Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Nanoscopic visualization of microgel-immobilized cytochrome P450 enzymes and their local activity†
Lukas
Schubert
 a,
Chiara
Nenninger
a,
Chiara
Nenninger
 b,
Maximilian
Nöth
b,
Thomke
Belthle
b,
Maximilian
Nöth
b,
Thomke
Belthle
 cd,
Robert Dirk
de Lange
cd,
Robert Dirk
de Lange
 a,
Andrij
Pich
a,
Andrij
Pich
 cde,
Ulrich
Schwaneberg
cde,
Ulrich
Schwaneberg
 bc and
Dominik
Wöll
bc and
Dominik
Wöll
 *a
*a
aInstitute of Physical Chemistry, RWTH Aachen University, Landoltweg 2, 52074 Aachen, Germany. E-mail: woell@pc.rwth-aachen.de; Tel: +49 241 80 98624
bInstitute of Biotechnology, RWTH Aachen University, Worringerweg 3, 52074 Aachen, Germany
cDWI – Leibniz-Institute for Interactive Materials, Forckenbeckstraße 50, 52074 Aachen, Germany
dFunctional and Interactive Polymers, Institute of Technical and Macromolecular Chemistry, RWTH Aachen University, Worringerweg 2, 52074 Aachen, Germany
eAachen Maastricht Institute for Biobased Materials (AMIBM), Maastricht University, Brightlands Chemelot Campus, Urmonderbaan 22, 6167 RD Geleen, The Netherlands
First published on 4th October 2024
Abstract
Microgels provide a controlled microenvironment for enzymes, protecting them from degradation while enhancing stability and activity. Their customizable and biocompatible structure allows for targeted delivery and controlled release, making them ideal for transporting and preserving enzyme function in various applications. For such applications, detailed knowledge of the distribution of enzymes and their activity within the microgels is essential. We present a combination of different localization-based super-resolution fluorescence microscopy measurements to localise single Cytochrome P450 BM3 enzymes and compare their local catalytic activity.
1 Introduction
Microgels are colloidal cross-linked 3D polymer networks swollen by water, which have emerged as promising materials for various applications, including (bio-)catalysis.1,2 Depending on the employed functional comonomers and synthesis method, microgels can be tailor-made for post-functionalization and physical interactions. These versatile and tunable soft polymeric gels offer a unique environment for enzymes, e.g., a porous structure and high water content, to enhance their stability, maintain their activity, and allow for efficient reusability. Furthermore, the 3D network allows immobilization of (bio-)catalysts not only on the microgel surface but also inside the polymer network, increasing the potential load and protection of active centres.3 The immobilization of enzymes within microgels is a promising approach to improve the catalytic efficiency due to the microgels’ ability to provide a protective and hydrated microenvironment that closely resembles natural conditions.4 Additionally, the meshsize, nano-structure, and surface properties of microgels can be engineered to optimise substrate diffusion, colloidal stability, interfacial activity, and enzyme–substrate interactions.5,6 This makes microgels highly suitable for applications in biocatalysis, ranging from industrial processes to environmental remediation and biomedical applications. The integration of enzymes into microgels not only enhances their performance but also facilitates easy recovery, separation, and reuse of the (bio-)catalysts, making the processes more cost-effective and sustainable.7 One important class of enzymes are P450 monooxygenases known for their remarkable ability to catalyze a wide range of oxidative reactions, including hydroxylation and epoxidation of fatty acids.8 These enzymes are highly efficient and versatile, as they combine a cytochrome P450 domain with a reductase domain allowing them to function independently of external electron donors and making them valuable in biotechnological applications.9,10 Despite the impressive potential for synthetic use of, in specific, P450 BM3, its small- and large-scale applicability is significantly hampered, amongst other factors, by its low operational stability with total turnover numbers (TTNs) in general below 40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000.8,11 Enzyme immobilization on a variety of carrier materials, e.g. agarose hydrogels,12 solid particles such as glutaraldehyde activated magnetic nanoparticles13 and microgels has become a universal method for improving enzyme activity, stability, and process metrics, especially reusability.14–16 Specifically, the characterization of P450 BM3 in PVCL/Vim microgels was reported previously, demonstrating that the enzyme retained full catalytic activity.17 This catalytic system enables the accessibility and profitability of enzymes for larger scale industrial processes, as the microgel can be easily recycled.17–19 However, the actual loading determination of enzymes in the microgels so far has been constrained to activity-based assay batch measurements.20,21 As a consequence, it is not possible to investigate whether the catalytic activity depends on the position of the catalyst within the microgel. Also, the spatial distribution of enzymes in the microgels affects the activity depending on whether the enzymes accumulate only in the shell, only in the core or evenly distributed throughout the microgel.18 As the core and shell region of microgels are in many cases associated with a difference in cross-linking density, the enzyme allocation may significantly alter their accessibility for substrates or flexibility of the active center. In addition, with the knowledge of immobilization distribution, processes can be improved in terms of decreased leaching, higher activity, or improved catalyst efficiency.22 In order to monitor enzyme immobilization within microgels, non-invasive analytical methods are required to generate knowledge on a nanoscopic level.
