Open Access Article
Open Access ArticleAnti-filarial efficacy of Centratherum anthelminticum: unravelling the underlying mechanisms through biochemical, HRAMS proteomics and MD simulation approaches†
Sunil Kumar,
Ayushi Mishra,
Surya Pratap Singh and
Anchal Singh
and
Anchal Singh *
*
Department of Biochemistry, Institute of Science, Banaras Hindu University, Varanasi, 221005, UP, India. E-mail: anchalsinghbhu@yahoo.com; anchalsingh@bhu.ac.in
First published on 12th August 2024
Abstract
Traditionally, Centratherum anthelminticum (CA) has been reported to be a potent anti-filarial, however no reports are available detailing its mechanism of action against filarial parasites. In this study, we have evaluated the anti-filarial activity of CA against lymphatic filarial parasites Setaria cervi using ex vivo biochemical, proteomics and in silico approaches. The motility and viability of the parasites decreased significantly after treatment with CA concentrations of ≥125 μg mL−1. An increase in lipid peroxidation (51.92%), protein carbonylation (48.99%), NADPH oxidase (88.88%) activity and decrease in the glutathione (GSH) (−39.23%), glutathione reductase (GR) (−60.17%), and glutathione S-transferase (GST) (−50.48%) activity was also observed after CA treatment. The proteomics analysis was performed by two-dimensional gel electrophoresis and high-resolution accurate mass spectrometry (HRAMS). In total, 185 proteins were differentially expressed (DEPs) following CA treatment. The major DEPs were mostly involved in tRNA processing, biosynthetic processes, metabolic activities, protein transport, the tricarboxylic acid cycle, protein translation, and stress response. The UPLC-ESI-MS/MS analysis of CA extract revealed the presence of 40 bioactive compounds. Further the docking analysis showed 10 CA bioactive compounds to have high binding affinity towards antioxidant proteins of filarial parasites. Additionally, MD simulation studies showed stable interactions (RMSF ≤ 10 Å) of 3-O-methylquercitin, quinic acid, gentisic acid, and vanillin with filarial antioxidant enzymes/proteins. To our knowledge, this is the first report detailing the molecular mechanism of anti-filarial activity of CA, which can be further evaluated for the development of new anti-filarial formulations.
1 Introduction
Lymphatic filariasis (LF) is a serious health problem caused by nematode parasites Wuchereria bancrofti, Brugia malayi, and Brugia timori. LF is prevalent in large parts of the tropical and subtropical regions of the world and more than 50 million people in 44 countries are infected while another 882 million people are at the risk of infection (https://www.who.int/news-room/fact-sheets/detail/lymphatic-filariasis). The World Health Organization aims to globally eradicate LF, through the implementation of a Mass Drug Administration (MDA) plan. This strategy entails providing pairs of anthelminthic medications (Albendazole with either Ivermectin or Diethylcarbamazine) to the entire population at risk.1 These drugs can reduce the microfilaria reservoir but cannot kill adult worms.2 Administration of Diethylcarbamazine (DEC) is often accompanied with serious adverse events such as fatal encephalopathy, also loss of vision can occur if DEC is given to persons with active loiasis and onchocerciasis.3 Furthermore, the use of Ivermectin (IVM) can result in severe encephalopathy and mortality in patients with a high burden of Loa loa infection.4 Furthermore, the development of drug resistance in helminths necessitates the discovery of novel and safer anti-filarial drugs.5 The use of medicinal plants for the treatment of parasitic diseases is becoming increasingly common in recent years as a method of avoiding the adverse effects of medication.6The seeds of Centratherum anthelminticum (L.) (CA) Kuntze (scientific synonyms: Veronia anthelmintica), commonly known as black cumin, are widely used as spices in tropical countries. The CA seeds have a variety of pharmacological properties, such as anti-viral, anti-microbial, anti-fungal, and anti-diabetic activities.7–9 For centuries, CA has been used as an efficacious anti-filarial and anti-helminthic remedy by ayurvedic practitioners in India. An earlier study has evaluated the effect of aqueous and alcoholic C. anthelminticum extracts on the filarial parasite Setaria cervi. The CA extracts inhibited spontaneous motility of S. cervi nerve-muscle preparations by decreasing the contraction amplitude and frequency.10 Although the anti-filarial effect of CA is known, there are no reports that provide a detailed explanation of its mechanism of action against the filarial parasites. Therefore, this work was conducted to evaluate the anti-filarial and adulticidal activity of CA extract against filarial parasite Setaria cervi using a combination of ex vivo biochemical, proteomics and in silico approaches.
2 Materials and methods
2.1. Parasites collection, culture and exposure to CA extract
The worms were procured as described previously11 and brought to the laboratory in Kreb's–Ringer bicarbonate buffer (KRB) supplemented with streptomycin, penicillin, glutamine and 0.5% glucose (KRB maintenance medium). Further worms were incubated in KRB maintenance medium (KRBM) at 37 °C in a water bath for one hour before further use.12 Equal numbers (N = 6) of adult female parasites were cultured in 20 mL of KRBM with varying doses of CA for 4 hours at 5% CO2 at 37 °C and 95% humidity. Worms incubated in KRBM with Dimethyl sulfoxide (DMSO) 0.37% served as vehicle control. The movement of the treated worms was visually inspected by an investigator who was blinded to the experiment, and the motility was assessed as either positive or negative at hourly interval for a period of four hours and marked as either positive or negative (+/−) accordingly. The motility analysis was based on the movement score; a score of “+++++” indicates that the parasites are very active, a score of “+” that they are not very active, and a score of “−” that they are not moving.11,13 In order to check the recovery, parasites were also transferred to new KRBM after 4 hours. The median lethal dose (LC50) was determined by using OriginPro 2024 software. All the experiments were carried out in triplicates.14Following treatment, the parasites were stored at −80 °C before subjecting them to further analysis.13
2.2. Effect of CA on parasite viability and production of reactive oxygen species (ROS)
The viability of control and CA treated S. cervi parasites were determined by MTT assay.15 S. cervi worms were incubated in Phosphate Buffer Saline (PBS) medium containing 0.5 mg mL−1 MTT (3-(4,5-dimethylthiazol-2yl)-2,5-diphenyl tetrazolium bromide) for 2 hours at 37 °C in dark. Next, the worms were transferred into 200 μL DMSO and formazan crystals were solubilized. After 1 (one) hour of incubation, medium was carefully aspirated and absorbance (OD) of the solution was measured at 540 nm in a microplate reader (BioRad). For ROS production, the method of Sim Choi et al.16 was followed with minor changes. The worms were incubated in 2% Nitro Blue Tetrazolium (NBT) solution for 1 hour at room temperature, followed by washing with PBS and methanol. In the next step, the formazan crystals were dissolved in 2 M KOH (prepared in DMSO) and the final absorbance was recorded at 620 nm in a microplate reader (BioRad).2.3. DNA fragmentation analysis
The worms were homogenized 20 mM Tris buffer pH 8.0, 50 mM EDTA, 0.5% SDS, 100 mM NaCl, 1% β-mercaptoethanol, and 0.1 mg mL−1 proteinase K, and then incubated at 55 °C for 3 hours. DNA was extracted using a 25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 24
24![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 mixture of phenol, chloroform, and isoamylalcohol, followed by centrifugation at 10
1 mixture of phenol, chloroform, and isoamylalcohol, followed by centrifugation at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm. Next the supernatant was treated with 3 M sodium acetate and 100% cold ethanol, the pellet was washed with 70% ethanol, and dissolved in 10 mM Tris–EDTA (TE) buffer (pH 8.0).17 The isolated DNA sample was separated on a 1.8% agarose gel containing ethidium bromide, and images were recorded in a GelDoc system (Biorad, Hercules CA).
000 rpm. Next the supernatant was treated with 3 M sodium acetate and 100% cold ethanol, the pellet was washed with 70% ethanol, and dissolved in 10 mM Tris–EDTA (TE) buffer (pH 8.0).17 The isolated DNA sample was separated on a 1.8% agarose gel containing ethidium bromide, and images were recorded in a GelDoc system (Biorad, Hercules CA).
2.4. Assessment of NADPH oxidase activity
Both the control and treatment groups were homogenized separately in 50 mM phosphate buffer (pH 7.2), 0.25% SDS, and centrifuged for 10 min. at 600 g at 4 °C. The resulting supernatant (100 μL) was combined with 1 mM MgCl2, 80 μM cytochrome c, and 2 mM sodium azide in a total volume of 1 mL. After adding 0.2 mM NADPH to start the reaction, the change in absorbance at 550 nm was measured.182.5. Determination of protein carbonylation (PC) and lipid peroxidation
Using 2,4-dinitrophenyl hydrazine (DNPH), protein carbonyl concentration was assessed in the control and CA treated worms.19 Equal volumes of 10% cytosolic extract and cold trichloroacetic acid (TCA) were mixed and centrifuged at 6000 g for 5 minutes at 4 °C. Next, the pellet was treated with DNPH (10 mM), and kept in dark at room temperature for one hour with occasional vortexing. After one hour the mixture was centrifuged at 6000 g for 5 min, and 20% TCA was added. The pellet was washed with a mixture of ethanol and ethyl acetate (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) until the yellow tint vanished. 6 M guanidine hydrochloride was added to the pellet and the mixture was centrifuged at 6000 rpm for 5 min. at 4 °C. The molar extinction coefficient of 22
1) until the yellow tint vanished. 6 M guanidine hydrochloride was added to the pellet and the mixture was centrifuged at 6000 rpm for 5 min. at 4 °C. The molar extinction coefficient of 22![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × 106 mM−1 cm−1 was used in calculations.
000 × 106 mM−1 cm−1 was used in calculations.
Assessment of lipid peroxidation of the control and treated worms was based on the levels of malondialdehyde (MDA). The reaction was started by adding 10% SDS to 300 μL of cytosolic extract to begin the reaction, which was then incubated at RT for 5 min. Next 600 μL of 20% acetic acid was added, followed by a second incubation at RT for 2 min, and finally 0.8% of 2-thiobarbituric acid (TBA) was added. In a water bath, the entire mixture was boiled for one hour.20 Next, the mixture was centrifuged at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 g for 5 min at 4 °C. The supernatant's absorbance was then measured at 532 nm to determine the amount of TBA reactive compounds. TBA was calculated using the molar extinction value of 1.53 × 105 M−1 cm−1.
000 g for 5 min at 4 °C. The supernatant's absorbance was then measured at 532 nm to determine the amount of TBA reactive compounds. TBA was calculated using the molar extinction value of 1.53 × 105 M−1 cm−1.
