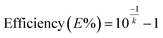Open Access Article
Open Access ArticleConvenient two-step method constructed silicon-based microfluidic chip for fast CYP2C19 SNPs detection†
Haobo Wang ,
Chi Yan,
Hua Tong,
Xiaojun Ye,
Xiao Yuan,
Cui Liu* and
Hongbo Li*
,
Chi Yan,
Hua Tong,
Xiaojun Ye,
Xiao Yuan,
Cui Liu* and
Hongbo Li*
School of Materials Science and Engineering, East China University of Science and Technology, Shanghai 200237, China. E-mail: liucui@ecust.edu.cn; lihongbo@ecust.edu.cn
First published on 19th August 2024
Abstract
The rapid detection of single nucleotide polymorphisms (SNPs) in the CYP2C19 gene is crucial for precise clopidogrel usage. Quantitative real-time polymerase chain reaction (qPCR), as a powerful amplification tool, has been widely employed for CYP2C19 SNPs detection. However, traditional qPCR suffers from long amplification times and high reagent consumption. To address these challenges, this work presents a microfluidic SNPs detection device based on on-chip qPCR. The device includes a rapid thermal cycling system, an optical detection system, a control system, and a complementary silicon-glass chip for CYP2C19 SNPs detection. Compared to commercial qPCR instruments that take 1 hour for testing, this device completes the test in just 15 minutes (40 PCR cycles). The resulting linearity is similar to that found using commercial qPCR instruments but with higher amplification efficiency. Additionally, compared with other silicon-based qPCR chips, this chip is constructed by using a convenient two-step method and offers low manufacturing costs, which potentially reduces single-test costs to an acceptable level. This makes our chip promising for point-of-care testing (POCT).
1 Introduction
The CYP2C19 gene plays a crucial role in the metabolism of several medications, including clopidogrel, S-mephenytoin, omeprazole, fluconazole, diazepam, and lorazepam. CYP2C19 genetic variation can lead to individual differences in enzyme activity, resulting in the emergence of ultrarapid metabolizers (UMs), extensive metabolizers (EMs), intermediate metabolizers (IMs), and poor metabolizers (PMs).1–3 The CYP2C19*2 (c.681G>A rs4244285), the CYP2C19*3 (c.636G>A rs4986893) and the CYP2C19*17 (c.-806C>A rs12248560) alleles are the most common and investigated polymorphism.4 Specifically, CYP2C19*2 results in a splicing defect, while CYP2C19*3 involves a premature stop codon mutation and CYP2C19*17 is a promoter variant.5 Notably, approximately 75–85% of PMs in East Asian populations are attributed to CYP2C19*2, while about 20–25% are due to CYP2C19*3. Furthermore, the CYP2C19*17 genotype is associated with UMs and may elevate the risk of bleeding in clopidogrel users.6 The efficacy and side effects of drugs metabolized by CYP2C19 exhibit variability based on patient genotype. Consequently, considering CYP2C19 genetic polymorphisms (SNPs) in drug selection holds critical clinical implications.Lab-on-a-chip (LoC) technology has emerged as a powerful platform for performing complex biological and chemical analyses.7–13 By integrating various functional units onto miniature material substrates, microfluidic devices offer several advantages, including rapid genetic testing, high throughput, low reagent consumption, and portability.14–16 These features make them particularly attractive for applications such as fast detection and POCT.17 When combined with qPCR technology, microfluidics provides a viable strategy to address challenges related to flexible small-scale analysis and rapid testing. Majeed et al.18 designed and manufactured a silicon-based microfluidic LoC system with rapid sensitivity and high specificity for SNP detection. This system utilizes silicon substrates and incorporates various fabrication processes, including mask deposition, photolithography, and dry etching. Integration with external pumps, heaters, and temperature control devices allows seamless single-sample purification, extraction, amplification, and detection. Zhang et al.19 developed a silicon-based integrated microsystem for detecting multiple SNPs directly from whole blood. This microsystem includes a blood lysis chamber, a cross-flow filter, a T-shaped mixer, and a microreactor for qPCR. Using this device, the successful detection of relevant SNPs in the human CYP2C9 gene was achieved on the chip. Despite these advancements, relevant microfluidic chips face certain limitations. The intricate design structures and high manufacturing costs hinder their widespread adoption for disposable use.20,21 Further research and engineering efforts are necessary to optimize these systems for practical clinical applications.