000.8,11 Enzyme immobilization on a variety of carrier materials, e.g. agarose hydrogels,12 solid particles such as glutaraldehyde activated magnetic nanoparticles13 and microgels has become a universal method for improving enzyme activity, stability, and process metrics, especially reusability.14–16 Specifically, the characterization of P450 BM3 in PVCL/Vim microgels was reported previously, demonstrating that the enzyme retained full catalytic activity.17 This catalytic system enables the accessibility and profitability of enzymes for larger scale industrial processes, as the microgel can be easily recycled.17–19 However, the actual loading determination of enzymes in the microgels so far has been constrained to activity-based assay batch measurements.20,21 As a consequence, it is not possible to investigate whether the catalytic activity depends on the position of the catalyst within the microgel. Also, the spatial distribution of enzymes in the microgels affects the activity depending on whether the enzymes accumulate only in the shell, only in the core or evenly distributed throughout the microgel.18 As the core and shell region of microgels are in many cases associated with a difference in cross-linking density, the enzyme allocation may significantly alter their accessibility for substrates or flexibility of the active center. In addition, with the knowledge of immobilization distribution, processes can be improved in terms of decreased leaching, higher activity, or improved catalyst efficiency.22 In order to monitor enzyme immobilization within microgels, non-invasive analytical methods are required to generate knowledge on a nanoscopic level.
Super-resolution microscopy and single molecule techniques have become a valuable tool to investigate soft matter and polymers in general23,24 and microgels in particular.25,26 Since the early stages of single molecule spectroscopy and microscopy, researchers were interested in analyzing single molecule enzyme dynamics.27,28 In parallel to the development of localization-based super-resolution methods,29 super-resolution maps of catalytic activity became accessible30 and revealed many new insights into the heterogeneity of catalytic activity in different systems.31–35 Roeffaers et al. called this approach nanometer accuracy by stochastic chemical reactions (NASCA).36 NASCA is not restricted to artificial catalytic systems, but can be also used to analyze biocatalysis.37 Resorufin is a suitable fluorophore for this approach since it can be catalytically produced from pro-fluorescent precursors,38 for example by the reduction of resazurin. The reduction of non-fluorescent resazurin to fluorescent resorufin has been used to visualise the local catalytic activity on metal nanoparticles,39,40 but it can be also applied to investigate the activity of cytochrome P450 oxidoreductase.41 In addition to determining the sites of catalysis, it is also important to visualise where the enzymes are located. This can be achieved with direct stochastic optical reconstruction microscopy (dSTORM).42 dSTORM has been used by different groups to analyze functionalization, deformation, cross-linking, inner structure and the local environment of microgels.26 This method can also be employed for the localization of enzymes, provided that the enzymes are labelled with an appropriate dye. A suitable way of labelling enzymes that was chosen herein, is the covalent reaction of amine groups present in the enzyme structure from corresponding amino acids with NHS esters from fluorescent dyes such as Alexa Fluor 647 (also referred to as Alexa 647 in the following).43 Furthermore, previous studies reported that the combination of dSTORM with points accumulation for imaging in nanoscale topography (PAINT) is possible in different imaging buffers.44 PAINT is a technique where temporal binding of probes to nanoscopic structures enables their super-resolved imaging.45,46 A comprehensive study of the fluorescence of Nile Red under different solvent conditions and the PAINT technique allowed Purohit et al. to visualise the polymer structure of microgels and connect it to the local nansocopic polarity inside them.47 In this context, super-resolution studies suggest that, if all regions can be fully accessed by the dye, a correlation between polymer density and number of localizations of non-covalently attached fluorescent probes can be assumed.48
In this paper, we report the investigation of the incorporation and the activity of P450 BM3 enzymes embedded in poly(N-vinylcaprolactam)-based microgels via electrostatic interaction14 with three different localization-based super-resolution methods. The microgels were functionalised with the comonomer N-vinylimidazole (Vim) to allow for a pH-dependent positive charge oppositely to the enzymes. The shape of microgels was determined with Nile Red PAINT, the positions of single enzymes in microgels were accessed with dSTORM measurements with Alexa Fluor™ 647 labelled enzymes, and the activity of single enzymes was analyzed with NASCA using the enzyme-catalyzed transition of non-fluorescent resazurin to fluorescent resorufin (see Fig. 1). The results and distributions obtained from the three different methods were combined to answer the question how the P450 BM3 enzymes are distributed inside the poly(VCL-co-Vim) microgels and whether catalytic activity depends on the respective enzyme location within the microgels.