2.6. Preparation of S. cervi homogenate
The 10% w/v homogenate of adult female S. cervi was prepared in 100 mM Tris–HCl, pH 7.0 containing, 1 mM EDTA, and 1 mM phenylmethylsulphonyl fluoride (PMSF) using a motor-driven homogenizer (REMI type RQ127A) at 4 °C. The homogenate was centrifuged at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 15 min at 4 °C. Next, the clear supernatant was stored at −20 °C in aliquots. The protein was quantified by the Bradford's method and Bovine serum albumin was used as a standard.13
000 rpm for 15 min at 4 °C. Next, the clear supernatant was stored at −20 °C in aliquots. The protein was quantified by the Bradford's method and Bovine serum albumin was used as a standard.13
2.7. 2D gel electrophoresis
With a few modifications, 2D gel electrophoresis was carried out as previously described.21,22 The S. cervi protein homogenate was treated with 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (acetone
1 (acetone![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) protein) volume of ice-chilled acetone and kept at −20 °C for 5 hours, followed by centrifugation at 10
protein) volume of ice-chilled acetone and kept at −20 °C for 5 hours, followed by centrifugation at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 10 minutes at 4 °C. 200 μl of the rehydration solution (7 M urea, 2 M thiourea, 2% w/v CHAPS, 15 mM DTT, 0.5% v/v IPG buffer pH 3–10) was used to collect and rehydrate the pellet. For improved resolution, 11 cm IPG strips with pI values 3–10 were used for better resolution of samples. The isoelectric focusing (IEF) at 20 °C was carried out in a Protean IEF Cell (BioRad, United States) as per: 150 μA per strip for 15 min, then quickly ramping up to 8000 V for 2 hours and 8000 V for 20
000 rpm for 10 minutes at 4 °C. 200 μl of the rehydration solution (7 M urea, 2 M thiourea, 2% w/v CHAPS, 15 mM DTT, 0.5% v/v IPG buffer pH 3–10) was used to collect and rehydrate the pellet. For improved resolution, 11 cm IPG strips with pI values 3–10 were used for better resolution of samples. The isoelectric focusing (IEF) at 20 °C was carried out in a Protean IEF Cell (BioRad, United States) as per: 150 μA per strip for 15 min, then quickly ramping up to 8000 V for 2 hours and 8000 V for 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 V for 7 hours (with a limit of 50 μA per strip). Following IEF, 40 mM Tris–HCl buffer (pH 8.8) containing 6 M urea, 25% w/v glycerol, 2% w/v SDS, 1% w/v DTT, and 2.5% iodoacetamide was used to equilibrate the strips.23 The second dimension was performed in 10% SDS PAGE. The gel was then stained with Coomassie Brilliant Blue G-250, (10% aluminum sulfate, 10% ethanol, 0.02% CBB G-250, and 2.5% orthophosphoric acid). Images of the gel were captured using a gel documentation system (Alpha Innotech, USA) and analyzed using PDQuest software (BioRad, USA). Three separate experiments were conducted to verify the reproducibility.24
000 V for 7 hours (with a limit of 50 μA per strip). Following IEF, 40 mM Tris–HCl buffer (pH 8.8) containing 6 M urea, 25% w/v glycerol, 2% w/v SDS, 1% w/v DTT, and 2.5% iodoacetamide was used to equilibrate the strips.23 The second dimension was performed in 10% SDS PAGE. The gel was then stained with Coomassie Brilliant Blue G-250, (10% aluminum sulfate, 10% ethanol, 0.02% CBB G-250, and 2.5% orthophosphoric acid). Images of the gel were captured using a gel documentation system (Alpha Innotech, USA) and analyzed using PDQuest software (BioRad, USA). Three separate experiments were conducted to verify the reproducibility.24
2.8. High resolution accurate mass spectrometry analysis
The samples were reduced with 10 mM dithiothreitol (DTT) for 1 h followed by treatment with 2% iodoacetamide (IDA), with 50 mM NH4HCO3/50% acetonitrile (ACN) thrice for 10–15 min with gentle vortexing and incubation in dark. The samples were then digested with Trypsin (Trypsin gold Promega, USA) and incubated at 37 °C for overnight. The extracted peptides were lyophilized, desalted and stored at −80 °C till further use.25 An Orbitrap Eclipse Tribrid Mass Spectrometer with nano-LC and UHPLC at Central Discovery Centre, Banaras Hindu University was used for peptide analysis. The samples were analyzed using a 120 min linear gradient of buffer B (80% Acetonitrile and 0.1% formic acid) at a flow rate of 0.300 μL min−1 and scanning was done in the range of 200–1600 m/z. The individual peptides MS/MS spectra were matched to the database sequence on Thermo Scientific™ Proteome Discoverer™ software. The samples were run in triplicates and abundance ratio value was set as ≥1.50 for upregulated and ≤0.667 for downregulated proteins respectively.25,26 The statistical significance was evaluated using T-tests and the significance index was computed based on the corresponding P value, where a default threshold of P < 0.05 was employed.252.9. Gene ontology analysis
The UniProt database, available at https://www.uniprot.org/, was utilized to facilitate the investigation of the Gene Ontology (GO) annotation proteome. The UniProt IDs were obtained by searching the UniProt database for the corresponding protein's accession number. By using GO annotation, major proteins were classified into categories according to their biological processes (BP), cellular components (CC), and molecular function (MF). The MF, CC, and BP of proteins were then visualized or formed into networks using Cytoscape (http://www.cytoscape.org, version 3.1.1). For this investigation, only the primary network-forming proteins were chosen. Excel was then used to create histograms for the classification and display of the MF, CC, and BP of proteins.2.10. Structure retrieval of filarial anti-oxidant proteins
Previously modelled structure of W. bancrofti glutathione S-transferase (GST) (5D73, DOI: https://doi.org/10.2210/Pdb5D73/pdb), W. bancrofti thioredoxin (TRx) (4FYUA, 10.2210/pdb4FYU/pdb) and B. malayi superoxide dismutase (SOD), (accession no. CTP82144.1) were previously constructed by our laboratory hence they were used as such for molecular docking.27 The 3 dimensional structure of filarial glutathione peroxidase (GPx) could not be located in any databases hence sequence of B. malayi GPx was retrieved (PM0077541). The structure was modeled with LOMETS and validated using PROCHECK and Rampage server.28 Further the 3D model for GPx was validated by ERRAT, ProSA, and ResProx server to determine its quality. The VADAR server was used to verify the hydrogen bond statistics and quality of the GPx models. The active site in the modelled 3D structure of GPx was predicted by Metapocket 2.0 server.292.11. C. anthelminticum extract preparation
The seeds of CA were purchased locally in Varanasi, Uttar Pradesh, India (between latitude 25.267878 and longitude 82.990494). Prof. Shashi Pandey, a taxonomist at the Botany Department, Institute of Science, Banaras Hindu University, made the botanical identification. 25 g of CA seeds were powdered under cold condition and defatted with n-hexane using a Soxhlet extractor. Thereafter the residue obtained was further fractionated with 250 mL of ethanol.30 The crude fractions were collected, filtered and concentrated to dryness under reduced pressure in a rotary evaporator (<40 °C). Before treatment the dried powder was solubilized in DMSO. The total percent of DMSO was always ≤0.37% of KRBM and an equal volume of DMSO was added to the control flasks also.2.12. FT-IR analysis of CA seed extract
A PerkinElmer Spectrum 65 Fourier Transform Infrared Spectrometer (FT-IR) was used to analyze the ethanolic extract of CA.31 The spectra were gathered between 4000 cm−1 and 400 cm−1 wavelength. Signal to noise ratio of spectra was improved by 100 interferograms with a special resolution of 4 cm−1 average. Additionally, background spectra were captured under the same circumstances and subtracted from the sample spectra. The experiment was done in triplicates, and OriginPro 8.0 was used to pick and integrate peaks, identify features and label them after importing the original FT-IR spectral files. Normalization and background removal was done to regulate the spectral quality.2.13. UPLC-ESI-MS/MS analysis of C. anthelminticum extract
UPLC-ESI-MS/MS analysis was performed on Acquity Ultra Performance Liquid Chromatography Electrospray Ionization Tandem Mass Spectrometry. Chromatographic separation of CA seed extracts was performed using an ACQUITY UPLC, BEH C18 column, 35 °C. The mobile phase has two phases: A phase, methanol and water (5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 95) and B phase methanol and water (95
95) and B phase methanol and water (95![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5) with 0.1% formic acid. Mass Lynx 4.1 software was used for data collection and processing. Phytochemical software equipped with RIKEN tandem mass spectral database (ReSpect) was utilized for detailed analysis of UPLC-ESI-MS/MS data.32
5) with 0.1% formic acid. Mass Lynx 4.1 software was used for data collection and processing. Phytochemical software equipped with RIKEN tandem mass spectral database (ReSpect) was utilized for detailed analysis of UPLC-ESI-MS/MS data.32
2.14. Retrieval of ligand structures
CA bioactive compounds were selected for docking analysis based on UPLC-ESI-MS/MS data. Using Biovia Discovery Studio 3.5 (https://discover.3ds.com/), the structures of the ligands were converted into PDB format which were retrieved from PubChem Database in SDF format.33 Drug like behavior of CA bioactive substances was predicted using the Lipinski filter.34 AdmetSAR server was used to forecast the Absorption, Distribution, Metabolism, Excretion, and Toxicity (ADMET) properties of CA bioactive substances.352.15. Docking analysis
YASARA and PatchDock server were employed to perform docking analysis of filarial antioxidant proteins with CA bioactive compounds. The PatchDock server's default setting for the RMSD value for protein and ligand complexes was 1.5. Discovery Studio 3.5 was used to visualize the docked complexes. The parameters GSC (geometric shape complementary) score and AI (approximate interface) area were obtained from PatchDock server,36 binding energy (kcal mol−1) and dissociation constant (μm) as given by YASARA (Yet Another Scientific Artificial Reality Application) server were used for data interpretation.372.16. Molecular dynamics simulation analysis
