 Open Access Article
Open Access ArticleDevelopment of an aptasensor for dibutyl phthalate detection and the elucidation of assay inhibition factors†
Hyerin Songab,
Hyun Jeong Limab and
Ahjeong Son *ab
*ab
aDepartment of Environmental Science and Engineering, Ewha Womans University, 52 Ewhayeodae-gil, Seodaemun-gu, Seoul 03760, Republic of Korea. E-mail: ason@ewha.ac.kr; ahjeong.son@gmail.com; Tel: +82(2)3277-3339
bCenter of SEBIS (Strategic Solutions for Environmental Blindspots in the Interests of Society), 52 Ewhayeodae-gil, Seodaemun-gu, Seoul 03760, Republic of Korea
First published on 28th June 2024
Abstract
We developed a fluorescence aptasensor (hereafter ‘SG-aptasensor’) using SYBR Green I, a newly truncated 20-mer aptamer, and probe DNA to detect dibutyl phthalate (DBP). The detection range of DBP was 0.1–100 ng L−1 with 0.08 ng L−1 as the limit of detection. To adapt the assay to environmental samples in the near future, possible inhibition factors (experimental and environmental) have been tested and reported. The experimental inhibitors included the incubation time, temperature, pH, and ionic strength. Consequently, temperature (2–25 °C) and pH (7.0–9.0) ranges did not significantly inhibit the assay. The incubation time required for sufficient reaction was at least 4 h, and a relative humidity <20% may have induced fluorescence quenching. Tris–HCl-based incubation buffer with excess ionic strength (more than 0.2 M NaCl) demonstrated an abnormal increase in fluorescence. Environmental inhibitors including cations (Mg2+, Ca2+, and Cu2+) and humic acids were tested. The fluorescence signal was significantly reduced (∼99%) by 100 mM Cu2+ compared to that by 0 mM Cu2+. In contrast, the reduction in fluorescence signal was marginal (<15%) when Mg2+ or Ca2+ ions were present. Inhibition of the assay was observed (∼28%) in the presence of 100 mg L−1 humic acids.
1. Introduction
Phthalic acid esters (PAEs) enhance flexibility and transparency as plasticizers for various industrial products such as electronics, plastics, and food packaging.1 Because PAEs can be easily released into the environment by leaching out from the products, their exposure to humans has increased. Subsequently, the endocrine disruption to humans by PAEs has been observed in various forms, such as developmental malformation, interference with reproduction in humans, and disturbances in the immune and nervous systems.2 At this juncture, the US EPA set the action plan to regulate PAEs including dibutyl phthalate (DBP), diisobutyl phthalate (DIBP), butyl benzyl phthalate (BBP), di(2-ethylhexyl) phthalate (DEHP), diisononyl phthalate (DINP), and diisodecyl phthalate (DIDP), di-n-pentyl phthalate, di-n-octyl phthalate.3 European Chemicals Agency of the European Commission added endocrine disrupting properties to DBP, DEHP, BBP, and DIBP in 2021.4Establishing PAE detection methods is important for preventing and mitigating potential hazards from chemicals that are not yet replaceable. In addition to reliable instrumental analysis, biosensor technologies have been developed to provide screening tools for these chemicals. Biosensors for environmental monitoring have demonstrated their advantages, including specificity, fast response times, low cost, and ease of use.5 Among the biological receptors, aptamers are short, single-stranded nucleotides. Several aptamers and related aptasensors have been developed for detecting PAEs (Table 1).
| Transducer type | Target | Characteristics of assay | Sensitivity of assay (LOQ & linearity range) | Selectivity of assay | References |
|---|---|---|---|---|---|
| a DMP: dimethyl phthalate, DEP: diethyl phthalate, DHP: dihexyl phthalate, DPP: dipentyl phthalate, MEHP: mono-2-ethylhexyl phthalate, PA: phthalic acid, PCB: polychlorinated biphenyl, AuFs: gold-nanoflowers, MCH: 6-mercapto-1-hexanol, DOP: dioctyl phthalate, DPHP: di(2-propylheptyl) phthalate, DNHP: di-n-hexyl phthalate, SMIPs: surface molecularly imprinted polymers, GCE: glassy carbon electrode, DVB: divinyl benzene, PPD: p-phenylenediamine, L-TRP: L-tryptophan, PEC: photoelectrochemical, MIT: molecular imprinted technology, SERS: surface-enhanced Raman spectroscopy, AgNCs: silver nanoclusters, TOTM: trioctyl trimellitate, UCNPs: upconversion nanoparticles. | |||||
| Optics-based (fluorescence and colorimetry) transducer | DBP | Aptamer-SYBR Green I (SG-aptasensor) | 0.0001–0.1 μg L−1 | DBP, nonylphenol ethoxylate, triclosan, bisphenol A (BPA), bisphenol S (BPS) | This study |
| DMP, DEP, DBP, DHP, DIBP, DINP, DPP, BBP, MEHP, DEHP, PA | AuNP-gQD aptasensor | 0.001–50 μg L−1 | DMP, DEP, DBP, DHP, DIBP, DINP, DPP, BBP, MEHP, DEHP, PA | Lim et al. (2022)6 | |
| Nonylphenol, benzoic acid, BPA, BPS, bisphenol F, DES, beta-estradiol | |||||
| PA, DMP, DEP, DBP, DIBP, BBP, DEHP | Non-equilibrium rapid replacement aptamer (NERRA) assay using aptamer and PoPo 3 dye | 0.1–200 μg L−1 (30 min) | 7 PAEs (PA, DMP, DEP, DBP, DIBP, BBP, DEHP), BPA, 4-nonylphenol | Kim et al. (2020)7 | |
| 1–100 μg L−1 (30 s) | |||||
