DOI:
10.1039/D4RA03003F
(Paper)
RSC Adv., 2024,
14, 18519-18527
Synthesis, antimicrobial activity and application of polymers of praseodymium complexes based on pyridine nitrogen oxide†
Received
22nd April 2024
, Accepted 27th May 2024
First published on 10th June 2024
Abstract
The traditional pyridine nitrogen oxide-based antimicrobial agents are often associated with health risks due to heavy metal enrichment. To mitigate this concern, we synthesized two novel complexes, Pr2(mpo)6(H2O)2 and Pr(hpo)(mpo)2(H2O)2, and integrated rare-earth salts, Hhpo (2-hydroxypyridine-N-oxide) and Nampo (2-mercapto-pyridine-N-oxide sodium salt). These complexes were characterized through infrared analysis, elemental analysis, thermogravimetric analysis, and X-ray crystallographic analysis. Our comparative analyses demonstrate that the synthesized rare-earth complexes exhibit stronger antimicrobial activity against Staphylococcus aureus (S. aureus ATCC6538) and Escherichia coli (E. coli ATCC25922) compared to the ligands and rare-earth salts alone. Quantitative results revealed the lowest inhibitory concentrations of the two complexes against S. aureus ATCC6538 and E. coli ATCC25922 at 3.125 μg mL−1, 6.25 μg mL−1, 3.125 μg mL−1 and 6.25 μg mL−1, respectively. Preliminary investigations indicated that the antibacterial mechanism of these complexes involved promoting intracellular substance exudation to achieve antibacterial effects. Incorporation of these complexes into polymeric antimicrobial films resulted in a potent antimicrobial effect, achieving a 100% inhibition rate against S. aureus ATCC6538 and E. coli ATCC25922 at a low addition level of 0.6 wt%. Our results suggest that nitrogen oxide-based praseodymium complexes have potential for various antimicrobial applications.
1. Introduction
Microorganisms are ubiquitous and pose significant threats to human beings.1 Rapid bacterial growth leads to the infection of other organisms, competition for resources and habitat, and the generation of toxic by-products that can harm both organisms and the ecosystem.2 Infections caused by microorganisms often lead to irreversible damage and immeasurable losses.3,4 Generally, the use of antimicrobial agents is an effective and important approach in combating microbial threats and controlling the growth of bacteria on the surfaces of materials. Bacteria can be categorized as Gram-positive (G+) or Gram-negative (G−) based on the Gram staining method. The method entails treating bacteria with the amethyst dye and iodine and then assessing their staining characteristics based on the presence of cytosolic polysaccharides and rhamnocytidylic acid in their cell walls. This classification divides bacteria into distinct categories, each displaying notable variances in growth attributes, resistance to drugs, and pathogenic tendencies.5 Hence, there is a global focus on the search for highly effective antimicrobial agents with broad-spectrum activity and minimal side effects.6–8
Pyridine nitrogen oxides are a class of common organic compounds containing pyridine, which are extensively utilized in pharmaceutical synthesis and other fields due to their diverse synthesis pathways, superior biological activity, ease of synthesis, and excellent chelating ability.9–21 Literature reports have demonstrated that complexes of pyridine nitrogen oxides and transition metals exhibit considerable anticancer and antibacterial activities,22,23 making them valuable in biomedicine and as antimicrobial materials.
The commonly available pyridine nitrogen oxide-based complex antimicrobials in the market are often detrimental to the environment and organisms as they contain heavy metals like copper and zinc.24,25 Advancements in the rare-earth industry have, however, propelled the active development of rare-earth-based antimicrobial materials,26–35 including various pyridine derivative complexes with metal ions;36–40 these rare-earth antimicrobials demonstrate favorable attributes of safety, environmental friendliness, and stability.41 In this study, we aimed to synthesize a new class of complexes with potential antimicrobial applications by substituting heavy metals with rare-earth ions coordinated with pyridine nitrogen oxides. The antimicrobial properties of the resulting products were assessed by incorporating them into polymeric antimicrobial packaging films. To achieve this, 2-hydroxypyridine-N-oxide (Hhpo) and 2-mercaptopyridine-N-oxide sodium salt (Nampo) were reacted with praseodymium chloride hexahydrate, yielding two metal complexes: Pr2(mpo)6(H2O)2 (1) and Pr(hpo)(mpo)2(H2O)2 (2). These complexes were then added into polylactic acid and polyethylene polymer to produce a series of antimicrobial packaging films. The antimicrobial activity of these complexes and films was assessed through in vitro resistance studies against S. aureus ATCC6538 and E. coli ATCC25922. Overall, this study demonstrates significant innovation and importance in the design of novel antimicrobial agents, the study of antimicrobial mechanisms, antimicrobial agent-modified polymer materials, and cross-disciplinary research (Scheme 1).
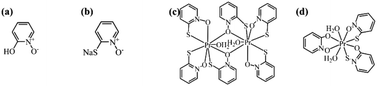 |
| | Scheme 1 The molecular structures of (a) Hhpo; (b) Nampo; (c) complex 1 Pr2(mpo)6(H2O)2; (d) complex 2 Pr(hpo)(mpo)2(H2O)2. | |
2. Experimental
2.1. Material
2-Hydroxypyridine-N-oxide (H-hpo) and 2-mercaptopyridine-N-oxide sodium salt (Na-mpo) were procured from Shanghai Bi De Pharmaceutical Technology Co. Ltd. Sodium hydroxide and praseodymium chloride hexahydrate were sourced from Aladdin Chemical Reagent Co. Ltd. Gram-negative bacterium (E. coli, ATCC25922) and Gram-positive bacterium (S. aureus, ATCC6538) were obtained from Beijing Preservation Biotechnology Co. Ltd. Blank drug-sensitive filter paper (sterilised, 5 mm diameter and 1 mm thickness), LB liquid medium (L1050), LB solid medium (L1010), 96-well plates (ABC796006) for bacterial culture, SCDLP medium (CM701), and NB medium (N8300) were purchased from Xiamen Hao he Biotechnology Co. Ltd. Polypropylene (PP, T30S) was sourced from China National Petroleum Co. Ltd. Polylactic acid (PLA, 601) was purchased from Anhui Feng yuan Co. Ltd. Deionized water, essential for all experiments, was prepared using a PSDK1-20-C ultrapure water system. All drugs employed in this study were of analytical purity.