Molecular dynamic simulation utilizing NAMD (Nanoscale Molecular Dynamics v 2.14) was used to assess the stability of the interaction between filarial antioxidant protein models and ligands.27 The Open Babel Chemical Format Converter (https://www.cheminfo.org/Chemistry/Cheminformatics/FormatConverter/index.html) was used to convert the PDB files of the CA compounds into Sybyl Mol2 files. Using the Sybyl Mol2 ligand modeler and the CHARMM-GUI input generator (https://www.charmmgui.org/input), PSF and forcefield parameter values of CA bioactive compounds were selected. The VMD dispdev command was used to produce complexes of proteins and CA bioactive substances. In protein and CA bioactive compound complexes, the complexes were solvated in the X, Y, and Z axis in an orthorhombic water model with a distance of 10 Å. The complexes was also neutralized with 0.15 M NaCl and solvated by a TIP3P water box with a 5 Å layer of water in each direction. The PARAM SHIVAY supercomputing facility of IIT BHU was used to simulate molecular dynamics. Under 3D periodic boundary conditions, an MD simulation was run at 310 K temperature, 1000 steps, energy minimization, and 50 ns time trajectory. Root mean square deviation (RMSD), root mean square fluctuation (RMSF), radius of gyration (Rg), and solvent accessible area analysis (SASA) fluctuations were calculated during the simulation run and the findings were visualized by VMD.2.17. Statistical analysis
Each experiment was run in triplicates and the data are shown as mean ± SD and were computed using the OriginPro 2023b (https://www.originlab.com/). Using GraphPad prism software 9.5.0, the Student's t-test was applied for the statistical significance between control and the CA-treated worms (*p < 0.05, **p < 0.01 and ***p < 0.001).253 Results and discussion
3.1. In vitro effect of CA treatment on motility and viability of adult S. cervi
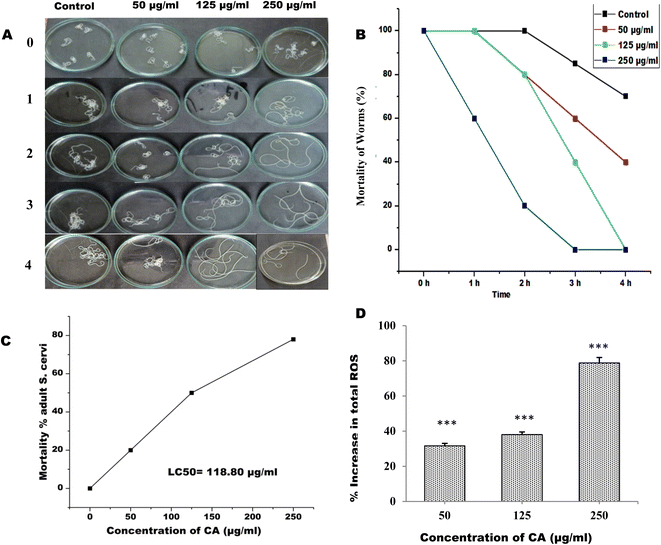
The adult female S. cervi were incubated in KRBM for 4 hours at 37 °C, with 5% CO2 and 95% humidity in a carbon dioxide incubator. It was observed that the S. cervi parasites treated with CA concentration of ≥125μg mL−1 were completely non-motile after 4 hours of incubation (Fig. 1A and B). The reduction in S. cervi motility was time and dose-dependent. After 4 hours of incubation, the adult parasites were transferred to fresh KRBM for 1 hour to check their recovery post CA treatment (Table 1). The worms treated with 50 μg mL−1 of CA were able to revive in the fresh medium, while the parasites treated with concentrations of 125 μg mL−1 and 250 μg mL−1 showed no evidence of recovery even after an hour of incubation. The lethal effect of CA appears to be of permanent nature at concentration >125 μg mL−1. The viability was decreased to 80%, 50.33%, and 22% after 4 hours of 250 μg mL−1, 125 μg mL−1, and 50 μg mL−1 CA treatments respectively. The 50% lethal concentration (LC50) was observed to be 118.80 μg mL−1 after 4 hours of treatment (Fig. 1C). The viability of S. cervi decreased as a function of concentration following CA treatment.| Sample | 0 h | 1 h | 2 h | 3 h | 4 h | Recovery |
|---|---|---|---|---|---|---|
| a Motility of the incubated parasites was evaluated as − (0%), no movement; + (20%), least active; ++ (40%), less active; +++ (60%), moderately active; ++++ (80%), highly active; and +++++ (100%), very high active. Worms were transferred into fresh medium after 4 h and motility recovery in treated group was compared with respect to the control. Results are from three independent experiments performed in duplicates. | ||||||
| Control | +++++ | +++++ | +++++ | ++++ | +++ | +++++ |
| 50 μg mL−1 | +++++ | +++++ | ++++ | +++ | ++ | ++++ |
| 125 μg mL−1 | +++++ | +++++ | ++++ | ++ | − | − |
| 250 μg mL−1 | +++++ | +++ | + | − | − | − |
3.2. CA treatment induces ROS production and DNA fragmentation in adult S. cervi
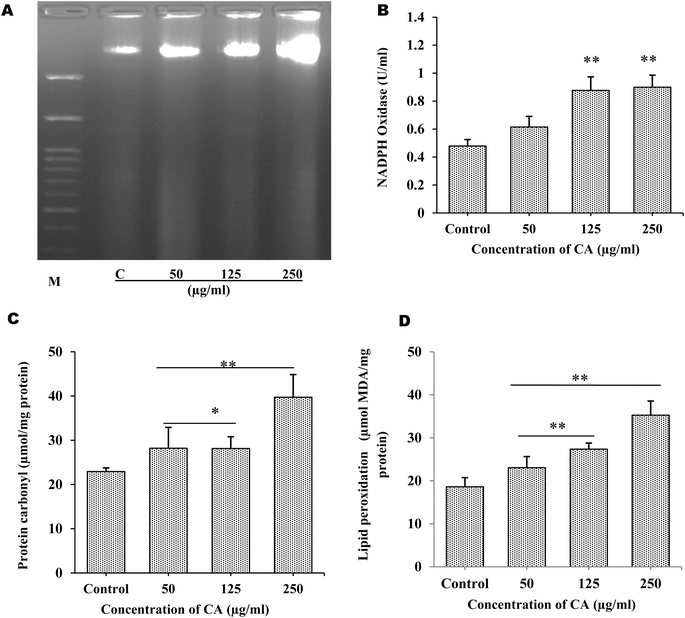
The production of ROS by S. cervi worms during CA treatment was estimated by NBT assay. The intracellular ROS was significantly higher in CA treated parasites as compared to control worms. The ROS production increased by 31.64% in 50 μg mL−1 (P-value ≤0.001), 38.04% in 125 μg mL−1 (P-value ≤0.001), and 78.78% in 250 μg mL−1 (P-value ≤0.001) in treated parasites as compared to the control group (Fig. 1D). The effect of elevated ROS level on cellular DNA was assessed by the DNA fragmentation assay. The DNA fragmentation analysis revealed dose-dependent nucleosomal DNA destruction and the maximal DNA laddering was seen at CA concentration of 250 μg mL−1 whereas 50 μg mL−1 concentrations, fragmentation was the least (Fig. 2A). Previously CA and its bioactive compound vernodalin have been shown to induce high levels of ROS in melanoma and breast cancer cells7,38 resulting in the apoptosis of the cancer cells. Since in our case too, the ROS was significantly higher after CA treatment causing a huge oxidative stress on the filarial parasites which could be a causative reason for the death of the parasites.3.3. CA treatment leads to increase in oxidative stress in S. cervi
The major hallmarks of programmed cell death are DNA fragmentation and increase in the cellular levels of ROS. Therefore the alterations in the oxidative stress indicators such as, protein carbonyl (PC) level, lipid peroxidation, and NADPH oxidase activity were also examined. Using 125 μg mL−1, and 250 μg mL−1 of CA seed extract, NADPH oxidase activity significantly increased by +82.05% (p ≤ 0.005), and +87.69% (p ≤ 0.005), respectively (Fig. 2B). Superoxide anions are produced, when active NADPH oxidase transfers electrons to oxygen, which in turn may cause production of H2O2 and other toxic reactive oxygen species leading to disruption of mitochondrial membrane. The oxidative damage production by elevated superoxide anions was assessed by examining the PC content and malondialdehyde levels. With CA treatment, PC content was shown to significantly increase by almost +27.01% (p ≤ 0.05), and +73.31% (p ≤ 0.005), in 125 μg mL−1 and 250 μg mL−1 respectively (Fig. 2C). Similar to the malondialdehyde levels, a rise in lipid peroxidation of about +47.1% (p ≤ 0.005), and +0.895% (p ≤ 0.005) fold change was observed in 125 μg mL−1 and 250 μg mL−1 respectively in CA treated worms in comparison to control parasites (Fig. 2D). The exposure of S. cervi to CA extract led to a significant increase in the lipid peroxidation and protein oxidation.3.4. Proteome profiling by 2D electrophoresis and HRAMS analysis
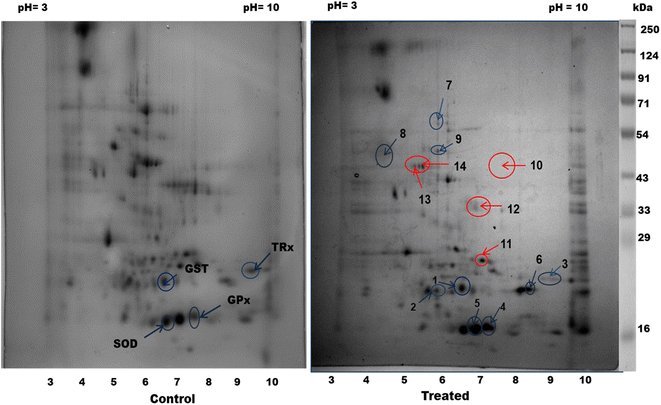
Next, proteomic profiling by 2D electrophoresis and HRAMS analysis was applied to investigate the effect of CA treatment on the filarial parasites. Upon exposure of S. cervi worms to 250 μg mL−1 CA extract, a significant alteration in the proteomic profile was observed with respect to the control worms. A total of 155 spots in control and 131 spots in CA treated parasite were observed in the proteome profiles after 2D gel electrophoresis. The PD-quest analysis identified 16 upregulated and 30 downregulated proteins (Fig. 3). The Pearson correlation coefficient between the treated and control samples were observed at 0.448.The HRAMS proteome profiling data was analyzed using the Thermo Scientific™ Proteome Discoverer™ software. The analysis of protein expression alteration was analyzed on the basis of abundance ratio. A threshold value of 0.67 was established for downregulated proteins, whereas a cut-off value of 1.5 was determined for upregulated proteins.23 A total of 185 proteins were identified as differentially expressed following, CA exposure, as indicated in Tables 2 and 3. Among these proteins, 97 were found to be considerably upregulated, while 88 were significantly downregulated.
| S. n. | Accession | Description | MW [kDa] | Score sequest HT | Abundance ratio: (treated)/(control) | Abundance ratio P-value: (treated)/(control) |
|---|---|---|---|---|---|---|
| 1 | A0A3P7FFD1 | Phosphoglucomutase (alpha-D-glucose-1,6-bisphosphate-dependent) | 62.5 | 16.6 | 25.911 | 6.88338 × 10−15 |
| 2 | J9EA55 | AV25 protein | 20.4 | 4.24 | 12.106 | 2.55534 × 10−9 |
| 3 | A0A8L7T780 | Transthyretin-like family protein | 15.9 | 60.78 | 10.112 | 3.28489 × 10−8 |
| 4 | E3UV59 | Glutathione S-transferase | 24.1 | 5.48 | 7.910 | 0.004939501 |
| 5 | A0A0J9Y0Q8 | BMA-HIP-1 | 38.9 | 19.14 | 6.427 | 9.12162 × 10−6 |
| 6 | A0A1I8EK35 | L-Lactate dehydrogenase | 35.7 | 156.75 | 4.506 | 0.000338363 |
| 7 | O97149 | Activation-associated secreted protein-1 | 24.6 | 13.85 | 4.493 | 0.000347![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 487 487 |
| 8 | A0A4E9FMP9 | Superoxide dismutase | 25.1 | 30.99 | 4.25 | 0.572008685 |
| 9 | J9APK4 | Glutathione peroxidase | 16.3 | 3.61 | 3.985 | 0.404530915 |
| 10 | J9EFL6 | Tropomyosin (fragment) | 9.4 | 45.79 | 3.835 | 0.001389045 |
| 11 | A0A1I8EE03 | Elongation factor 1-alpha | 50.8 | 212.39 | 3.543 | 0.002641362 |
| 12 | Q04009 | Myosin heavy chain | 225.9 | 48.52 | 3.515 | 0.002810523 |
| 13 | A0A8L7YQ50 | Alanine transaminase | 60.8 | 8.34 | 3.312 | 0.004447048 |
| 14 | A0A1I8EW65 | Succinate–CoA ligase [ADP/GDP-forming] subunit alpha, mitochondrial | 37.8 | 49.34 | 3.262 | 0.004990625 |
| 15 | A0A3P7G595 | Thioredoxin domain-containing protein | 9.3 | 16.85 | 2.971 | 0.009770672 |
| 16 | J9E6J2 | Transthyretin-like family protein | 20.2 | 44.08 | 2.945 | 0.010383163 |
| 17 | A0A3P7DHN6 | 60S ribosomal protein L27a | 28.6 | 4.05 | 2.904 | 0.011454856 |
| 18 | A0A4E9FP34 | Peptidyl-prolyl cis–trans isomerase | 18.5 | 44.37 | 2.809 | 0.014346473 |
| 19 | J9ETG6 | UMP-CMP kinase | 22.2 | 10.53 | 2.69 | 0.019053637 |
| 20 | J9EPU8 | RNA transcription, translation and transport factor protein | 28.5 | 8.4 | 2.678 | 0.019571524 |
| 21 | A0A8L7T3Z0 | BMA-ERP-1, isoform d | 28.7 | 43.95 | 2.559 | 0.026113795 |
| 22 | A0A3P7DF31 | Myosin tail domain-containing protein | 127.7 | 136.72 | 2.431 | 0.035547965 |
| 23 | A0A4E9FKG6 | Tropomyosin family protein | 20.5 | 315.12 | 2.415 | 0.037035642 |
| 24 | A0A1I8EKE6 | Elongation factor 1-alpha | 50.7 | 417.89 | 2.41 | 0.037422667 |
| 25 | A0A1I8EC27 | DB domain-containing protein | 22.4 | 9.92 | 2.357 | 0.042620241 |
| 26 | A0A0K0JX89 | Tubulin alpha chain | 45.1 | 3.8 | 2.333 | 0.045151395 |
| 27 | J9EYX9 | 30S ribosomal protein S19e | 16.9 | 9.65 | 2.312 | 0.047586267 |
| 28 | J9EKD7 | 50S ribosomal protein L31e | 12.9 | 17.96 | 2.289 | 0.050293391 |
| 29 | S6FMC3 | Triosephosphate isomerase | 27.1 | 229.44 | 2.28 | 0.051511685 |
| 30 | A0A0K0J057 | BMA-CYC-2.2 | 12.2 | 62.71 | 2.207 | 0.061510051 |
| 31 | A0A1I8ENA1 | ATP-dependent RNA helicase | 81 | 15.94 | 2.19 | 0.064165527 |
| 32 | A0A4E9FD82 | S-methyl-5′-thioadenosine phosphorylase | 31.6 | 38.6 | 2.182 | 0.065381791 |
| 33 | A0A3P7DLL1 | Glutamate dehydrogenase [NAD(P)(+)] | 60.5 | 590.15 | 2.178 | 0.06616905 |
| 34 | J9B9B8 | SWIB/MDM2 domain-containing protein | 34.2 | 4.79 | 2.122 | 0.075918113 |
| 35 | A0A4E9FEL1 | Aconitate hydratase, mitochondrial | 84.7 | 8.72 | 2.121 | 0.076134949 |
| 36 | A0A1I9G417 | Bm5160, isoform b | 9 | 154.89 | 2.076 | 0.084986192 |
| 37 | A0A0H5S2M8 | Bm3307 (fragment) | 228.9 | 81.5 | 2.072 | 0.085738954 |