In this study, we demonstrate a rapid and low-cost qPCR detection device for SNP analysis of the CYP2C19 gene. Utilizing a convenient two-step process reported previously, we fabricated a microfluidic chip on a silicon substrate. By circumventing the complexities associated with standard photolithography techniques, this fabrication approach reduces production costs and enhances suitability for disposable use. A custom-built thermal cycling module ensures precise and rapid temperature cycling, while an integrated optical detection module captures fluorescence images after each qPCR cycle. Software analysis extracts fluorescence intensity curves, providing real-time data. Utilizing TaqMan probe-based assays, we successfully performed CYP2C19 SNPs detection directly within a short timeframe of 15 minutes (40 PCR cycles). The results obtained using our microfluidic device were consistent with those obtained using commercial qPCR instruments. Overall, this innovative system offers substantial advantages in terms of reduced testing time, cost of consumables and maintained accuracy, making it promising for POCT applications.
2 Materials and methods
2.1 Chip design and fabrication
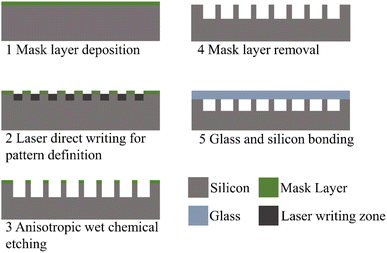
The substrate of the chip is standard polished N-type (100) silicon wafers (Rice-semi Co. Ltd, Suzhou, China), which have a diameter of 4 inches and a thickness of 500 ± 10 μm. On the wafer surface, S-shaped microchannels are fabricated to function as the reaction channels. The microchannel fabrication process is depicted in Fig. 1. Initially, a protective layer is deposited on the wafer surface. To define the microchannel pattern, laser direct writing is employed to create etching windows. Subsequently, anisotropic wet etching is employed to form the microchannels. The detailed fabrication process has been documented in prior reports.22 The KOH solution is used for wet etching, and the HPM solution is used for microchannel cleaning before and after etching. Then, a volume fraction of 10% of the HF solution is used for mask layer removal. After another HPM cleaning step, glass (Schott BF33) is anodically bonded to the wafer, serving as a cover plate to hermetically seal the microchannels. Following this, the bonded wafer is precisely cut into 10 × 10 mm2 chips. After fabrication, the channel morphology and dimensions are characterized using a 3D surface profiler (Keyence, Japan).2.2 Optical detection module
The optical detection module includes an LED light source, filters, and a high-resolution camera for signal collection. Filters of different light wavelengths are installed behind the light source and in front of the camera lens. The structure diagram is shown in Fig. 2(a). The entire system is placed in a small darkroom. During operation, the light emitted by the LED light source passes through the filters, transforming into excitation light corresponding to the excitation wavelength of fluorescent molecules. This excitation light reaches the interior of the chip channel, exciting FAM, ROX, or VIC molecules to produce fluorescence. The fluorescence then passes through the filters and reaches the camera and the fluorescence intensity is read by software.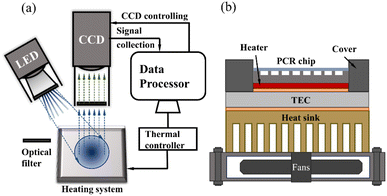 | ||