| DEHP, DBP, BBP | Aptamer-AuNP-based colorimetric assay | 0.003–10 μg L−1 (mixture) | Mixture (DEHP, DBP, BBP), Cd2+, atrazine, PCB77, PCB126, estrone, estradiol, ethinylestradiol, glucose, L-histidine, humic acids | Chen et al. (2021)8 | |
| DEP, DBP, DEHP | DNA-modified AuNPs based colorimetric sensor | 421–1661 μg L−1 (DEP) | DEP, DBP, DEHP, Fe2+, Ni2+, Zn2+, Na+, K+, Cu2+, CO32−, NO3−, PO43−, CH3COO− | Guo et al. (2021)9 | |
| 321–701 μg L−1 (DBP) | |||||
| 841–3322 μg L−1 (DEHP) | |||||
| Electrochemical-based transducer | DEHP | Signaling-probe displaced electrochemical aptamer-based biosensor (SD-EAB) | 0.0039–39 μg L−1 | DEHP, Hg2+, Cr3+, Cd2+, ethyl acetate, benzoic acid, PA, kanamycin, sulfadimethoxine | Han et al. (2017)10 |
| DEHP | AuFs-methylene blue | 0.0005–0.001 μg L−1 | DEHP, DMP, DEP, DINP, DIBP, BBP, DIBP | Lee et al. (2022)11 | |
| DEHP | DNA junction-aptamer-MCH-capture probe-Au electrode | 0.1–5000 μg L−1 | DEHP, DOP, DPHP, BBP, DBP, DNHP | Chen et al. (2022)12 | |
| DBP | Coating SMIPs on the surface of modified GCE | 0.1–10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 μg L−1 000 μg L−1 |
DBP, BPA, DVB, PPD, L-TRP | Wang et al. (2022)13 | |
| PEC-based transducer | DBP | MIT using metal organic framework and Cu2O heterostructure | 0.000028–0.278 μg L−1 | DBP, NH4+, K+, Na+, Ca2+, Mg2+, SO42−, Cl−, and NO3− | Yu et al. (2023)14 |
| SERS-based transducer | DEHP | AgNCs–SiO2-NH2 | 0.0032–72.8 μg L−1 | DEHP, DEP, DBP, DINP, DIDP, BBP, TOTM | Tu et al. (2019)15 |
| DBP | UCNPs decorated with AuNPs-aptamer | 0.001–100 μg L−1 | DBP, DEHP, BBP, ethyl acetate, PA, Na+, Mg2+, Ca2+, K+, Fe2+ | Rong et al. (2021)16 | |
Analyzing environmental samples using biosensors can be challenging due to the complexity of their nature. Environmental samples contain a wide range of potentially interfering substances, such as organic and inorganic compounds, microbes, and particulate matter, making it difficult to detect specific targets.17 Experimental conditions, such as temperature and pH, also change dynamically, causing fluctuations in biosensor signals.18
As summarized in Table 2, previous studies have indicated that inhibition occurs during biosensor-based analyses. Wang et al.19 and Jin et al.20 described that the assay was inhibited by Mg2+, possibly through the mechanism of DNA aggregation, followed by disruption of DNA hybridization. Zhou et al.21 reported a reduction in the fluorescence signal due to water hardness from Ca2+ and Mg2+ ions. Zhan et al.22 indicated the interference of fluorescence resonance energy transfer (FRET) process by sodium vanadate. Wu et al.23 showed that an assay using nanomaterials was inhibited by butyrylcholinesterase, causing the aggregation of AuNPs. Kim et al.24 and Jin et al.20 developed an inhibition resistance assay based on DNA hybridization and quantum dot nanoparticles.
| Biosensor type | Components | Target | Inhibition factors | Inhibition type (effects) | References |
|---|---|---|---|---|---|
| a N/S-CDs: nitrogen/sulfur co-doped carbon dots, DPA: diaminophenazine, PPi: pyrophosphate, GFET: graphene field-effect transistors, PDMS: polydimethylsiloxane, XOD: xanthine oxidate. | |||||
| Fluorescence aptasensor | NanoGene assay MB-QD-probe & signaling probe DNA | E. coli O157![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H7 (bacteria) H7 (bacteria) |
Humic acids, Ca2+, SDS, ethanol | Compared to PCR, MB-QD assay is resistant to the presence of inhibitors | Kim et al. (2011)24 |
| Pseudomonas putida (bacteria) | Mg2+ | DNA aggregation | Wang et al. (2018)25 | ||
| Microcystis aeruginosa (bacteria) | Mg2+ | Prevent disrupting DNA hybridization using electrical discharge treatment | Jin et al. (2020)20 | ||
| SYBR Green I & Ag+ specific oligonucleotides | Ag+ | Ca2+ & Mg2+ water hardness, hypochlorite | The instability of silver hypochlorite formed by silver ions and hypochlorite and the oxidation of hypochlorite, which would cause an unstable DNA | Zhou et al. (2020)21 | |
| Fluorescence immunosensor (FRET) | CQDs & AuNps | Paraoxon (insecticide) | Butyrylcholinesterase (BChE) | Causing the aggregation of AuNPs and the corresponding recovery of FRET-quenched fluorescence emission | Wu et al. (2017)26 |
| N/S-CDs & 2,3-DPA | Alkaline phosphatase (ALP) | Sodium vanadate (Na3VO4) | NA3VO4 inhibited the process of ALP hydrolysis of PPi. (PPi and free Cu2+ form a stable complex, which cannot form DPA, in the absence of ALP) | Zhan et al. (2021)22 | |
| Electrochemical aptasensor | GFETs sensor with PDMS | 17β-estradiol (E2) (EDCs) | pH, ionic strength | pH & ionic strength value in the environment (tap water) could fluctuate with time | Li et al., (2019)27 |
| Sonic Hedgehog/aptamer complexes | Sonic Hedgehog (protein) | Exonuclease III | Inhibiting cleavage of aptamers by exonuclease III via the steric hindrance effect to yield the displacement strands | Chen et al. (2023)28 | |
In this study, we developed an aptasensor to detect DBP (SG-aptasensor) to investigate the possible inhibition effects of various factors in environmental samples. The tested inhibition factors included experimental (incubation time, temperature, pH, and ionic strength) and environmental factors (divalent cations of Mg2+, Ca2+, Cu2+, and humic acids).
