
All animal experiments were conducted in accordance with the Guidelines on the Care and Use of Laboratory Animals (National Institutes of Health Publication No. 85-23, revised 1996) and approved by the Xinhua Hospital Ethics Committee affiliated with Shanghai Jiaotong University School of Medicine (XHEC-NSFC-2019-288).
Conclusions
To detect the small molecule (H2O2), GQDs with disulfide bonds were designed and synthesized. The resulting SGQDs show a decrease in fluorescent intensity after reacting with H2O2 because of the oxidation of disulfide bonds into sulfoxide and sulfone. Using PEG5 to connect SGQDs and Gd3+, the paramagnetic SGQDs-PEG5-Gd exhibits T1 reduction after reacting with H2O2. The r1 of SGQDs-PEG5-Gd increases from 30.72 to 45.75 L mmol−1 s−1. Such an acceleration in relaxation is mainly attributed to the low energy barrier in the PEG5 tube and leads to fast proton transport near Gd3+. Ultimately, the SGQDs-PEG5-Gd was applied in the in vitro fluorescent imaging to differentiate healthy and senescent HUVECs as well as the in vivo MRI to show the senescent region in 1-year-old rat. More importantly, this work not only overcomes the difficulty of small molecule sensing using the MRS probe but also inspires the design of carbon nanostructures with the ability to transport protons.Author contributions
Y. Li: conceptualization, investigation, methodology, writing – original draft, visualization, funding acquisition; H. Wang: validation, writing – original draft; C. Ye: formal analysis; visualization; X. Wang: methodology, resources; Peng He: validation; S. Yang: conceptualization, methodology, writing – review and editing, funding acquisition; H. Dong: validation, writing – review and editing, funding acquisition, supervision; G. Ding: supervision.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was financially supported by Science and Technology Commission of Shanghai Municipality grants 21ZR1482800 (S. Y.) and 23YF1455800 (Y. L.), Mobility Programme of the Sino-German Center for Research Promotion grant M-0022 (H. D.), and Shanghai Post-doctoral Excellence Program grant 2022675 (Y. L.), National Key R&D Program of China grant 2022YFA1203400 (C. Y.), Guangdong Provincial Key Laboratory of Computational Science and Material Design grant 2019B030301001 (C. Y.). S. Yang also wants to acknowledge the support from the Youth Innovation Promotion Association of CAS and the Xinweizhixing Project of SIMIT. Computing resources were supported by the Center for Computational Science and Engineering at Southern University of Science and Technology.References
- L. Josephson, J. M. Perez and R. Weissleder, Angew. Chem., Int. Ed., 2001, 40, 3204–3206 CrossRef CAS PubMed.
- J. M. Perez, L. Josephson, T. O'Loughlin, D. Hogemann and R. Weissleder, Nat. Biotechnol., 2002, 20, 816–820 CrossRef CAS PubMed.
- Z. C. Xu, C. Liu, S. J. Zhao, S. Chen and Y. C. Zhao, Chem. Rev., 2019, 119, 195–230 CrossRef CAS PubMed.
- X. Q. Chi, D. T. Huang, Z. H. Zhao, Z. J. Zhou, Z. Y. Yin and J. H. Gao, Biomaterials, 2012, 33, 189–206 CrossRef CAS PubMed.
- X. L. Huang, Y. J. Liu, B. Yung, Y. H. Xiong and X. Y. Chen, ACS Nano, 2017, 11, 5238–5292 CrossRef CAS PubMed.
- Y. F. Shen, F. Jia, Y. W. He, Y. C. Fu, W. H. Fang, J. P. Wang and Y. B. Li, Biosens. Bioelectron., 2022, 213, 114437 CrossRef CAS PubMed.
- J. P. Wen, L. Q. Ren, Q. F. He, J. W. Bao, X. W. Zhang, Z. X. Pi and Y. P. Chen, Biosens. Bioelectron., 2023, 219, 114790 CrossRef CAS PubMed.
