Open Access Article
Open Access ArticleAutomated chemoenzymatic modular synthesis of human milk oligosaccharides on a digital microfluidic platform†
Yiran Wu‡
abc,
Yunze Sun‡abc,
Caixia Peiabd,
Xinlv Pengabc,
Xianming Liue,
Eika W. Qian d,
Yuguang Du*ab and
Jian-Jun Li
d,
Yuguang Du*ab and
Jian-Jun Li *ab
*ab
aState Key Laboratory of Biochemical Engineering, Institute of Process Engineering, Chinese Academy of Sciences, Beijing 100190, China. E-mail: jjli@ipe.ac.cn; ygdu@ipe.ac.cn
bKey Laboratory of Biopharmaceutical Preparation and Delivery, Institute of Process Engineering, Chinese Academy of Sciences, Beijing 100190, China
cSchool of Chemical Engineering, University of Chinese Academy of Sciences, Beijing 100049, China
dGraduate School of Bio-Applications and Systems Engineering, Tokyo University of Agriculture and Technology, Nakacho 2-24-16, Koganei, Tokyo 184-8588, Japan
eKey Laboratory of Separation Science for Analytical Chemistry, Dalian Institute of Chemical Physics, Chinese Academy of Sciences, Dalian 116023, China
First published on 29th May 2024
Abstract
Glycans, along with proteins, nucleic acids, and lipids, constitute the four fundamental classes of biomacromolecules found in living organisms. Generally, glycans are attached to proteins or lipids to form glycoconjugates that perform critical roles in various biological processes. Automatic synthesis of glycans is essential for investigation into structure–function relationships of glycans. In this study, we presented a method that integrated magnetic bead-based manipulation and modular chemoenzymatic synthesis of human milk oligosaccharides (HMOs), on a DMF (Digital Microfluidics) platform. On the DMF platform, enzymatic modular reactions were conducted in solution, and purification of products or intermediates was achieved by using DEAE magnetic beads, circumventing the intricate steps required for traditional solid-phase synthesis. With this approach, we have successfully synthesized eleven HMOs with highest yields of up to >90% on the DMF platform. This study would not only lay the foundation for OPME synthesis of glycans on the DMF platform, but also set the stage for developing automated enzymatic glycan synthesizers based on the DMF platform.
1 Introduction
Glycans, along with oligonucleotides, oligopeptides, and lipids, are recognized as the four fundamental biomacromolecules of life, with glycans being the most abundant biopolymers in nature.1 As significant biological macromolecules, glycans are ubiquitous in all living organisms, encompassing animals, plants, and microorganisms.2 It is now well established that glycans play important roles in a variety of physiological and pathological processes, including cell growth and proliferation, immune responses, angiogenesis and tumor cell metastasis, toxin interaction, protein folding and degradation, cell–cell communications, and cell–pathogen interactions. Human milk oligosaccharides (HMOs), important components in human milk, have been found to be present in colostrum at concentrations as high as 20 g L−1.3 In fact, as early as in the late 19th century, HMOs were recognized for their significant impact on infant survival.4–6 As research into HMOs has progressed over time, structures of more than 200 HMOs have been determined. HMOs are generally classified as fucosylated, sialylated or acidic oligosaccharides, as well as non-fucosylated neutral oligosaccharides. The proportions of fucosylated, sialylated, and nonfucosylated neutral HMOs in term breast milk are approximately 35–50%, 12–14%, and 42–55%, respectively.7 A range of biological functions of HMOs have been investigated. For example, HMOs can act as soluble decoy receptors to prevent pathogenic attachment, reducing the risk of infection, promoting brain development in infants, serving as metabolic substrates for infant gut microbes to promote healthy gastrointestinal function, and so on.8–10 Being almost the sole prebiotic source for breastfed infants, HMOs shape the gut flora and stimulate bifidobacteria growth. In fact, some HMOs have been added commercially to infant formula. However, so far the exact roles of many single HMOs are still unknown due to unavailability, necessitating the need to explore their structure–function relationship by using pure single HMO isomers. Unfortunately, the low abundance of HMOs in milk makes their isolation and purification exceedingly challenging and almost impossible. Thus, synthesis remains the optimal route to obtain structurally well-defined HMOs for functional studies.11In recent years, an array of synthetic strategies have been explored for the assembly of HMOs, including chemical approach, chemo-enzymatic one, whole-cell fermentation or cell factory, and enzymatic synthesis. Pure chemical synthesis has been used to prepare structurally well-defined HMOs, such as lacto-N-tetraose (LNT), lacto-N-neohexaose (LNnT), and 2′-FL (2′-fucosyllactose).12 Notably, Glycom A/S has even established a chemical approach to obtaining kilogram-scale 2′-FL with high purity.13,14 However, the chemical synthetic route generally requires multi-step protection and deprotection steps, and reaction conditions are significantly more demanding, including high or low temperatures, toxic metal-based catalysts, special reaction equipment, and organic solvents, etc. Instead, the whole-cell fermentation approach offers the advantage of non-toxicity and low cost, and can achieve large-scale production. For instance, synthetic biology techniques utilizing E. coli as the microorganism chassis have facilitated the industrial-scale production of 2′-FL, 6′-sialyllactose (6′-SL), 3′-sialyllactose (3′-SL), and LNnT, etc.15–17 However, the technical barriers of whole-cell fermentation are typically high due to the complicated anabolism steps, the challenging coordination of multi-step metabolism, the regulation of metabolic flow, and select of appropriate transporters. Furthermore, this method is limited in its ability to produce more complex and branched HMOs, which need special transporters for product export15. In comparison, glycosyltransferase-based enzymatic synthesis is highly efficient and specific, making it the optimal choice for synthesizing HMOs. To streamline the process of enzymatic synthesis, the one-pot multi-enzyme (OPME) method, combining the biosynthetic process of sugar nucleotides with glycosyltransferase-catalyzed reactions, was developed for the efficient synthesis of oligosaccharides,18 which used specified enzymatic modules to extend specific monosaccharides with specific configurations and glycosidic linkages.