2. Materials and methods
2.1. Preparation of aptamer and probe DNA
The schematic in Fig. 1A shows the interaction of the aptamer and complementary probe DNA with DBP and SYBR Green I. The aptamer, 5′-TCT GTC CTT CCG TCA CAG GT-3′ (20-mer) was truncated from the previous study's aptamer (27-mer),6 which has shown the specific binding to PAEs. The probe DNA, 5′-TGT GAC GGA A-3′, was designed as complementary to the aptamer sequence in the previous study6 (Fig. 1B). All oligonucleotides used in this study were commercially synthesized and purified using high-performance liquid chromatography (Bioneer Co., Daejeon, Korea). The aptamer and probe DNA were mixed with TE buffer (Tris–HCl 10 mM of pH 8.0 and EDTA 1 mM, Bioneer Co.), making their concentrations 100 μM and 200 μM, respectively. The final concentration of the aptamer and probe DNA in the reaction was 2.5 μM after dilution with Tris–HCl buffer. Tris–HCl buffer (pH 8.0) comprises 0.02 M of Tris–HCl (Dyne Bio Inc., Gyeonggi-do, Korea), 0.02 M of MgCl2·6H2O (Daejung, Gyeonggi-do, Korea), 0.04 M of KCl (Duksan, Gyeonggi-do, Korea), and 0.1 M of NaCl (Junsei, Tokyo, Japan). All experiments were performed in triplicates unless otherwise stated.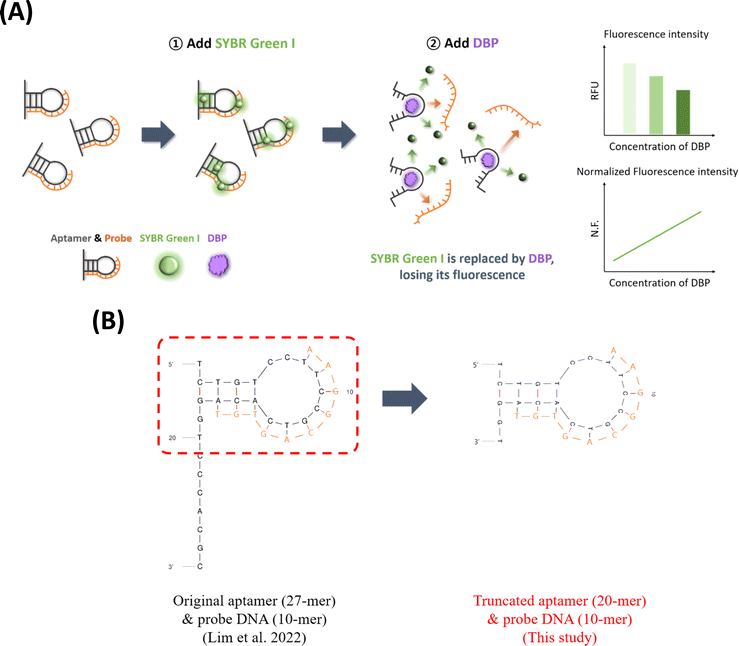 | ||
| Fig. 1 (A) The schematic of SG-aptasensor for DBP detection and (B) secondary structures of the original aptamer, truncated aptamer, and probe DNA. | ||
2.2. Fluorescence measurement
SYBR Green I (Invitrogen, Carlsbad, CA, USA) is a fluorescence dye that intercalates double-stranded nucleic acids. The stock solution (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000×) was prepared with 1 mL of dimethyl sulfoxide, and the final SYBR Green I concentration used for the SG-aptasensor was 1×, which was serially diluted with Tris–HCl buffer.
000×) was prepared with 1 mL of dimethyl sulfoxide, and the final SYBR Green I concentration used for the SG-aptasensor was 1×, which was serially diluted with Tris–HCl buffer.
The fluorescence was measured at λex = 265 nm and λem = 525 nm using a SpectraMax M2 spectrofluorometer (Molecular Devices, San Jose, CA, USA). The fluorescence signal was converted to normalized fluorescence based on eqn (1) to minimize the background signal, which changed in every reaction.
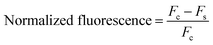 | (1) |
2.3. Quantification of DBP using the SG-aptasensor
The reaction (total 200 μL) of the assay was prepared with Tris–HCl buffer (70 μL), the aptamer (100 μL, 5 μM), probe DNA (10 μL, 50 μM), and SYBR Green I (20 μL, 10×). Subsequently, target analyte DBP (20 μL, various concentrations) was added. The samples were then incubated for 4 h at ambient temperature (25 °C) with gentle mixing at 300 rpm (MixMate Shaker, Eppendorf, Hamburg, Germany). For the quantification experiment, 100 mL DBP stock solution was prepared to 1000 mg L−1 by adding 0.1 g of DBP (99% purity, Junsei) to methanol (LC-MS Grade, Thermo Fisher Scientific, Waltham, MA, USA). DBP was serially diluted with deionized water to obtain final concentrations of 0.1, 0.5, 1, 5, 10, 50, 100, 500, 1000, 5000, and 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 ng L−1.
000 ng L−1.
For the selectivity experiment, four endocrine-disrupting or potentially endocrine-disrupting compounds were selected for comparison with DBP.29,30 The details of four chemicals are listed in Table S1.† All compounds affect the human endocrine system. Nonylphenol ethoxylate (NPE) is commonly used as a surfactant in various products.31 Triclosan (TCS) has been used as an antibacterial agent for personal products and is known for disrupting the thyroid hormone.32,33 Bisphenol A (BPA) and bisphenol S (BPS) are chemical analogs and well-known endocrine-disrupting chemicals that are used in plastics, receipts, and food packaging.34 DBP, NPE (70% in H2O, Sigma-Aldrich, St Louis, MO, USA), TCS (>98%, TCI Co., Tokyo, Japan), BPA (>99%, Daejung), and BPS (≥98%, Sigma-Aldrich) were first prepared to 1000 mg L−1 stock solution in methanol (LC-MS grade, Thermo Fisher Scientific) and subsequently diluted with Tris–HCl buffer. Each chemical (20 μL, 1 ng L−1) was subjected to SG-aptasensor reaction, including the Tris–HCl buffer (70 μL), aptamer (100 μL), and probe (10 μL).
2.4. Assay inhibition experiments
To investigate the assay inhibition factors, four experimental factors (incubation time, temperature, pH, and ionic strength of the Tris–HCl buffer) and four environmental factors (Mg2+, Ca2+, Cu2+, and humic acids) were selected.Temperature experiments were conducted in a manner similar to that of the incubation time experiment, as described above. The temperature experiment occurred at 2 °C, 13 °C, 25 °C, and 37 °C using a refrigerator, incubator (Wise Cube, Daihan Scientific, Gangwondo, Korea), or oven (HB-500 Minidizer™, Ultra-Violet Products Ltd, Cambridge, UK). At each incubation, the humidity was measured to determine the effect of relative humidity.