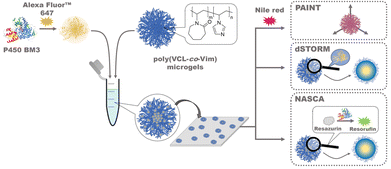 | ||
| Fig. 1 Schematic depiction of the experimental set-up for the immobilization of fluorescent labelled P450 BM3 enzymes (crystal structure from PDB: 1BVY) in poly(VCL-co-Vim) microgels and their analysis using points accumulation for imaging in nanoscale topography (PAINT), direct stochastic optical reconstruction microscopy (dSTORM), and nanometer accuracy by stochastic chemical reactions (NASCA). | ||
2 Experimental
2.1 Chemicals and reagents
All chemicals were purchased from Sigma Aldrich (Darmstadt, Germany), Carl Roth GmbH & Co KG (Karlsruhe, Germany), AppliChem GmbH (Darmstadt, Germany) or Ambinter (Orléans, France) with analytical purity unless stated differently. The microgel crosslinker N,N′-methylenebis(acrylamide) (BIS; Sigma-Aldrich, 99%) and initiator 2,2′-azobis(2-methyl-propionamidine) dihydrochloride (AMPA; Sigma-Aldrich; 97%) were used as received. N-Vinylcaprolactam (VCL; Sigma-Aldrich, 98.0%) was distilled and recrystallised from n-hexane before use. The comonomer N-vinylimidazole (Vim; Sigma-Aldrich, 99%) was purified by basic aluminium oxide column filtration to remove the stabilizer. Primers for polymerase chain reaction (PCR) were obtained from Eurofins genomics (Ebersberg, Germany). Microtiter plates were purchased from Greiner Bio-One GmbH (Frickenhausen, Germany). Microscopic supplies were purchased from VWR International GmbH (Darmstadt, Germany). Nile Red was purchased from Sigma Aldrich (Darmstadt, Germany) and purified via recrystallization. Alexa dyes and resazurin were purchased from Thermo Fisher Scientific (Schwerte, Germany) and used as received.2.2 Synthesis of poly(VCL-co-Vim) microgels
Poly(VCL-co-Vim) microgels were prepared by free radical precipitation polymerization in water. The main monomer VCL (661.8 mg, 90 mol%), the crosslinker BIS (24.4 mg, 3 mol%) and the comonomer Vim (47.7 μL, 10 mol%) were dissolved in 47 mL ultrapure water in a 50 mL-roundbottom flask. This mixture was heated to 70 °C and flushed with N2 for 30 min while stirring. The initiator AMPA (8.6 mg, 0.6 mol%) was separately dissolved in 3 mL ultrapure water and purged with N2 for 30 min. The polymerization was started by addition of the AMPA solution to the reaction flask and removal of the N2 source. The reaction was allowed to proceed for 2 h at 70 °C while being stirred. Afterwards, the microgel dispersion was purified by dialysis in a regenerated cellulose tube against distilled H2O (MWCO 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000–14
000–14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 Da) with daily exchange of the H2O for 4 days. The microgels have a hydrodynamic radius of approx. 428 nm (see DLS part of the ESI†).
000 Da) with daily exchange of the H2O for 4 days. The microgels have a hydrodynamic radius of approx. 428 nm (see DLS part of the ESI†).
2.3 Expression and purification of P450 BM3
P450 BM3 was expressed in E. coli BL21 Gold (DE3) cells as described previously.49 The cells were harvested by centrifugation using a Sorvall RC 6 centrifuge (ThermoFisher, Dreieich, Germany) for 30 min at 6340 RCF and 4 °C. The supernatant was removed, and the cell pellets were frozen at −20 °C until further use.P450 BM3 was purified via ion exchange chromatography using a 5 mL HiTrap Capto Q chromatography column (Cytiva, Freiburg, Germany), and the ÄKTA pure™ chromatography system (Cytiva, Freiburg, Germany) for UV-detection. Prior purification, the cell pellet from 200 mL of expression culture was resuspended in 30 mL of binding buffer (100 mM Tris HCl, pH 7.8) and sonicated for 4 min (15 s on + 15 s off, 70% amplitude) using the Sonicator Vibra-Cell VCX130 from Sonics & Materials (Newtown, USA) to lyse the cells. The cell lysate was centrifuged (15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 RCF, 4 °C, for 30 min) and the supernatant was afterwards filtered using a 0.2 μM mixed cellulose membrane filter from Cytiva (Freiburg, Germany).
000 RCF, 4 °C, for 30 min) and the supernatant was afterwards filtered using a 0.2 μM mixed cellulose membrane filter from Cytiva (Freiburg, Germany).
For the ion exchange chromatography, the binding buffer (100 mM Tris HCl, pH 7.8), elution buffer (100 mM Tris HCl, 1 M NaCl, pH 7.8), 20% EtOH, and ultrapure H2O were prepared, sterile filtered using a 0.2 μM mixed cellulose membrane filter from Cytiva (Freiburg, Germany) and degassed. The HiTrap Capto Q chromatography column (Cytiva, Freiburg, Germany) was connected to the chromatography system and pre-washed with 5 column volumes (cv) of ultrapure water followed by 5 cv of binding buffer (100 mM Tris HCl, pH 7.8). After pre-washing, the protein sample was loaded onto the column. The column was then washed with 5 cv of 23% of elution buffer (100 mM Tris HCl, 1 M NaCl, pH 7.8). Protein elution was performed using a gradient from 23% to 40% of elution buffer over 6 cv. The fractions were collected and analyzed via sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) using a 10% Mini-PROTEAN® TGXTM precast protein gel from Bio-Rad Laboratories GmbH (Feldkirchen, Germany). The fractions containing P450 BM3 were pooled, concentrated, and buffer exchanged to potassium phosphate buffer (100 mM, pH 7.5) using an Amicon (Merck Millipore, Darmstadt, Germany) Ultra-15 centrifugation tube (100 kDa cut-off). Samples were stored at 4 °C until further use or freeze dried in liquid nitrogen and lyophilised in an Alpha 1-2 LC plus freeze dryer (Christ, Osterode am Harz, Germany).