| Fig. 2 (a) Diagram of the optical detection system and control system. (b) Diagram of thermocycling system structure. | ||
2.3 Thermocycling module
We independently assemble a rapid thermal cycling system, which includes two thermocouples, a heating pad, two phase-change materials (PCM, Honeywell, America), a thermal electronic cooler (TEC), a fan, and a custom-made copper heat sink, as shown in Fig. 2(b). The heating pad is positioned beneath the silicon-based microfluidic chip to achieve rapid chip heating. The TEC maintains a constant temperature, ensuring consistent environmental conditions for each experiment. Two thermocouples are affixed near the heating pad and the TEC to provide real-time temperature measurements of the chip. The TEC interfaces with the heat sink and the heating pad via PCM, ensuring rapid heat transfer.2.4 Control system
The whole system is under computer control. The computer controls the activation of the LED light source filter position and adjusts both the intensity and wavelength of the excitation light. At the end of each cycle, the camera captures images of fluorescence in the channel by configuring the exposure time.2.5 PCR kits and thermal cycling protocol
Primers and probes for three polymorphisms of human CYP2C19 gene: CYP2C19*17, CYP2C19*3, CYP2C19*2, and internal reference primer RnaseP were provided by General Biol (Anhui, China) Co., LTD. The sequence information of primers is shown in Table 1.| Allele | Primer | Sequence |
|---|---|---|
| CYP2C19*17 rs12248560 (c.-806C>A) | rs12248560-zyFAM | FAM-5′-ATCAGAGATACTTTGAGAAC-3′-MGB |
| rs12248560-zyVIC | VIC-5′-ATCAGAGATGCTTTGAGAA-3′-MGB | |
| rs12248560-zyF | 5′-CTGATACCCATCGTGGCG-3′ | |
| rs12248560-zyR | 5′-ACTAAGGTTTGGAAGTTGTTTTGT-3′ | |
| CYP2C19*3 rs4986893 (c.636G>A) | rs4986893-zyFAM | FAM-5′-CACCCCCTGAATC-3′-MGB |
| rs4986893-zyVIC | VIC-5′-CACCCCCTGGATC-3′-MGB | |
| rs4986893-zyF | 5′-ATTGAATGAAAACATCAGGATTGTA-3′ | |
| rs4986893-zyR | 5′-ATACAGAATTTTGGATTTCCCAG-3′ | |
| CYP2C19*2 rs4244285 (c.681G>A) | rs4244285-FAMTaq | FAM-5′-CCCGGGAACCC-3′-MGB |
| rs4244285-VICTaq | VIC-5′-TCCCAGGAACCC-3′-MGB | |
| rs4244285taq-F | 5′-ATGCAATAATTTTCCCACTATCAT-3′ | |
| rs4244285taq-R | 5′-TATCACTTTCCATAAAAGCAAGGT-3′ | |
| RNaseP | RNAseP-ytaq | ROX-5′-TTCTGACCTGAAGGCTCTGCGCG-3-MGB |
| RNAseP-yF | 5′-AGATTTGGACCTGCGAGCG-3′ | |
| RNAseP-yR2 | 5′-GGCAACTCACGGTGAGCG-3′ |
The qPCR amplification diagram is shown in Fig. 3.
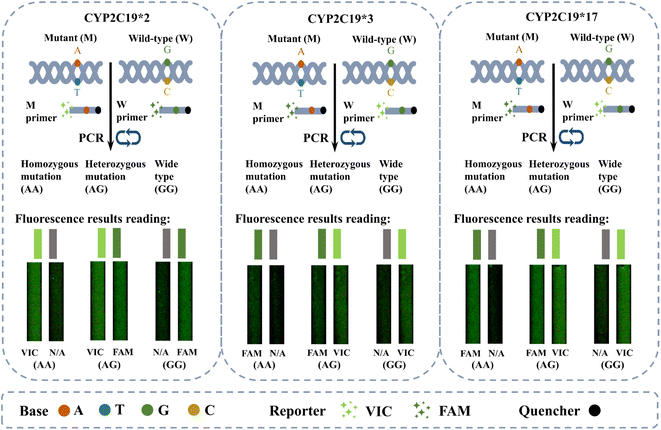 | ||
| Fig. 3 Schematic diagram of the on-chip qPCR for CYP2C19 SNPs detection. The figure shows the genotypes corresponding to the different fluorescence results after amplification. | ||
For on-chip qPCR, the total volume of the reaction solution is 5 μL, including 5× GUeasy Taq DNA polymerase buffer (General Biol Co. LTD Anhui, China), 10 mM dNTP mixture (Vazyme International LLC. Nanjing, China), 25 mM MgCl2 (Takara Bio Inc. Dalian, China), 10 μg μL−1 BSA (Perlong Inc. Beijing, China), 5 U μL−1 of Taq DNA polymerase (General Biol Co. LTD Anhui, China), 10 μM primers and probes of CYP2C19*2, *3 and *17 and 10 μM RnaseP primers and probe and ddH2O. The reaction mix was injected respectively into different channels in the chips for detection.