The inhibition effect of the pH and ionic strength of the Tris–HCl buffer was examined. The tested pHs were 6.0, 7.0, 9.0, and 10.0. Solutions of various pH values were prepared with Tris–HCl (pH 8.0) buffer by adding HCl (0.02 M, Sigma-Aldrich) or NaOH (0.01 M, pH 12.0, Duksan). pH was measured using a PB-10 pH meter (Sartorius Co., Göttingen, Germany).
Tris–HCl buffer as an incubation buffer included 0.02 M MgCl2·6H2O, 0.04 M KCl, and 0.1 M NaCl. To examine the effect of ionic strength, various NaCl concentrations (0.01, 0.05, 0.1, 0.2, and 0.5 M) were added to the Tris–HCl buffer, and DBP quantification was conducted at various ionic strengths. The total ionic strengths were 0.11, 0.15, 0.2, 0.3, and 0.6 M, respectively, based on eqn (2):35
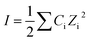 | (2) |
The 1 M stock solution of cations (Mg2+, Ca2+, and Cu2+ ions) was prepared by dissolving magnesium chloride hexahydrate (MgCl2·6H2O, 10.165 g, Daejung), magnesium sulfate heptahydrate (MgSO4·7H2O, 12.324 g, Daejung), calcium chloride dihydrate (CaCl2·2H2O, 14.701 g, Junsei), and copper(II) sulfate pentahydrate (CuSO4·5H2O, 24.968 g, Daejung) in 100 mL of deionized water. The stock solutions were serially diluted to 0.01, 0.1, 1, 10, and 100 mM using deionized water. Each Mg2+, Ca2+, and Cu2+ ion concentration in 20 μL was added to the reaction. When conducting cations experiments, the added volumes of ion solution and Tris–HCl buffer were 20 μL and 50 μL, respectively.
Humic acids (Suwannee River Humic Acid Standard II 2S101H, 200 mg) was obtained from the International Humic Substances Society (Denver, CO, USA). The humic acids stock solution was prepared by dissolving humic acids (4 mg) in 10 mL of Tris–HCl buffer (pH 8.0). After shaking overnight (shaking incubator, Wise Cube) to ensure complete dissolution, the stock solution was serially diluted to 0.001, 0.01, 0.1, 1, 10, and 100 mg L−1 in Tris–HCl buffer. The 20 μL of humic acids was subjected to the reaction (200 μL total) with 50 μL of Tris–HCl buffer.
3. Results and discussion
3.1. Assay configuration of SG-aptasensor
In the developed SG-aptasensor configuration (Fig. 1A), SYBR Green I was intercalated between the base pairs of double-stranded DNA, which were formed by the aptamer and probe DNA. Once the aptamer bound to the DBP, the probe DNA and SYBR Green I dissociate owing to the conformational change of the aptamer binding with the target. SYBR Green I loses its fluorescence owing to fluorescence quenching by surrounding water molecules.7 The reduced signal of SYBR Green I is inversely proportional to the amount of DBP. A truncated aptamer was used in this study instead of the original aptamer6 because of its smaller standard deviation for DBP detection (Fig. S1†).3.2. Sensitivity and selectivity of the SG-aptasensor for DBP detection
The fluorescence intensity gradually decreased with increasing DBP concentration, as shown in the emission spectra (Fig. 2A). The range of quantification for DBP was three orders of magnitude (0.1–100 ng L−1 or ppt) (Fig. 2B). In Fig. 2B, the linear regression equation is y = 0.0357 log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) x + 0.0535 (r2 = 0.70) and the limit of detection (LOD) is 0.08 ng L−1 based on eqn (3a) and (3b).37
x + 0.0535 (r2 = 0.70) and the limit of detection (LOD) is 0.08 ng L−1 based on eqn (3a) and (3b).37| LOB = meanblank + 1.645 (SDblank) | (3a) |
| LOD = LOB + 1.645 (SDlow concentration sample) | (3b) |
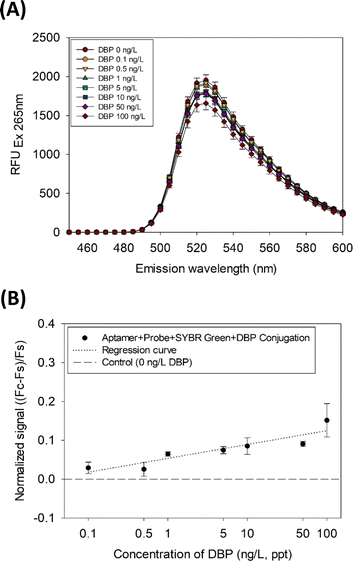 | ||
| Fig. 2 (A) SYBR Green I emission spectra measured with DBP concentration variation and (B) DBP quantification results via the SG-aptasensor. | ||
Based on the limit of quantification of this study (LOQ = 0.0001 μg L−1), the sensitivity of this assay is considered excellent as compared to the previous similar aptasensors for the detection of DBP, which ranges from 0.000028 to 321 μg L−1 (Table 1). However, the limitation of this assay may be its lower linearity, because r2 was similar to or lower than that in other studies, where the linearity ranged from 0.71 to 0.99.
As shown in Fig. 3, the selectivity of the SG-aptasensor is demonstrated in the presence of other endocrine-disrupting compounds (NPE, TCS, BPA, and BPS). DBP showed a significant decrease in fluorescence between 0 and 1 ng L−1 DBP (dotted box in Fig. 3A, p-value = 0.0058) compared to the other four chemicals (p-values of 0.274, 0.204, 0.259, and 0.488 for NPE, TCS, BPA, and BPS, respectively) (Table S2†). The selective quantification of DBP was clearly demonstrated by normalized fluorescence (Fig. 3B, red bar for DBP).