| 38 | A0A3P7FDU5 | 60S ribosomal protein L7a | 31.1 | 35.42 | 2.069 | 0.086395097 |
| 39 | A0A1I8EUR5 | Malate dehydrogenase | 38.4 | 163.96 | 2.053 | 0.08993189 |
| 40 | A0A4E9FPQ9 | Moesin/ezrin/radixin homolog 1 | 67.2 | 9.32 | 2.032 | 0.094605644 |
| 41 | A0A4E9FDM3 | Hypothetical RNA-binding protein T28D9.2 in chromosome II, putative | 23.6 | 20.84 | 2.018 | 0.098165902 |
| 42 | A0A4E9EPZ8 | Troponin family protein | 32 | 95.48 | 2.001 | 0.102260576 |
| 43 | A0A0K0J070 | 60S ribosomal protein L38 | 8.2 | 91.98 | 1.958 | 0.113584457 |
| 44 | A0A4E9FA37 | Triosephosphate isomerase | 27.1 | 277.16 | 1.923 | 0.123993316 |
| 45 | A0A3P7DR94 | Cysteine rich repeat family protein | 137.9 | 4.07 | 1.89 | 0.134409097 |
| 46 | A0A4E9EZP7 | Arginine kinase | 40.5 | 15.23 | 1.881 | 0.13745321 |
| 47 | A0A3P7FEU7 | Aminotransferase class I/classII domain-containing protein | 47.4 | 26.82 | 1.876 | 0.139157966 |
| 48 | A0A4E9FZS3 | Sodium/potassium-transporting ATPase subunit alpha | 111.1 | 20.35 | 1.87 | 0.141310621 |
| 49 | J9EHH9 | Uncharacterized protein | 134.4 | 5.05 | 1.855 | 0.146485992 |
| 50 | J9ELW9 | Chaperonin GroL | 61.4 | 750.65 | 1.854 | 0.14701626 |
| 51 | A0A4E9FT05 | Chloride intracellular channel exc-4(excretory canal abnormal protein4), putative | 33.9 | 11.42 | 1.849 | 0.148646905 |
| 52 | A0A4E9ESS7 | Methionine aminopeptidase 2 | 46.7 | 28.36 | 1.841 | 0.151566548 |
| 53 | J9BDB6 | Uncharacterized protein | 13.6 | 36.76 | 1.814 | 0.162075474 |
| 54 | A0A0K0JCL5 | Bm3963, isoform b | 12.6 | 2.01 | 1.814 | 0.161884262 |
| 55 | J9FAQ8 | Cation-transporting P-type ATPase N-terminal domain-containing protein | 10.5 | 2.17 | 1.809 | 0.163845709 |
| 56 | J9ASR6 | Mlp/crp family protein 1 | 14.5 | 38.31 | 1.807 | 0.164843254 |
| 57 | A0A5S6PN68 | Fumarate hydratase | 54.3 | 639.19 | 1.804 | 0.166166117 |
| 58 | A0A1I8ESR7 | Glutathione-disulfide reductase | 52.6 | 7.02 | 1.785 | 0.173902686 |
| 59 | A0A3P7DIY7 | Glyceraldehyde-3-phosphate dehydrogenase | 36.2 | 503.05 | 1.785 | 0.173814354 |
| 60 | A0A3P7EAK0 | Ribosome maturation protein SBDS | 33.5 | 13.53 | 1.783 | 0.17465467 |
| 61 | A0A1I8ERE7 | Protein disulfide-isomerase | 59 | 26.5 | 1.764 | 0.183044436 |
| 62 | J9ENJ4 | Ribosomal protein L37ae | 12.7 | 10.28 | 1.762 | 0.183809632 |
| 63 | A0A1P6BM73 | Succinate dehydrogenase [ubiquinone] iron–sulfur subunit, mitochondrial | 31.7 | 24.6 | 1.76 | 0.184836825 |
| 64 | A0A1I8EAU9 | Ndr family protein | 39 | 16.16 | 1.758 | 0.18598488 |
| 65 | J9AQV1 | Adenylate kinase isoenzyme 1 | 22.8 | 139.74 | 1.720 | 0.89073951 |
| 66 | A0A1I8EX81 | Galectin | 36.7 | 63.54 | 1.712 | 0.208012151 |
| 67 | A0A1I8F0A6 | Vacuolar proton pump subunit B | 57.6 | 10.7 | 1.708 | 0.209815911 |
| 68 | J9B374 | Sorting nexin-12 | 19 | 19.5 | 1.701 | 0.213465697 |
| 69 | A0A3P7E0Z2 | MICOS complex subunit MIC60 | 79.8 | 12.91 | 1.689 | 0.219699941 |
| 70 | A0A1P6BMC5 | Ribonucleoprotein | 14 | 19.75 | 1.683 | 0.223170446 |
| 71 | J9EY80 | Translation elongation factor Tu | 54 | 22.13 | 1.673 | 0.22834361 |
| 72 | A0A0H5SBF4 | Bm3026 | 15.4 | 22.27 | 1.671 | 0.22962774 |
| 73 | A0A4E9EUM6 | Methionine aminopeptidase | 43.3 | 5.66 | 1.644 | 0.245122576 |
| 74 | A0A0M4FXK5 | Phosphoglycerate kinase (fragment) | 29 | 257.43 | 1.644 | 0.245077256 |
| 75 | A0A4E9FW13 | Adenylosuccinate synthetase | 52.7 | 26.35 | 1.639 | 0.248281406 |
| 76 | A0A4E9FV29 | Tubulin gamma chain | 49.2 | 17.68 | 1.631 | 0.252845192 |
| 77 | A0A0J9XNT3 | 40S ribosomal protein S27, putative; BMA-RPS-27 | 9.5 | 20.5 | 1.63 | 0.253758622 |
| 78 | Q6H323 | Protein disulfide-isomerase (fragment) | 53.9 | 19.63 | 1.625 | 0.256455645 |
| 79 | A0A3P7G9Q2 | 26S proteasome complex subunit dss-1 | 62.4 | 14.76 | 1.619 | 0.260540116 |
| 80 | A0A4E9FND0 | Transthyretin-like family protein | 15.3 | 156.07 | 1.619 | 0.26040714 |
| 81 | A0A4E9FP97 | DUF19 domain-containing protein | 40.8 | 8.39 | 1.608 | 0.267531713 |
| 82 | A0A3P7FCC0 | Peptidase S1 domain-containing protein | 31.5 | 18.54 | 1.601 | 0.272003697 |
| 83 | A0A0K0JWH8 | BMA-HMG-1.1 | 10.3 | 92.09 | 1.6 | 0.272838833 |
| 84 | A0A5S6PN83 | Ubiquitin carboxyl-terminal hydrolase 7 | 127.3 | 10.1 | 1.593 | 0.277634590 |
| 85 | A0A4E9FGM3 | Calponin-homology (CH) domain-containing protein | 15.5 | 482.63 | 1.590 | 0.710268997 |
| 86 | J9ES30 | Cytoplasmic tRNA 2-thiolation protein 1 (fragment) | 27.8 | 5.37 | 1.577 | 0.288359931 |
| 87 | J9FJW2 | 60S ribosomal protein L12 | 31.1 | 93.71 | 1.576 | 0.288749514 |
| 88 | A0A4E9FAX4 | Hypothetical RNA-binding protein T28D9.2 in chromosome II, putative | 45.5 | 8.2 | 1.576 | 0.288804578 |
| 89 | A0A4E9FMS4 | TATA-binding protein interacting (TIP20) domain-containing protein | 142.8 | 10.07 | 1.564 | 0.297346175 |
| 90 | A0A1I8EJ18 | BAR domain-containing protein | 34 | 77.51 | 1.563 | 0.297723468 |
| 91 | J9FEN6 | Succinate–CoA ligase [ADP-forming] subunit beta, mitochondrial | 47.4 | 15.9 | 1.544 | 0.3118908 |
| 92 | A0A4E9ESV3 | Guanine nucleotide-binding protein subunit gamma | 7.5 | 4.99 | 1.542 | 0.313185891 |
| 93 | A0A1I8ETH8 | GDP-L-fucose synthase | 54.4 | 12.87 | 1.537 | 0.317223493 |
| 94 | A0A4E9FBF2 | Peripheral subunit-binding (PSBD) domain-containing protein | 35.5 | 10.11 | 1.527 | 0.324770277 |
| 95 | A0A4E9ER74 | Uncharacterized protein | 226 | 61.89 | 1.527 | 0.324530557 |
| 96 | A0A5S6PLZ5 | FAD_binding_2 domain-containing protein | 56.8 | 76.37 | 1.518 | 0.331946859 |
| 97 | A0A0I9NBF1 | BMA-SNR-2 | 18.1 | 17.01 | 1.517 | 0.332302623 |
| S. n. | Accession | Description | MW [kDa] | Score sequest HT | Abundance ratio: (treated)/(control) | Abundance ratio P-value: (treated)/(control) |
|---|---|---|---|---|---|---|
| 1 | J9FES9 | Proteasome subunit alpha type (fragment) | 24.7 | 187.5 | 0.669 | 0.317621517 |
| 2 | J9FGQ7 | MPN domain-containing protein | 38.2 | 2.44 | 0.669 | 0.318789073 |
| 3 | Q962A2 | Translationally-controlled tumor protein homolog | 20.8 | 85.57 | 0.66 | 0.302533179 |
| 4 | J9DX04 | RRM domain-containing protein (fragment) | 6.7 | 5.6 | 0.658 | 0.29902988 |
| 5 | A0A4E9F9C9 | SGS domain containing protein | 23 | 52.1 | 0.658 | 0.30006964 |
| 6 | A0A3P7GA46 | SH3 domain-containing protein | 73.4 | 16.78 | 0.655 | 0.294743553 |
| 7 | A0A1I8EEX0 | Skp1-related protein | 25.6 | 9.09 | 0.653 | 0.291820357 |
| 8 | J9AKD6 | 26S protease regulatory subunit 8 | 29.8 | 42.72 | 0.652 | 0.289955506 |
| 9 | A0A3P7GHM4 | Vesicle-fusing ATPase | 91.6 | 687.71 | 0.649 | 0.283644745 |
| 10 | A0A1I8EG93 | RuvB-like helicase | 47.4 | 13.63 | 0.646 | 0.278619261 |
| 11 | A0A8L7TJD2 | UNC-52/perlecan, putative | 375 | 10.96 | 0.641 | 0.27153754 |
| 12 | A0A3P7FIZ2 | Proteasome subunit alpha type | 29 | 37.22 | 0.634 | 0.259568518 |
| 13 | A0A4E9EWP4 | ATP-dependent 6-phosphofructokinase | 89.6 | 32.66 | 0.633 | 0.258764632 |
| 14 | J9EVC3 | Protein serine/threonine phosphatase 2C C-terminal domain-containing protein (fragment) | 12.4 | 35.48 | 0.633 | 0.258401234 |
| 15 | A0A4E9FBQ2 | Trans-ketolase, putative | 67.2 | 159.55 | 0.631 | 0.254455621 |
| 16 | A0A3P7DP86 | Uncharacterized protein | 8.5 | 64.05 | 0.63 | 0.25304265 |
| 17 | A0A5S6PC29 | VWFA domain-containing protein | 530.4 | 2.39 | 0.628 | 0.251074583 |
| 18 | J9ENW2 | Uncharacterized protein | 13.8 | 6.32 | 0.623 | 0.241800228 |
| 19 | J9FG14 | Heat shock 70 protein (fragment) | 67.8 | 847.76 | 0.623 | 0.242147498 |
| 20 | A0A4E9FKH9 | TPR domain containing protein | 30.5 | 49.27 | 0.616 | 0.231755568 |
| 21 | J9FBW7 | Small heat shock protein | 17.8 | 20.23 | 0.613 | 0.226798658 |
| 22 | J9EFE8 | Profilin | 14.1 | 9.32 | 0.612 | 0.225932411 |
| 23 | A0A1I8EI05 | Twitchin | 752.9 | 31.93 | 0.599 | 0.206227775 |
| 24 | A0A0H5S9A3 | Dihydrolipoyllysine-residue succinyl transferase component of 2-oxoglutarate dehydrogenase complex, mitochondrial | 51.2 | 18.03 | 0.597 | 0.203409966 |
| 25 | A0A0K0J9J7 | 60S ribosomal protein L35a | 14.2 | 14.19 | 0.596 | 0.202731388 |
| 26 | A0A4E9EZ61 | Ribosomal protein L10e/L16 domain-containing protein | 24.7 | 10.63 | 0.593 | 0.198052286 |
| 27 | A0A4E9FBN8 | Cytoplasmic intermediate filament protein, putative | 67.8 | 262.21 | 0.59 | 0.19310016 |
| 28 | A0A1I9G5N0 | Bm898 (fragment) | 4.3 | 11.34 | 0.577 | 0.176269485 |
| 29 | A0A0J9Y2D9 | BMA-SEM-5 | 23.5 | 21.62 | 0.569 | 0.165095268 |
| 30 | A0A4E9EXP0 | Uncharacterized protein | 47.8 | 86.79 | 0.569 | 0.165994255 |
| 31 | A0A5S6PR17 | BMA-SRAP-1 | 211.8 | 13.27 | 0.565 | 0.159960888 |
| 32 | A0A3P7DJL2 | SHSP domain-containing protein | 19.9 | 16.54 | 0.564 | 0.159719255 |
| 33 | A0A0K0J064 | Mitochondrial import inner membrane translocase subunit | 10.6 | 36.55 | 0.556 | 0.149230289 |
| 34 | J9BHI4 | Prefoldin | 18.2 | 2.95 | 0.554 | 0.14646369 |
| 35 | A0A1I9G512 | Bm2039, isoform c | 50.8 | 14.3 | 0.552 | 0.144422143 |
| 36 | A0A8L7SNZ6 | Transcriptional activator protein Pur-alpha | 29.4 | 14.53 | 0.551 | 0.142684051 |
| 37 | A0A4E9FBQ6 | NADP-dependent oxidoreductase domain-containing protein | 36.3 | 7.59 | 0.546 | 0.137456776 |
| 38 | A0A1I8EXK7 | Oxoglutarate dehydrogenase (succinyl-transferring) | 112.5 | 5.65 | 0.544 | 0.134605963 |
| 39 | A0A4E9FE28 | V-type proton ATPase subunit F | 13.6 | 9.24 | 0.543 | 0.134175691 |
| 40 | A0A8L7SX06 | Fatty acid synthase | 138.4 | 8.64 | 0.543 | 0.133977568 |
| 41 | A0A0J9XPL7 | BMA-LSM-7, isoform a | 11.3 | 6.38 | 0.53 | 0.119384074 |
| 42 | A0A0J9XYB9 | BMA-DNJ-13, isoform c | 36.8 | 20.03 | 0.527 | 0.115927318 |
| 43 | A0A0J9XRU7 | 60S ribosomal protein L35 | 19 | 9.51 | 0.524 | 0.113361363 |
| 44 | A0A1I8EDE6 | Proteasome endopeptidase complex | 26.1 | 18.22 | 0.523 | 0.112421588 |
| 45 | J9FF58 | Laminin subunit gamma-1 (fragment) | 183.1 | 10.78 | 0.521 | 0.110046266 |
| 46 | A0A0J9XLH0 | Bm9133 | 26.6 | 21.79 | 0.521 | 0.109601854 |
| 47 | A0A7I4NJV0 | ATP-dependent (S)-NAD(P)H-hydrate dehydratase | 34.2 | 29.92 | 0.515 | 0.104291209 |
| 48 | J9EGA5 | Uncharacterized protein (fragment) | 8.8 | 99.86 | 0.511 | 0.099662931 |
| 49 | A8Q043 | cAMP-dependent protein kinase regulatory chain, putative | 7.2 | 34.74 | 0.501 | 0.091034522 |
| 50 | J9EJZ2 | Proliferating cell nuclear antigen | 29.1 | 24.21 | 0.501 | 0.091036289 |
| 51 | J9F0I0 | Clathrin light chain | 22.8 | 16.91 | 0.486 | 0.077156737 |
| 52 | A0A4E9EVU8 | Uncharacterized protein | 58 | 17.39 | 0.485 | 0.077007995 |
| 53 | A0A8L7SQJ2 | Glutamine synthetase | 41.2 | 19.22 | 0.478 | 0.071305005 |
| 54 | A0A4E9FH92 | RRM domain-containing protein | 42.2 | 10.62 | 0.475 | 0.06861159 |
| 55 | A0A4E9FEZ1 | Vitellogenin domain-containing protein | 361.1 | 10.63 | 0.473 | 0.06696446 |
| 56 | A0A4E9FBY7 | Proteasome alpha-type subunits domain-containing protein | 27.7 | 67.83 | 0.472 | 0.066088914 |
| 57 | A0A0K0JIQ0 | Bm5388, isoform a | 19.4 | 1.89 | 0.466 | 0.061851423 |
| 58 | J9FCT2 | Mannose-6-phosphate isomerase | 45 | 3.59 | 0.452 | 0.052220282 |