- X. Wang, S. P. Ni and Y. N. Wang, Appl. Magn. Reson., 2021, 52, 1561–1580 CrossRef CAS.
- Y. Dong, R. Chen, L. Wu, X. Wang, F. Jiang, Z. Fan, C. Huang and Y. Chen, Biosens. Bioelectron., 2022, 207, 114127 CrossRef CAS PubMed.
- C. X. Huang, J. P. Zhao, R. S. Lu, J. Wang, S. R. Nugen, Y. P. Chen and X. H. Wang, Food Chem., 2023, 400, 134035 CrossRef CAS PubMed.
- P. Caravan, Chem. Soc. Rev., 2006, 35, 512–523 RSC.
- X. B. Qi, Z. L. Wang, R. S. Lu, J. W. Liu, Y. Li and Y. P. Chen, Food Chem., 2021, 338, 127837 CrossRef CAS PubMed.
- C. Wang, C. Yan, L. An, H. Zhao, S. Song and S. Yang, J. Mater. Chem. B, 2021, 9, 7734–7740 RSC.
- T. A. Meyer, C. Zhang, G. Bao and Y. G. Ke, Nano Lett., 2020, 20, 2799–2805 CrossRef CAS PubMed.
- Z. Wang, X. Xue, H. Lu, Y. He, Z. Lu, Z. Chen, Y. Yuan, N. Tang, C. A. Dreyer, L. Quigley, N. Curro, K. S. Lam, J. H. Walton, T.-y. Lin, A. Y. Louie, D. A. Gilbert, K. Liu, K. W. Ferrara and Y. Li, Nat. Nanotechnol., 2020, 15, 482–490 CrossRef CAS PubMed.
- Y. Q. Li, Y. Xiao, Q. Tao, M. M. Yu, L. Zheng, S. W. Yang, G. Q. Ding, H. Dong and X. M. Xie, Chin. Chem. Lett., 2021, 32, 3926–3931 Search PubMed.
- J. Yu, F. Zhao, W. L. Gao, X. Yang, Y. M. Ju, L. Y. Zhao, W. S. Guo, J. Xie, X. J. Liang, X. Y. Tao, J. Li, Y. Ying, W. C. Li, J. W. Zheng, L. Qiao, S. B. Xiong, X. Z. Mou, S. L. Che and Y. L. Hou, ACS Nano, 2019, 13, 10002–10014 CrossRef CAS PubMed.
- L. Đorđević, F. Arcudi, M. Cacioppo and M. Prato, Nat. Nanotechnol., 2022, 17, 112–130 CrossRef PubMed.
- S. Yang, Y. Li, L. Chen, H. Wang, L. Shang, P. He, H. Dong, G. Wang and G. Ding, Small, 2023, 19, 2205957 CrossRef CAS PubMed.
- M. Zheng, F. Åslund and G. Storz, Science, 1998, 279, 1718–1722 CrossRef CAS PubMed.
- F. Li, Y. Y. Li, X. Yang, X. X. Han, Y. Jiao, T. T. Wei, D. Y. Yang, H. P. Xu and G. J. Nie, Angew. Chem., Int. Ed., 2018, 57, 2377–2382 CrossRef CAS PubMed.
- J. M. Holt and G. K. Ackers, in Methods in Enzymology, Academic Press, 2009, vol. 455, pp. 193–212 Search PubMed.
- G. C. Li, M. D. Brady and G. J. Meyer, J. Am. Chem. Soc., 2018, 140, 5447–5456 CrossRef CAS PubMed.
- Y. Q. Li, H. Dong, Q. Tao, C. C. Ye, M. M. Yu, J. P. Li, H. F. Zhou, S. W. Yang, G. Q. Ding and X. M. Xie, Biomaterials, 2020, 250, 120056 CrossRef CAS PubMed.
- M. M. Yu, Q. Tao, H. Dong, T. Huang, Y. Q. Li, Y. Xiao, S. W. Yang, B. Gao, G. Q. Ding and X. M. Xie, J. Magn. Reson., 2020, 317, 106775 CrossRef CAS PubMed.