However, it is slow, laborious work to manually synthesize oligosaccharide at any scale, and labour-intensive and time-consuming purification protocols have still been an impediment for the implementation of these approaches. Automated enzymatic synthesis of oligosaccharides is, therefore, an emerging and still developing technology. In 2010, Nishimura and co-workers developed an artificial Golgi apparatus that utilized high-performance liquid chromatography for the fully automated synthesis of oligosaccharides.19 More innovatively, in 2018 Wang et al. realized machine-driven enzymatic oligosaccharide synthesis by modifying a commercially available peptide synthesizer.20 In 2019, Li et al. created an easy-to-install sulfonate tag to establish an automated platform for enzyme-mediated oligosaccharide synthesis based on catch and release of tagged oligosaccharides on diethylaminoethyl (DEAE) resin by using a liquid handling system. Each strategy has its own pros and cons as discussed recently.21
Recently, digital microfluidics (DMF) has emerged as a new portable droplet processing platform that employs electrode arrays to maneuver discrete pico-to micro-liters of droplets (Fig. 1).22 On a DMF platform, droplets can be manipulated to move, dispense, mix, separate, and merge. Compared to bulkly systems, microfluidics technology offers numerous benefits, including rapid mass and heat transport, low reagent consumption, easy automation, and precise manipulation of fluids at the microscale.23,24 DMF has been widely utilized in biomedical analysis, chemical synthesis and preparation of biochemical materials, such as for PCR (Polymerase Chain Reaction), single-cell analysis, immunoassay, chemical synthesis of a pentapeptide25 and macrocyclic peptides,26 preparation of microfibers with core–shell structures27 and multi-component particles of various structures,28 etc. In 2009, the Linhardt group reported that heparan sulfate immobilized on magnetic nanoparticle was enzymatically modified in a DMF device by recombinant D-glucosaminyl 3-O-sulfotransferase with a yield of ∼5%, highlighting the challenge in enzymatically modifying immobilized saccharide chains.29 However, until now enzymatic synthesis of HMOs on the DMF platform hasn't been reported.
Since magnetic carriers can simplify the implementation of automated processes, thus allowing the automation of complex multistep enzymatic cascade reaction. In this study, we reported on the integration of a digital microfluidic platform and magnetic beads-based purification with a one-pot multi-enzyme strategy to automatically synthesize various human milk oligosaccharides, including fucosylated, sialylated ones, as well as non-fucosylated neutral ones. This study would set the foundation for the creation of modular (chemo)enzymatic glycan synthesizers based on the DMF platform in future.
2 Materials and methods
2.1 Reagents and chemicals
Unless specified, all chemicals were of analytical grade and purchased from Sigma (St. Louis, Missouri, USA), Beijing BioDee (Beijing, China), Sinopharm (Shanghai, China), Aladdin (Shanghai, China) or Beijing Solarbio Science & Technology Co. Ltd (Beijing, China). Tag (7-(2-(2-(N-methyl-aminooxy)ethoxy)ethoxy)naphthalene-1,3-disulfonic acid) was synthesized by Shanghai Nafu Biotechnology Co. Ltd according to the published procedure (Shanghai, China).21 In addition, all solutions were prepared with double-distilled water.2.2 Establishment of the DMF device
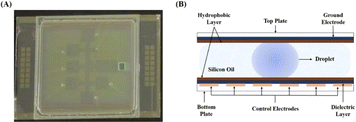
On the two-plate DMF device used for enzymatic reactions, 32 actuation electrodes were fabricated on the bottom plate, which were covered with 8 μm thick SU-8 as the dielectric layer to prevent hydrolysis of the electrodes and 50 nm thick Teflon to render hydrophobility and smooth actuation of droplets, while the top plate was also covered with the same hydrophobic layer. One square hole and seven smaller through holes were fabricated on the top plate via laser scribing for droplet loading and collecting (or sampling) with a pipette. The device was assembled by adhering the top and bottom plate with a spacer formed by adhesive double-side tape with a thickness of 0.15 mm approximately. Accordingly, the volume of one droplet on one electrode is around 2–2.5 μL.2.3 Enzymes
All enzymatic modules were constructed, expressed and purified following the published procedures. Enzyme abbreviations: BiNahK, Bifidobacterium infantis N-acetylhexosamine-1-kinase;30 PmGlmU, Pasteurella multocida N-acetylglucosamine 1-phosphate uridylyltransferase;31 NmLgtA, Neisseria meningitidis β1,3 N-acetylglucosaminyltransferase;32 BLUSP: Bifidobacterium longum UDP-sugar pyrophosphorylase;33 BfFKP, a bifunctional enzyme from Bacteroides fragilis that has both L-fucokinase and GDP-fucose pyrophosphorylase activities;34 Hp1,3FT, Helicobacter pylori α1,3-fucosyltransferase;35 Hp1,2FT, Helicobacter pylori α1,2-fucosyltransferase;36 Pd2,6ST, Photobacterium damselae α2,6-sialyltransferase;37 GalK, galactokinase from E. coli;38 β1,3GalT, β1,3-galactosyltransferase from;39 NmLgtB, Neisseria meningitidis β1,4-galactosyltransferase;40 NmCSS, Neisseria meningitidis CMP-sialic acid synthetase;41 PmST1 (M144D), Pasteurella multocida 2,3-sialyltransferase;42 PpA, Escherichia coli pyrophosphatase.432.4 Droplet operation on the DMF device
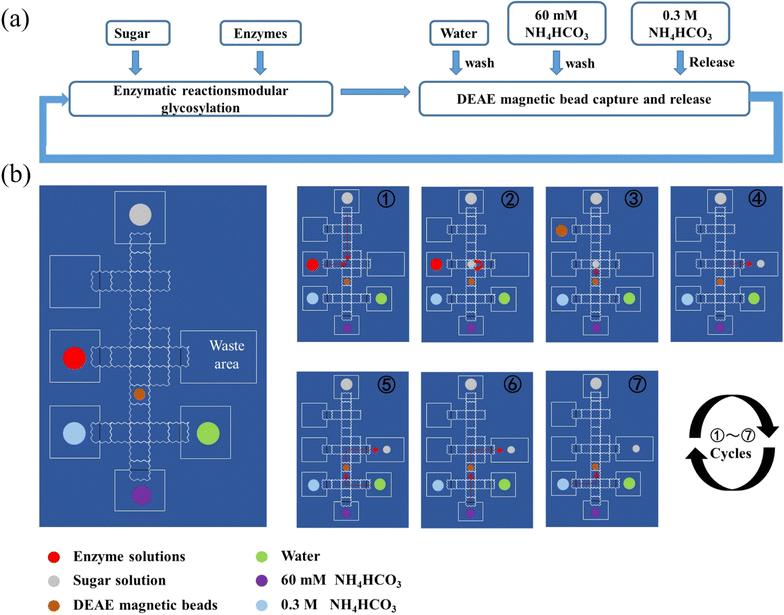