| 59 | A0A3P7E5V5 | Integrin beta N-terminal domain-containing protein | 13.7 | 18.4 | 0.449 | 0.05058465 |
| 60 | J9F5C4 | Mitochondria bc1 complex core subunit 1 (fragment) | 50.3 | 5.48 | 0.447 | 0.049436044 |
| 61 | J9ARA6 | 40S ribosomal protein S8 (fragment) | 17.2 | 80.97 | 0.438 | 0.044081236 |
| 62 | A0A0J9XNT1 | Bm255 | 9.2 | 52.04 | 0.436 | 0.042833626 |
| 63 | A0A4E9F8W1 | UBC core domain-containing protein | 19 | 7.45 | 0.414 | 0.031326666 |
| 64 | A0A1I9GCP6 | Bm9018 | 138.4 | 7.98 | 0.4 | 0.025577555 |
| 65 | A0A5S6PPU7 | BMA-ALX-1 | 75.9 | 19.01 | 0.399 | 0.025295669 |
| 66 | A0A0J9Y905 | BMA-TLN-1, isoform a | 278.1 | 38.09 | 0.398 | 0.024679215 |
| 67 | A0A1I8EBP1 | RRM domain-containing protein | 40.7 | 21.66 | 0.366 | 0.014543053 |
| 68 | J9BBS8 | NADAR domain-containing protein | 36.5 | 28.47 | 0.366 | 0.014527273 |
| 69 | A0A3P7EB04 | Uncharacterized protein | 28 | 12.76 | 0.364 | 0.013891842 |
| 70 | A0A4E9FSQ9 | Leucine rich repeat family protein | 27.6 | 9.56 | 0.358 | 0.012376684 |
| 71 | J9FDW3 | Transketolase | 67.3 | 133.19 | 0.348 | 0.010169759 |
| 72 | J9DT68 | Uncharacterized protein | 10.5 | 8.39 | 0.348 | 0.01018951 |
| 73 | A0A8L7SNS8 | Adenosylhomocysteinase | 48.1 | 21.63 | 0.346 | 0.009749431 |
| 74 | A0A5S6PIB0 | BMA-PQN-22 | 84.6 | 11.12 | 0.342 | 0.009029136 |
| 75 | A0A5S6P7N8 | Uncharacterized protein | 91 | 5.47 | 0.325 | 0.006333444 |
| 76 | A0A1I8EWK5 | BSD domain-containing protein | 38.7 | 2.41 | 0.304 | 0.003846617 |
| 77 | A0A0H5S5L6 | BMA-ALP-1 | 67 | 5.34 | 0.3 | 0.003448479 |
| 78 | A0A5S6PX95 | Bm8873, isoform c | 100.3 | 21.58 | 0.297 | 0.003182232 |
| 79 | A0A3P7ETZ6 | PDZ domain-containing protein | 44.7 | 3.84 | 0.289 | 0.002555259 |
| 80 | J9E3C5 | Uncharacterized protein | 7 | 5.69 | 0.284 | 0.002213946 |
| 81 | A0A4E9FE79 | Proteasome subunit beta type 2, putative | 17.9 | 4.54 | 0.27 | 0.001482822 |
| 82 | A0A5S6PAI6 | Uncharacterized protein | 24.6 | 5.79 | 0.244 | 0.000627717 |
| 83 | A0A3P7FJZ7 | Uncharacterized protein (fragment) | 50.6 | 8.01 | 0.132 | 9.35986 × 10−7 |
| 84 | A0A1I8EP56 | 60S ribosomal protein L30 | 12.3 | 38.81 | 0.013 | 1 × 10−17 |
| 85 | A0A3P7DU85 | Coatomer subunit beta | 107.2 | 2.38 | 0.01 | 1 × 10−17 |
| 86 | A0A4E9EXG9 | RWD domain-containing protein | 29.9 | 2.84 | 0.01 | 1 × 10−17 |
| 87 | J9EMX1 | Eukaryotic translation initiation factor 3 subunit K | 18.8 | 3.34 | 0.01 | 1 × 10−17 |
| 88 | A0A3P7DVD5 | Activator of Hsp90 ATPase AHSA1-like N-terminal domain-containing protein | 40.3 | 1.76 | 0.01 | 1 × 10−17 |
After CA treatment the levels of detoxifying enzymes such as glutathione S-transferase (GST), superoxide dismutase (SOD), thioredoxin, glutathione peroxidase and glutathione reductase were significantly increased. These enzymes play a crucial role in scavenging oxidants and serve as the parasites' primary defense mechanism. The enzymes GST and SOD have a role in the metabolism of xenobiotics and their overexpression indicates an enhanced requirement for detoxification in CA treated parasites.
The expression of key components of the cytoskeletal structure, tropomyosin, myosin family proteins, tubulin, and moesin/ezrin/radixin (MER) homolog-1 was increased in the filarial worms treated with CA. Myosin is the molecular component responsible for the contraction of sarcomeres and has the ability to convert chemical energy into mechanical energy. Moesin/ezrin/radixin homolog-1 facilitates the interaction of plasma membrane and filamentous actin, thus facilitating the cell cortex stability. The MERs control the signaling pathway by binding transmembrane receptors and connecting them to downstream signaling components and the overexpression of these proteins could be correlated to significant alterations in the cytoskeleton of the parasite.39
The glycolytic enzymes enolase, triose phosphate isomerase, glyceraldehyde 3 phosphate dehydrogenase, and phosphoglycerate kinase were identified among the major upregulated proteins. Several enzymes involved in the energy metabolism such as phosphoglucomutase, L-lactate dehydrogenase, succinate CoA ligase subunit alpha, triose phosphate isomerase, BMA-CYC-2.2, aconitate hydratase, malate dehydrogenase, glyceraldehyde-3-phosphate dehydrogenase, succinate dehydrogenase, phosphoglycerate kinase, were significantly upregulated after CA treatment. Some of these enzymes are part of TCA cycle and glycolysis while BMA-CYC-2.2 is a component of the oxidative phosphorylation machinery. The upregulation could be due to increased demands for energy in CA treated parasites. Another highly upregulated protein was the transthyretin-like family protein molecular weight 15.9 and 20.2, which is involved in the apoptotic process of corps engulfment. The transthyretin-like family protein has been shown to have neuroprotective role as it protects dopaminergic neurons against degradation caused by oxidative stress.40
The major protein degradation pathways involves ubiquitin proteasome system involving proteasome subunit alpha type fragment (J9FES9), proteasome subunit alpha type subunit (A0A4E9FBY7), proteasome endopeptidase complex, proteasome alpha-type subunits domain-containing protein, proteasome subunit beta type 2, and RWD domain-containing protein was highly downregulated. This system is responsible for degradation of more than 80% of the cellular proteins and is also actively involved in other cellular processes like apoptosis, control of cell-cycle progression and metabolic regulation.41,42
Harnessing the proteasome's destructive force to selectively degrade the drivers of human disease, has opened up a new and fascinating field of drug discovery. For example, targeted immunoproteasome inhibition has excellent clinical efficacy for autoimmune disease and inflammation and proteasome inhibitors could be used as innovative therapies for malaria and other microbes.43 Also the heat shock proteins SHSP domain-containing protein and activator of Hsp90 ATPase AHSA1-like N-terminal domain-containing protein were highly downregulated. In another study, similar downregulation of HSPs was correlated with the death of filarial parasites.
The versatile central factor Proliferating Cell Nuclear Antigen (PCNA) was highly downregulated after treatment with CA seed extract in filarial parasites. The downregulation of PCNA after CA treatment could be one of the major factors for death of the filarial parasites. The PCNA encircles DNA, and act as proclivity factor in DNA replication.44 PCNA forms the protein complexes in base excision repair, nucleotide excision repair, mismatch repair, homologous recombination, and cell cycle progression. Several researchers have established the fact that inhibition of PCNA could be a successful therapeutic strategy for treatment of cancer.45
The CA treated worms showed reduced expression of coatomer subunit β (abundance ratio P-value 0.01), low levels of coatomer leads to the fragmentation of Golgi apparatus, suppression of autophagy and cell death. It was also observed that many crucial enzymes such as adenosylhomocysteinase, transketolase, mannose-6-phosphate isomerase, and fatty acid synthase were significant downregulated, thus severely affecting the survival of the filarial worms.
3.5. Gene ontology and functional classification of differentially expressed proteins
Gene ontology annotation analysis for the most significant Differentially Expressed Proteins (DEPs), categorized by their molecular function, cellular components and biological process is shown in Fig. 4. Regarding molecular function, the main DEPs were involved in ATP binding, metal ion binding, GTP binding, ATP hydrolysis activity, actin monomer binding, oxidoreductase activity, protein folding, chaperone binding, RNA binding, transferase activity, and cytoskeletal motor activity as structural constituents of chromatin. The major DEPs in the biological processes category were mostly involved in tRNA processing, biosynthetic processes, metabolic activities, protein transport, tricarboxylic acid cycle, reaction to stimulus, glutamine biosynthesis processes, SCF complex assembly, translation process, and stress response. The biological components that showed substantial enrichment were the cell surface, nucleus, plasma membrane, cytoplasm, mitochondria, ribonucleoprotein complex, nucleosome, ribosomal protein, extracellular matrix, and spliceosome complex. Proteomic analysis showed that treating filarial worms with CA led to the suppression of many proteins involved in energy metabolism, signal transduction, stress response, chaperone proteins, and highly antigenic proteins.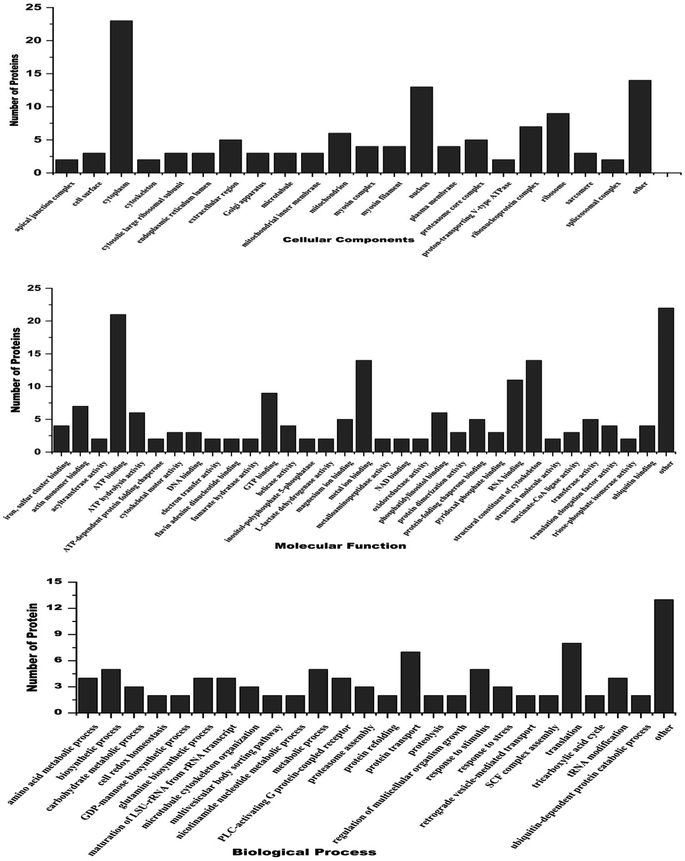 | ||
| Fig. 4 Gene ontology analysis of differentially expressed proteins belonging to 3 major classes i.e. cellular component, molecular function and biological processes. | ||
3.6. FT-IR spectral analysis of CA ethanolic extract
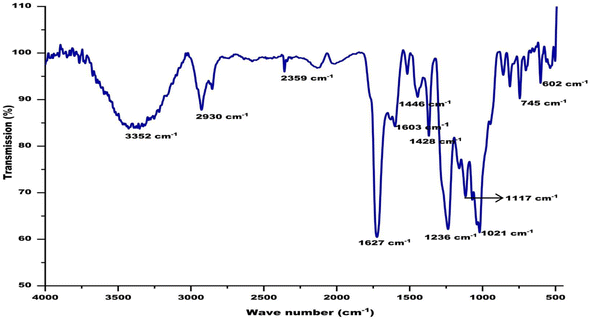
FT-IR spectra of biological samples are typically performed in the range of 4000 to 400 cm−1 to identify functional groups of the active components by observing the emitted peaks in the infrared radiations. The spectrum pattern from the CA seed extract was observed at 602, 745, 1021, 1117, 1236, 1428, 1446, 1603, 1627, 2359, 2930, and 3352 cm−1, respectively. The small sharp peak at 602 corresponds to the aromatic H- out of plane bending. Several small peaks in the range of 729–759 correspond to the C–C in the CA extract. The sharp peak at 1021 corresponds to present of phosphate ion in the extract. The phenolic groups involved in ion replacement reactions are placed in the 1250–1270 cm−1 and 1485–1620 cm−1 spectrum of the plant extract. The peak at 1627 represents N–H bending in amide group and the peak at 2930 is accredited to asymmetric stretching of sp3 carbon atoms. The broad peak found between 3280–3495 cm−1 is assigned to the stretching of the (–NH) aliphatic secondary groups present in the extract (Fig. 5).3.7. UPLC-ESI MS/MS analysis of CA ethanolic seed extract
The UPLC-ESI-MS/MS analysis was performed for the identification of bioactive compounds of CA ethanolic seed extract. The bioactive compounds were identified based on molecular mass and retention time with database ResPect phytochemical software. The UPLC-ESI MS/MS analysis of both positive and negative ions modes was performed, and in total 40 compounds were identified. The detected compounds are listed in Table 4 along with their retention periods, molecular weights, molecular formula, and amounts (ppm). The robust antioxidative defense system of filarial parasites aids in evading the host oxidative attack mechanism. For the discovery of novel pharmaceuticals, molecular docking is a more expanding and cost effective alternative to the laborious in vitro drug screening procedure. The 25 most abundant CA bioactive compounds were selected for in silico screening against the forementioned filarial anti-oxidant proteins/enzymes. Based on binding energy values the 10 top scoring bioactive compounds namely 3-O-methylquercitin, 4-methoxycinnamoyloleanolic acid methyl ester, carbenicillin, podorhizol-β-D glucoside, RU5135, soraphen A, beta-obscurine, carbenicillin, vanillin, gentisic acid, and quinic acid were chosen for further in silico studies (Table 5 and Fig. 6).| S. n. | Name of compound | Retention time (min) | Theoretical mass | Molecular formula | DB diffa (ppm) |