- W. J. Wu, Y. F. Li, J. D. Liu, J. T. Wang, Y. K. He, K. Davey and S. Z. Qiao, Adv. Mater., 2018, 30, 1707516 CrossRef PubMed.
- B. B. Shi, H. Wu, J. L. Shen, L. Cao, X. Y. He, Y. Ma, Y. Li, J. Z. Li, M. Z. Xu, X. L. Mao, M. Qiu, H. B. Geng, P. F. Yang and Z. Y. Jiang, ACS Nano, 2019, 13, 10366–10375 CrossRef CAS PubMed.
- H. Z. Dou, M. Xu, B. Y. Wang, Z. Zhang, D. Luo, B. B. Shi, G. B. Wen, M. Mousavi, A. P. Yu, Z. Y. Bai, Z. Y. Jiang and Z. W. Chen, Angew. Chem., Int. Ed., 2021, 60, 5864–5870 CrossRef CAS PubMed.
- W. J. Wu, Z. F. Zhou, Y. Wang, Y. T. Zhang, Y. Wang, J. T. Wang and Y. C. Zou, Nano Res., 2022, 15, 4124–4131 CrossRef CAS.
- M. A. Saadiah, H. M. Tan and A. S. Samsudin, Bull. Mater. Sci., 2020, 43, 203 CrossRef CAS.
- Z. Zheng, Q. Zhou, M. Li and P. Yin, Chem. Sci., 2019, 10, 7333–7339 RSC.
- L. Partridge, M. Fuentealba and B. K. Kennedy, Nat. Rev. Drug Discovery, 2020, 19, 513–532 CrossRef CAS PubMed.
- A. J. Covarrubias, R. Perrone, A. Grozio and E. Verdin, Nat. Rev. Mol. Cell Biol., 2021, 22, 119–141 CrossRef CAS PubMed.
- C. D. Wiley and J. Campisi, Nat. Metab., 2021, 3, 1290–1301 CrossRef CAS PubMed.
- N. Basisty, A. Kale, O. H. Jeon, C. Kuehnemann, T. Payne, C. Rao, A. Holtz, S. Shah, V. Sharma, L. Ferrucci, J. Campisi and B. Schilling, PLoS Biol., 2020, 18, e3000599 CrossRef PubMed.
- A. Hernandez-Segura, T. V. de Jong, S. Melov, V. Guryev, J. Campisi and M. Demaria, Curr. Biol., 2017, 27, 2652–2660 CrossRef CAS PubMed.
- A. Hernandez-Segura, J. Nehme and M. Demaria, Trends Cell Biol., 2018, 28, 436–453 CrossRef CAS PubMed.
- J. P. Perdew, K. Burke and M. Ernzerhof, Phys. Rev. Lett., 1996, 77, 3865–3868 CrossRef CAS PubMed.
- G. Kresse and J. Furthmuller, Phys. Rev. B: Condens. Matter Mater. Phys., 1996, 54, 11169–11186 CrossRef CAS PubMed.
- G. Kresse and J. Furthmuller, Comput. Mater. Sci., 1996, 6, 15–50 CrossRef CAS.
- H. J. Monkhorst and J. D. Pack, Phys. Rev. B: Solid State, 1976, 13, 5188–5192 CrossRef.
- S. Grimme, J. Comput. Chem., 2006, 27, 1787–1799 CrossRef CAS PubMed.
- G. Henkelman, B. P. Uberuaga and H. Jonsson, J. Chem. Phys., 2000, 113, 9901–9904 CrossRef CAS.
- P. Samakkarnthai, D. Saul, L. Zhang, Z. Aversa, M. L. Doolittle, J. G. Sfeir, J. Kaur, E. J. Atkinson, J. R. Edwards, G. G. Russell, R. J. Pignolo, J. L. Kirkland, T. Tchkonia, L. J. Niedernhofer, D. G. Monroe, N. K. Lebrasseur, J. N. Farr, P. D. Robbins and S. Khosla, Aging, 2023, 15, 3331–3355 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3nr05053j |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2024 |