2.2. Characterization
The crystal structure of the complexes was determined using a Bruker single-crystal diffractometer (XRD, Bruker D8 Venture). Surface morphology analysis of the complex samples was conducted with a scanning electron microscope (SEM, Apreo S Lo Vac). Fourier transform infrared spectroscopy (FTIR) spectra were obtained in ATR mode utilizing an infrared spectrometer (Thermo Nicolet is50 spectrometer) with a resolution of 4 cm−1, scanning in the range of 4000–525 cm−1. Praseodymium content in the complexes was quantitated using an inductively coupled plasma emission spectrometer (ICP-OES, JY-Horiba ICP-OES Ultima 2), while the C, H, O, N, and S contents were determined using an elemental analyzer (EA, Elementar Vari EL Cube). The thermal stability of the samples was assessed using a thermogravimetric analyzer (STA, Mettler-Toledo TGA/DSC 1) under an argon atmosphere, employing a heating rate of 10 K min−1 within a temperature range of 30–800 °C. To prepare antimicrobial plastic plates, samples were mixed on a double screw extruder (TSE-20, Co-rotating Twin Screw Extruder) and subsequently processed using a desktop casting machine (FDSI, Cast Film Testing Machine). Antimicrobial experimental procedures were conducted within an ultra-clean bench (ZHJH-C1109B). Bacterial suspension concentrations were measured using a spectrophotometer (Nano-300, Micro-spectrophotometer). Bacterial cultures were maintained in a constant temperature oscillator (SHZ-82A) and a biochemical incubator (SHP-080).
2.3. Synthesis of complex 1
Nampo (0.67 g, 4.5 mmol) was dissolved in 15 mL of H2O to create solution 1. Praseodymium chloride hexahydrate (0.53 g, 1.5 mmol) was dissolved in 15 mL of H2O to form solution 2. A U-shaped tube was utilized, and 30 mL of H2O was introduced into it. Solution 1 was slowly added to the left end of the tube, while solution 2 was carefully dropped into the right end. The tube's opening was tightly sealed with sealing film. The U-tube was left at room temperature for five days. After this period, green transparent crystals precipitated. The crystals were separated via filtration, washed repeatedly with deionized water, and vacuum-dried. The yield was 76.95%. Anal. calc. for C30H28N6O8Pr2S6: Pr, 26.240; C, 33.525; H, 2.608; O, 11.920; N, 7.823; S, 17.880. Found: Pr, 26.123; C, 33.126; H, 2.578; O, 12.572; N, 7.929; S, 17.678. IR (cm−1): 3209s, 1593s, 1538s, 1452w, 1413m, 1262s, 1214w, 1192m, 1157s, 1140m, 1083m, 1033s, 952s, 839m, 823m, 753w, 701w, 594w, 579m, 541m.
2.4. Synthesis of complex 2
Nampo (0.45 g, 3 mmol) and Hhpo (0.17 g, 1.5 mmol) were dissolved in 15 mL of H2O, and the pH was adjusted to 7, forming solution 3. Praseodymium chloride hexahydrate (0.53 g, 1.5 mmol) was dissolved in 15 mL of H2O to create solution 4. A U-shaped tube was employed, and 30 mL of H2O was introduced into it. Solution 3 was slowly added along the left end of the tube, while solution 4 was introduced along the right end. The U-tube's opening was securely sealed with sealing film. The sealed U-tube was allowed to stand at room temperature; after seven days, dark green opaque crystals precipitated. The crystals were isolated by filtration, washed multiple times with deionized water, and vacuum-dried. The yield was 78.03%. Anal. calc. for C15H16N3O6PrS2: Pr, 26.146; C, 33.401; H, 2.969; O, 17.814; N, 7.794; S, 11.876. Found: Pr, 25.520; C, 33.368; H, 2.946; O, 18.136; N, 7.805; S, 12.162. IR (cm−1): 3103s, 1673s, 1616s, 1595s, 1520w, 1455w, 1412m, 1361s, 1267s, 1199w, 1146w, 1119s, 1088m, 1034s, 973s, 924s, 882s, 830w, 786m, 755w, 740w, 708w, 608m, 592w, 544w.
2.5. X-ray crystallography
The diffraction data for complex 1 and complex 2 were collected on a Bruker single crystal diffractometer at 200 K, which was equipped with a graphite-monochromated MoKα (λα = 0.71073 Å) radiation. The structure factors were obtained after Lorentz and polarization. An empirical absorption correction based on “multi-scan” was applied to the data. The positions of some of the heavy atoms, including the praseodymium atoms, were located by the direct method or Patterson method of the SHELXS program, while the remaining atoms were found in a series of alternating difference Fourier maps and least-square refinements. The hydrogen atoms were added using the HADD command and refined as riding atoms. Basic information pertaining to crystal parameters and structure refinement was summarized in Table S1.† The important bond lengths and bond angles are listed in Tables S2–S7.† The CCDC (2292292 for complex 1, 2292293 for complex 2) contains the supplementary crystallographic data for this study.†
2.6. Filter paper disc diffusion
We evaluated the antimicrobial properties using the filter paper disc diffusion method.42 The two ligands, two complexes, and rare earth salt ions were each dissolved in dimethyl sulfoxide (DMSO) to prepare a 0.5 mg mL−1 solution. The LB solid medium (L1010) was dissolved in water, sterilized through boiling, and poured into a suitable plate. Upon agar solidification, 120 μL of the tested strains (S. aureus ATCC6538 or E. coli ATCC25922) at a concentration of 106 CFU mL−1 were uniformly inoculated onto the agar's surface. Specific antimicrobial drug-sensitive filter paper (sterilised, 5 mm diameter and 1 mm thickness), containing antimicrobial compounds, was positioned at five predetermined spots on the agar plate. These treated agar plates were inverted and placed in an incubator at 37 °C for 24 h. The antimicrobial performance was evaluated by measuring the size of the five inhibition zones. Furthermore, the antimicrobial activities of the rare earth complexes were assessed at various concentrations (50, 100, 150, 200, and 250 μg mL−1) using the same filter paper disc diffusion method.