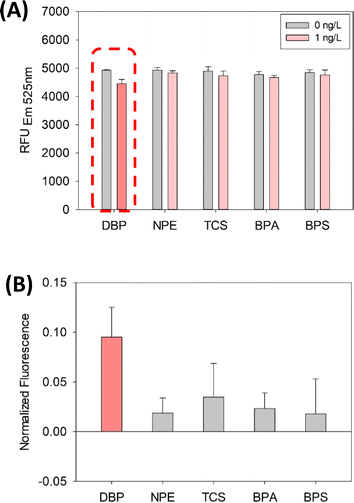 | ||
| Fig. 3 Selectivity results of the SG-aptasensor for DBP detection with non-phthalate compounds: (A) fluorescence intensity and (B) normalized fluorescence. | ||
3.3. Experimental inhibitors
The results pertaining to the experimental inhibitors (incubation time, temperature, pH, and ionic strength) are shown in Fig. 4. Various incubation times (0.5, 2, 4, 6, 8, 10, and 12 h) were tested for DBP detection using the SG-aptasensor and the results are presented in Fig. 4A and B. The fluorescence intensity for 0.5 h and 2 h are 5197 ± 1157 and 5300 ± 338 in the presence of DBP, respectively (Fig. 4A). The relatively higher standard deviations of 0.5 h and 2 h incubation indicate that the reaction might not have been completed. Afterwards, the standard deviations decreased after a 4 h incubation period, as depicted by the red arrow in Fig. 4A. Therefore, DBP detection by the SG-aptasensor requires an incubation time of at least 4 h. Additionally, a mild photobleaching effect of SYBR Green I was observed over an incubation period of 4 h (Fig. S2†). Therefore, the fluorescence decreased by ∼10% after 12 h. The signal decrease over time might have been due to exposure of the fluorescent dye to light or air.38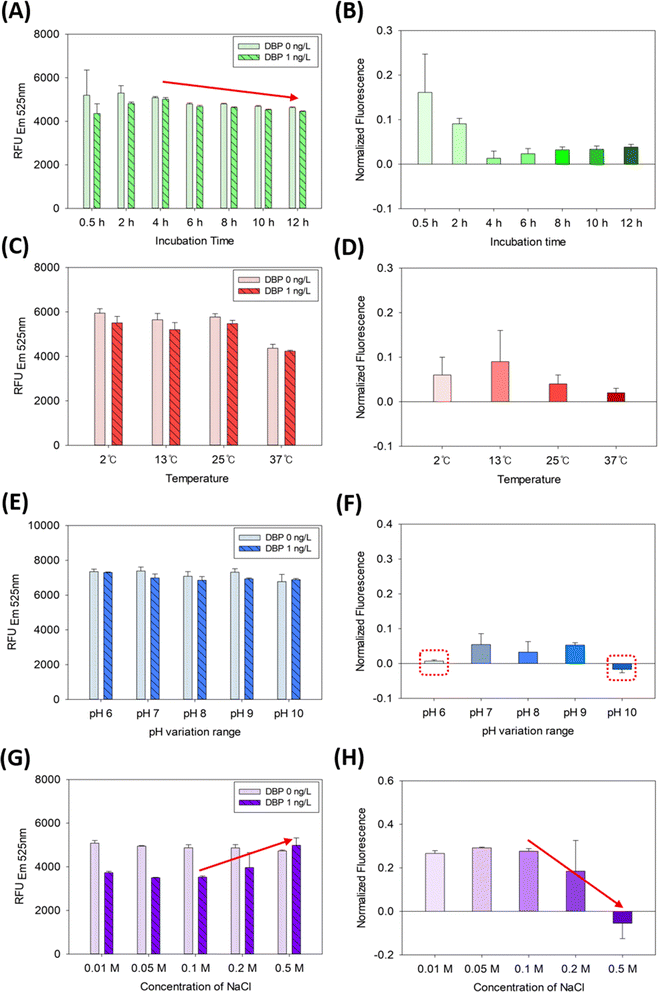 | ||
| Fig. 4 DBP quantification using the SG-aptasensor in the presence of experimental inhibitors: (A) and (B) incubation time, (C) and (D) temperature, (E) and (F) pH, (G) and (H) ionic strength. | ||
The effect of temperature on DBP detection by the SG-aptasensor was tested and is depicted in Fig. 4C and D. The temperature effect was somewhat interesting, as it did not follow the optimal conditions (i.e., 37–42 °C) for general DNA hybridization. Unlike the typical pattern of higher temperatures providing better results for DNA hybridization, lower temperatures resulted in a higher normalized fluorescence in the SG-aptasensor platform (Fig. 4C). However, the individual comparison of 2 °C, 13 °C, and 25 °C each using a t-test indicates that they are not significantly different (all p-values > 0.05) (Table S3†). This indicates that the actual temperature change did not significantly influence the assay results.
Conversely, 37 °C provides the lowest normalized fluorescence as compared to the other three temperatures (Fig. 4D). After measuring the relative humidity of each temperature incubation setting, the 37 °C setting had a markedly lower relative humidity of <20%. In contrast, the humidity in the other three temperature settings ranged from 30% to 75%. As indicated in a previous study, microliter-scale volumes in microarrays are vulnerable to inadequate humidity, causing incomplete hybridization and degradation of the fluorescent dye.39 The lower humidity can be the reason for the lower fluorescence signal. However, more studies may be required to elucidate the actual mechanism.
A pH range (6.0–10.0) of Tris–HCl buffer for DBP detection was tested and the results are presented in Fig. 4E and F. As shown in Fig. 4E, the fluorescence values are similar for all pH values. However, the Tris–HCl buffer with a pH lower than 7.0, or higher than 9.0, may inhibit the assay, because the normalized fluorescence was smaller than other pH values (depicted by dotted red boxes in Fig. 4F). Acidic or basic buffers can affect the assay by either protonating or deprotonating SYBR Green I and DNA.40,41
The effect of ionic strength of the Tris–HCl buffer was also investigated, as shown in Fig. 4G and H. As shown on the right side of the columns (DBP 1 ng L−1, indicated by the red arrow) in Fig. 4G, the fluorescence increases corresponding to the excess NaCl concentrations added, whereas the fluorescence of the negative control (DBP 0 ng L−1) is unchanged over NaCl concentrations. In the same manner, the normalized fluorescence at 0.5 M of NaCl was significantly reduced (−0.05 ± 0.07) as compared to 0.1 M of NaCl (0.27 ± 0.01) (red arrow in Fig. 4H). More than 0.2 M NaCl might have provided over-stringency of the pH buffer for aptamer-DBP binding or an imbalance of charges in the solution. This result is in line with that of Hianik et al.42 DNA molecules have negatively charged phosphate groups in the backbone. At higher salt concentrations, the positively charged Na+ ion can preferentially bind with the negatively charged phosphate group, reducing the repulsive forces between DNA molecules and facilitating DNA hybridization.43 Therefore, the effect may cause unnecessary binding between the aptamer and probe DNA, where the detachment of probe DNA is required for DBP detection.
