Open Access Article
Open Access ArticleStructure-dependent detection of polyphenols using crown ether-immobilized gold nanoparticles†
Yuto Yamakia,
Hiroki Seob,
Akihiko Hatano c,
Manabu Suzukid and
Kenichi Niikura
c,
Manabu Suzukid and
Kenichi Niikura *ab
*ab
aDepartment of Applied Chemistry, Faculty of Fundamental Engineering, Nippon Institute of Technology, Japan
bGraduate School of Environmental Symbiotic System Major, Nippon Institute of Technology, Minamisaitama-Gun Saitama 345-8501, Japan. E-mail: niikura.kenichi@nit.ac.jp
cDepartment of Materials Science and Engineering, Shibaura Institute of Technology, Fukasaku, Minuma-ku, Saitama-City, Saitama 337-8570, Japan
dResearch & Development Center for Advanced Materials and Technology, Nippon Institute of Technology, Japan
First published on 24th May 2024
Abstract
Gold nanoparticles functionalized with 18-crown 6-ether (18C6-AuNPs) can be used for detection of tannic acid, epigallocatechin gallate, and epicatechin gallate by color change in the μg mL−1 range. 18C6-AuNPs were insensitive to L-ascorbic acid and L-tyrosine unlike conventional detection methods, such as Folin & Ciocalteu assay, whose principle is based on the redox reaction of polyphenols. Although 18C6-AuNPs did not respond to some polyphenols, such as gallic acid and epicatechin, if the polyphenols of interest are responsive to this approach, these are expected to be effective nanomaterial for simple sensing of polyphenols.
Polyphenols are a group of molecules that are widely found in plants and have multiple hydroxyl groups in their aromatic rings. Polyphenols are obtained from a variety of plant-derived foods and beverages, such as fruits, tea, coffee, red wine, vegetables, chocolate, and cereals.1 Polyphenol molecules remove oxygen radicals generated in the body; thus, polyphenols have been reported to have potential benefits for human health through their antioxidant properties.1–7 Further, the receptor proteins for green tea-derived polyphenols have already been identified,8 and research into the effects of these polyphenols on living organisms is progressing. In recent years, drug carriers have also been developed by focusing on the unique intermolecular forces and antioxidant effects of polyphenol molecules.9,10 In addition to foods and beverages, it is important to clarify the amount of polyphenols remaining in biological samples such as blood and urine.
As research on polyphenols increases, methods for sensing the amount and structure of polyphenols are becoming increasingly important. Polyphenol molecules contain aromatic rings, therefore they are detectable by High Performance Liquid Chromatography (HPLC) with an UV detector. For example, polyphenols contained in beverage, plant and biological samples have been separated and analyzed using HPLC or LC-MS.11–16 Recently, quantitative nuclear magnetic resonance spectroscopy was applied to polyphenol detection.17
Many polyphenol molecules have a phenol, 1,2-,1,3-dihydroxylbenzene (catechol or resorcinol) or 1,2,3-trihydroxylbenzene (pyrogallol) structure. The detection of polyphenols relies on redox reactions involving these structures, which can be monitored by electrochemical techniques such as cyclic voltammetry.18–20 For example, the detection of phenol and catechol at low concentrations (10−8 M) has been achieved using nanocarbon electrodes.21 Simultaneously detection of dihydroxybenzene (DHB) isomers were also achieved by electrochemical methods.22,23 Polyphenol oxidase-immobilized quantum dots (QDs) can be used as sensing elements based on the principle of fluorescence quenching due to charge transfer from quinone to quantum dots.24 However, electrochemical methods require special equipment, and the signals obtained are not easy to interpret.
A colorimetric detection method that utilizes the specific chemical bond formation between polyphenols and boronic acids has also been reported. The formation of boronic esters causes a decrease in pH, resulting in a color change in the pH indicator.25 The Folin & Ciocalteu method (F–C method) is a well-known practical method for quantifying total polyphenols.