2.7. Minimum inhibitory concentration (MIC)
The MIC of the rare earth complexes was determined using a stepwise twofold dilution method.43 The bacterial suspension, prepared at a concentration of 106 CFU mL−1 using LB liquid medium, served as the test medium. A stock solution of the complex, prepared at 0.5 mg mL−1 concentration using water as the solvent, was mixed with the bacterial suspension in a test tube. The mixture comprised 180 μL of the bacterial stock solution and 20 μL of the sample stock solution. The mixture was well-shaken, and 200 μL of this co-culture solution was dispensed into the first well of a 96-well plate. Subsequent dilutions followed a twofold dilution scheme, with 100 μL of the diluted bacterial solution added to each successive well, except for the starting wells. This dilution process continued until the eleventh well. A control group was included where sterile purified water replaced the sample solution. The 96-well plate was then placed in a constant temperature incubator at 37 °C for 24 h, and the MIC values were subsequently observed and recorded.
2.8. Preparation of antimicrobial films
To evaluate the antimicrobial properties of antimicrobial plastics, the lamination method was employed. The two complexes were incorporated into polypropylene (PP) and polylactic acid (PLA) at varying concentrations: 0 wt%, 0.2 wt%, 0.4 wt%, and 0.6 wt%. The mixtures were processed into uniform flakes using a double screw extruder and desktop casting machine. The specific formulations of various antimicrobial films are shown in Table 2. Fig. S1† shows the schematic process of LC1/0.2% cast film. After drying PLA at 80 °C for 12 h, the rare earth compounds and PLA were weighed according to the ratio in Table 2, and uniformly mixed, and then pelletized using a twin-screw extruder, with the temperatures of zones set at 180 °C, 180 °C, 180 °C, 185 °C, 185 °C, and 180 °C, respectively, and the screw rotating speed being 200 rpm, and the charging speed being 10 rpm. The cast film was prepared using a desktop casting machine, with temperatures of 165 °C, 180 °C, and 190 °C for barrel zones, 195 °C for the transition zone, and 200 °C, 200 °C, 205 °C, and 205 °C for mold zones, respectively. The screw speed was 30 rpm, the line speeds of the casting rollers and the haul-off rollers were 1 rpm, and the tension of the take-up rollers was 25%. Other antimicrobial films were prepared in a similar manner. Specimens of size 50 mm × 50 mm were cut from the sheet for testing using the film affixing method.
2.9. Antibacterial activity on plastics surfaces
In the film affixing tests, the prepared antimicrobial plastic specimens were placed in appropriately sized sterile Petri dishes (90 cm in diameter) with the test surfaces facing upwards. A bacterial solution, with a concentration of 106 CFU mL−1 achieved by dilution with NB medium, was prepared. Subsequently, 0.4 mL of the bacterial solution was added to the center of each specimen's surface. A sterile cling film measuring 40 mm × 40 mm was placed over the solution to allow the solution to overspread without overflowing to the edges. The Petri dishes were covered and incubated at 37 °C with a relative humidity of at least 90% for 24 h. After incubation, the specimens were gently removed from the plastic wrap. The test surface was rinsed with 10 mL of SCDLP culture solution. Next, the specimens were taken out, and the culture solution was diluted using sterile water in a ten-fold gradient. Subsequently, 30 μL of the solution was added to a 6 cm diameter Petri dish containing culture medium dropwise. The plate was evenly coated with glass beads, inverted, and incubated for 24 h. After incubation, the antimicrobial effect of the different specimens was assessed by counting the number of colonies on the plate with the same dilution gradient.
2.10. Nucleic acid leaching
Note that 10 mL of saline containing E. coli ATCC25922 and S. aureus ATCC6538 at a concentration of 108 CFU mL−1 were treated with 2.5 mg of complex 1 and complex 2, respectively. The mixture was incubated at 37 °C for 24 h. After incubation, the supernatant was obtained by centrifugation at 4000 rpm for 2 min. The concentration of released nucleic acids was determined by measuring the UV absorbance of the sample at 260 nm, which was then converted in the supernatant to DNA and RNA concentrations using Beer–Lambert law. Control experiments were conducted using cells immersed in saline solutions without complex treatment.
2.11. Electron microscope morphology
Note that 2.5 mg of complex 1 and complex 2 were added separately to 10 mL of saline solution containing S. aureus ATCC6538 and E. coli ATCC25922 at a concentration of 108 CFU mL−1. The mixtures were incubated for 24 h. Subsequently, the complexes were separated by centrifugation at 4000 rpm for 2 min. To eliminate the complexes, the remaining mixtures were washed three times with a PBS buffer. The control group consisted of bacterial cells cultured in saline without the addition of complexes. Following complex removal, the three bacterial groups were fixed with 5 mL of 2.5 wt% glutaraldehyde at 4 °C for 24 h. After three times washing with PBS, the bacterial cells underwent a sequential dehydration process using varying concentrations of ethanol (30 vol%, 50 vol%, 70 vol%, 90 vol%, 100 vol%) for 15 min. Finally, the bacterial cells were suspended in anhydrous ethanol (100 vol%, AR). A volume of 0.2 mL of the cell suspension was added dropwise to a glass slide and allowed to air dry. The morphology of the cells was subsequently examined using scanning electron microscopy (SEM).
3. Results and discussion
3.1. Characterization of antimicrobial agents
The rare earth complexes were synthesized using a low concentration diffusion method. complex 1, formed by adding Na-mpo and PrCl3·6H2O in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 ratio, and complex 2, created by combining H-hpo, Na-mpo and PrCl3·6H2O in a 1
3 ratio, and complex 2, created by combining H-hpo, Na-mpo and PrCl3·6H2O in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio, were characterized through elemental analysis and X-ray single crystal analysis, revealing structures in the triclinic space group of P
1 ratio, were characterized through elemental analysis and X-ray single crystal analysis, revealing structures in the triclinic space group of P![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) and P1, respectively, as shown in Fig. 1(a). The crystal structure of complex 1 exhibits a distorted-octahedral geometry with each Pr(III) ion coordinated by three sulfur atoms, two oxygen atoms, two μ2-oxygen atoms from mpo ligands, and one coordinated water molecule.44 Weak π–π stacking interactions between pyridine-ring of mpo ligands are observed as shown in Fig. 1(b). Complex 2 displays a distorted dodecahedral geometry, with Pr(III) ion coordinated by two sulfur atoms, two oxygen atoms from mpo ligands, two oxygen atoms from hpo ligand, and two coordinated water molecules, shown in Fig. 1(c). Two-dimensional layered structures are formed with alternative weak π–π interactions, shown in Fig. 1(d).