3.4. Environmental inhibitors
The results pertaining to environmental inhibitors (divalent cations of Mg2+, Ca2+, Cu2+, and humic acids) are presented in Fig. 5. DBP (1 ng L−1) detection in the presence of various Mg2+ ion showed a pattern similar to that of the negative control (0 ng L−1 DBP), except for 100 mM Mg2+ (Fig. 5A). This is somewhat different from the results of several studies that have demonstrated a range of Mg2+ ion inhibition in DNA hybridization-based assays. For example, Jin et al.20 indicated that the Mg2+ ion of 0.01–0.1 mM caused under-estimation of quantification, whereas the Mg2+ ion of 1–1000 mM caused over-estimation of quantification. Conversely, the SG-aptasensor did not demonstrate the serious inhibition in the presence of Mg2+. However, 100 mM Mg2+ ion (dotted red box in Fig. 5A) appeared to over-estimate the quantification (i.e., by reducing the fluorescence signal [∼13%] of 1 ng L−1 DBP compared to the negative control of 0 mM Mg2+ ion). This probably is due to the specific role (‘shield effect’) of the Mg2+ ion, contributing to the secondary structure of the DNA.43,44 Based on the DNA folding form (mfold) calculation,45 the Gibbs free energy (ΔGf) of the aptamer secondary structure formation changed from −1.07 to −1.47 when 100 mM of the Mg2+ ion was added to the reaction as compared to the negative control (0 mM Mg2+ ion added). This indicates that a certain amount of Mg2+ ion can reinforce the formation of the secondary structure of the ssDNA aptamer, preventing it from reverting to its original structure via electrostatic binding with negatively charged DNA. This may explain why more SYBR Green I dye was released from the aptamer-probe DNA hybrid in the presence of 100 mM Mg2+. Because of the overestimation of the quantity compared to the negative control (0 mM Mg2+ ion), it can be considered as inhibition of the assay. However, the concentration of the Mg2+ ion in the environment is 4–528 mg L−1, which is equivalent to 0.17–21.72 mM.19,46,47 Because 100 mM Mg2+ ion is beyond the environmentally relevant concentration, the inhibition by the Mg2+ ion may not be the major concern for the aptasensor-based applications.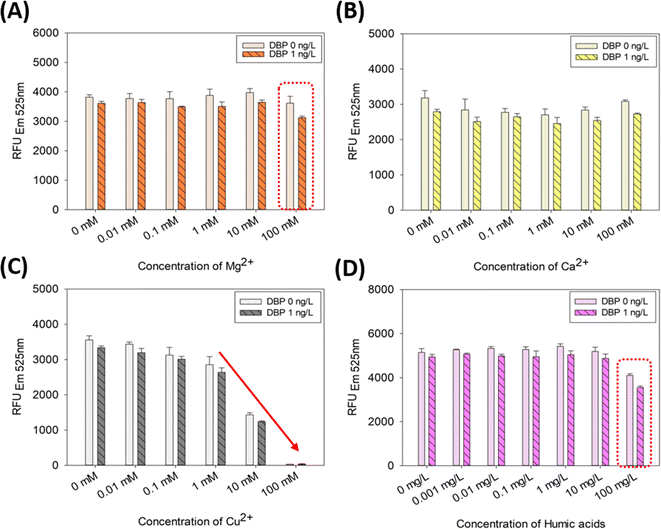 | ||
| Fig. 5 DBP quantification using the SG-aptasensor in the presence of environmental inhibitors: (A) Mg2+ ion, (B) Ca2+ ion, (C) Cu2+ ion, and (D) humic acids. | ||
DBP detection in the presence of various Ca2+ ion showed no remarkable inhibition (Fig. 5B). This result is consistent with that of Jin et al.20 The ubiquity of Ca2+ ion is accentuated by its concentration in lake, river, or soil samples and ranges from 0.109 to 127 mg L−1 (0.0027 to 3.17 mM).46,47 Therefore, the concentration range of Ca2+ ion in the environment is acceptable for the SG-aptasensor.
DBP detection in the presence of various Cu2+ ion showed a dramatic inhibitive change at concentrations of 10 and 100 mM Cu2+ (Fig. 5C). In the presence of 100 mM Cu2+, ∼99% of the fluorescence intensity disappeared compared to that of the negative control (0 mM Cu2+). This result can be deduced from the chemical nature of the Cu2+ ion. The Cu2+ ion is classified as a transition metal ion and an inherent fluorescence quenching ion because it suppresses the fluorescence emission by interfering with the process of the Jablonski diagram.36,48 In Zhao et al.,49 the fluorescence was quenched approximately 88% in the presence of 100 μM Cu2+ ion. Furthermore, the environmentally relevant concentration range of Cu2+ ion was <0.033 mM in water and 0.11–64.7 mM in soils and sediments.50–52 Therefore, the aptasensor assay can exhibit the inhibition by the Cu2+ ion that is environmentally relevant, when working with real environmental samples (e.g., soils and sediments).