The on-chip qPCR thermal cycling scheme is shown in Table 2.
| Stage | Temperature (°C) | Time (s) | Fluorescence signal collection |
|---|---|---|---|
| Pre-denaturation | 95 | 30 | — |
| Denaturation | 95 | 5 | — |
| Annealing/extension | 60 | 10 | Collecting signal at 9 s |
| Cycles | 40 |
In addition, as a comparison and verification of genotype results, the amplification system with the same formula was amplificated in a commercial qPCR amplification instrument (Lepgen-96, Beijing, China), the reaction liquid volume was 20 μl, and the thermal cycle scheme was 95 °C 5 min, 95 °C 10 s, and 60 °C 30 s, 45 cycles. As a matter of fact, 40 cycles are enough for CYP2C19 gene SNPs detection.
For comparing the system's linearity of on-chip qPCR and commercial qPCR instrument, the PTGS1 gene was amplificated on a qPCR chip and commercial instrument. The sequence information of primers and probe are shown in Table 3.
| Allele | Primer | Sequence |
|---|---|---|
| PTGS1 | PTGS1-zyF2 | 5′-GCCTTCCGATAACTGAGCAC-3′ |
| PTGS1-zyR2 | 5′-CATCCCAGAGGGTGGTTGTA-3′ | |
| PTGS1-zyFAMA | FAM-5′-ATGCTGGACACTGC-3′-MGB |
The total volume of the reaction solution is 5 μL on qPCR chips and 20 μL on commercial qPCR instrument (Quantstudio, Thermo Fisher Scientific Inc., America), respectively. The reaction reagents include 5×GUeasy Taq DNA polymerase buffer, 10 mM dNTP, 25 mM MgCl2 10 μM PTGS1 primer probe, 10 mM BSA, gradient diluted DNA template, and ddH2O. The on-chip qPCR and commercial qPCR instrument thermal cycling scheme are shown in Table 4.
| On-chip qPCR | Commercial qPCRinstrument | ||||
|---|---|---|---|---|---|
| Stage | Temperature (°C) | Time (s) | Stage | Temperature (°C) | Time (s) |
| Pre-denaturation | 95 | 30 | Pre-denaturation | 95 | 300 |
| Denaturation | 95 | 6 | Denaturation | 95 | 10 |
| Annealing/extension | 60 | 12 | Annealing/extension | 60 | 30 |
| Cycles | 45 | Cycles | 45 | ||
3 Results and discussion
3.1 Fabrication results of qPCR chips
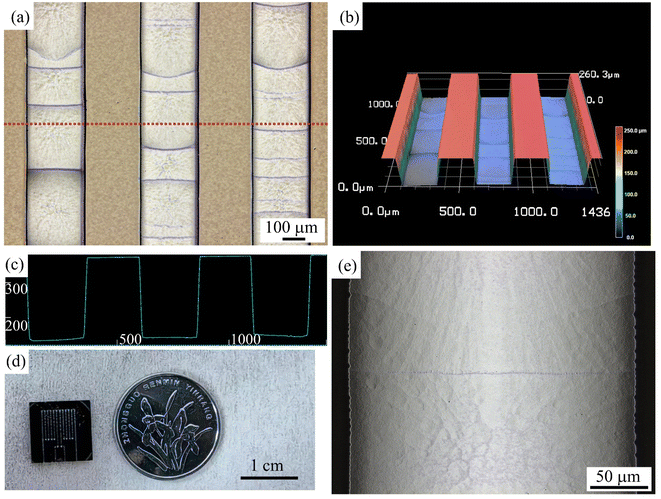
The fabricated microfluidic chip consists of two S-shaped reaction channels. The micrographs of microchannels are depicted in Fig. 4(a), with smooth channel edges. Within the microchannel, black horizontal lines are observed resulting from the etching processes. The 3D display in Fig. 4(b) reveals a rectangular cross-section of the microchannel. The cross-sectional profile of the microchannel, as depicted in Fig. 4(c), exhibits an aspect ratio of approximately 0.87 (see in ESI†). The vertical sidewalls effectively utilize the limited chip area and avoid reflection interference.23 It is worth noting that the aspect ratio of the microchannels can be freely and flexibly adjusted according to laser and etching conditions to meet different liquid handling needs. Fig. 4(d) shows a size comparison between the chip and an ordinary coin. Fig. 4(e) presents a high-magnification micrograph of the interior of the microchannel, revealing a relatively smooth channel inside the surface with a measured roughness (Ra) of 2.3 μm. The interior of the channel exhibits hydrophilicity (the water contact angle data is shown in ESI†), minimizing the occurrence of bubbles during sample injection. Bubbles within the microchannel are detrimental to maintaining temperature stability during qPCR processes and may lead to sample evaporation, resulting in failed amplification.23–25 During detection, capillaries are connected to the inlet and outlet of the microchannel and thermally sealed. Samples are injected into the chip through the capillary and then thermally sealed.30 chips can be rapidly produced on standard 4-inch wafers using the convenient two-step method to meet the needs of injection and qPCR reactions, without standard lithography processes. It makes the material cost of a single chip only $2, which is expected to meet the cost of a single test.