and P1, respectively, as shown in Fig. 1(a). The crystal structure of complex 1 exhibits a distorted-octahedral geometry with each Pr(III) ion coordinated by three sulfur atoms, two oxygen atoms, two μ2-oxygen atoms from mpo ligands, and one coordinated water molecule.44 Weak π–π stacking interactions between pyridine-ring of mpo ligands are observed as shown in Fig. 1(b). Complex 2 displays a distorted dodecahedral geometry, with Pr(III) ion coordinated by two sulfur atoms, two oxygen atoms from mpo ligands, two oxygen atoms from hpo ligand, and two coordinated water molecules, shown in Fig. 1(c). Two-dimensional layered structures are formed with alternative weak π–π interactions, shown in Fig. 1(d).
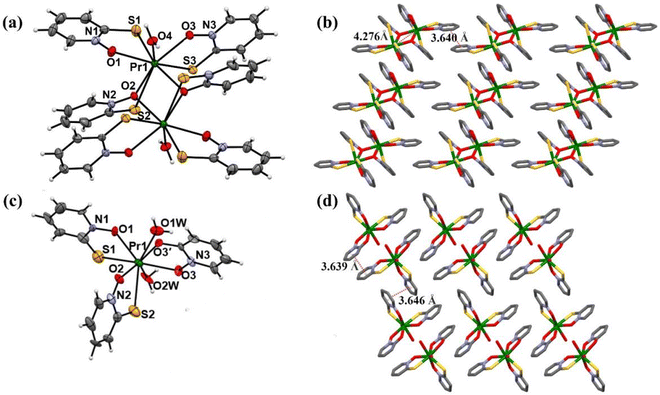 |
| | Fig. 1 (a) Thermal ellipsoids for complex 1; (b) molecular packing structure of complex 1, viewed along the b-axis; (c) thermal ellipsoids for complex 2; (d) molecular packing structure of complex 2, viewed along the a-axis. | |
Infrared spectroscopy highlights the interaction between rare earth metal ions and ligands, revealing distinct shifts in vibration bands. The stretching modes of the pyridine ring (1600–1500 cm−1) and N–O group (1020–825 cm−1) were identified as shown in Fig. 2(a). Comparing Nampo in complex 1 and complex 2, shifts in the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O of Hhpo from 1627 cm−1 to 1615 cm−1 and N–O from 892 cm−1 to 830 cm−1 suggest the occurrence of coordination reactions. Fig. 2(b) presenting a thermal stability analysis under argon indicates a three-stage mass loss process for both complexes of 1 and 2, with complex 1 exhibiting 7.7 wt% coordinated water weight loss (37 °C to 179 °C) and decomposition initiating at 261 °C, meeting polymer processing requirements for PLA and PP.
O of Hhpo from 1627 cm−1 to 1615 cm−1 and N–O from 892 cm−1 to 830 cm−1 suggest the occurrence of coordination reactions. Fig. 2(b) presenting a thermal stability analysis under argon indicates a three-stage mass loss process for both complexes of 1 and 2, with complex 1 exhibiting 7.7 wt% coordinated water weight loss (37 °C to 179 °C) and decomposition initiating at 261 °C, meeting polymer processing requirements for PLA and PP.
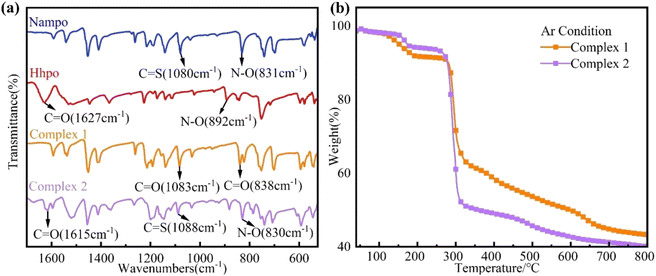 |
| | Fig. 2 (a) Infrared spectra of complexes 1 and 2 in the wavenumber range of 1700–525 cm−1; (b) thermogravimetric analysis (TG) of rare earth complexes 1 and 2. | |
3.2. Antimicrobial activity of agents
To evaluate antimicrobial performance, Nampo ligand was used as a ref. 45. The antimicrobial properties of different substances were compared based on the size of their inhibition circles, as shown in Fig. 3(a) and (b). The ligand Nampo showed strong resistance to both strains, and this result was compared with the antimicrobial performance of other experimental drugs. The rare earth salts show no resistance against E. coli ATCC25922 and S. aureus ATCC6538. Hhpo does not exhibit resistance to E. coli ATCC25922; however, it demonstrates a significant inhibition against S. aureus ATCC6538. The rare earth complexes show strong resistance against E. coli ATCC25922 and S. aureus ATCC6538. Furthermore, the antimicrobial activities of the rare earth complexes were assessed at various concentrations (50, 100, 150, 200, and 250 μg mL−1) using the same filter paper disc diffusion method. As shown in Fig. 4(a) and (b), tests conducted at different concentrations showed that the inhibitory effect of the complexes on both bacteria increased with increasing concentration, and the trend was more pronounced especially at higher concentrations. We then quantitatively characterize the antimicrobial properties (MIC) of the complexes through stepwise dilution. Both complexes exhibit significant resistance against both bacteria, with MICs of 6.25 μg mL−1 for E. coli ATCC25922 and 3.125 μg mL−1 for S. aureus ATCC6538, as shown in Table 1.