The Folin & Ciocalteu assay (F–C assay) has been used as a quantitative method for the determination of total phenolic content based on color change.26,27 In fact, ISO Standards to determine total polyphenol content in tea is based on the F–C assay.28 The strong reducing power of polyphenols reduces phosphomolybdic acid, turning the solution blue. By examining the absorbance at 765 nm, the total phenolic content can be determined in units of gallic acid equivalents. Recently, total polyphenol amounts in biological samples have been determined using the F–C assay.29 However, the F–C reagent can also react with various reducing molecules in the assay sample other than polyphenols, such as ascorbic acid and tyrosine, resulting in an overestimation of the total polyphenol contents.29
In this study, we propose a simpler detection method that focuses on molecular recognition of polyphenols rather than their redox reactions. A specific interaction between phenol and 18-crown-6 ether (18C6) in the gas phase has been reported.30 It is further reported that catechol and 18C6 form a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
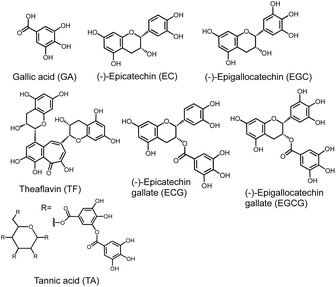
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 complex in cyclohexane, leading to an increase in the catechol fluorescence intensity.31 This allows fluorescence-based detection of catechol in the organic solvent. Recently, we found that 18C6 binds specifically to tannic acid (TA), which has five pyrogallol units (see Fig. 5), resulting in a turbid solution in buffer (pH 7.4), with K+ ions promoting the aggregation of 18C6-TA.32 Here, we applied the specific interaction of 18C6 with (poly)hydroxybenzenes to polyphenol sensing using 18C6-immobilized gold nanoparticles. Gold nanoparticles (referred to as AuNPs) undergo a clear color change when aggregated from monodispersion, making them useful as a material for colorimetric sensing.33–35 Many polyphenols contain multiple phenol, catechol, and pyrogallol structures, and these binding sites draw the gold nanoparticles closer together, causing a color change from red to blue. Importantly, AuNP-based polyphenol sensing is not hindered by reducing molecules such as ascorbic acid and tyrosine. We demonstrate that 18C6-immobilized AuNPs are excellent sensing elements for the convenient detection of tannic acid, epigallocatechin gallate, and epicatechin gallate, well-known polyphenol molecules.
1 complex in cyclohexane, leading to an increase in the catechol fluorescence intensity.31 This allows fluorescence-based detection of catechol in the organic solvent. Recently, we found that 18C6 binds specifically to tannic acid (TA), which has five pyrogallol units (see Fig. 5), resulting in a turbid solution in buffer (pH 7.4), with K+ ions promoting the aggregation of 18C6-TA.32 Here, we applied the specific interaction of 18C6 with (poly)hydroxybenzenes to polyphenol sensing using 18C6-immobilized gold nanoparticles. Gold nanoparticles (referred to as AuNPs) undergo a clear color change when aggregated from monodispersion, making them useful as a material for colorimetric sensing.33–35 Many polyphenols contain multiple phenol, catechol, and pyrogallol structures, and these binding sites draw the gold nanoparticles closer together, causing a color change from red to blue. Importantly, AuNP-based polyphenol sensing is not hindered by reducing molecules such as ascorbic acid and tyrosine. We demonstrate that 18C6-immobilized AuNPs are excellent sensing elements for the convenient detection of tannic acid, epigallocatechin gallate, and epicatechin gallate, well-known polyphenol molecules.