The droplets were actuated with an alternative wave generator in a sinusoid waveform (150 V RMS with a frequency of 1 kHz) and loaded onto the DMF device by a micropump. The actuation of droplets started from loading deionized water to test the actuation versatility of the DMF device. The reagents used on the DMF device included saccharides, salts, enzymes and magnetic beads. For saccharides, 10 mM glucose, 10 mM galactose, 10 mM lactose and 5 mg mL−1 lacto-N-tetraose were tested respectively. For salts, 50 mM NaAc pH = 5.5 buffer, 60 mM NH4HCO3, 1 M Tris–HCl pH = 8 buffer and 100 mM MgCl2 were tested respectively. For enzymes, each enzyme mentioned above was tested respectively. Before loading, Pluronic F127 surfactant was added at a concentration of 0.05% (w/v) to prevent biofouling caused by non-specific adsorption of proteins on the surface of DMF chips.2.5 General procedure for automatic enzymatic synthesis of tagged HMOs on the DMF device
The movement of droplets and addition of all reagents were automatically manipulated by a predefined program which had been scripted by a computer. Droplets were manually removed from the DMF platform by pipette and the magnet bar was also maneuvered manually.① Lactose or GlcNAc modified at the reducing end with the tag at a concentration of 3 mM was loaded in the first droplet and transferred onto the DMF device by a micropump. Enzymatic module, 10 mM MgCl2, 50 mM NH4HCO3, 3 mM monosacharide, 3.6 mM CTP or 3.6 mM ATP and 3.6 mM UTP/GTP were loaded in another droplet and transferred onto the DMF device by a micropump controlled by a program.
② On the DMF platform, two droplets were mixed evenly by applying power to the electrodes below two droplets and left for 1 h for enzymatic synthesis.
③ The reaction mixture was mixed with DEAE magnetic beads in one droplet by applying power to the electrodes below specified droplets. The DEAE magnetic beads were suspended in the droplet to capture products modified with tag.
④ DEAE magnetic beads and supernatants were separated by a magnet controlled manually, and the supernatant including enzymes, excess (sugar) nucleotides, MgCl2 and NH4HCO3 was taken out and discarded.
⑤ DEAE magnetic beads were washed with water.
⑥ DEAE magnetic beads were washed with 60 mM NH4HCO3.
⑦ Captured products with tag on DEAE magnetic beads were released by washing with 0.3 M NH4HCO3, and the eluent obtained was mixed evenly with the third droplet and left for 1 hour for the next enzymatic synthesis reaction. Like the first cycle, DEAE magnetic beads were used for capturing synthesized products. Finally, the eluent washed by 0.3 M NH4HCO3 was transferred by a pipette and characterized by MS.
2.6 Removal of sulfonate tag after enzymatic synthesis on the DMF device
The eluent containing product modified at the reducing end with the tag was mixed with a droplet (2 μL) of trifluoroacetic acid (0.25%, v/v) on the DMF platform. The reaction mixture was agitated by applying power to the electrode below droplet at room temperature for 2 h.21 After the reaction, the product was mixed with a droplet of 50 mM NH4HCO3 to adjust pH. Then, DEAE magnetic beads were used for capturing sulfonate tags removed from products, and the supernatant including target oligosaccharides was taken out by a pipette and characterized by MS.2.7 MS analysis of products in droplets
Analysis of products was performed on UltiMate-3000-ISQ-EM (ThermoFisher Scientific). The m/z scanning range was set to 150 to 1200. MS was performed in positive and negative mode. The droplets were diluted to 100 μL before analysis. Samples were loaded directly without columns. The mobile phase was water and the flow rate was 0.1 mL min−1.Since it's not possible and realistic to purify the synthesized oligosaccharides from a droplet of 2 μL and traditional methods such as HPLC (high performance liquid chromatography) or NMR (nuclear magnetic resonance) are not suitable for quantification, the yields of enzymatic synthesis of oligosaccharides on the DMF platform were determined as follows. It is worth pointing out that capture of tagged substrates or products by the magnetic beads reached >90% according to our experimental results (data not shown). Since the staring substrate and the final product contain the same tag which could be captured by magnetic beads, in theory their probabilities captured by magnetic beads are very close. In addition, considering the fact that tagged oligosaccharides can be easily ionized and give strong MS signals, when the samples were analyzed by MS, ion peaks corresponding to molecular weights of tagged starting materials or expected products were first searched. If only ion peaks corresponding to molecular weights of tagged expected products were observed and no ion peaks corresponding to molecular weights of tagged starting materials were found, we roughly assumed that the yields of expected products were >90%. If ion peaks corresponding to molecular weights of both tagged starting materials and expected products were observed simultaneously, the yields of expected products were estimated based on their relative abundance. In addition, it has been demonstrated the TFA (trifluoroacetic acid)-cleavage step is nearly 100%.21 Therefore, if the tagged starting substrate wasn't completely converted into the tagged product, both untagged starting substrate and product cleaved off by TFA could be detected by MS. Since saccharides have close ionization capabilities in the process of MS analysis, the relative MS signal intensities of substrates and/or products were roughly used to define yield.