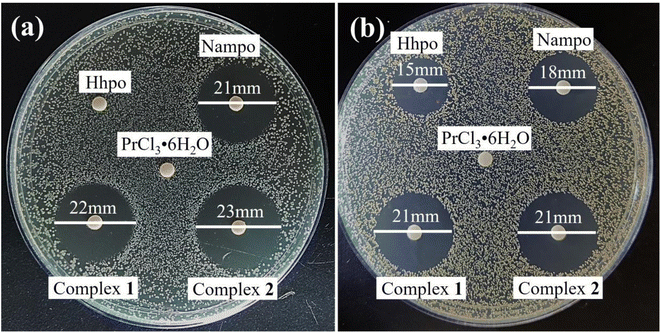 |
| | Fig. 3 Comparison of antimicrobial activity of ligands Nampo and Hhpo, rare earth salts and complexes against (a) E. coli ATCC25922 and (b) S. aureus ATCC6538. | |
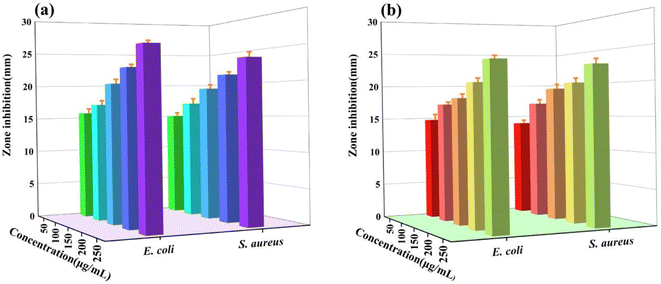 |
| | Fig. 4 (a) Complex 1 and (b) complex 2 antibacterial activity against S. aureus ATCC6538 and E. coli ATCC25922. | |
Table 1 MIC values of metal complexes against various pathogens
| Pathogen |
Minimum inhibiting concentrations (μg mL−1) |
| Complex 1 |
Complex 2 |
| E. coli ATCC25922 |
6.250 |
6.250 |
| S. aureus ATCC6538 |
3.125 |
3.125 |
Table 2 Formulation of films
| Films |
PLA (g) |
PP (g) |
Complex 1 (g) |
Complex 2 (g) |
| PLA |
400 |
0 |
0 |
0 |
| LC1/0.2 wt% |
399.2 |
0 |
0.8 |
0 |
| LC1/0.4 wt% |
398.4 |
0 |
1.6 |
0 |
| LC1/0.6 wt% |
397.6 |
0 |
2.4 |
0 |
| LC2/0.2 wt% |
399.2 |
0 |
0 |
0.8 |
| LC2/0.4 wt% |
398.4 |
0 |
0 |
1.6 |
| LC2/0.6 wt% |
397.6 |
0 |
0 |
2.4 |
| PP |
0 |
400 |
0 |
0 |
| PC1/0.2 wt% |
0 |
399.2 |
0.8 |
0 |
| PC1/0.4 wt% |
0 |
398.4 |
1.6 |
0 |
| PC1/0.6 wt% |
0 |
397.6 |
2.4 |
0 |
| PC2/0.2 wt% |
0 |
399.2 |
0 |
0.8 |
| PC2/0.4 wt% |
0 |
398.4 |
0 |
1.6 |
| PC2/0.6 wt% |
0 |
397.6 |
0 |
2.4 |
3.3. Antimicrobials in plastics
Bacterial films of polymerized PLA and PP are currently successfully prepared by blending, granulation, and hot pressing. According to Fig. 5, the addition of compounds to PP and PLA at a concentration of 0.2 wt% exhibits significant antibacterial effects, whereas a concentration of 0.6 wt% completely inhibits bacterial growth. Furthermore, Fig. 6 demonstrates that the amount of compound addition does not significantly affect the transparency of the film. Despite the complete inhibition of bacterial growth, the resulting antimicrobial film still maintained a high level of transparency.
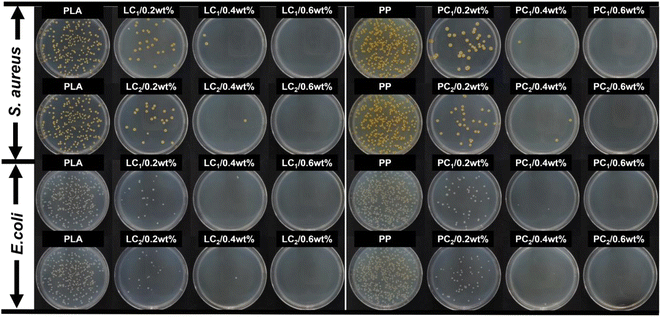 |
| | Fig. 5 Surface resistance to S. aureus ATCC6538 and E. coli ATCC25922 of composite antimicrobial films prepared with different complex additions (0 wt%, 0.2 wt%, 0.4 wt%, 0.6 wt%) in PLA and PP. | |
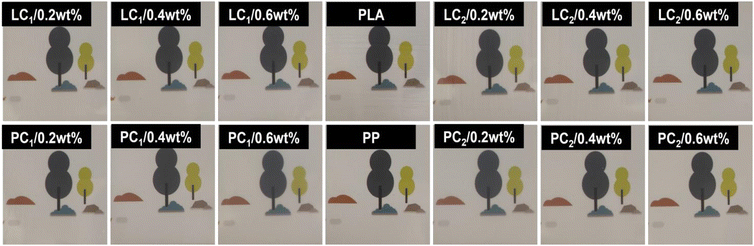 |
| | Fig. 6 Transparency illustration of different antimicrobial films with complex additives (0 wt%, 0.2 wt%, 0.4 wt%, 0.6 wt%) in PLA and PP. | |
3.4. Antimicrobial mechanism
Based on above discussion, the antimicrobial mechanism was proposed. The findings can partially be attributed to chelation interactions,46 as chelation reduces metal ion polarity, enhancing lipophilicity for better cell membrane penetration, as illustrated in Fig. 9. The complex under investigation disrupts the vital cellular components, impairing essential bacterial processes, and ultimately causing cell death. This process is supported by literature.47 In the control group, the two bacterial strains exhibit their expected normal structures. However, treatment with the complex induces profound morphological alterations, including wrinkling, deformation, membrane rupture, and the formation of cavities with varying degrees of severity, as shown in Fig. 7. These findings strongly suggest that the antimicrobial activity of the complex primarily arises from its interference with normal physiological processes within living organisms, mainly by compromising the integrity of the cell membrane. The nucleic acid leakage experiments, depicted in Fig. 8, further substantiate these observations. After 24 h of static incubation, the addition of antimicrobial agents leads to a considerably higher release of DNA and RNA into the supernatants of the two bacterial strains compared to the control group. This result indicates that the introduction of these complexes accelerates the apparent release of cytogenetic material into extracellular environment. The consistency between these nucleic acid assay results and the SEM observations reinforces the understanding that irreversible cell membrane damage leads to the loss of essential cellular components.