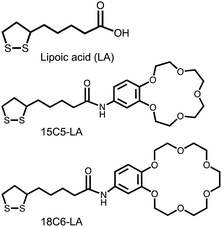
DL-α-Lipoic acid (LA) (see the chemical structure in Fig. 1) has been used as surface ligand enabling stable dispersion of AuNPs in water.36 A disulfide moiety is anchored to the surface of the AuNPs, and the carboxylate anion affords good dispersibility to the AuNPs in water. AuNPs were synthesized using citric acid and TA as reducing agents.37 Then, AuNPs were coated with LA ligands according to the literature,36 with some modifications (details in ESI†). Briefly, AuNPs were incubated with LAs in an aqueous solution under a basic condition (approx. pH 11.5) for two weeks. Scanning transmission electron microscopy (STEM) images of LA-AuNPs showed the spherical shape of the gold nanoparticles with a diameter of approximately 20 nm (Fig. 2). The hydrodynamic diameter of lipoic acid-coated-AuNPs (LA-AuNPs) in this basic solution was 20.8 nm, indicating that they were monodispersed (Fig. S5†) in the solution. Successful coating of LA molecules can be confirmed by the color in the buffer solution. While untreated gold nanoparticles aggregated in blue color, LA-AuNPs maintained red color in phosphate buffer (Fig. S6†). For Fig. S6,† high concetration of AuNPs was used to cleraly show the color change.
LA-AuNPs were concentrated 33 times by centrifugation in this solution. The two ligands shown in Fig. 1, 18C6-LA and 15C5-LA, were synthesized by a one-step condensation between lipoic acid and an amine-functionalized crown ether. 15C5-LA is a molecule known in the literature.38 Synthesis details, 1H-NMR and FTIR spectra are shown in ESI (Fig. S1–S3).† The purity of these two ligands was high at approximately 97% as judged by HPLC analysis (Fig. S4†). A methanolic solution of the 18C6 ligand was added to the concentrated basic solution of LA-AuNPs to produce 18C6-LA-immobilized AuNPs (18C6-AuNPs) via a partial ligand exchange reaction from LA to 18C6-LA on the AuNP surface. After purification by centrifugation, 18C6-AuNPs were dispersed in phosphate buffer (100 mM, pH 7.4) and used as the sensor solution. The 15C5-AuNPs were prepared using the same procedure. The zeta potentials of the LA-AuNPs and 18C6-AuNPs in phosphate buffer (pH 7.4) were −37.4 mV and −33.8 mV, respectively, suggesting that the exchange rate from the surface LA ligand to 18C6-LA is low at less than 10%. A similar increase of approximately 3 mV was observed for 15C5-AuNPs after ligand exchange. However, these low exchange rate allowed good dispersion of AuNPs in the buffer solution. Indeed, the hydrodynamic diameter of 18C6-AuNPs is about 19.5 nm, which is almost the same as that of LA-AuNPs (Fig. S5†).
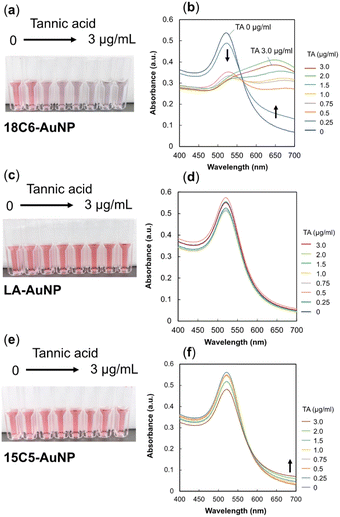
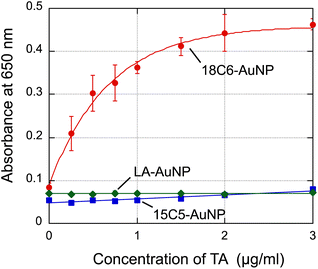
First, we tested tannic acid, which has five pyrogallol moieties, as a representative polyphenol. Fig. 3a shows a photograph of changes when tannic acid (0–3 μg mL−1 in each cuvette) was added to 18C6-AuNPs. The sensor solution was allowed to stand for 20 minutes until the color changed completely. Interestingly, TA concentrations as low as a few μg mL−1 induced a color change from red to purple. The spectral change in Fig. 3b shows an increase in absorbance at wavelengths longer than 600 nm, indicating that the gold nanoparticles aggregated.33 As the absorbance at 650 nm increases with aggregation (Fig. 3b), the increase in absorbance at 650 nm was used as an indicator of aggregation. The addition of TA was also performed for LA-AuNPs and 15C5-AuNPs (Fig. 3c–f). In these cases, no color change was observed. In the spectra for 15C5-AuNPs (Fig. 3f), a slight increase in absorbance at 650 nm was observed for 15C5, but the increase was much smaller than that for 18C6. The absorbance changes observed at 650 nm for the three types of AuNPs upon addition of TA are summarized in Fig. 4, indicating that 18C6-AuNPs show a specific response to TA. Importantly, the detection sensitivity of 18C6-AuNPs to TAs is much lower than the concentration (sub mg mL−1) at which a mixed solution of free 18-crown 6-ether and TA becomes cloudy, meaning that 18C6-LA AuNPs are a higher affinity to TA than free 18C6 molecules.