|---|---|---|---|---|---|
| a (ppm) parts per million. | |||||
| 1 | CDP-DG (12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/12 0/12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0) 0) |
15.875 | 840.3915 | C36H65N3O15P2 | 1187.08 |
| 2 | Quinic acid | 1.202 | 192.0596 | C7H12O6 | 19.47 |
| 3 | Gentisic acid | 2.028 | 154.0236 | C7H6O4 | 19.45 |
| 4 | 2-Acetylthiophene | 4.724 | 126.0116 | C6H6OS | 18.86 |
| 5 | Trans-chlorogenic acid | 3.526 | 354.0897 | C16H18O9 | 15.19 |
| 6 | Vanillin | 5.639 | 152.0453 | C8H8O3 | 13.41 |
| 7 | Soraphen A | 14.703 | 520.2982 | C29H44O8 | 10.36 |
| 8 | 3-Acetyl-6-methoxybenzaldehyde | 6.897 | 178.0612 | C10H10O3 | 9.85 |
| 9 | IAA/3-indoleacetic acid | 7.011 | 175.0617 | C10H9NO2 | 9.44 |
| 10 | Irisolidone 7-O-glucuronide | 5.948 | 490.1064 | C23H22O12 | 9.62 |
| 11 | Flavine mononucleotide (FMN) | 5.999 | 456.1006 | C17H21N4O9P | 8.85 |
| 12 | 4-Methoxycinnamoyloleanolic acid methyl ester | 18.566 | 630.4232 | C41H58O5 | 8.29 |
| 13 | 3-Carboxyethenyl-3,5-cyclohexadiene-1,2-diol | 8.308 | 182.0564 | C9H10O4 | 8.22 |
| 14 | 3-Methylindolepyruvate | 10.651 | 217.0724 | C12H11NO3 | 7 |
| 15 | 3-O-Methylquercetin | 9.393 | 316.0562 | C16H12O7 | 6.54 |
| 16 | 4-Dodecylbenzenesulfonic acid | 19.63 | 326.1895 | C18H30O3S | 6.4 |
| 17 | Annotemoyin 1 | 20.124 | 564.4722 | C35H64O5 | 6.39 |
| 18 | PG(16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1(9Z)/16 1(9Z)/16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0) 0) |
19.087 | 720.4896 | C38H73O10P | 6.26 |
| 19 | Theasapogenol E | 19.641 | 504.342 | C30H48O6 | 6.18 |
| 20 | Dihydroxy-epoxyoctadecanoate | 9.902 | 330.2386 | C18H34O5 | 6.18 |
| 22 | Podorhizol beta-D-glucoside | 7.221 | 578.1964 | C28H34O13 | 6.14 |
| 23 | 15-O-demethyl-dideoxydihydro-striatin C | 15.039 | 434.2644 | C25H38O6 | 5.57 |
| 24 | Ascorbyl stearate | 10.851 | 442.2906 | C24H42O7 | 5.52 |
| 25 | Avocadene 2-acetate | 12.222 | 328.2596 | C19H36O4 | 5.5 |
| 26 | Stypandrol | 10.921 | 430.1393 | C26H22O6 | 5.48 |
| 27 | RU 5135 | 13.253 | 304.2135 | C18H28N2O2 | 5.3 |
| 28 | Beta-obscurine | 16.561 | 272.1877 | C17H24N2O | 4.41 |
| 29 | MG(15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/0 0/0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/0 0/0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0) 0) |
14.437 | 316.26 | C18H36O4 | 4.38 |
| 30 | Carbenicillin | 1.275 | 378.0869 | C17H18N2O6S | 4.32 |
| 31 | Dibutyl decanedioate | 13.252 | 314.2444 | C18H34O4 | 4.32 |
| 32 | LysoPE(18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1(11Z)/0 1(11Z)/0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0) 0) |
18.424 | 479.2995 | C23H46NO7P | 3.61 |
| 33 | N-undecylbenzenesulfonic acid | 18.137 | 312.1748 | C17H28O3S | 3.51 |
| 34 | LysoPE(0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0/18 0/18![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2(9Z,12Z)) 2(9Z,12Z)) |
15.105 | 477.2839 | C23H44NO7P | 3.4 |
| 35 | 2-(Methylthiomethyl)-3-phenyl-2-propenal | 3.746 | 192.0603 | C11H12OS | 2.98 |
| 36 | Isopetasoside | 15.16 | 396.2142 | C21H32O7 | 1.63 |
| 37 | N-adenylyl-L-phenylalanine | 1.276 | 494.131 | C19H23N6O8P | 1.09 |
| 38 | S-nitroso-L-glutathione | 9.159 | 336.0738 | C10H16N4O7S | 0.64 |
| 39 | Mytilin A | 5.385 | 332.1219 | C13H20N2O8 | 0.27 |
| 40 | Remifentanil | 10.823 | 376.1997 | C20H28N2O5 | 0.2 |
| S. n. | Compounds name | RT (min) | Formula | MW | Fragmentation profile (m/z) | DB diffa (ppm) |
|---|---|---|---|---|---|---|
| a (ppm) parts per million. | ||||||
| 1 | 3-O-methylquercetin | 9.393 | C16H12O7 | 316.05 | 207.0644 | 6.54 |
| 243.0273 | ||||||
| 255.0285 | ||||||
| 271.0234 | ||||||
| 300.0251 | ||||||
| 301.0295 | ||||||
| 315.0483 | ||||||
| 329.2307 | ||||||
| 395.0819 | ||||||
| 2 | 4-Methoxycinnamoyloleanolic acid methyl ester | 18.566 | C41H58O5 | 630.42 | 325.1836 | 8.29 |
| 689.4342 | ||||||
| 690.434 | ||||||
| 719.4857 | ||||||
| 3 | Podorhizol β D-glucoside | 7.221 | C28H34O13 | 578.19 | 160.839 | 6.14 |
| 162.8346 | ||||||
| 195.8088 | ||||||
| 255.0482 | ||||||
| 4 | RU5135 | 13.253 | C18H28N2O2 | 304.21 | 129.0904 | 5.30 |
| 183.138 | ||||||
| 295.2262 | ||||||
| 296.2199 | ||||||
| 313.2369 | ||||||
| 314.2401 | ||||||
| 5 | Soraphen A | 14.703 | C19H44O8 | 520.29 | 277.2167 | 10.36 |
| 313.2366 | ||||||
| 403.2242 | ||||||
| 6 | Vanillin | 5.639 | C8H8O3 | 152.04 | 108.0196 | 13.41 |
| 109.0253 | ||||||
| 137.0221 | ||||||
| 151.0373 | ||||||
| 187.095 | ||||||
| 197.8061 | ||||||
| 262.065 | ||||||
| 7 | Quinic acid | 1.202 | C7H12O6 | 192.39 | 191.0524 | 19.47 |
| 192.0555 | ||||||
| 193.0577 | ||||||
| 195.0473 | ||||||
| 317.0493 | ||||||
| 377.0802 | ||||||
| 379.0777 | ||||||
| 539.1314 | ||||||
| 8 | Gentisic acid | 2.028 | C7H6O4 | 154.02 | 109.0266 | 19.45 |
| 110.0305 | ||||||
| 153.0165 | ||||||
| 175.0571 | ||||||
| 218.1004 | ||||||
| 282.0811 | ||||||
| 9 | Beta-obscurine | 16.561 | C17H24N2O | 272.1877 | 331.201 | 4.41 |
| 332.2001 | ||||||
| 333.2019 | ||||||
| 367.1791 | ||||||
| 368.1794 | ||||||
| 369.1708 | ||||||
| 370.176 | ||||||
| 10 | Carbenicillin | 1.275 | C17H18N2O6 S | 377.08 | 191.0508 | 4.32 |
| 192.0493 | ||||||
| 377.0772 | ||||||
| 379.0766 | ||||||
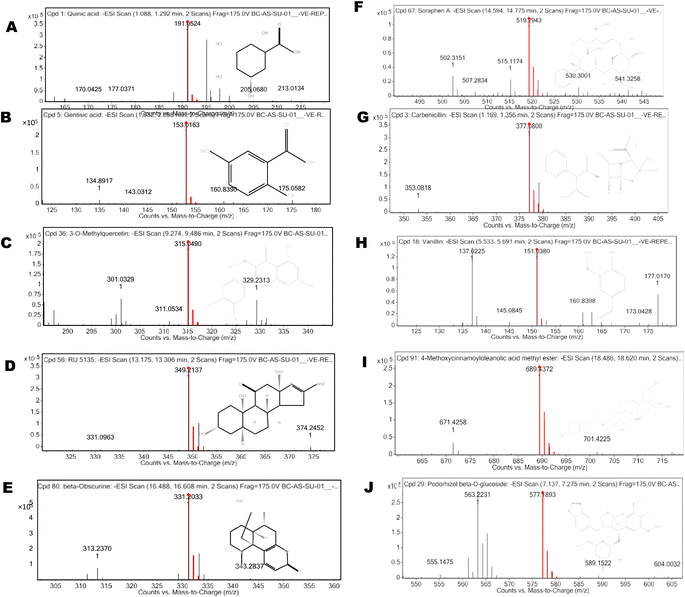 | ||
| Fig. 6 Graphical representation of LC-MS/MS spectra and fragmentation profile of CA bioactive compounds. | ||
3.8. Molecular docking of CA bioactive compounds
The drug-likeness properties of bioactive compounds were checked by the Lipinski filter and their Absorption, Distribution, Metabolism, Excretion, and Toxicity properties were analyzed by admetSAR server (Table 6). Out of the 10 bioactive compounds of CA, only five confirmed to be safe and non-toxic as predicted by the admetSAR values. The bioactive compounds 3-O-methylquercetin, vanillin, gentisic acid, and quinic acid only were used for further simulation studies. The 3D interactions of these CA bioactive compounds with the antioxidant proteins were visualized through BIOVIA Discovery Studio (Fig. 7).| Parameter absorption | 3-O-methylquercetin | 4-Methoxycinnamoyloleanolic acid methyl ester | Albendazole | DEC | Beta-obscurine | RU 5135 | Soraphen A | Carbenicillin | Gentisic acid | Quinic acid | Vanillin | Podorhizol beta-D-glucoside |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Blood brain barrier | BBB− | BBB+ | BBB+ | BBB+ | BBB+ | BBB+ | BBB− | BBB− | BBB+ | BBB+ | BBB+ | BBB− |
| Human intestinal absorption | HIA+ | HIA− | HIA+ | HIA+ | HIA+ | HIA+ | HIA− | HIA− | HIA+ | HIA+ | HIA+ | HIA− |
| Coco-2 permeability | Caco2+ | Caco2+ | Caco2− | Caco2+ | Caco2+ | Caco2− | Caco2+ | Caco2− | Caco2+ | Caco2− | Caco2+ | Caco2− |
| P-glycoprotein substrate | Substrate | Substrate | No substrate | Substrate | Substrate | Substrate | Substrate | Substrate | No substrate | No substrate | No substrate | Substrate |
| hERG | Weak inhibitor | Weak inhibitor | Weak inhibitor | Weak inhibitor | Weak inhibitor | Weak inhibitor | Weak inhibitor | Weak inhibitor | Weak inhibitor | Weak inhibitor | Weak inhibitor | Weak inhibitor |
| AMES toxicity | Non AMES toxic | Non AMES toxic | Non AMES toxic | Non AMES toxic | Non AMES toxic | Non AMES toxic | Non AMES toxic | Non AMES toxic | Non AMES toxic | Non AMES toxic | Non AMES toxic | Non AMES toxic |
| Carcinogens | Non carcinogen | Non carcinogen | Non carcinogen | Non carcinogen | Non carcinogen | Non carcinogen | Non carcinogen | Non carcinogen | Non carcinogen | Non carcinogen | Non carcinogen | Non carcinogen |
| Acute oral toxicity | III | III | III | III | III | III | III | IV | III | III | III | III |
| Rat acute toxicity | 2.6388 LD50 mol kg−1 | 2.0343 LD50 mol kg−1 | 2.0752 LD50 mol kg−1 | 2.2639 LD50 mol kg−1 | 2.9623 LD50 mol kg−1 | 2.6484 LD50 mol kg−1 | 3.0223 LD50 mol kg−1 | 1.4399 LD50 mol kg−1 | 2.1788 LD50 mol kg−1 | 1.7528 LD50 mol kg−1 | 1.9642 LD50 mol kg−1 | 2.6524 LD50 mol kg−1 |
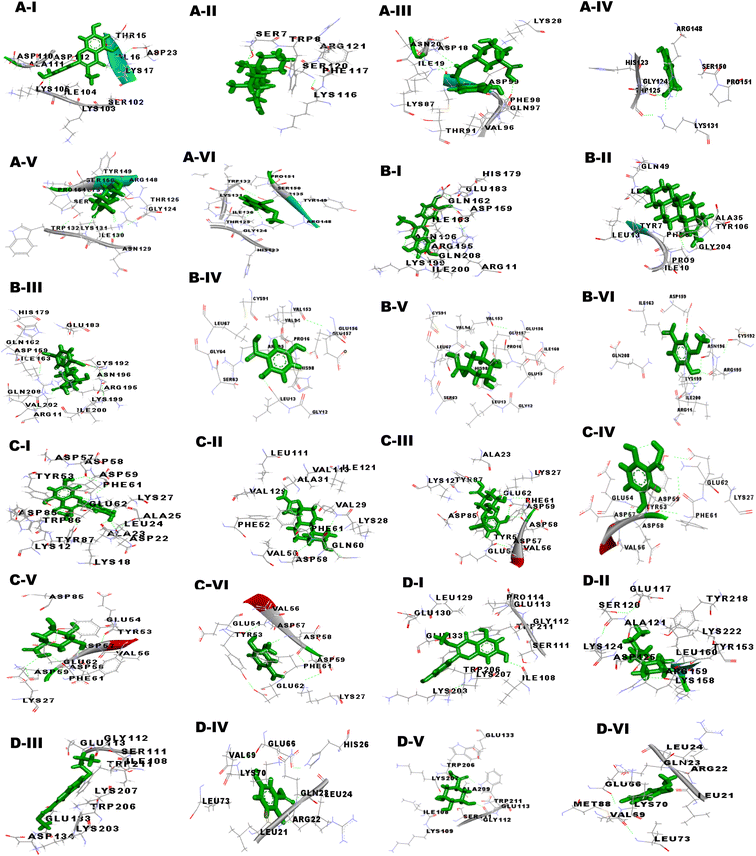
Our studies targeted the antioxidant proteins i.e. glutathione-S-transferase (GST), thioredoxin (TRx), glutathione peroxidase (GPx), and superoxide dismutase (SOD). The GPx model was validated by the RAMPAGE server, as well as the PDBsum and ProCheck servers showed that none of the amino acids were located in the disallowed region. Further, the quality of the 3D model was examined by ERRAT, ProSA and RAMPAGE servers. Metapocket 2.0 server was used to predict the binding site of GPx and the top 3 binding sites were considered as the active sites of protein (S Table 1†). The PatchDock server and YASARA tool were used to investigate the docking characteristics of CA bioactive compounds with filarial antioxidant proteins. The following parameters were studied in this work: (a) interacting amino acid residue, (b) interacting residue active site number, (c) CA bioactive compounds and antioxidant proteins involved in the H-bonding, (d) binding energy, (e) dissociation constant, (f) GSC score, and (g) AI area. Interacting residues were identified using the YASARA programme and the PatchDock server, and notable binding sites were predicted with the Metapocket 2.0 server and Discovery Studio 3.5. The retrieved docked complexes were screened for the highest binding energy, lowest dissociation constant, maximum hydrogen bonding, higher GSC score, AI area, and docking within the top three binding sites of anti-oxidant proteins, GST, GPx, SOD, and TRx, with only the best complex being chosen for further analysis. On the basis of docking studies CA bioactive compounds 4-methoxycinnamoyloleanolic acid methyl ester, 3-O-methylquercetin, Podorhizol β D-glucoside, and soraphen A had the highest computed binding energies. The binding energies of these compounds were much higher than anti-filarial drugs Albendazole and DEC (Table 7). The docking analysis also showed that CA bioactive compounds and antioxidant proteins could form ample hydrogen bonds with one another. Soraphen A, 3-O-methylquercetin and quinic acid showed maximum hydrogen bonding among all CA bioactive compounds, forming 8, 7 and 5 bonds with GST, TRx and GR respectively. Also, the interacting amino acid residues were mostly found in the predicted binding sites of the antioxidant proteins.