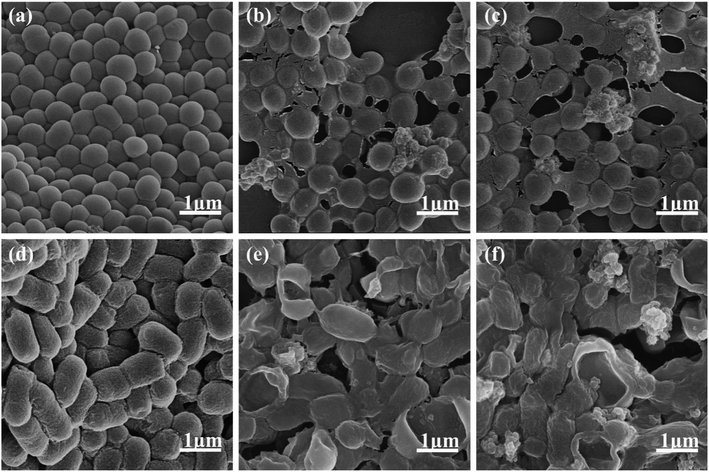 |
| | Fig. 7 SEM image at 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000× magnification of (a) S. aureus ATCC6538; (b) co-incubation of S. aureus ATCC6538 with complex 1; (c) co-incubation of S. aureus ATCC6538 with complex 2; (d) E. coli ATCC25922; (e) co-incubation of E. coli ATCC25922 with complex 1; (f) co-incubation of E. coli ATCC25922 with complex 2. 000× magnification of (a) S. aureus ATCC6538; (b) co-incubation of S. aureus ATCC6538 with complex 1; (c) co-incubation of S. aureus ATCC6538 with complex 2; (d) E. coli ATCC25922; (e) co-incubation of E. coli ATCC25922 with complex 1; (f) co-incubation of E. coli ATCC25922 with complex 2. | |
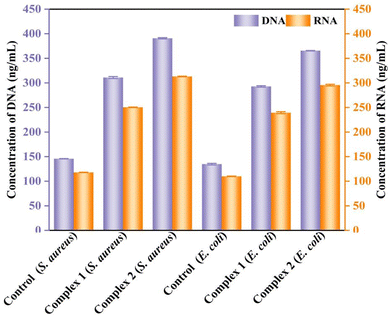 |
| | Fig. 8 The leaching of nucleic acid in the co-culture solution of S. aureus ATCC6538 and E. coli ATCC25922 was examined: the control group and the groups with the addition of complex 1 and complex 2. | |
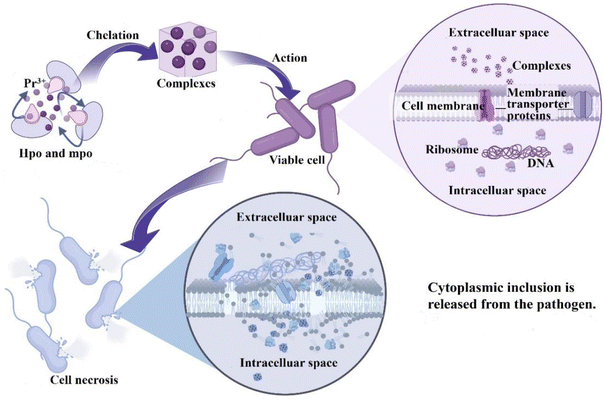 |
| | Fig. 9 The proposed antimicrobial mechanism of complexes. | |
4. Conclusions
In conclusion, we succeeded in synthesizing two praseodymium-based complexes using pyridine nitrogen oxide ligand and Pr3+ ions via the U-tube diffusion method. Both complexes show strong inhibitory activities against the Gram-negative bacterium E. coli ATCC25922 and the Gram-positive bacterium S. aureus ATCC6538 even in low concentrations. The results of these studies showed that the prepared complexes containing mpo ligands were superior to the original ligands H-hpo and rare earth salts in terms of antimicrobial properties. Specifically, their antibacterial effects against S. aureus ATCC6538 and E. coli ATCC25922 were enhanced, and the susceptibility of H-hpo to the Gram-negative bacterium E. coli ATCC25922 was effectively solved, thus expanding the antibacterial spectrum of H-hpo. The synthesized complexes currently exhibit desirable thermal stability, as confirmed through TG-DTG analysis, making them suitable for applications in polymer processing and related fields. The composites prepared from PLA/PP and the films show good antimicrobial effects against S. aureus ATCC6538 and E. coli ATCC25922. In our future research, we will also address existing limitations in methodology, sample size, mechanistic understanding, and practical applications. This will involve continual optimization of our methods, expansion of our research scope, and deeper exploration of antimicrobial mechanisms. Additionally, we recognize the importance of conducting a rigorous contact safety assessment, which will be invaluable for the development of transparent PLA and PP packaging materials endowed with high antimicrobial properties.
Conflicts of interest
There are no conflicts to declare.
Acknowledgements
This work has been supported by the STS Program of Chinese Academy of Sciences in Fujian Province, No. 2023T3016, 2023T3083, 2023T3061.
Notes and references
- K. Tram, P. Kanda, B. J. Salena, S. Y. Huan and Y. F. Li, Angew. Chem., Int. Ed., 2014, 53, 12799–12802 CrossRef CAS PubMed.
- F. Nakonechny and M. Nisnevitch, Adv. Funct. Mater., 2021, 31, 24 CrossRef.
- X. J. Li, N. X. Gu, Y. R. Ye, H. F. Lan, F. Peng and G. Y. Peng, Front. Microbiol., 2023, 13, 8 Search PubMed.
- C. Yang, Y. Luo, H. Lin, M. Ge, J. L. Shi and X. L. Zhang, ACS Nano, 2021, 15, 1086–1099 CrossRef CAS PubMed.
- J. Yuan, H. Zhao, M. Zhang and J. Zhang, J. Chin. Comput. Syst., 2021, 42, 678–684 Search PubMed.
- C. Nastasa, D. C. Vodnar, I. Ionut, A. Stana, D. Benedec, R. Tamaian, O. Oniga and B. Tiperciuc, Int. J. Mol. Sci., 2018, 19, 18 Search PubMed.
- A. Sanmugam, D. Vikraman, H. J. Park and H. S. Kim, Nanomaterials, 2017, 7, 14 CrossRef.