3 Results
3.1 Establishment and operation of digital microfluidic device
Based on previous research, a microfluidic device was set up (Fig. 1).44 The DMF chip includes two functional plates: a teflon-coated ITO glass as the top plate and a serrated square electrode array as the bottom plate, which drove droplets to move, as depicted in Fig. 1. The top plate was a perforated ITO conductive glass that acted as a ground electrode.45 The holes present in the top plate were utilized for introducing or removing droplets on the DMF device. The bottom plate housed 32 control electrodes that were responsible for manipulating droplets. The dielectric layer, covering the base plate, created an interfacial force in electrowetting-on-dielectrics necessary for liquid manipulation. Both top and bottom plates were uniformly coated with a hydrophobic material, facilitating direct contact with the reagents. This hydrophobic layer was crucial in addressing issues such as electrode surface contamination and sample loss. Additionally, to ensure smooth movement of droplets and minimize evaporation of droplets, the top and bottom electrode plates were bonded and the space between them was filled with silicone oil to further assist droplet transportation.Initially, different droplets were maneuvered on the DMF platform to test the feasibility of enzymatic synthesis of oligosaccharides, including saccharides, enzymes, buffers, metal ions, etc., involved in HMOs synthesis. The results proved that droplets containing salts (50 mM NaAc pH 5.5 buffer, 60 mM NH4HCO3 and 1 M Tris-HCl pH 8.0 buffer) or saccharides (10 mM glucose, 10 mM galactose, 10 mM lactose, 5 mg mL−1 lacto-N-tetraose, etc.) can be actuated smoothly in both air and silicone oil (cSt 1.5). However, those containing enzymes such as β1,3 N-acetylglucosaminyltransferase can only be actuated in silicone oil even after being doped with 0.05% (w/v) pluronic F127. Our experimental results also demonstrated that enzymatic activities were not notably affected by the presence of 0.05% pluronic F127 and silicone oil (data not shown). Therefore, in consideration of compatibility with the DMF platform, the droplets for enzymatic reactions were conducted in the presence of 0.05% pluronic F127 in silicone oil in the following experiments, unless indicated otherwise.
3.2 Design and validation of the strategy for enzymatic synthesis and magnetic bead-based purification of HMOs
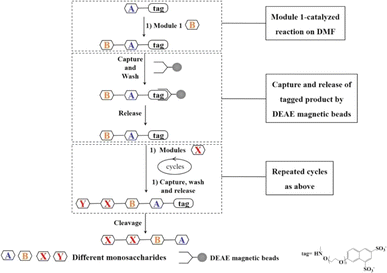
In order to achieve automatic synthesis of HMOs, a strategy integrating enzymatic modular assembly and magnetic beads-based purification after enzymatic reactions on the DMF platform was designed (Fig. 2). The sulfonate tags bearing two sulfonate groups, which can be attached to the reducing ends of starting saccharides, were captured onto anion (DEAE) exchange resin, and released by 0.3 M NH4HCO3.21 The sulfonated tags have been demonstrated not to interfere with enzymatic reactions.21Starting from the monosaccharides or cheap and commercially available disaccharides such as lactose, the sulfonate tag was attached to the reducing end of the first monosaccharide or disaccharide by a simple and successful chemical coupling reaction as a starting material for oligosaccharide synthesis (Fig. 3).21 After the enzymatic glycosylation reaction catalyzed by one enzyme module, the disaccharide or trisaccharide-tag was generated. Then DEAE magnetic beads were introduced to capture the disaccharide or trisaccharide product with the sulfonate tag by charge interaction between tag and DEAE. With the help of a magnet, the impurities including buffer and other components which couldn't be captured onto the DEAE magnetic beads were washed off in turn with water and 60 mM NH4HCO3 solution. Finally the product was released from DEAE magnetic beads with 0.3 M NH4HCO3 solution, which was used as the acceptor for the next enzymatic glycosylation step. The pH value of the oligosaccharide solution was adjusted and the reaction was continued with the addition of new reaction components required for the next glycosylation reaction catalyzed by another enzyme module to produce the trisaccharide or tetrasaccharide-tag, followed by capture-release process on the DEAE magnetic beads. In a similar fashion, different enzyme modules were used to produce different tagged oligosaccharides depending on the target oligosaccharide structure to be synthesized. Finally, since low concentration and short duration of trifluoroacetic acid (TFA) treatment have no significant effects on the oligosacharide structure, fucosylated and sialylzted oligosaccharides in particular, the tag was removed using low concentration of TFA21 and the target oligosaccharides were obtained. The capture and release of the tagged product by DEAE magnetic beads ensures the homogeneous synthesis of the oligosaccharides in solution and quick separation and purification (several minutes), while separation and purification steps of traditional oligosaccharide synthesis need several hours.
Based on structural characteristics of HMOs, we constructed 7 enzymatic modules for synthesis of them. Because lactose is inexpensive and abundant in nature, it can be used as a starting material to synthesize HMOs by using different enzymatic modules (Table 1). For each step, the enzymes in the current study which were commonly used and worked best in published studies were chosen (Table 1).