2.4 Fluorescence Labelling of P450 BM3
1 mg of Alexa 647 NHS ester was dissolved in 0.1 mL of PBS labelling buffer (pH 8.3) and vortexed for 1 min. This Alexa 647 NHS ester solution was then slowly added to a stirred solution of 10 mg of P450 BM3 enzymes in 1.29 mL of PBS labelling buffer (pH 8.3). The final concentrations of enzyme and dye were 0.061 mM and 0.58 mM, respectively. The reaction mixture was kept under continuous stirring at room temperature for 2 h. Subsequently, the labelled enzymes were purified using Zeba spin columns (150 μL per column, 4 columns in total). The remaining enzyme solution underwent multiple rounds of dialysis against 1 L of potassium phosphate buffer (100 mM, pH 7.5) at 5 °C until no significant amount of free dye could be detected by UV Vis absorption or fluorescence measurements of the dialysis liquid. Afterwards, the solution was stored at 5 °C.2.5 Immobilization of P450 BM3 in microgels
Before immobilization, lyophilised microgels were reconstituted to a final concentration of 10–40 mg mL−1 with ultrapure H2O and washed with the respective buffer solutions by centrifugation and redispersion. For the latter, microgels (100–400 μL per immobilization reaction) were centrifuged (20 °C, 9391–21![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 130 RCF, for 5–10 min) in an Eppendorf 5424 R centrifuge (Eppendorf, Hamburg, Germany). The supernatant was removed and the microgels were redispersed in the initial volume of buffer solution. Centrifugation time and RCF were adapted depending on the colloidal stability of the microgel sample to ensure proper pellet formation and subsequent re-dispersion.
130 RCF, for 5–10 min) in an Eppendorf 5424 R centrifuge (Eppendorf, Hamburg, Germany). The supernatant was removed and the microgels were redispersed in the initial volume of buffer solution. Centrifugation time and RCF were adapted depending on the colloidal stability of the microgel sample to ensure proper pellet formation and subsequent re-dispersion.
For immobilization, lyophilised P450 BM3 was dissolved in potassium phosphate buffer (50 mM, pH 6–8) to a final concentration of 0.6–3.0 mg mL−1. An equal volume of pre-washed microgel pellet was redispersed in a P450 BM3 solution and incubated for 30 min on ice. After incubation, unbound P450 BM3 was removed by two consecutive wash steps. Each wash step consisted of a centrifugation step (20 °C, 9391 RCF, for 5 min). After centrifugation, the supernatant was discarded and the P450 BM3 μ-gelzymes were redispersed in the respective buffer solutions and volume. After the final washing step, the P450 BM3 μ-gelzymes were redispersed in the respective buffer solution and volume. The amounts of immobilised P450 BM3 were quantified employing the Pierce™ BCA Protein Assay kit from Thermo Fisher Scientific (Rockford, USA), following the manufacturer's protocol. The activity of soluble and microgel-immobilised P450 BM3 was determined by the 7-benzoxy-3-carboxycoumarin ethyl ester (BCCE) assay as described elsewhere.50 The immobilization of Cytochrome P450 BM3 reduces the hydrodynamic radius of the microgels to 359 nm as determined by DLS (see the ESI†).
2.6 Quantification of P450 BM3 microgel loading and activity after immobilization
The protein concentration of immobilised P450 BM3 was quantified by employing the Pierce™ BCA Protein Assay kit from Thermo Fisher Scientific (Rockford, USA), following the manufacturers instructions. 25 μL of a bovine serum albumin (BSA) standard (2–0.125 mg mL−1) was mixed with 25 μL microgel sample and 200 μL working solution in a 96-well transparent flatbottom microtiter plate (MTP). The samples were incubated at 37 °C for 30 min and the absorbance was quantified afterwards at 562 nm using a Tecan Infinite M1000 Pro plate reader (Tecan Group, Männedorf, Switzerland). All measurements were performed in triplicates.For samples containing microgels, the solution was centrifuged (20 °C, 21![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 130 RCF, for 1 min) after the incubation step at 37 °C for 30 min to remove microgels from the reaction mixture and prevent light scattering during the absorption measurement. After centrifugation, the supernatant was transferred to a fresh microtiter plate, and the absorbance at 562 nm was measured using a Tecan Infinite M1000 Pro plate reader (Tecan Group, Männedorf, Switzerland).
130 RCF, for 1 min) after the incubation step at 37 °C for 30 min to remove microgels from the reaction mixture and prevent light scattering during the absorption measurement. After centrifugation, the supernatant was transferred to a fresh microtiter plate, and the absorbance at 562 nm was measured using a Tecan Infinite M1000 Pro plate reader (Tecan Group, Männedorf, Switzerland).
The activity of soluble and microgel-immobilised P450 BM3 was determined via BCCE assay.50 The assay was performed in a black flat bottom 96-well MTP. For activity determination of immobilised P450 BM3, each well contained 188 μL potassium phosphate buffer (50 mM, pH 6–8), 2 μL BCCE (2 mM in DMSO), and 10 μL of P450 BM3 μ-gelzymes. The MTP was incubated for 5 min at 600 RPM before the reaction was started by adding 50 μL NADPH (1 mM in ultrapure H2O). The fluorescent signal (Ex: 400 nm/Em: 440 nm) was monitored using a Tecan Infinite M1000 Pro plate reader (Tecan Group, Männedorf, Switzerland). To quantify P450 BM3 activity before and after immobilization, similar concentrations of soluble and immobilised enzyme were used. The activity of soluble P450 BM3 at different pH values was assessed using the BCCE assay and 600 nM of P450 BM3. Buffer conditions included sodium acetate buffer (50 mM, pH 4/5), potassium phosphate buffer (50 mM, pH 6–8), Tris HCl buffer (50 mM, pH 9), and glycine NaOH buffer (50 mM, pH 10). All measurements were performed in triplicates and the product concentration was calculated using a standard curve generated with BCCE in potassium phosphate buffer (50 mM, pH 6–8).