- E. Yousif, A. Majeed, K. Al-Sammarrae, N. Salih, J. Salimon and B. Abdullah, Arabian J. Chem., 2017, 10, S1639–S1644 CrossRef CAS.
- A. Daolio, A. Pizzi, M. Calabrese, G. Terraneo, S. Bordignon, A. Frontera and G. Resnati, Angew. Chem., Int. Ed., 2021, 60, 20723–20727 CrossRef CAS PubMed.
- Y. H. Dong, S. Q. Xie, P. Zhang, Q. Q. Fan, X. Y. Du, H. J. Sun, X. Y. Li, O. Fuhr and D. Fenske, Inorg. Chem., 2021, 60, 4551–4562 CrossRef CAS PubMed.
- J. Gao, K. Y. Luo, X. H. Wei, H. Wang, H. Liu and Y. Zhou, Org. Lett., 2023, 25, 3341–3346 CrossRef CAS PubMed.
- T. Ikai, M. Ando, M. Ito, R. Ishidate, N. Suzuki, K. Maeda and E. Yashima, J. Am. Chem. Soc., 2021, 143, 12725–12735 CrossRef CAS PubMed.
- J. M. Lipshultz and A. T. Radosevich, J. Am. Chem. Soc., 2021, 143, 14487–14494 CrossRef CAS PubMed.
- M. Schlegel, S. R. Qian and D. A. Nicewicz, ACS Catal., 2022, 12, 10499–10505 CrossRef CAS PubMed.
- S. Tamba, M. Nitani, T. Seo, H. Nitta, H. Tanaka, K. Hagiya, Y. Aso and Y. Ie, Org. Lett., 2022, 24, 3792–3796 CrossRef CAS PubMed.
- G. Tobajas-Curiel, Q. Q. Sun, J. K. M. Sanders, P. Ballester and C. A. Hunter, Chem. Sci., 2023, 14, 6226–6236 RSC.
- M. Toya, T. Omine, F. Ishiwari, A. Saeki, H. Ito and K. Itami, J. Am. Chem. Soc., 2023, 145, 11553–11565 CrossRef CAS PubMed.
- B. Wang, C. A. Pettenuzzo, J. Singh, G. E. McCabe, L. Clark, R. Young, J. Z. Pu and Y. M. Deng, ACS Catal., 2022, 12, 10441–10448 CrossRef CAS.
- M. S. Xie, M. Shan, N. Li, Y. G. Chen, X. B. Wang, X. Cheng, Y. Tian, X. X. Wu, Y. Deng, G. R. Qu and H. M. Guo, ACS Catal., 2022, 12, 877–891 CrossRef CAS.
- G. Z. Xu, Z. J. Liu, X. K. Wang, T. B. Lu, R. L. DesJarlais, T. Thieu, J. Zhang, Z. H. Devine, F. Y. Du, Q. Li, C. M. Milligan, P. Shaffer, P. E. Cedervall, J. C. Spurlino, C. F. Stratton, B. Pietrak, L. M. Szewczuk, V. C. R. Wong, R. A. Steele, W. Bruinzeel, M. Chintala, J. Silva, M. D. Gaul, M. J. Macielag and R. Nargund, J. Med. Chem., 2022, 65, 10419–10440 CrossRef CAS PubMed.
- Y. Zhu, G. Q. Liu, R. Zhao, H. Gao, X. N. Li, L. C. Sun and F. Li, Chem. Sci., 2022, 13, 4955–4961 RSC.
- Z. Iyigundogdu, B. Basar, R. Couvreur, S. Tamrakar, J. Yoon, O. G. Ersoy, F. Sahin, D. Mielewski and A. Kiziltas, Composites, Part B, 2022, 242, 8 CrossRef PubMed.
- X. Pei, J. C. Cui, S. L. Shen, H. X. Qiu, G. Z. Yang and J. Li, Composites, Part B, 2023, 251, 9 CrossRef.
- X. Li, S. Ru, H. Tian, S. Zhang, Z. Lin, M. Gao and J. Wang, Sci. Total Environ., 2021, 797, 149131 CrossRef CAS PubMed.
- D. Shin, Y. Choi, M. Kim, M.-C. Jang, J.-Y. Seo, J.-H. Kang, K. Shin and J.-H. Jung, Ecotoxicol. Environ. Saf., 2023, 253, 114653 CrossRef CAS PubMed.
- M. Gu, W. Li, L. Jiang and X. Li, Acta Biomater., 2022, 148, 22–43 CrossRef CAS PubMed.
- F. Guo, F. Pan, W. Zhang, T. Liu, F. Zuber, X. Zhang, Y. Yu, R. Zhang, M. Niederberger and Q. Ren, ACS Appl. Mater. Interfaces, 2022, 14, 44158–44172 CrossRef CAS PubMed.
- M. Luo, K. Shaitan, X. Qu, A. P. Bonartsev and B. Lei, Appl. Mater. Today, 2022, 26, 101304 CrossRef.
- V. Syrvatka, A. Rabets, O. Gromyko, A. Luzhetskyy and V. Fedorenko, Trends Biotechnol., 2022, 40, 1088–1101 CrossRef CAS PubMed.
- X. Tong, Y. Han, R. Zhou, W. Jiang, L. Zhu, Y. Li, S. Huang, J. Ma, C. Wen and J. Lin, Acta Biomater., 2023, 155, 684–702 CrossRef CAS PubMed.
- C. Wang, Y. Liu, Y. Zhang, Y. Song, Q. Chen, A. Cai, H. Guo and P. Zhang, Ceram. Int., 2023, 49, 11378–11392 CrossRef CAS.
- J. Wei, H. Zhao, L. Zhang, S. Chai, H. Liu, Y. Wang and J. Xue, Carbohydr. Polym., 2022, 297, 119967 CrossRef CAS PubMed.
- J. Wu, Y.-q. Chi, Y.-j. Yan, M.-z. Ji, X. Chen, X.-q. Yang, Y. Gao, Q. Zou, L. Zou and X.-y. Li, Nanoscale, 2023, 15, 3940–3951 RSC.
- L. Wu, F. Yang, Y. Xue, R. Gu, H. Liu, D. Xia and Y. Liu, Mater. Today Bio, 2023, 19, 100595 CrossRef CAS PubMed.