| Module | Name | References |
|---|---|---|
| (1) β-1,4-gal glycosylation | Escherichia coli galactose kinase (GalK) | 38 |
| Bifidobacterium longum UDP-sugar synthase (BLUSP) | 33 | |
| Neisseria meningitidis β1,4-galactosyltransferase (NmLgtB) | 40 | |
| Escherichia coli pyrophosphatase (PpA) | 43 | |
| (2) α-2,6-sialyltransferase | Neisseria meningitidis CMP-sialic acid synthetase (NmCSS) | 41 |
| Photobacterium damselae α2,6-sialyltransferase (Pd2,6ST) | 37 | |
| Escherichia coli pyrophosphatase (PpA) | 43 | |
| (3) α-2,3-sialylation | Neisseria meningitidis CMP-sialic acid synthetase (NmCSS) | 41 |
| Pasteurella multocida α2,3-sialyltransferase M144D mutant (PmST1) | 42 | |
| Escherichia coli pyrophosphatase (PpA) | 43 | |
| (4) α-1,3-fucosylation | Bacteroides fragilis both L-fucokinase and GDP-fucose pyrophosphorylase (BfFKP) | 34 |
| Helicobacter pylori α1,3FucT (Hp1,3FT) | 35 | |
| Escherichia coli pyrophosphatase (PpA) | 43 | |
| (5) α-1,2-fucosylation | Bacteroides fragilis both L-fucokinase and GDP-fucose pyrophosphorylase BfFKP) | 34 |
| Helicobacter pylori α1,2FucT (Hp1,2FT) | 36 | |
| Escherichia coli pyrophosphatase (PpA) | 43 | |
| (6) β-1,3 N-acetylglucosaminylation | Bifidobacterium infantis N-acetylhexosamine-1-kinase (BiNahK) | 30 |
| Pasteurella multocida N-acetylglucosamine 1-phosphate uridylyltransferase (PmGlmU) | 31 | |
| Neisseria meningitidis β1,3 N-acetylglucosaminyltransferase (LgtA) | 32 | |
| Escherichia coli pyrophosphatase (PpA) | 43 | |
| (7) β-1,3-gal glycosylation | Escherichia coli K-12 galactose kinase (GalK) | 38 |
| Bifidobacterium longum UDP-sugar synthase (BLUSP) | 33 | |
| Campylobacter jejuni β1,3-galactosyltransferase (WbgO) (β1,3GalT) | 39 | |
| Escherichia coli pyrophosphatase (PpA) | 43 |
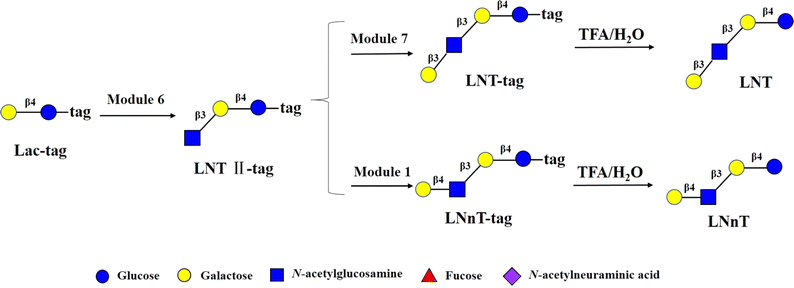
To validate the feasibility of above strategy, modular enzymatic synthesis and magnetic bead-based purification of Lacto-N-triose II (LNT II) starting from lactose was used as an example. The lactose-tag was synthesized following the published procedure.21 An enzymatic module 6 (β1,3 N-acetylglucosaminylation) was used to convert lactose-tag into LNT II-Tag on the DMF platform. One droplet containing Lac-tag, ATP and UTP and another one containing enzymatic module 6 (including NahK, GlmU, LgtA, PpA), MgCl2, buffer and GlcNAc were mixed and incubated for 1 h. Then, lacto-N-triose II-tag (LNT II-tag), the formed product with the sulfonate Tag was quantitatively captured onto DEAE magnetic beads and the rest reaction components including enzymes and excess monosaccharides, NTPs, etc. were separated with magnetic beads by a magnet bar. Then, the captured intermediate was washed in turn with H2O and 60 mM NH4HCO3, and was finally released by washing with 0.3 M NH4HCO3. The product LNT II-tag was characterized by MS. LNT II-tag was clearly observed (Fig. S3†). Finally, the tag was removed under mild acidic condition to give LNT II as the final product having a free reducing end, which was confirmed by MS (Fig. S4†). The results proved that LNT II was successfully synthesized on the DMF platform through an automatic process.
3.3 Enymatic modular synthesis of HMOs on the DMF platform
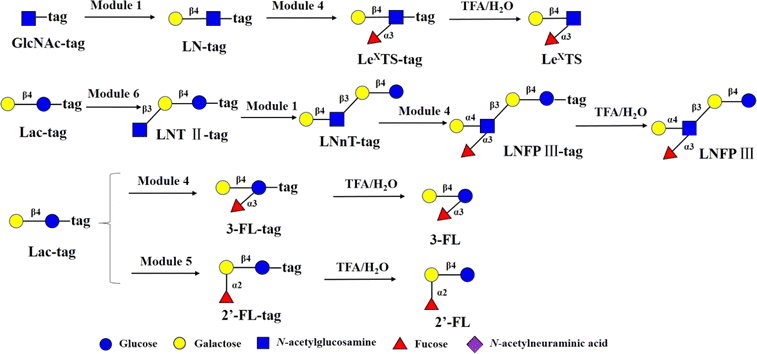
Encouraged by the above results, to test robustness of the technology, chemoenzymatic module-catalyzed synthesis of other HMOs on the DMF platform was also attempted, including neutral non-fucosylated oligosaccharides: LNT, LNnT, fucosylated HMOs: 2′-FL, 3-FL (3-fucosyllactose), lacto-N-fucopentaose III (LNFP III), sialylated HMOs: 6′-SL, 3′-SL, 6′-sialyl-N-acetyllactosamine (6′-SLN), 3′-sialyl-N-acetyllactosamine (3′-SLN), sialyllacto-N-tetraose a (LSTa), and Lewis X antigen triaose (LeX triaose).Neutral non-fucose-based HMOs serve as the core structure or backbone for all HMOs, such as LNT and LNnT. It has been reported that LNT is a carbon source by most bifidobacteria, and can reduce the cytotoxicity of protozoan parasites in tissues.46 LNnT is utilized as a carbon source for certain bifidobacteria and has been shown to possess immunosuppressive and inflammation-reducing effects in humans.47,48 Some studies have reported that higher LNnT concentrations in the milk of HIV-infected individuals are associated with reduced postnatal transmission through breastfeeding,49 making LNnT a potential target for the development of prebiotic and infectious therapies. LNT-tag and LNnT-tag were prepared from LNT II-tag by module 7 and module 1 following the synthetic procedure of LNT II-tag on the DMF platform with the yields of >81% (Fig. 4). Synthesis of LNT-tag and LNnT-tag was confirmed by MS. Finally, the tag was removed by 0.25% TFA (Fig. 4), and the final product was proved by MS.