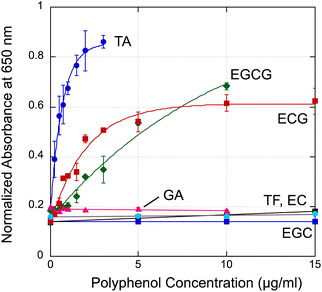
Next, we tested six types of polyphenols; gallic acid hydrate (GA), (−)-epigallocatechin gallate (EGCG), (−)-epigallocatechin (EGC), (−)-epicatechin (EC), (−)-epicatechin gallate (ECG), and theaflavin (TF) (see chemical structures in Fig. 5). EGCG, EGC, EC, ECG are polyphenols found in green tea and theaflavin is found in black tea.39 To eliminate errors caused by differences in the concentration of gold nanoparticles between each assay solution, the absorbance at 520 nm of a 18C6–AuNP solution without polyphenols was normalized to 1. Fig. 6 shows the change in normalized absorbance at 650 nm. Typical absorbance spectra upon addition of each sample are shown in Fig. S7.† 18C6-AuNPs responded to three (TA, ECG, EGCG) of the seven types of polyphenols tested this time.
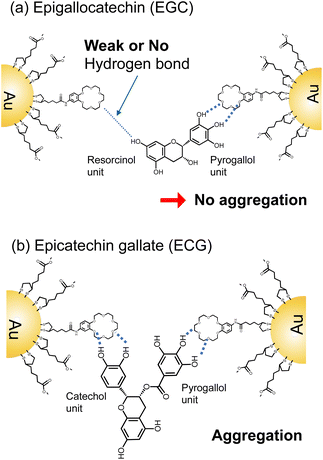
The speculated binding model is summarized in Fig. 7. At least two binding sites are essential to bring AuNPs closer together. In this illustration using EGC and ECG as an example. There are no responses observed for EG, EGC, and TF, therefore it means the resorcinol units do not induce the aggregation of 18C6-AuNPs due to weak or no hydrogen bond (Fig. 7a). Thus, we assumed that the catechol and pyrogallol units in epicatechin gallate (ECG) were each attached to 18C6 through hydrogen bonds, resulting in the aggregation of two gold nanoparticles (Fig. 7b). ECG, having a catechol and pyrogallol units, induced aggregation of 18C6-AuNPs similarly to EGCG having two pyrogallol units. This suggests that catechol and pyrogallol unit have similar affinity to 18-crown 6-ether. Tannic acid is thought to exhibit the strongest aggregation because it has five pyrogallols. Although only hydrogen bonds are shown here, 18-crown 6-ether and the benzene ring of polyphenols also contribute to the specific interaction.30,31 Currently, to test these hypotheses, we are currently synthesizing molecules with a single isomer of catechol or resorcinol structure.