- L. Wu, M. Yang, L. Yao, Z. He, J.-x. Yu, W. Yin and R.-a. Chi, ACS Appl. Mater. Interfaces, 2022, 14, 53947–53959 CrossRef CAS PubMed.
- M. A. A. Hay, R. W. W. Gable and C. Boskovic, Dalton Trans., 2023, 52, 3315–3324 RSC.
- G. Tan, R.-Q. Jia, X. Zhao, Y.-Q. Guo, L. L. Zhang, X.-H. Wang, J.-G. Wang, X. Feng, B. Li and L.-Y. Wang, Inorg. Chem., 2022, 61, 11866–11878 CrossRef CAS PubMed.
- H. Yan, H. Ni, Y. Yang, C. Shan, X. Yang, X. Li, J. Cao, W. Wu, W. Liu and Y. Tang, Chin. Chem. Lett., 2020, 31, 1792–1796 CrossRef CAS.
- M. Zhang, C. Zhao, J.-Q. Song, W.-Q. He, P. Zhao and J. Liu, Mater. Today Bio, 2022, 7, 160–167 Search PubMed.
- M. Zhou, N. Ren and J. Zhang, Acta Phys.-Chim. Sin., 2021, 37, 2004071 Search PubMed.
- T. Wakabayashi, A. Ymamoto, A. Kazaana, Y. Nakano, Y. Nojiri and M. Kashiwazaki, Biol. Trace Elem. Res., 2016, 174, 464–470 CrossRef CAS PubMed.
- J. Tang, X. Huang, M.-H. Cao, Z. Wang, Z. Yu, Y. Yan, J.-P. Huang, L. Wang and S.-X. Huang, Front. Chem., 2022, 9, 812564 CrossRef PubMed.
- S. M. Jang, M. P. Pai, A. R. Shaw and B. A. Mueller, Crit. Care Med., 2019, 47, E863–E871 CrossRef CAS PubMed.
- J. Li, Q. Zhu, Y. Wu, F. Lin, L. Liu, L. Chen, S. Wang and L. Song, ACS Appl. Bio Mater., 2024, 7, 104–113 CrossRef CAS PubMed.
- J. K. Aulakh, T. S. Lobana, H. Sood, D. S. Arora, V. A. Smolinski, C. E. Duff and J. P. Jasinski, J. Inorg. Biochem., 2018, 178, 18–31 CrossRef CAS PubMed.
- L. Thunus and R. Lejeune, Coord. Chem. Rev., 1999, 184, 125–155 CrossRef.
- K. Andiappan, A. Sanmugam, E. Deivanayagam, K. Karuppasamy, H.-S. Kim and D. Vikraman, Int. J. Biol. Macromol., 2019, 124, 403–410 CrossRef CAS PubMed.
|
| This journal is © The Royal Society of Chemistry 2024 |
Click here to see how this site uses Cookies. View our privacy policy here.  Open Access Article
Open Access Article abcd,
Wayne Hsu
abcd,
Wayne Hsu bcd,
Shenglong Wangbcd,
Fenglong Linbcd,
Yincai Wu
bcd,
Shenglong Wangbcd,
Fenglong Linbcd,
Yincai Wu bcd,
Yimin Fange,
Jinglin Chen
bcd,
Yimin Fange,
Jinglin Chen *a and
Lijun Song
*a and
Lijun Song *bcd
*bcd

![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 ratio, and complex 2, created by combining H-hpo, Na-mpo and PrCl3·6H2O in a 1
3 ratio, and complex 2, created by combining H-hpo, Na-mpo and PrCl3·6H2O in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2
2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio, were characterized through elemental analysis and X-ray single crystal analysis, revealing structures in the triclinic space group of P
1 ratio, were characterized through elemental analysis and X-ray single crystal analysis, revealing structures in the triclinic space group of P![[1 with combining macron]](https://www.rsc.org/images/entities/char_0031_0304.gif) and P1, respectively, as shown in Fig. 1(a). The crystal structure of complex 1 exhibits a distorted-octahedral geometry with each Pr(III) ion coordinated by three sulfur atoms, two oxygen atoms, two μ2-oxygen atoms from mpo ligands, and one coordinated water molecule.44 Weak π–π stacking interactions between pyridine-ring of mpo ligands are observed as shown in Fig. 1(b). Complex 2 displays a distorted dodecahedral geometry, with Pr(III) ion coordinated by two sulfur atoms, two oxygen atoms from mpo ligands, two oxygen atoms from hpo ligand, and two coordinated water molecules, shown in Fig. 1(c). Two-dimensional layered structures are formed with alternative weak π–π interactions, shown in Fig. 1(d).
and P1, respectively, as shown in Fig. 1(a). The crystal structure of complex 1 exhibits a distorted-octahedral geometry with each Pr(III) ion coordinated by three sulfur atoms, two oxygen atoms, two μ2-oxygen atoms from mpo ligands, and one coordinated water molecule.44 Weak π–π stacking interactions between pyridine-ring of mpo ligands are observed as shown in Fig. 1(b). Complex 2 displays a distorted dodecahedral geometry, with Pr(III) ion coordinated by two sulfur atoms, two oxygen atoms from mpo ligands, two oxygen atoms from hpo ligand, and two coordinated water molecules, shown in Fig. 1(c). Two-dimensional layered structures are formed with alternative weak π–π interactions, shown in Fig. 1(d).
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O of Hhpo from 1627 cm−1 to 1615 cm−1 and N–O from 892 cm−1 to 830 cm−1 suggest the occurrence of coordination reactions. Fig. 2(b) presenting a thermal stability analysis under argon indicates a three-stage mass loss process for both complexes of 1 and 2, with complex 1 exhibiting 7.7 wt% coordinated water weight loss (37 °C to 179 °C) and decomposition initiating at 261 °C, meeting polymer processing requirements for PLA and PP.
O of Hhpo from 1627 cm−1 to 1615 cm−1 and N–O from 892 cm−1 to 830 cm−1 suggest the occurrence of coordination reactions. Fig. 2(b) presenting a thermal stability analysis under argon indicates a three-stage mass loss process for both complexes of 1 and 2, with complex 1 exhibiting 7.7 wt% coordinated water weight loss (37 °C to 179 °C) and decomposition initiating at 261 °C, meeting polymer processing requirements for PLA and PP.