The main chain structures of HMOs can be fucosylated through α1,2-, α1,3- or α1,4 glycosidic bonds 2′-FL is the most abundant HMO in human milk. 2′-FL and 3-FL were shown to selectively promote the growth of bifidobacteria, and high levels of 2′-FL in mother's milk corresponded to lower occurrences of campylobacter diarrhea of the infants.50 In addition, in the rat brain, 2′-FL induces long-term enhancement involved in learning and memory.51 The immunomodulating function of fucosylated HMOs was represented by LewisX-type LNFP III, which was shown to have immunosuppressive functions. 2′-FL and 3-FL were synthesized from Lac-tag by module 5 and module 4 and followed by treatment with 0.25% TFA on the DMF platform (Fig. 5). For 3-FL, the total yield was >90%. In the case of 2′-FL, since Lac-tag was detected in MS of 2′-FL-tag, the total yield of 2′-FL was around 63%. LeX tiraose was prepared from GlcNAc-tag through sequential module 1 and module 4, and followed by 0.25% TFA treatment with yield of >81% (Fig. 5). In the case of LNFP III,52 it was assembled from Lac-tag by sequential module 6, module 1 and 4, and followed by 0.25% TFA treatment (Fig. 5). The peak corresponding to m/z [LNnT-tag]2- was observed (Fig. S15†). Combined with enzymatic synthesis of LNnT-tag, this result indicated that in the three-step consecutive reactions of Lac-tag → LNFP III-tag on the DMF platform, the conversion rate of Lac-tag → LNT II-tag → LNnT-tag exceeded 90%, while the conversion rate from LNnT-tag to LNFP III-tag was approximately 70%. Thus the total yield of LNFP III was 51%.
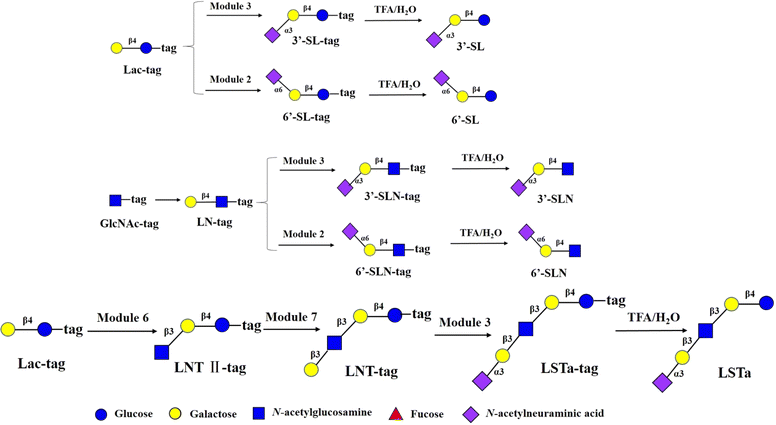
Sialic acid can modify the main chain structure of HMOs with α2,6 and α2,3- glycosidic bonds to form sialylated HMOs. The prebiotic, antiadhesive antimicrobial, and immunomodulating activities of sialylated HMOs, as well as their nutritional value for infant brain development, have been demonstrated. One study found53 that sialylated HMOs such as 3′-SL (∼23%), 6′-SL (∼3%), 3′-SLN (∼14%) and 6′-SLN (∼23%) in the plasma of partially breastfed infants were in greater amounts than in formula-only fed infants. These findings support the hypothesis that sialylated HMOs are transported to the small intestinal or colonic lumen of newborns and infants and then absorbed into the plasma.53 6′-SL and 3′-SL were synthesized from Lac-tag by module 2 and module 3 and followed by treatment with 0.25% TFA on the DMF platform with the total yields of >90% (Fig. 6). 6′-SLN and 3′-SLN were prepared from GlcNAc-tag through sequential module 1 and module 2/3, and followed by 0.25% TFA treatment with yields of >81% respectively. LSTa was assembled from Lac-tag by sequential module 6, module 7 and module 3 and followed by 0.25% TFA treatment the total yield of >72.9% (Fig. 6).
4 Discussion
Reliable and rapid access to oligonucleotides and oligopeptides by commercial automated synthesizers has greatly altered biological research. In recent years, many oligosaccharides have been successfully prepared by automated chemical synthesizers. However, the application of automated chemical synthesis of oligosaccharides has been hindered by some limitations, such as tedious protection/deprotection manipulations, control of regio- and stereo-specificity, etc.Glycosyltransferases-based oligosaccharide synthesis provides an attractive alternative to chemical synthesis. However, labour-intensive and time-consuming purification procedures of intermediates are the main limitation for the preparation of large collections of glycans by enzymatic approaches. The current automation platform could greatly speed up glycan preparation by removing tedious purification procedures.
A key feature of the current study is to integrate modular chemoenzymatic assembly of oligosaacharide, DEAE magnetic beads-based purification and DMF. In our protocol, DEAE-modified resins, which were typically used for the purpose of product purification in synthesis of oligosaccharides,21 were replaced by magnetic beads, which promoted automation level when combined with programmed actuation of droplets in DMF chips. Besides, compared with other automatic platforms for enzymatic synthesis of oligosaccharides, the DMF platform demonstrated the following advantages as shown in this study, including simple-to-implement programmability, free bacterial enzymes and free substrates used for glycan synthesis, which can make sure that all substrates can be enzymatically transformed into products. The main limitation of the DMF platform is that reactions can only be carried out in droplets of 2 μL, which can be addressed by multiple parallel reactions in greater number of droplets on high-throughput platform in the future, such as active matrix engaged DMF devices.25,54
We believe that the scope of the current methodology can be expanded to prepare highly complex and structurally differing oligosaccharides such as gangliosides, blood group O, A, and B antigens, poly-LacNAc, and N-linked glycans.