DBP detection in the presence of various humic acids showed a significant decrease at 100 mg L−1 humic acids (red dotted box in Fig. 5D). The fluorescence signal decreased by 20% (without DBP) and 28% (with DBP) in the presence of 100 mg L−1 humic acids compared with that of the negative control (0 mg L−1 humic acids). The t-test also indicated a significant inhibition of the quantification results in the presence of 100 mg L−1 (p-values were 0.00071 without DBP and 0.00006 with DBP). In Kim et al.,53 humic acids were found to interfere with DNA hybridization by causing random nonspecific binding between humic acids and DNA. The quantification capability of the assay was inhibited by approximately 50% by humic acids in the range of 0.001–1000 mg L−1. The reduction in gene quantity was 20–50% in the presence of 100 mg L−1 humic acids. This result is in line with the present study, which showed a 20–28% decrease in the presence of 100 mg L−1 humic acids. In the previous studies regarding the occurrence of humic acids in the environment, the environmentally relevant concentration ranged from ∼0.1 to 1970 mg L−1.54–56 Therefore, 100 mg L−1 of humic acids is still environmentally relevant concentration and it may act as an inhibition factor of SG-aptasensor applications to environmental samples.
4. Conclusions
An SG-aptasensor was developed to detect DBP. The quantification range of DBP was 0.1–100 ng L−1 with a LOD of 0.08 ng L−1. Environmental samples are complex and contain various constituents that can inhibit aptasensor experiments. Therefore, potential inhibition factors of the SG-aptasensor were investigated. The experimental inhibitors were tested for time, temperature, pH, and ionic strength. Our findings indicated that the assay could detect DBP in a more stable manner for at least 4 h. Various temperature ranges (2 °C, 13 °C, 25 °C) and pH buffers (7.0 to 9.0) had no significant influence. Excess ionic strength (above 0.2 M of NaCl) of the Tris–HCl buffer may cause an inhibitive increase in the fluorescence signal because SYBR Green I can bind to aggregated DNA. Environmental inhibitors such as divalent cations (Mg2+, Ca2+, and Cu2+) and humic acids were tested. Cu2+ ion can significantly inhibit the assay, resulting in the reduction of 99% of the fluorescence signal in 100 mM Cu2+ ion, whereas the inhibition by Mg2+ and Ca2+ ion is marginal (<15%). A relatively high but environmentally relevant concentration of humic acids (100 mg L−1) could also inhibit the assay. These findings underscore the importance of considering potential inhibitors when using SG-aptasensor for detecting DBP in environmental samples. The robustness of the proposed aptasensor with field samples will require further characterization in the future. This will allow us to observe its response in the presence of potential inhibitors as well as interference species.Data availability
The data supporting this article have been included as part of the ESI.†Author contributions
AS and HL performed the experimental design. HS performed the experimental setup and measurements. AS, HL, and HS analyzed the data. HS drafted the manuscript. AS and HL reviewed and revised the manuscript. All authors have approved the final manuscript.Conflicts of interest
The authors declare that they have no competing financial interests or personal relationships that may have influenced this study.Acknowledgements
This study was supported by the National Research Foundation of Korea (RS-2023-00217228) as part of the research conducted by the Convergence Research Center (CRC) at Ewha Womans University, Seoul, Korea.Notes and references
- H. C. Erythropel, M. Maric, J. A. Nicell, R. L. Leask and V. Yargeau, Appl. Microbiol. Biotechnol., 2014, 98, 9967–9981 CrossRef CAS PubMed.
- M. A. Kamrin, J. Toxicol. Environ. Health, Part B, 2009, 12, 157–174 CAS.
- U. S. E. P. Agency, Phthalates Action Plan, 2012 Search PubMed.
- E. C. (EC), Commission Regulation (EU) 2021/2045 of 23 November 2021 amending Annex XIV to Regulation (EC) No 1907/2006 of the European Parliament and of the Council concerning the Registration, Evaluation, Authorisation and Restriction of Chemicals (REACH) (Text with EEA relevance), European Commission, Off. J. Eur. Union, 2021, 6–10 Search PubMed.
- M. J. Dennison and A. P. Turner, Biotechnol. Adv., 1995, 13, 1–12 CrossRef CAS PubMed.
- H. J. Lim, H. Jin, B. Chua and A. Son, ACS Appl. Mater. Interfaces, 2022, 14, 4186–4196 CrossRef CAS PubMed.
- D. Kim, H. J. Lim, Y. G. Ahn, B. Chua and A. Son, Talanta, 2020, 219, 121216 CrossRef CAS PubMed.
- Y. Chen, Z. Wang, S. Liu and G. Zhao, J. Hazard. Mater., 2021, 412, 125174 CrossRef CAS PubMed.
- R. H. Guo, C. C. Shu, K. J. Chuang and G. B. Hong, Mater. Lett., 2021, 293, 129756 CrossRef CAS.
- Y. Han, D. Diao, Z. Lu, X. Li, Q. Guo, Y. Huo, Q. Xu, Y. Li, S. Cao, J. Wang, Y. Wang, J. Zhao, Z. Li, M. He, Z. Luo and X. Lou, Anal. Chem., 2017, 89, 5270–5277 CrossRef CAS PubMed.
- K. Lee, N. G. Gurudatt, W. Heo, K. A. Hyun and H. I. Jung, Sens. Actuators, B, 2022, 357, 131381 CrossRef CAS.
- Q. Chen, M. J. Du and X. Q. Xu, J. Electroanal. Chem., 2022, 914, 116300 CrossRef CAS.
- S. Wang, M. Pan, K. Liu, X. Xie, J. Yang, L. Hong and S. Wang, Food Chem., 2022, 381, 132225 CrossRef CAS PubMed.
- L. Y. Yu, Y. Z. Shen, P. W. Gao, Q. Zhang, X. Y. Hu and Q. Xu, Chem. Eng. J., 2023, 472, 144925 CrossRef CAS.
- D. Tu, J. T. Garza and G. L. Cote, RSC Adv., 2019, 9, 2618–2625 RSC.
- Y. W. Rong, S. Ali, Q. Ouyang, L. Wang, H. H. Li and Q. S. Chen, J. Food Compos. Anal., 2021, 100, 103929 CrossRef CAS.
- H. Brammer and F. O. Nachtergaele, Int. J. Environ. Stud., 2015, 72, 56–73 CrossRef.
- K. R. Rogers, Biosens. Bioelectron., 1995, 10, 533–541 CrossRef CAS.
- X. F. Wang, H. Kweon, S. Lee, H. Shin, B. Chua, M. R. Liles, M. K. Lee and A. Son, Soil Biol. Biochem., 2018, 125, 300–308 CrossRef CAS.
- H. Jin, Y. Yoon, M. R. Liles, B. Chua and A. Son, Analyst, 2020, 145, 6846–6858 RSC.