As the F–C assay is not specific for polyphenols, reducing compounds other than polyphenols, such as ascorbic acid, tyrosine, can also cause color changes, potentially overestimating the total polyphenol content.29 The effects of ascorbic acid and tyrosine on the AuNP sensor were, therefore, investigated. The addition of ascorbic acid or tyrosine at concentrations as high as 100 μg mL−1 had no effect on the adsorption of 18C6-AuNPs (Fig. 8). Conversely, in the F–C assay, these reagents increased the absorbance at 765 nm by about one-half that of GA (Fig. S8†). A major advantage of our sensing system is that it is not affected by reducing substances, such as ascorbic acid.
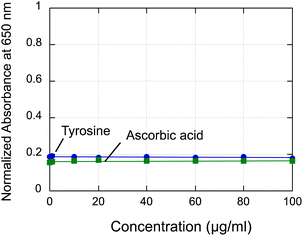 | ||
| Fig. 8 Change in normalized absorbance at 650 nm of sensor solutions with increasing L-tyrosine and L-ascorbic acid concentrations. | ||
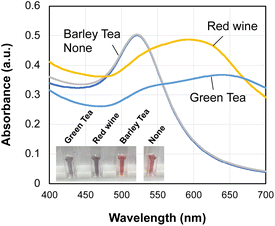
Finally, we tested three commercially available beverages: red wine, green tea, and barley tea. Green tea and barley tea were brewed before assay, and red wine was purchased from the market and used as is. An aliquot (20 μL) of the beverage was added to the sensor solution (1 mL) and allowed to stand for 20 minutes. UV-vis spectra and pictures are shown in Fig. 9. A color change in the gold nanoparticles was observed for green tea and red wine, but not for barley tea. It is reported that green tea and red wine, but not for barley tea, contain abundant polyphenols.40 The major polyphenols in green tea are ECG and EGCG,2,41 and these polyphenols are thought to induce the aggregation of AuNPs. Red wine has the highest polyphenol content among the beverages tested, and condensed tannin molecules with multiple catechol and pyrogallol units42 may be bound to gold nanoparticles. Crown ether is a representative molecule used in supramolecular chemistry for the specific recognition of ammonia and alkali metal ions.43–45 Our results further expand the use of crown ethers and demonstrate their effectiveness as a new and simple structure-dependent detection method for polyphenols.
In summary, we detected polyphenols, such as TA, ECG and EGCG, in a structure-dependent manner through color change using 18-crown 6-ether-functionalized gold nanoparticles. This method is based on supramolecular chemistry in which polyphenols recognize 18C6, and is a simple and new detection method that differs in principle from conventional redox-based assays. Our method has the advantage of not being affected by non-polyphenol antioxidants such as ascorbic acid. Polyphenol-rich beverages such as red wine and green tea responded to 18C6-AuNP sensor. This indicates that the 18C6-AuNP sensor is expected to be an effective nanomaterial for easy sensing of polyphenols in beverages without pretreatment.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This research was financially supported by JSPS KAKENHI Grant Number 22K04878. For the ESI-MS measurements, we thank Ms S. Kado of the Center for Analytical Instrumentation, Chiba University.Notes and references
- A. Scalbert, I. T. Johnson and M. Saltmarsh, Am. J. Clin. Nutr., 2005, 81, 215S CrossRef CAS PubMed.
- M. Kobayashi, T. Kawano, Y. Ukawa, Y. M. Sagesaka and I. Fukuhara, Food Funct., 2016, 7, 498 RSC.
- A. Rana, M. Samtiya, T. Dhewa, V. Mishra and R. E. Aluko, J. Food Biochem., 2022, 46, e14264 CrossRef CAS PubMed.
- U. Etxeberria, A. Fernandez-Quintela, F. I. Milagro, L. Aguirre, J. A. Martinez and M. P. Portillo, J. Agric. Food Chem., 2013, 61, 9517 CrossRef CAS PubMed.
- H. Cory, S. Passarelli, J. Szeto, M. Tamez and J. Mattei, Front. Nutr., 2018, 5, 87 CrossRef PubMed.