In conclusion, we proposed a strategy for the synthesis of HMOs based on the DMF platform and DEAE magnetic beads-based purification. We successfully achieved the chemoenzymatic modular sequential synthesis of HMOs on the DMF platform incorporating DEAE magnetic beads-based purification. A total of eleven HMOs were synthesized in a maximum of four sequential steps with highest yields up to >90%. The current methodology would be extended to synthesize other oligosaccharides. This study represents a proof-of-concept demonstration that modular chemoenzymatic synthesis of HMOs can be achieved in an automated manner by integrating DEAE-based magnetic beads on a DMF platform. This work will enable the development of (chemo)enzymatic module-based glycan synthesizers in the future.
Author contributions
Conceptualization: J-JL, YD, XL, EWQ; methodology, validation, investigation, formal analysis, data curation: YW, YS, CP, XP; writing–original draft, review and editing: J-JL, YW; supervision: J-JL, YD; project administration: J-JL; funding acquisition: YD, XL.Conflicts of interest
The authors declare no competing interest.Acknowledgements
This research was funded by National Natural Science Foundation of China (grant number 21877114 and 31927802).References
- A. Smith, S. Moxon and G. Morris, Biomater, 2016, 2, 261–287 Search PubMed.
- J. Qin, H. Y. Wang, D. Zhuang, F. C. Meng, X. Zhang, H. Huang and G. P. Lv, Int. J. Biol. Macromol., 2019, 136, 341–351 CrossRef CAS PubMed.
- D. Viverge, L. Grimmonprez, G. Cassanas, L. Bardet and M. Solere, J. Pediatr. Gastroenterol. Nutr., 1990, 11, 361–364 CAS.
- C. Kunz, Adv. Nutr., 2012, 3, 430s–439s CrossRef CAS PubMed.
- L. Bode, Glycobiology, 2012, 22, 1147–1162 CrossRef CAS PubMed.
- A. Kobata, Proc. Jpn. Acad. Ser. B Phys. Biol. Sci., 2010, 86, 731–747 CrossRef CAS PubMed.
- S. M. Donovan and S. S. Comstock, Ann. Nutr. Metab., 2016, 2(69), 42–51 Search PubMed.
- L. Bode, N. Contractor, D. Barile, N. Pohl, A. R. Prudden, G. J. Boons, Y. S. Jin and S. Jennewein, Nutr. Rev., 2016, 74, 635–644 CrossRef PubMed.
- J. B. German, S. L. Freeman, C. B. Lebrilla and D. A. Mills, Nestle Nutr. Inst. Workshop Ser., 2008, 62, 218–222 Search PubMed 205-218.
- P. Thomson, D. A. Medina and D. Garrido, Food Microbiol., 2018, 75, 37–46 CrossRef CAS PubMed.
- C. Sabater, M. Prodanov, A. Olano, N. Corzo and A. Montilla, Food Chem., 2016, 194, 6–11 CrossRef CAS.
- J. Zheng, H. Xu, J. Fang and X. Zhang, Carbohydr. Polym., 2022, 291, 119564 CrossRef.
- K. Agoston, M. J. Hederos, I. Bajza and G. Dekany, Carbohydr. Res., 2019, 476, 71–77 CrossRef CAS PubMed.
- M. D. Bandara, K. J. Stine and A. V. Demchenko, Carbohydr. Res., 2019, 486, 107824 CrossRef CAS.
- K. Bych, M. H. Mikš, T. Johanson, M. J. Hederos, L. K. Vigsnæs and P. Becker, Curr. Opin. Biotechnol., 2019, 56, 130–137 CrossRef CAS PubMed.
- G. A. Sprenger, F. Baumgärtner and C. Albermann, J. Biotechnol., 2017, 258, 79–91 CrossRef CAS.
- W. Zhou, H. Jiang, L. Wang, X. Liang and X. Mao, ACS Synth. Biol., 2021, 10, 447–458 CrossRef CAS PubMed.
- D. Horton, Adv. Carbohydr. Chem. Biochem., 2009, 62, xi–xiii CrossRef PubMed.
- T. Matsushita, I. Nagashima, M. Fumoto, T. Ohta, K. Yamada, H. Shimizu, H. Hinou, K. Naruchi, T. Ito, H. Kondo and S. Nishimura, J. Am. Chem. Soc., 2010, 132, 16651–16656 CrossRef CAS PubMed.
- J. Zhang, C. Chen, M. R. Gadi, C. Gibbons, Y. Guo, X. Cao, G. Edmunds, S. Wang, D. Liu, J. Yu, L. Wen and P. G. Wang, Angew Chem. Int. Ed. Engl., 2018, 57, 16638–16642 CrossRef CAS.
- T. Li, L. Liu, N. Wei, J. Y. Yang, D. G. Chapla, K. W. Moremen and G. J. Boons, Nat. Chem., 2019, 11, 229–236 CrossRef CAS PubMed.
- B. Wu, S. von der Ecken, I. Swyer, C. Li, A. Jenne, F. Vincent, D. Schmidig, T. Kuehn, A. Beck, F. Busse, H. Stronks, R. Soong, A. R. Wheeler and A. Simpson, Angew Chem. Int. Ed. Engl., 2019, 58, 15372–15376 CrossRef CAS PubMed.
- W. Li, L. Zhang, X. Ge, B. Xu, W. Zhang, L. Qu, C. H. Choi, J. Xu, A. Zhang, H. Lee and D. A. Weitz, Chem. Soc. Rev., 2018, 47, 5646–5683 RSC.
- L. Shang, Y. Cheng and Y. Zhao, Chem. Rev., 2017, 117, 7964–8040 CrossRef CAS.
- Y. Xing, Y. Liu, R. Chen, Y. Li, C. Zhang, Y. Jiang, Y. Lu, B. Lin, P. Chen, R. Tian, X. Liu and X. Cheng, Lab Chip, 2021, 21, 1886–1896 RSC.
- M. J. Jebrail, A. H. Ng, V. Rai, R. Hili, A. K. Yudin and A. R. Wheeler, Angew Chem. Int. Ed. Engl., 2010, 49, 8625–8629 CrossRef CAS.