The optimization of silicon chips can still be further enhanced by considering the impact of minuscule bubbles in the reaction. The combination of various optimization conditions is still in the process of exploration and whether the coating is needed in the channel and the specific influence of the surface structure of the channel on the qPCR reaction need to be further explored.
3.2 Thermal cycling performance
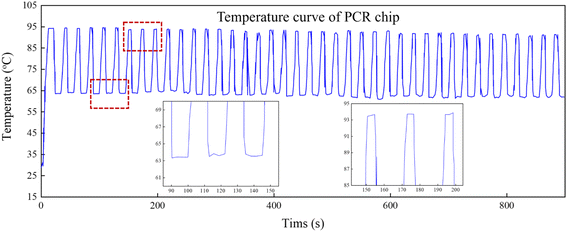
The equipment described can rapidly complete temperature cycling with precise temperature control. The temperature of the qPCR chip can transition from 60 °C to 90 °C within 3 s. The average heating rate from 60 °C to 90 °C is 10 °C s−1, while the average cooling rate from 90 °C to 60 °C is 8 °C s−1. The temperature profile during the qPCR process is depicted in Fig. 5. The qPCR process comprises an initial pre-denaturation at 95 °C for 30 s, followed by denaturation at 95 °C for 5 s and extension at 60 °C for 10 s. Throughout the entire qPCR process, temperature overshoot is minimal due to the well-defined temperature points.The result of rapid temperature cycling is closely related to the structure of the thermal cycling system because the heating pad has a small heat capacity, it can carry the silicon chip to quickly achieve heating and cooling. At the same time, TEC keeps the whole system at a constant temperature so that temperature overshoot rarely occurs during the thermal cycle.
3.3 Fast CYP2C19 gene SNPs detection
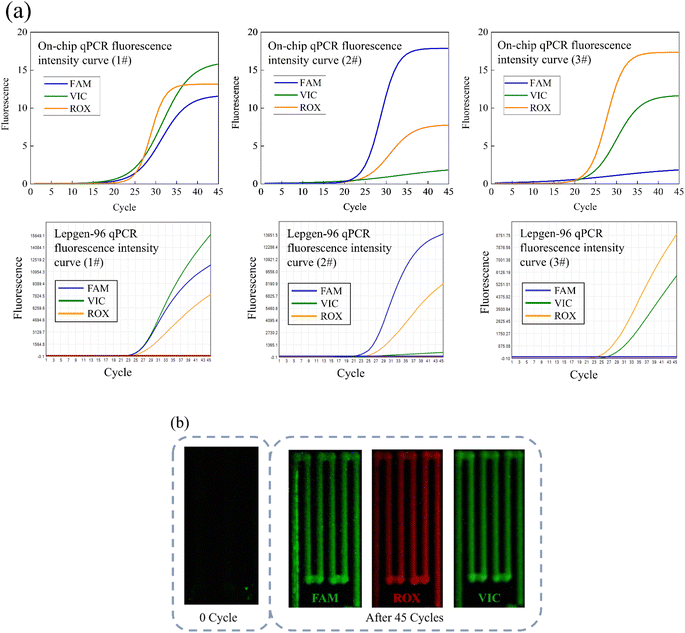
Three CYP2C19 gene templates of unknown genotypes, as samples 1#, 2#, and 3# respectively, were detected on the chip, in order to assess the accuracy of on-chip qPCR detection. The thermal cycling conditions for on-chip amplification are described above. Additionally, the same sample was amplified using a commercial qPCR instrument, and the results of the amplification serve as the standard for genotype interpretation. The thermal cycling process in the commercial machine is set according to kit requirements, taking approximately 80 minutes in total. Fig. 6 displays the results of the amplification curves. The top half of picture 6(a) represents the fitted on-chip qPCR fluorescence curve, while the bottom half of picture 6(a) corresponds to the fitted fluorescence curve of a commercial qPCR instrument. It is worth noting that it has significant difference in the fluorescence intensity obtained from on-chip and commercial instruments, which makes it difficult to compare the fluorescence intensity between the two devices. The main reason for this difference is that on-chip qPCR instruments and commercial instruments have different fluorescence collection methods.The ROX fluorescence curve demonstrates successful amplification of the internal control gene RnaseP both on-chip and in commercial instruments. This result shows the capability of our chip and reagent for on-chip qPCR with small reaction volume and rapid temperature cycling conditions. Furthermore, the FAM and VIC fluorescence curves reveal the expected resolution capabilities of this device. The 1# sample labeled CYP2C19*2 probes, exhibiting FAM and VIC fluorescence, indicating the presence of the CYP2C19*2 AG genotype according to the results reading instructions presented above. At the same time, the 2# labeled CYP2C19*3 probes, only showed strong FAM fluorescence and weak VIC fluorescence, indicating the reading result was CYP2C19*3 AA genotype. Lastly, the 3# of the sample labeled CYP2C19*17 probe, which showed VIC fluorescence and weak FAM fluorescence, indicating the interpretation result was CYP2C19*17 GG genotype.