- N. A. M. Toriigahara, M. Shibata, T. Ishii, T. Nakayama and N. Osakabe, J. Funct. Foods, 2014, 10, 355 CrossRef.
- L. Xing, H. Zhang, R. Qi, R. Tsao and Y. Mine, J. Agric. Food Chem., 2019, 67, 1029–1043 CrossRef CAS PubMed.
- H. Tachibana, K. Koga, Y. Fujimura and K. Yamada, Nat. Struct. Mol. Biol., 2004, 11, 380 CrossRef CAS.
- J. Zhou, Z. Lin, Y. Ju, M. A. Rahim, J. J. Richardson and F. Caruso, Acc. Chem. Res., 2020, 53, 1269 CrossRef CAS PubMed.
- C. Liu, W. Shen, B. Li, T. Li, H. Chang and Y. Cheng, Chem. Mater., 2019, 31, 1956 CrossRef CAS.
- M. Kanlayavattanakul, M. Khongkow, W. Klinngam, P. Chaikul, N. Lourith and P. Chueamchaitrakun, Sci. Rep., 2024, 14, 2253 CrossRef CAS PubMed.
- J. Bayer and P. Högger, J. Pharm. Biomed. Anal., 2024, 239, 115914 CrossRef CAS.
- L. Lian, Y. Cui, M. Wen, D. Granato, Y.-Q. Xu, Y.-Q. Fu, Z. Jiang, J. Ke, X. Wan and L. Zhang, Food Biosci., 2024, 57, 103480 CrossRef CAS.
- R. J. Pachulicz, L. Yu, B. Jovcevski, V. Bulone and T. L. Pukala, Food Funct., 2022, 13, 8585 RSC.
- M. F. Alves, A. Katchborian-Neto, P. C. P. Bueno, F. Carnevale-Neto, R. Casoti, M. S. Ferreira, M. Murgu, A. C. C. de Paula, D. F. Dias, M. G. Soares and D. A. Chagas-Paula, RSC Adv., 2024, 14, 10481 RSC.
- L. A. Orozco-Flores, E. Salas, B. Rocha-Gutiérrez, M. D. R. Peralta-Pérez, G. González-Sánchez and L. Ballinas-Casarrubias, ACS Omega, 2024, 9, 196 CrossRef CAS PubMed.
- D.-Y. Shi, Y. Zheng, Q.-S. Guo, C. Gong, X. Xu and J.-P. Gao, Anal. Methods, 2023, 15, 6561 RSC.
- M. Guo, Z. Wang, Z. Wang and L. Zhang, Anal. Methods, 2023, 15, 1378 RSC.
- P. A. Raymundo-Pereira, T. A. Silva, F. R. Caetano, L. Ribovski, E. Zapp, D. Brondani, M. F. Bergamini, L. H. Marcolino Jr, C. E. Banks, O. N. Oliveira Jr, B. C. Janegitz and O. Fatibello-Filho, Anal. Chim. Acta, 2020, 198 CrossRef CAS PubMed.
- H. S. Ali and Y. Yardım, Food Chem., 2024, 441, 138262 CrossRef PubMed.
- L. Jiang, I. Santiago and J. Foord, Commun. Chem., 2018, 1, 43 CrossRef.
- R. Thangaraj, N. Manjula and A. S. Kumar, Anal. Methods, 2012, 4, 2922 RSC.
- N. Vishnu, M. Gandhi, S. Badhulika and A. S. Kumar, Anal. Methods, 2018, 10, 2327 RSC.
- U. S. Akshath, L. R. Shubha, P. Bhatt and M. S. Thakur, Biosens. Bioelectron., 2014, 57, 317 CrossRef CAS.
- Y. Hu, L. Shao, Y. Fan, L. Lu, C. Zhou, H. Fu and Y. She, Dyes Pigm., 2022, 203, 110326 CrossRef CAS.
- M. Y. Peker, Ö. K. Koç, A. Üzer and R. Apak, ACS Appl. Polym. Mater., 2024, 6, 1864 CrossRef.