- X. Zhou, A. G. Memon, W. Sun, F. Fang and J. Guo, Biosensors, 2020, 11, 6 CrossRef PubMed.
- Y. J. Zhan, S. T. Yang, L. F. Chen, Y. B. Zeng, L. Li, Z. Y. Lin, L. H. Guo and W. Xu, ACS Sustainable Chem. Eng., 2021, 9, 12922–12929 CrossRef CAS.
- X. L. Wu, Y. Song, X. Yan, C. Z. Zhu, Y. Q. Ma, D. Du and Y. H. Lin, Biosens. Bioelectron., 2017, 94, 292–297 CrossRef CAS PubMed.
- G. Y. Kim, X. Wang and A. Son, J. Environ. Monit., 2011, 13, 1344–1350 RSC.
- X. Wang, H. Kweon, S. Lee, H. Shin, B. Chua, M. R. Liles, M.-k. Lee and A. Son, Soil Biol. Biochem., 2018, 125, 300–308 CrossRef CAS.
- X. Wu, Y. Song, X. Yan, C. Zhu, Y. Ma, D. Du and Y. Lin, Biosens. Bioelectron., 2017, 94, 292–297 CrossRef CAS PubMed.
- Y. Li, Y. Zhu, C. Wang, M. He and Q. Lin, Biosens. Bioelectron., 2019, 126, 59–67 CrossRef CAS PubMed.
- Q. R. Chen, H. H. Gao, J. L. Yao, B. Y. Jiang, R. Yuan and Y. Xiang, Sens. Actuators, B, 2023, 385, 133702 CrossRef CAS.
- A. Soares, B. Guieysse, B. Jefferson, E. Cartmell and J. N. Lester, Environ. Int., 2008, 34, 1033–1049 CrossRef CAS PubMed.
- J. Xue, Q. Wu, S. Sakthivel, P. V. Pavithran, J. R. Vasukutty and K. Kannan, Environ. Res., 2015, 137, 120–128 CrossRef CAS PubMed.
- A. R. Kim, S. H. Kim, D. Kim, S. W. Cho, A. Son and M. Y. Yoon, Int. J. Mol. Sci., 2019, 21, 208 CrossRef PubMed.
- A. Son, I. M. Kennedy, K. M. Scow and K. R. Hristova, Presented in Part at the WEFTEC 2009, 2009 Search PubMed.
- K. B. Paul, J. M. Hedge, M. J. Devito, and K. M. Crofton, Triclosan and Endocrine Disruption: Evidence for Alterations in Thyroid Hormone Homeostasis, United States Envrionmental Protection Agency, 2007 Search PubMed.
- H. J. Lim, E. H. Lee, S. D. Lee, Y. Yoon and A. Son, Chemosphere, 2018, 211, 72–80 CrossRef CAS PubMed.
- G. N. Lewis and M. Randall, J. Am. Chem. Soc., 1921, 43, 1112–1154 CrossRef CAS.
- S. S. Tan, S. J. Kim and E. T. Kool, J. Am. Chem. Soc., 2011, 133, 2664–2671 CrossRef CAS PubMed.
- D. A. Armbruster and T. Pry, Clin. Biochem. Rev., 2008, 29, S49–S52 Search PubMed.
- C. L. Tiller and K. D. Jones, Environ. Sci. Technol., 1997, 31, 424–429 CrossRef CAS.
- J. R. Choi, J. Hu, S. Feng, W. A. Wan Abas, B. Pingguan-Murphy and F. Xu, Biosens. Bioelectron., 2016, 79, 98–107 CrossRef CAS PubMed.
- E. Tervola, K. N. Truong, J. S. Ward, A. Priimagi and K. Rissanen, RSC Adv., 2020, 10, 29385–29393 RSC.
- G. Jiang, Y. Ma, J. Ding, J. Liu, R. Liu and P. Zhou, Chem.–Eur. J., 2023, 29, e202300625 CrossRef CAS PubMed.
- T. Hianik, V. Ostatna, M. Sonlajtnerova and I. Grman, Bioelectrochemistry, 2007, 70, 127–133 CrossRef CAS PubMed.
- K. L. Wong and J. Liu, Biotechnol. J., 2021, 16, e2000338 CrossRef PubMed.
- J. Anastassopoulou, J. Mol. Struct., 2003, 651, 19–26 CrossRef.
- M. Zuker, Nucleic Acids Res., 2003, 31, 3406–3415 CrossRef CAS PubMed.
- A. Amadi, J. Yisa, I. Ogbonnaya, M. Dan-Hassan, J. Jacob and Y. Alkali, J. Geogr. Geol., 2012, 4, 13–21 Search PubMed.
- A. Potasznik and S. Szymczyk, J. Elem., 2015, 20, 677–692 Search PubMed.
- A. W. Varnes, R. B. Dodson and E. Wehry, J. Am. Chem. Soc., 1972, 94, 946–950 CrossRef CAS PubMed.
- H. Zhao and M. L. Zastrow, Biochemistry, 2022, 61, 494–504 CrossRef CAS PubMed.
- Y. Liu, H. Wang, Y. Cui and N. Chen, Int. J. Environ. Res. Public Health, 2023, 20, 3885 CrossRef CAS PubMed.
- D. G. Heijerick, P. A. Van Sprang and A. D. Van Hyfte, Environ. Toxicol. Chem., 2006, 25, 858–864 CrossRef CAS PubMed.
- T. F. Rozan and G. Benoit, Geochim. Cosmochim. Acta, 1999, 63, 3311–3319 CrossRef CAS.
- G. Y. Kim, X. F. Wang, H. Ahn and A. Son, Environ. Sci. Technol., 2011, 45, 8873–8880 CrossRef CAS PubMed.
- M. Ali and W. Mindari, Presented in Part at the MATEC Web of Conferences, 2016 Search PubMed.
- A. H. Jasim and K. A. Alghrebawi, Dysona, Appl. sci., 2020, 1, 88–95 Search PubMed.
- L. Tripolskaja, A. Kazlauskaite-Jadzevice, E. Baksiene and A. Razukas, Agriculture, 2022, 12, 488 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4ra03045a |
| This journal is © The Royal Society of Chemistry 2024 |