| Receptor | S. n. | Ligand | Binding energy (kcal mol−1) | Dissociation constant (μm) | Score | Area | ACE |
|---|---|---|---|---|---|---|---|
| Glutathione peroxidase | 1 | 4-Methoxycinnamoyloleanolic acid methyl ester | 7.540 | 2.967 | 5448 | 647.70 | −174.14 |
| 2 | 3-O-methylquercetin | 6.026 | 8.272 | 3864 | 500.00 | −243.56 | |
| 3 | RU 5135 | 4.312 | 246.392 | 4576 | 555.70 | −219.07 | |
| 4 | Podorhizol beta-D-glucoside | 6.443 | 18.932 | 5060 | 748.80 | −430.03 | |
| 5 | Vanillin | 4.782 | 312.426 | 2945 | 372.1 | −123.40 | |
| 6 | Soraphen A | 5.801 | 55.951 | 3764 | 401.6 | −30.84 | |
| 7 | Gentisic acid | 5.944 | 23.684 | 3010 | 310.5 | −125.61 | |
| 8 | Quinic acid | 5.427 | 105.1855 | 3066 | 322.4 | −115.59 | |
| 9 | Carbenicillin | 5.443 | 112.013 | 3129 | 298.3 | −124.41 | |
| 10 | Albendazole | 5.016 | 210.486 | 3698 | 397.90 | −165.80 | |
| 11 | DEC | 4.189 | 850.006 | 3708 | 454.50 | −212.10 | |
| 12 | Beta-obscurine | 4.012 | 910.020 | 3001 | 129.70 | −80.01 | |
| Glutathione-S-transferase | 1 | 4-Methoxycinnamoyloleanolic acid methyl ester | 8.756 | 0.381 | 5830 | 848.9 | −68.31 |
| 2 | 3-O-methylquercetin | 6.907 | 8.651 | 3746 | 450.5 | −97.94 | |
| 3 | RU 5135 | 4.109 | 135.196 | 3488 | 492.5 | −192.03 | |
| 4 | Vanillin | 4.757 | 325.891 | 3134 | 355.69 | −105.40 | |
| 5 | Podorhizol beta-D-glucoside | 7.309 | 4.389 | 4802 | 599.3 | −138.33 | |
| 6 | Soraphen A | 6.472 | 18.028 | 3730 | 486 | −109.89 | |
| 7 | Carbenicillin | 6.243 | 32.013 | 3629 | 298.3 | −64.41 | |
| 8 | Gentisic acid | 6.019 | 11.727 | 2706 | 305.3 | −86.24 | |
| 9 | Quinic acid | 5.869 | 49.884 | 2790 | 327.7 | −84.51 | |
| 10 | Albendazole | 5.025 | 207.312 | 3540 | 466.6 | −140.15 | |
| 11 | DEC | 4.215 | 13.512 | 3314 | 414.1 | −151.11 | |
| 12 | Beta-obscurine | 3.929 | 928.020 | 3101 | 239.70 | −94.21 | |
| Thioredoxin transferase | 1 | 4-Methoxycinnamoyloleanolic acid methyl ester | 7.786 | 1.962 | 5324 | 587.3 | 2.92 |
| 2 | 3-O-methylquercetin | 6.727 | 7.723 | 3920 | 423.6 | −177.16 | |
| 3 | RU 5135 | 3.987 | 113.250 | 3432 | 377.5 | −45.57 | |
| 4 | Beta-obscurine | 4.204 | 112.187 | 2994 | 331.7 | −50.98 | |
| 5 | Soraphen A | 6.728 | 11.703 | 3520 | 383.7 | −117.43 | |
| 6 | Carbenicillin | 6.518 | 16.681 | 3992 | 454.3 | −163.83 | |
| 7 | Vanillin | 4.975 | 225.567 | 3092 | 394.1 | −91.2 | |
| 8 | Gentisic acid | 5.605 | 7.889 | 4290 | 493.8 | −8.56 | |
| 9 | Quinic acid | 5.379 | 114.061 | 2208 | 277 | −80.32 | |
| 10 | Albendazole | 5.250 | 141.807 | 3374 | 363.3 | −98.38 | |
| 11 | DEC | 4.521 | 485.358 | 3000 | 320.2 | −95.42 | |
| 12 | Podorhizol beta-D-glucoside | 7.491 | 4.389 | 4792 | 569.3 | −128.33 | |
| Superoxide dismutase | 1 | 4-Methoxycinnamoyloleanolic acid methyl ester | 8.420 | 0.6730 | 5480 | 629.9 | −50.26 |
| 2 | 3-O-methylquercetin | 6.177 | 9.661 | 3352 | 415.4 | −111.54 | |
| 3 | RU 5135 | 4.035 | 111.565 | 3154 | 361.9 | −100.79 | |
| 4 | Beta-obscurine | 3.595 | 114.648 | 2968 | 320.6 | 29.8 | |
| 5 | Carbenicillin | 6.048 | 36.877 | 3934 | 475.5 | −212.64 | |
| 6 | Vanillin | 4.503 | 500.330 | 2948 | 226.1 | −21.4 | |
| 7 | Soraphen A | 5.728 | 91.703 | 3520 | 383.7 | −57.43 | |
| 8 | Gentisic acid | 4.967 | 8.633 | 2282 | 252.3 | 6.44 | |
| 9 | Quinic acid | 5.726 | 63.501 | 2316 | 255.9 | 27.02 | |
| 10 | Albendazole | 5.358 | 118.177 | 3648 | 413.9 | −109.75 | |
| 11 | DEC | 4.252 | 764.262 | 2936 | 342.3 | −4.55 | |
| 12 | Podorhizol beta-D-glucoside | 7.191 | 6.389 | 4692 | 499.3 | −138.33 |
3.9. Molecular dynamics simulation analysis
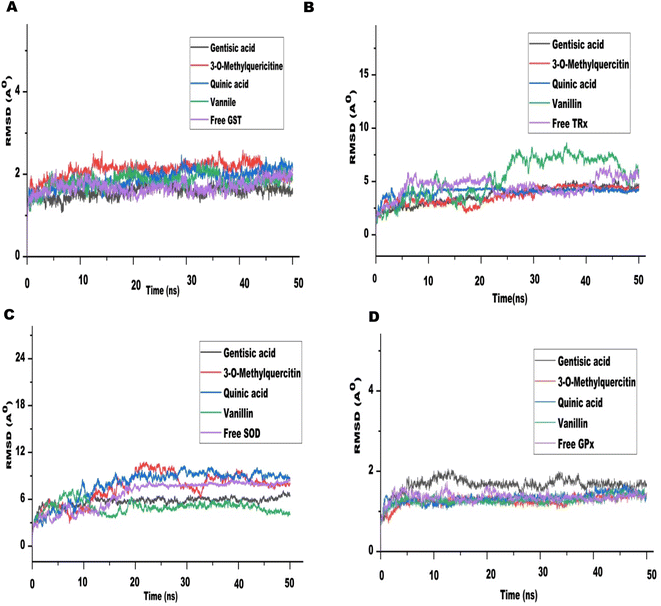
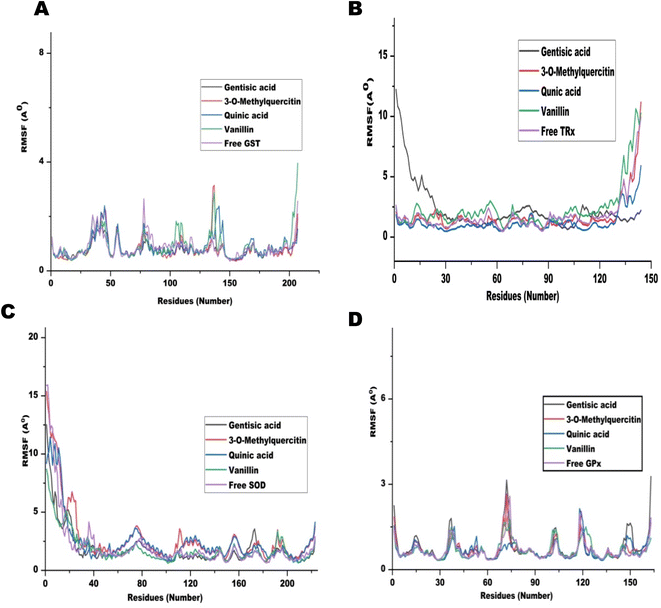
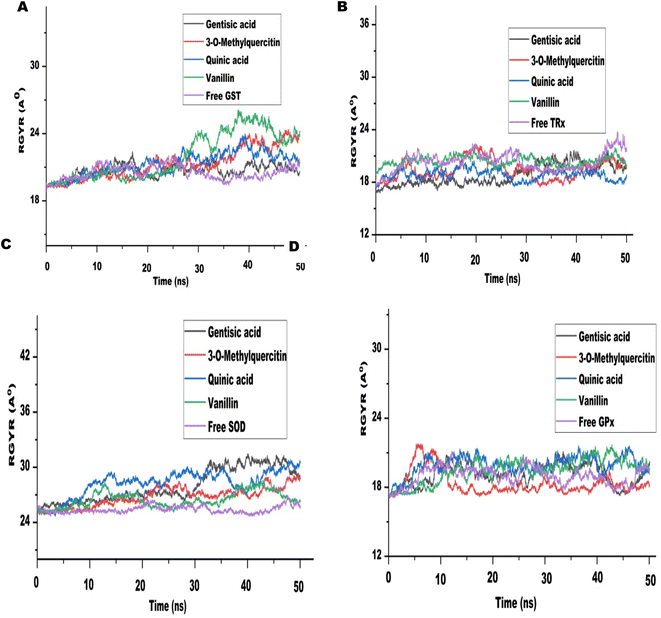
The MD run took place in an isothermal, isobaric (NPT) ensemble (310 K and 1 bar) for 50 nanoseconds (ns). Due to their stronger interactions and comparatively higher binding energies in molecular docking, we chose 3-O-methylquercetin, quinic acid, gentisic acid, and vanillin for the MD run. For each target, the MD simulations of individual filarial GST, TRx, SOD, and GR with water were used as control.The Root Mean Square Deviation (RMSD) of the protein ligand complexes of filarial antioxidant proteins and CA bioactive compounds is shown in Fig. 8. In the entire, MD simulation run, RMSD of GST ranged from 1.316 Å to 2.022 Å and was lowest with gentisic acid. The RMSD for TRx's interaction with 3-O-methylquercetin was in the range of 2.03–4.743 Å whereas for SOD it was 4.39–10.75 Å. The anti-oxidant protein GPx formed most stable complexes with all CA bioactive compounds and the variation in RMSD was less than 1 Å with 3-O-methylquercetine. Upon comparison of average RMSD values for protein-ligand complexes, 3-O-methylquercetin and gentisic acid formed most stable complexes with filarial antioxidant proteins. During the entire run, the total energy, potential energy, and temperature remained constant and the RMSD of each docked complex was below 10 Å.46 The interaction between the ligand and protein residue was demonstrated by the Root Mean Square Fluctuation (RMSF).11 The graph of antioxidant proteins with CA bioactive compounds is represented in Fig. 9. The attachment stability of binding with the amino acids sequence over a given time period, such as the ligand, can be established using RMSF analysis. In comparison to other locations fluctuation were more frequent at the N- and C-terminal regions in all the complexes. The RMSF graph of GST showed minor deviations in amino acid residues at positions 136 to 144 during the simulation run. The RMSF of TRx complex with CA bioactive compounds shows fluctuation between 132 to 144 amino acid residues. The RMSF of SOD complexes initially fluctuated between 1 to 20 amino acid residues, but later less pronounced oscillations were seen throughout the complete run. The minor fluctuations of GPx complex with CA bioactive compounds was observed in between 69 to 76 amino acid residues. The compactness of CA bioactive compounds was analyzed by radius of gyration (Rg) plots and depicted in Fig. 10. The average radius of gyration of GPx with 3-O-methylquercetin was least (18.359) among all the bioactive compounds. Quinic acid complexes with GST and TRx had most stable complex structure with an average radius of gyration values of 21.080 Å and 19.085 Å respectively. Furthermore SOD complex with vanillin had the lowest Rg value of 26.95 Å.