2.7 Super-resolution fluorescence microscopy
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 frames were recorded with 20 ms exposure time.
000 frames were recorded with 20 ms exposure time.
Subsequently, for the combined PAINT and dSTORM measurements, the channel was flushed with an imaging buffer solution (bidest. water with KPi buffer pH 6 50 mM, Cysteamine 50 mM, Nile Red in MeOH (1%), cend approx. 5 × 10−11 M). The sample was illuminated simultaneously with both the 532 nm and 640 nm laser. 60![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 frames were taken with an exposure time of 20 ms. The image was spectrally split using the CAIRN OptoSplit II described above to distinguish Alexa 647 and Nile Red blinking events. Inner filter effects can be excluded due to the low concentration of (active) dyes and the small path length of light traveling through the microgels due to their sub-micron size. The images were further processed using ThunderSTORM analysis including a correction for drifts of the sample during the measurement time.51 From the microgels observed in the field of view, a previously published algorithm was used to chose the most representative microgels.52
000 frames were taken with an exposure time of 20 ms. The image was spectrally split using the CAIRN OptoSplit II described above to distinguish Alexa 647 and Nile Red blinking events. Inner filter effects can be excluded due to the low concentration of (active) dyes and the small path length of light traveling through the microgels due to their sub-micron size. The images were further processed using ThunderSTORM analysis including a correction for drifts of the sample during the measurement time.51 From the microgels observed in the field of view, a previously published algorithm was used to chose the most representative microgels.52
3 Results and discussion
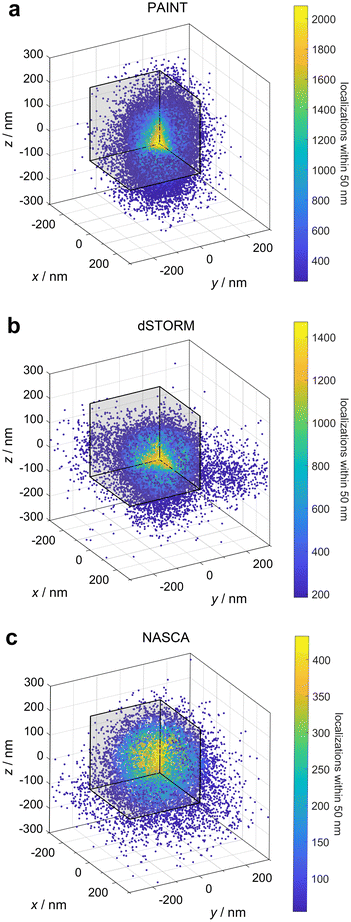
The main objective of this research was the analytical, non-invasive allocation of negatively charged P450 BM3 enzymes embedded into positively charged poly(VCL-co-Vim) microgels by electrostatic interaction. Further, the enzymatic activity of the enzymes was studied in dependence of their position within the microgels, as well as the local polymer density. For that purpose, we performed three different types of super-resolution fluorescence measurements: PAINT to visualise the microgels, dSTORM to show the positions of enzymes and NASCA to determine the local activity of the enzymes. The results of these measurements can be found in section 2 of the ESI.† Overlay of the representative microgels according to a previously developed analysis method52 yields the point clouds shown in Fig. 2.Fig. 2a presents the results of the PAINT measurements. During PAINT, the solvatochromic dye Nile Red temporarily binds to the polymer chains of the microgels and becomes fluorescent, due to the change from the polar aqueous environment to the more apolar polymer surrounding. Based on our previous experience with PAINT measurements on microgels47 and on measurements with non-covalently bound rhodamine 6G of Wrede et al.,48 we assume that the localization density profiles obtained from the PAINT experiments correlate with the polymer density profiles of the poly(VCL-co-Vim) microgels. The radial point density distribution (see Fig. 3, red squares) derived from the PAINT point cloud data in Fig. 2a represents a high polymer density in the microgel center that gradually decreases towards the periphery. This observation is in line with the known polymerization kinetics of the main monomer VCL and cross-linker BIS employed here, which is characterised by an early consumption of the cross-linking agent, resulting in a dense polymer core.54–56 It must be noted that the microgel radius represented in Fig. 3 differs from the hydrodynamic radius reported in Table S1 (see the ESI†). As the microgels are sensitive to small changes in ionic strength, their hydrodynamic radius in the buffer solution used for dynamic light scattering (DLS) measurements is larger than the radius determined after several washing steps with buffer, as required for the PAINT sample preparation. In addition, DLS analysis of charged microgels gives a size that also includes a significant hydration shell of the colloids.