- X. Y. Du, Q. Li, G. Wu and S. Chen, Adv. Mater., 2019, 31, e1903733 CrossRef PubMed.
- Y. K. Jo and D. Lee, Small, 2020, 16, e1903736 CrossRef PubMed.
- J. G. Martin, M. Gupta, Y. Xu, S. Akella, J. Liu, J. S. Dordick and R. J. Linhardt, J. Am. Chem. Soc., 2009, 131, 11041–11048 CrossRef CAS.
- H. Li, X. Liu, F. Zhu, D. Ma, C. Miao, H. Su, J. Deng, H. Ye, H. Dong, X. Bai, Y. Luo, B. Lin, T. Liu and Y. Lu, Biosens. Bioelectron., 2022, 215, 114557 CrossRef CAS.
- D. Grissom and P. Brisk, Proceedings of the 50th Annual Design Automation Conference, 2013, pp. 1–9 Search PubMed.
- E. Jantscher-Krenn, T. Lauwaet, L. A. Bliss, S. L. Reed, F. D. Gillin and L. Bode, Br. J. Nutr., 2012, 108, 1839–1846 CrossRef CAS PubMed.
- S. Duboux, M. Golliard, J. A. Muller, G. Bergonzelli, C. J. Bolten, A. Mercenier and M. Kleerebezem, Sci. Rep., 2021, 11, 7236 CrossRef CAS PubMed.
- E. M. Moya-Gonzálvez, A. Rubio-Del-Campo, J. Rodríguez-Díaz and M. J. Yebra, Sci. Rep., 2021, 11, 23328 CrossRef PubMed.
- L. Bode, L. Kuhn, H. Y. Kim, L. Hsiao, C. Nissan, M. Sinkala, C. Kankasa, M. Mwiya, D. M. Thea and G. M. Aldrovandi, Am. J. Clin. Nutr., 2012, 96, 831–839 CrossRef CAS PubMed.
- L. Bode, Early Hum. Dev., 2015, 91, 619–622 CrossRef CAS PubMed.
- G. M. Ruiz-Palacios, L. E. Cervantes, P. Ramos, B. Chavez-Munguia and D. S. Newburg, J. Biol. Chem., 2003, 278, 14112–14120 CrossRef CAS PubMed.
- C. Chen, Y. Zhang, M. Xue, X. W. Liu, Y. Li, X. Chen, P. G. Wang, F. Wang and H. Cao, Chem. Commun., 2015, 51, 7689–7692 RSC.
- L. R. Ruhaak, C. Stroble, M. A. Underwood and C. B. Lebrilla, Anal. Bioanal. Chem., 2014, 406, 5775–5784 CrossRef CAS.
- Y. Li, H. Yu, Y. Chen, K. Lau, L. Cai, H. Cao, V. K. Tiwari, J. Qu, V. Thon and P. G. Wang, Molecules, 2011, 16, 6396–6407 CrossRef CAS.
- Y. Chen, V. Thon, Y. Li, H. Yu, L. Ding, K. Lau, J. Qu, L. Hie and X. Chen, Chem. Commun., 2011, 47, 10815–10817 RSC.
- H. Yu, K. Lau, V. Thon, C. A. Autran, E. Jantscher-Krenn, M. Xue, Y. Li, G. Sugiarto, J. Qu and S. Mu, Angew. Chem., Int. Ed., 2014, 53, 6687–6691 CrossRef CAS.
- M. M. Muthana, J. Qu, Y. Li, L. Zhang, H. Yu, L. Ding, H. Malekan and X. Chen, Chem. Commun., 2012, 48, 2728–2730 RSC.
- W. Yi, X. Liu, Y. Li, J. Li, C. Xia, G. Zhou, W. Zhang, W. Zhao, X. Chen and P. G. Wang, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 4207–4212 CrossRef CAS PubMed.
- G. Sugiarto, K. Lau, J. Qu, Y. Li, S. Lim, S. Mu, J. Ames, A. Fisher, X. Chen, X. Song, H. Yu, X. Chen, Y. Lasanajak, M. M. Tappert, G. M. Air, V. K. Tiwari, H. Cao, H. A. Chokhawala, H. Zheng, R. D. Cummings and D. F. Smith, J. Biol. Chem., 2011, 286, 31610–31622 CrossRef.
- D. B. Stein, Y. N. Lin and C. H. Lin, Adv. Synth. Catal., 2008, 350, 2313–2321 CrossRef CAS.
- M. Sun, Y. Li, H. A. Chokhawala, R. Henning and X. Chen, Biotechnol. Lett., 2008, 30, 671–676 CrossRef CAS PubMed.
- X. Chen, J. Fang, J. Zhang, Z. Liu, J. Shao, P. Kowal, P. Andreana and P. G. Wang, J. Am. Chem. Soc., 2001, 123, 2081–2082 CrossRef CAS PubMed.
- J. B. McArthur, H. Yu and X. Chen, ACS Catal., 2019, 9, 10721–10726 CrossRef CAS PubMed.
- K. Lau, V. Thon, H. Yu, L. Ding, Y. Chen, M. M. Muthana, D. Wong, R. Huang and X. Chen, Chem. Commun., 2010, 46, 6066–6068 RSC.
- H. Yu, H. Yu, R. Karpel and X. Chen, Bioorg. Med. Chem., 2004, 12, 6427–6435 CrossRef CAS PubMed.
- J. Cheng, H. Yu, K. Lau, S. Huang, H. A. Chokhawala, Y. Li, V. K. Tiwari and X. Chen, Glycobiology, 2008, 18, 686–697 CrossRef CAS PubMed.
- L. Li, Y. Liu, Y. Wan, Y. Li, X. Chen, W. Zhao and P. G. Wang, Org. Lett., 2013, 15, 5528–5530 CrossRef CAS PubMed.
- S. Anderson, B. Hadwen and C. Brown, Lab Chip, 2021, 21, 962–975 RSC.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4ra01395f |
| ‡ These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2024 |