- M. Pérez, I. Dominguez-López and R. M. Lamuela-Raventós, J. Agric. Food Chem., 2023, 71, 17543 CrossRef.
- ISO 14502-1:2005. Determination of Substances Characteristic of Green and Black Tea — Part 1: Content of Total Polyphenols in Tea — Colorimetric Method using Folin – Ciocalteu Reagent Search PubMed.
- I. Dominguez-López, M. Pérez and R. M. Lamuela-Raventós, Crit. Rev. Food Sci. Nutr., 2023 DOI:10.1080/10408398.2023.2220031.
- R. Kusaka, Y. Inokuchi, T. Haino and T. Ebata, J. Phys. Chem. Lett., 2012, 3, 1414 CrossRef CAS PubMed.
- F. Morishima, R. Kusaka, Y. Inokuchi, T. Haino and T. Ebata, J. Phys. Chem. B, 2015, 119, 2557 CrossRef CAS PubMed.
- S. Endo, M. Sarai, A. Hatano and K. Niikura, Chem. Lett., 2023, 52, 455 CrossRef CAS.
- V. Chegel, O. Rachkov, A. Lopatynskyi, S. Ishihara, I. Yanchuk, Y. Nemoto, J. P. Hill and K. Ariga, J. Phys. Chem. C, 2012, 116, 2683 CrossRef CAS.
- R. Iida, H. Mitomo, Y. Matsuo, K. Niikura and K. Ijiro, J. Phys. Chem. C, 2016, 120, 15846 CrossRef CAS.
- K. Saha, S. S. Agasti, C. Kim, X. Li and V. M. Rotello, Chem. Rev., 2012, 112, 2739 CrossRef CAS PubMed.
- J. Tournebize, A. Boudier, A. Sapin-Minet, P. Maincent, P. Leroy and R. Schneider, ACS Appl. Mater. Interfaces, 2012, 4, 5790 CrossRef CAS PubMed.
- G. Tsutsui, S. Huang, H. Sakaue, S. Shingubara and T. Takahagi, Jpn. J. Appl. Phys., 2001, 40, 346 CrossRef CAS.
- I. Turcu, I. Zarafu, M. Popa, M. C. Chifiriuc, C. Bleotu, D. Culita, C. Ghica and P. Ionita, Nanomaterials, 2017, 7, 43 CrossRef CAS PubMed.
- L. K. Leung, Y. Su, R. Chen, Z. Zhang, Y. Huang and Z. Y. Chen, J. Nutr., 2001, 131, 2248 CrossRef CAS PubMed.
- Y. Fukushima, T. Ohie, Y. Yonekawa, K. Yonemoto, H. Aizawa, Y. Mori, M. Watanabe, M. Takeuchi, M. Hasegawa, C. Taguchi and K. Kondo, J. Agric. Food Chem., 2009, 57(4), 1253 CrossRef CAS PubMed.
- D. Botten, G. Fugallo, F. Fraternali and C. Molteni, J. Phys. Chem. B, 2015, 119, 12860 CrossRef CAS PubMed.
- B. Teng, Y. Hayasaka, P. A. Smith and K. A. Bindon, J. Agric. Food Chem., 2021, 69, 4804 CrossRef CAS PubMed.
- S. D. P. Fielden, D. A. Leigh, C. T. McTernan, B. Pérez-Saavedra and I. J. Vitorica-Yrezaba, J. Am. Chem. Soc., 2018, 140, 6049 CrossRef CAS PubMed.
- S. Li, L. Lin, W. Wang, X. Yan, B. Chen, S. Jiang, S. Liu, X. Ma, H. Tian and X. Yu, Chem. Commun., 2020, 56, 5552 RSC.
- G. W. Gokel, W. M. Leevy and M. E. Weber, Chem. Rev., 2004, 104, 2723 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Syntheses of ligands, supporting spectra. See DOI: https://doi.org/10.1039/d4ra02182g |
| This journal is © The Royal Society of Chemistry 2024 |