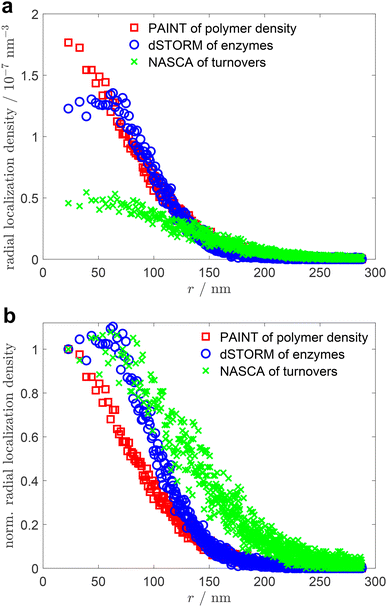 | ||
| Fig. 3 Comparison of the radial localization densities of the point clouds of the three different super-resolution fluorescence microscopy results presented in Fig. 2; (a) absolute localization densities combined from several microgels; (b) radial localization densities normalised to the value in the center for a direct comparison of the functions. | ||
The immobilization of P450 BM3 within the microgels can be determined with dSTORM measurements. The enzymes were covalently labelled with the fluorophore Alexa 647 and subsequently embedded into microgels by electrostatic interaction. The latter was achieved by the cationic charges of the microgels resulting from functionalization with the comonomer Vim. As depicted in Fig. 2b, the enzymes are distributed within the entire microgel. Additionally, Fig. 3 (blue circles) shows that in the periphery, at distances larger than 80 nm from the microgel center, the density of enzyme labels drops similar to the density of the polymer with increasing distance from the core as determined by the PAINT measurements. In the microgel core region, however, a constant enzyme density is observed. This indicates that either there is insufficient space or charge for more enzymes to be immobilised, or that the mesh size is so dense that only a few paths are available for the enzyme to reach the centre.
With the knowledge that enzymes can be immobilised within the entire microgel volume, the question arises whether their activity depends on the position in the microgel. The activity of the enzymes was addressed with the super-resolution technique NASCA, where the non-fluorescent substrate resazurin is reduced to fluorescent resorufin by the present enzymes with NADPH as co-factor. The positions of resorufin molecules were determined and their distribution is presented in Fig. 2c. The corresponding radial analysis (green crosses in Fig. 3) exhibits a similar radial distribution function as the one obtained for the enzymes. A detailed comparison between the localization of enzymes and the enzyme turnover, however, indicates differences. When the both radial distribution functions are normalised to their values in the center of the microgels, as presented in Fig. 3b, it becomes obvious that the enzyme activity is significantly higher in the periphery compared to the microgel center. This observation indicates that either the accessibility for the substrate resazurin to the enzymes or the enzyme activity itself is increased in the periphery compared to the center. Based on our previous experience with diffusion measurements in analogous microgels, we can infer that the diffusion of small molecules is not significantly impeded. Consequently, we can conclude that the activity of the enzymes in the periphery is significantly increased. The reason for this may be attributed to the lower polymer density observed in the outer regions of the microgels, which resembles a more natural aqueous environment for the enzyme. In contrast, the central region of the microgels appears to cause some degree of inhibition.
4 Conclusions
We presented a combination of super-resolution fluorescence microscopy techniques to analyse sub-diffraction sized polymeric gels with enzymes embedded by electrostatic interaction. In contrast to bulk methods, we were able to localise single enzymes embedded into microgels and to compare the local activity of substrate conversion at different distances to the microgel centre. Our results show that the enzyme Cytochrome P450 BM3 can be embedded within the entire volume of the microgels and that its activity is in general preserved within the microgel environment, however, with a small but significant trend of higher enzyme activity towards the periphery. Our conclusions can be transferred to similar micro- and hydrogel systems with enzymes of comparable size. They indicate that microgels are excellent carrier systems for (bio-)catalysts with 3D capacity compared to solid supports where immobilization is only possible at the particle surface. The combination of methods can be extended to other systems if suitable pro-fluorescent substrates are available or can be developed.Author contributions
D. W., U. S. and A. P. designed the project within the framework of collaborative research center SFB 985. T. B. provided the functionalised microgels. M. N. and C. N. provided the enzymes and helped with the enzyme labelling of microgels. L. S. performed all super-resolution measurements. L. S. and D. W. analysed the super-resolution measurements. R. D. d. L. performed and analysed light scattering measurement. L. S., C. N., T. B., R. D. d. L. and D. W. wrote the manuscript which was approved by all authors.Data availability
Data are stored according to the guidelines of the DFG. Additional data is available upon request.The data supporting this article have been included as part of the ESI.†
Conflicts of interest
There are no conflicts to declare.Acknowledgements
We gratefully acknowledge funding from the German Research Foundation (DFG) via project C5 of the Collaborative Research Center SFB 985 “Functional Microgels and Microgel Systems”.References
- A. Pich and W. Richtering, in Microgels by Precipitation Polymerization: Synthesis, Characterization, and Functionalization, ed. A. Pich and W. Richtering, Springer Berlin Heidelberg, 2011, vol. 234, book section 70, pp. 1–37 Search PubMed.
- M. Karg, A. Pich, T. Hellweg, T. Hoare, L. A. Lyon, J. J. Crassous, D. Suzuki, R. A. Gumerov, S. Schneider, I. Potemkin and W. Richtering, Langmuir, 2019, 35, 6231–6255 CrossRef CAS PubMed.
- S. Walta, D. V. Pergushov, A. Oppermann, A. A. Steinschulte, K. Geisel, L. V. Sigolaeva, F. A. Plamper, D. Wöll and W. Richtering, Polymer, 2017, 119, 50–58 CrossRef CAS.