4 Conclusion
In the present work, CA seed extract was prepared using a sustainable, efficient, and easily reproducible approach. The seed extract from CA significantly increased the levels of ROS, antioxidant proteins/enzymes, thus disrupting the redox balance of the filarial parasites. Overall, the CA treatment had a huge impact on the metabolism and survival of S. cervi filarial parasites, demonstrating excellent efficacy even at extremely low doses. Further the HRAMS proteomics results demonstrated that the parasites' exposures to CA extract led to the disruption of crucial signaling and metabolic pathways. The bioactive components in CA such as sterols, tannin, terpenes, fatty acids, lactones, phenolics, tetrahydroxyflavone, flavonoids, cyclic polyol, and benzoic acid are known to have several biological activities. The in silico studies proved that CA bioactive compounds like 3-O-methylquercetin, quinic acid, vanillin, and gentisic acid could stably interact with the parasites anti-oxidant proteins GPx, TRx, SOD, and GST. Hence, on the basis of biochemical, HRAMS, molecular docking, and in silico simulation studies, it seems imperative to integrate CA as a future treatment modality for Lymphatic Filariasis.Ethical statement
Indian water buffaloes (Bubalus bubalis) are a part of the non-vegetarian diet in India. S. cervi is a bovine filarial parasite that is found in the peritoneal folds of freshly slaughtered Indian water buffaloes. S. cervi worms are routinely discarded as waste in the slaughter houses. For this study S. cervi was collected and brought to the laboratory in KRB that was supplemented with streptomycin, penicillin, glutamine.Data availability
All data supporting the finding of this study are available within the paper and its ESI.†Author contributions
Sunil Kumar: conceptualization, methodology, data curation, formal analysis, investigation, writing – original draft, review & editing. Ayushi Mishra: methodology, formal analysis, investigation, review & editing. Surya Pratap Singh: review & editing. Anchal Singh: supervision, project administration, validation, writing – original draft, review & editing. All authors have read and agreed to the published version of the manuscript.Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
The author(s) reported there is no funding associated with the work featured in this article. Sunil Kumar is thankful to the Banaras Hindu University for UGC-NET Research Fellowship (BHU Res. Sch.) 2021-22/34927. Ayushi Mishra is grateful to the Council of Scientific and Industrial Research (CSIR), India (09/013(0832)/2018-EMR-I) for providing a Senior Research Fellowship (SRF). We acknowledge the center for Bioinformatics, School of Biotechnology for facilitating YASARA and DBT-BHU Interdisciplinary School of Life Sciences, Banaras Hindu University for providing laboratory space and Nanodrop and Gel documentation, Central Discovery Center BHU provided HRAMS facility. Prof. Shashi Pandey Dept. of Botany Inst. of Science BHU is acknowledged for providing botanical identification. The authors are also thankful to PARAM Shivay supercomputing facility, IIT, BHU, Varanasi.References
- D. J. Tisch, E. Michael and J. W. Kazura, Lancet Infect. Dis., 2005, 5, 514–523 CrossRef CAS PubMed.
- R. K. Prichard, M.-G. Basanez, B. A. Boatin, J. S. McCarthy, H. H. Garcia, G.-J. Yang, B. Sripa and S. Lustigman, PLoS Neglected Trop. Dis., 2012, 6, e1549 CrossRef PubMed.
- J. N. Sangshetti, D. B. Shinde, A. Kulkarni and R. Arote, RSC Adv., 2017, 7, 20628–20666 RSC.
- O. Ojurongbe, A. A. Akindele, M. A. Adeleke, M. O. Oyedeji, S. A. Adedokun, J. F. Ojo, C. A. Akinleye, O. S. Bolaji, O. A. Adefioye and O. A. Adeyeba, PLoS Neglected Trop. Dis., 2015, 9, e0003633 CrossRef PubMed.
- R. Prichard, Human and Animal Filariases: Landscape, Challenges, and Control, 2022, pp. 283–305 Search PubMed.
- A. Singh, A. Mishra, R. Chaudhary and V. Kumar, J. Sci. Res., 2020, 64, 50–58 Search PubMed.
- C. Y. Looi, B. Moharram, M. Paydar, Y. L. Wong, K. H. Leong, K. Mohamad, A. Arya, W. F. Wong and M. R. Mustafa, BMC Complementary Altern. Med., 2013, 13, 1–14 CrossRef PubMed.
- S. Sharma and B. Mehta, J. Hyg., Epidemiol., Microbiol., Immunol., 1991, 35, 157–161 CAS.
- A. Arya, C. Y. Looi, S. C. Cheah, M. R. Mustafa and M. A. Mohd, J. Ethnopharmacol., 2012, 144, 22–32 CrossRef PubMed.
- K. Singhal, S. Sharma and B. Mehta, Indian J. Exp. Biol., 1992, 30, 546–548 CAS.
- V. Kumar, A. Mishra and A. Singh, RSC Adv., 2022, 12, 22542–22554 RSC.
- A. Singh and S. Rathaur, Biochem. Biophys. Res. Commun., 2005, 331, 1069–1074 CrossRef CAS PubMed.
- S. Sharma, F. Ahmad, A. Singh and S. Rathaur, Vet. Parasitol., 2021, 290, 109357 CrossRef CAS.
- S. Yadav, S. Sharma, F. Ahmad and S. Rathaur, J. Drug Delivery Sci. Technol., 2020, 56, 101557 CrossRef CAS.
- T. Mosmann and T. Fong, J. Immunol. Methods, 1989, 116, 151–158 CrossRef CAS PubMed.
- H. Sim Choi, J. Woo Kim, Y. N. Cha and C. Kim, J. Immunoassay Immunochem., 2006, 27, 31–44 CrossRef PubMed.
- A. Nayak, P. Gayen, P. Saini, N. Mukherjee and S. P. Sinha Babu, Parasitol. Res., 2012, 111, 1173–1186 CrossRef.
- W. M. Nauseef and R. A. Clark, NADPH Oxidases: Methods and Protocols, 2019, pp. 3–16 Search PubMed.
- E. Shacter, J. A. Williams, M. Lim and R. L. Levine, Free Radicals Biol. Med., 1994, 17, 429–437 CrossRef CAS PubMed.
- B. Halliwell and S. Chirico, Am. J. Clin. Nutr., 1993, 57(5), 715S–724S CrossRef CAS PubMed.
- S. Rathaur, M. Yadav, N. Singh and A. Singh, J. Proteomics, 2011, 74, 1595–1606 CrossRef CAS PubMed.
- S. Tiwari, M. Wadhawan, N. Singh and S. Rathaur, J. Proteomics, 2015, 113, 435–446 CrossRef CAS.
- M. Wadhawan, F. Ahmad, S. Yadav and S. Rathaur, Protein J., 2022, 41, 613–624 CrossRef CAS PubMed.
- V. Kumar, A. Mishra, A. K. Yadav, S. Rathaur and A. Singh, Plos One, 2022, 17(7), e0270635 CrossRef CAS.
- A. Mishra, S. Kumar and A. Singh, RSC Adv., 2024, 14, 5893–5906 RSC.
- H. Birla, C. Keswani, S. S. Singh, W. Zahra, H. Dilnashin, A. S. Rathore, R. Singh, M. Rajput, P. Keshri and S. P. Singh, ACS Chem. Neurosci., 2021, 12, 4319–4335 CrossRef CAS.
- A. Mishra, V. Kumar and A. Singh, Pharm. Biol., 2022, 60, 2237–2252 CrossRef CAS PubMed.
- L. Willard, A. Ranjan, H. Zhang, H. Monzavi, R. F. Boyko, B. D. Sykes and D. S. Wishart, Nucleic Acids Res., 2003, 31, 3316–3319 CrossRef CAS.
- B. Huang, OMICS: J. Integr. Biol., 2009, 13, 325–330 CrossRef CAS.
- F. Amir and K. Y. Chin, Int. J. PharmTech Res., 2011, 3, 1772–1779 CAS.
- A. B. D. Nandiyanto, R. Oktiani and R. Ragadhita, Indones. J. Sci. Technol., 2019, 4, 97–118 CrossRef.
- Y. Sawada, R. Nakabayashi, Y. Yamada, M. Suzuki, M. Sato, A. Sakata, K. Akiyama, T. Sakurai, F. Matsuda and T. Aoki, Phytochemistry, 2012, 82, 38–45 CrossRef CAS PubMed.
- S. Kim, J. Chen, T. Cheng, A. Gindulyte, J. He, S. He, Q. Li, B. A. Shoemaker, P. A. Thiessen and B. Yu, Nucleic Acids Res., 2021, 49, D1388–D1395 CrossRef CAS.
- C. Lipinski and A. Hopkins, Nature, 2004, 432, 855–861 CrossRef CAS PubMed.
- F. Cheng, W. Li, Y. Zhou, J. Shen, Z. Wu, G. Liu, P. W. Lee and Y. Tang, J. Chem. Inf. Model., 2012, 52(11), 3099–3105 CrossRef CAS PubMed.
- D. Schneidman-Duhovny, Y. Inbar, R. Nussinov and H. J. Wolfson, Nucleic Acids Res., 2005, 33, W363–W367 CrossRef CAS.
- E. Krieger and G. Vriend, Bioinformatics, 2014, 30, 2981–2982 CrossRef CAS PubMed.
- C. Y. Looi, A. Arya, F. K. Cheah, B. Muharram, K. H. Leong, K. Mohamad, W. F. Wong, N. Rai and M. R. Mustafa, PLoS One, 2013, 8, e56643 CrossRef CAS PubMed.
- H. C. Santiago, S. Bennuru, A. Boyd, M. Eberhard and T. B. Nutman, J. Allergy Clin. Immunol., 2011, 127, 479–486 CrossRef CAS PubMed.
- J. Magalhães, J. Eira and M. A. Liz, Cell. Mol. Life Sci., 2021, 78, 6105–6117 CrossRef PubMed.
- L. Beld, H. Jung, C. A. Bulman, B. A. Rosa, P. U. Fischer, J. W. Janetka, S. Lustigman, J. A. Sakanari and M. Mitreva, Pathogens, 2022, 11, 707 CrossRef CAS PubMed.
- P. M. Cromm and C. M. Crews, ACS Cent. Sci., 2017, 3, 830–838 CrossRef CAS PubMed.
- B. Dahlmann, BMC Biochem., 2007, 8, 1–12 CrossRef PubMed.
- M. D. Bartolowits, J. M. Gast, A. J. Hasler, A. M. Cirrincione, R. J. O'Connor, A. H. Mahmoud, M. A. Lill and V. J. Davisson, ACS Omega, 2019, 4, 15181–15196 CrossRef CAS PubMed.
- S. O. Wendel, J. A. Snow, L. Gu, N. S. Banerjee, L. Malkas and N. A. Wallace, J. Med. Virol., 2023, 95, e29244 CrossRef CAS.
- C. B. M. Platania and C. Bucolo, Allostery: Methods and Protocols, 2021, pp. 245–254 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4ra03461a |
| This journal is © The Royal Society of Chemistry 2024 |