- Q. Wang, Q. Wu, T. Ye, X. Wang, H. Qiu, J. Xie, Y. Wang, S. Zhou and W. Wu, ACS Macro Lett., 2021, 26–32 CAS.
- F. A. Plamper and W. Richtering, Acc. Chem. Res., 2017, 50, 131–140 CrossRef CAS PubMed.
- L. Sigolaeva, D. Pergushov, M. Oelmann, S. Schwarz, M. Brugnoni, I. Kurochkin, F. Plamper, A. Fery and W. Richtering, Polymers, 2018, 10, 791 CrossRef PubMed.
- N. C. Dubey, D. Gaur and B. P. Tripathi, J. Polym. Sci., 2023, 61, 1730–1748 CrossRef CAS.
- C. J. Whitehouse, S. G. Bell and L. L. Wong, Chem. Soc. Rev., 2012, 41, 1218–1260 RSC.
- L. Zhang, Z. Xie, Z. Liu, S. Zhou, L. Ma, W. Liu, J. W. Huang, T. P. Ko, X. Li, Y. Hu, J. Min, X. Yu, R. T. Guo and C. C. Chen, Nat. Commun., 2020, 11, 2676 CrossRef CAS PubMed.
- G. C. Schröder, M. S. Smit and D. J. Opperman, Curr. Opin. Green Sustainable Chem., 2023, 39, 100734 CrossRef.
- D. J. Cook, J. D. Finnigan, K. Cook, G. W. Black and S. J. Charnock, in Chapter Five - Cytochromes P450: History, Classes, Catalytic Mechanism, and Industrial Application, ed. C. Z. Christov, Academic Press, 2016, vol. 105, pp. 105–126 Search PubMed.
- V. M. Martínez, G. D. Cremer, M. B. J. Roeffaers, M. Sliwa, M. Baruah, D. E. De Vos, J. Hofkens and B. F. Sels, J. Am. Chem. Soc., 2008, 130, 13192–13193 CrossRef PubMed.
- A. Bahrami, A. Garnier, F. Larachi and M. C. Iliuta, Can. J. Chem. Eng., 2018, 96, 2227–2235 CrossRef CAS.
- M. Nöth, E. Gau, F. Jung, M. D. Davari, I. El-Awaad, A. Pich and U. Schwaneberg, Green Chem., 2020, 22, 8183–8209 RSC.
- L. Cai, Y. Chu, X. Liu, Y. Qiu, Z. Ge and G. Zhang, Microb. Cell Fact., 2021, 20, 37 CrossRef CAS PubMed.
- Y. Wang, X. Luo, X. Sun, J. Hu, Q. Guo, B. Shen and Y. Fu, Colloids Surf., B, 2022, 216, 112604 CrossRef CAS PubMed.
- M. Nöth, L. Hussmann, T. Belthle, I. El-Awaad, M. D. Davari, F. Jakob, A. Pich and U. Schwaneberg, Biomacromolecules, 2020, 21, 5128–5138 CrossRef PubMed.
- L. Schoonen and J. C. M. van Hest, Adv. Mater., 2016, 28, 1109–1128 CrossRef CAS PubMed.
- M. Faulde, J. Tonn and A. Jupke, Chem. Eng. Technol., 2019, 43, 137–142 CrossRef.
- N. Welsch, A. Wittemann and M. Ballauff, J. Phys. Chem. B, 2009, 113, 16039–16045 CrossRef CAS PubMed.
- S. Schachschal, H.-J. Adler, A. Pich, S. Wetzel, A. Matura and K.-H. van Pee, Colloid Polym. Sci., 2011, 289, 693–698 CrossRef CAS.
- C. Mateo, J. M. Palomo, G. Fernandez-Lorente, J. M. Guisan and R. Fernandez-Lafuente, Enzyme Microb. Technol., 2007, 40, 1451–1463 CrossRef CAS.
- D. Wöll and C. Flors, Small Methods, 2017, 1700191 CrossRef.
- A. Aloi and I. K. Voets, Curr. Opin. Colloid Interface Sci., 2018, 34, 59–73 CrossRef CAS.
- F. Scheffold, Nat. Commun., 2020, 11, 4315 CrossRef CAS PubMed.
- O. Nevskyi and D. Wöll, Annu. Rev. Phys. Chem., 2023, 74, 391–414 CrossRef CAS PubMed.
- H. P. Lu, L. Xun and X. S. Xie, Science, 1998, 282, 1877–1882 CrossRef CAS PubMed.
- X. S. Xie, Science, 2013, 342, 1457–1459 CrossRef CAS PubMed.
- E. Betzig, G. H. Patterson, R. Sougrat, O. W. Lindwasser, S. Olenych, J. S. Bonifacino, M. W. Davidson, J. Lippincott-Schwartz and H. F. Hess, Science, 2006, 313, 1642–1645 CrossRef CAS PubMed.
- M. B. J. Roeffaers, B. F. Sels, H. Uji-i, F. C. De Schryver, P. A. Jacobs, D. E. De Vos and J. Hofkens, Nature, 2006, 439, 572–575 CrossRef CAS PubMed.
- G. De Cremer, M. B. J. Roeffaers, E. Bartholomeeusen, K. Lin, P. Dedecker, P. P. Pescarmona, P. A. Jacobs, D. E. De Vos, J. Hofkens and B. F. Sels, Angew. Chem., Int. Ed., 2010, 49, 908–911 CrossRef CAS PubMed.
- M. B. J. Roeffaers, B. F. Sels, H. Uji-i, B. Blanpain, P. L'hoëst, P. A. Jacobs, F. C. De Schryver, J. Hofkens and D. E. De Vos, Angew. Chem., Int. Ed., 2007, 46, 1706–1709 CrossRef CAS PubMed.
- X. Zhou, N. M. Andoy, G. Liu, E. Choudhary, K. S. Han, H. Shen and P. Chen, Nat. Nanotechnol., 2012, 7, 237–241 CrossRef CAS PubMed.
- K. P. Janssen, G. De Cremer, R. K. Neely, A. V. Kubarev, J. Van Loon, J. A. Martens, D. E. De Vos, M. B. Roeffaers and J. Hofkens, Chem. Soc. Rev., 2014, 43, 990–1006 RSC.
- N. M. Andoy, X. Zhou, E. Choudhary, H. Shen, G. Liu and P. Chen, J. Am. Chem. Soc., 2013, 135, 1845–1852 CrossRef CAS PubMed.
- M. B. J. Roeffaers, G. De Cremer, J. Libeert, R. Ameloot, P. Dedecker, A.-J. Bons, M. Bückins, J. A. Martens, B. F. Sels, D. E. De Vos and J. Hofkens, Angew. Chem., Int. Ed., 2009, 48, 9285–9289 CrossRef CAS PubMed.
- R. Ye, X. Mao, X. Sun and P. Chen, ACS Catal., 2019, 9, 1985–1992 CrossRef CAS.
- L. Tian, H. Feng, Z. Dai and R. Zhang, J. Mater. Chem. B, 2021, 9, 53–79 RSC.
- G. Chen, N. Zou, B. Chen, J. B. Sambur, E. Choudhary and P. Chen, ACS Cent. Sci., 2017, 3, 1189–1197 CrossRef CAS PubMed.
- R. F. Hamans, M. Parente and A. Baldi, Nano Lett., 2021, 21, 2149–2155 CrossRef CAS PubMed.
- T. Laursen, A. Singha, N. Rantzau, M. Tutkus, J. Borch, P. Hedegard, D. Stamou, B. L. Moller and N. S. Hatzakis, ACS Chem. Biol., 2014, 9, 630–634 CrossRef CAS PubMed.
- M. Heilemann, S. van de Linde, M. Schüttpelz, R. Kasper, B. Seefeldt, A. Mukherjee, P. Tinnefeld and M. Sauer, Angew. Chem., Int. Ed., 2008, 47, 6172–6176 CrossRef CAS PubMed.
- Y. Zhang, K. Y. Park, K. F. Suazo and M. D. Distefano, Chem. Soc. Rev., 2018, 47, 9106–9136 RSC.
- C. K. Spahn, M. Glaesmann, J. B. Grimm, A. X. Ayala, L. D. Lavis and M. Heilemann, Sci. Rep., 2018, 8, 14768 CrossRef PubMed.
- A. Sharonov and R. M. Hochstrasser, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 18911–18916 CrossRef CAS PubMed.
- R. Jungmann, C. Steinhauer, M. Scheible, A. Kuzyk, P. Tinnefeld and F. C. Simmel, Nano Lett., 2010, 10, 4756–4761 CrossRef CAS PubMed.
- A. Purohit, S. P. Centeno, S. K. Wypysek, W. Richtering and D. Wöll, Chem. Sci., 2019, 10, 10336–10342 RSC.
- O. Wrede, S. Bergmann, Y. Hannappel, T. Hellweg and T. Huser, Soft Matter, 2020, 16, 8078–8084 RSC.
- A. Dennig, J. Marienhagen, A. J. Ruff, L. Guddat and U. Schwaneberg, ChemCatChem, 2012, 4, 771–773 CrossRef CAS.
- A. J. Ruff, A. Dennig, G. Wirtz, M. Blanusa and U. Schwaneberg, ACS Catal., 2012, 2, 2724–2728 CrossRef CAS.
- M. Ovesný, P. Křížek, J. Borkovec, Z. Švindrych and G. M. Hagen, Bioinformatics, 2014, 30, 2389–2390 CrossRef PubMed.
- R. Azad, P. Lenßen, Y. Jia, M. Strauch, B. A. Bener, D. Merhof and D. Wöll, Nano Lett., 2024, 24, 4447–4453 CrossRef CAS PubMed.
- R. L. Thorndike, Psychometrika, 1953, 18, 267–276 CrossRef.
- J. Meyer-Kirschner, M. Kather, A. Pich, D. Engel, W. Marquardt, J. Viell and A. Mitsos, Appl. Spectrosc., 2016, 70, 416–426 CrossRef CAS PubMed.
- F. A. L. Janssen, M. Kather, A. Ksiazkiewicz, A. Pich and A. Mitsos, ACS Omega, 2019, 4, 13795–13807 CrossRef CAS PubMed.
- H. J. M. Wolff, M. Kather, H. Breisig, W. Richtering, A. Pich and M. Wessling, ACS Appl. Mater. Interfaces, 2018, 10, 24799–24806 CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4nr03435j |
| This journal is © The Royal Society of Chemistry 2024 |