The fluorescence curves obtained from the same sample after amplification using the commercial qPCR instrument exhibit consistency with those generated by on-chip qPCR, thereby enabling us to obtain identical reading results. Consequently, our device can accurately detect the CYP2C19 genotype based on qPCR fluorescence. Moreover, owing to the utilization of fewer reagents and composite silicon chips, our on-chip SNP detection time is reduced to 15 min (40 PCR cycles), compared to the 80 min required by commercial instruments, providing a significant time advantage. Fig. 6(b) provides the fluorescence photographs of 1 and 45 cycles.
3.4 Comparison of the system's linearity
In order to assess the linearity of samples, on-chip qPCR was performed using a positive DNA template with a copy number concentration ranging from 2.128 to 2.128 × 103 copies per μL. Following amplification, Ct values were obtained for each sample and a standard curve was plotted. The linear correlation coefficient R2 and amplification efficiency were calculated. The calculation of R2 involves the ratio of the explained sum of squares (ESS) to the total sum of squares (TSS). Additionally, the amplification efficiency is related to the Ct value standard curve, and the equation for calculating efficiency is shown in the following equation.where k is the slope of the standard curve of Ct value. For comparison, the same DNA template was amplified using a commercial qPCR instrument. Both on-chip qPCR and commercial instrument qPCR thermal cycling parameters were set according to the conditions described above. Notably, the faster temperature ramp rates of on-chip qPCR devices allowed for faster completion of the entire process compared to commercial qPCR instruments.
The system linearity results for both on-chip qPCR instrument and commercial qPCR instrument are depicted in Fig. 7. Ct values were calculated and summarized in the Table 5 of each fluorescence curve. By constructing the Ct value standard curve, an R2 value of 0.9463 was obtained for the on-chip qPCR device, indicating good linear correlation in the detection results across various concentrations of samples. In contrast, the commercial qPCR equipment achieved a higher R2 value of 0.9962.
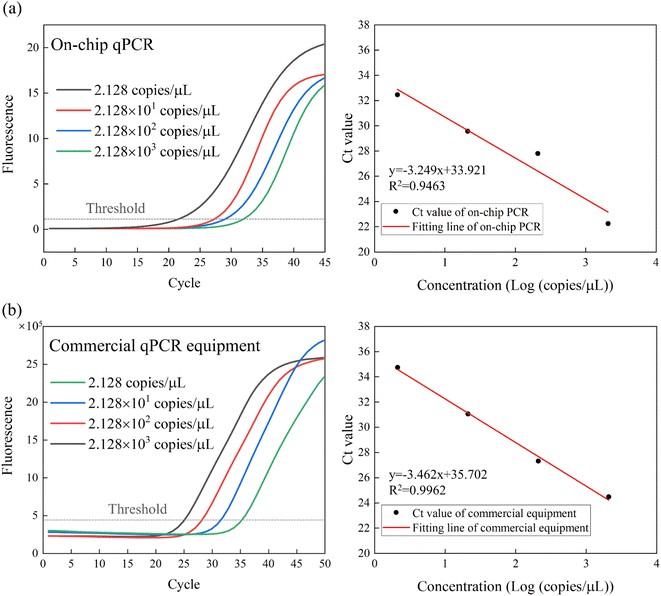 | ||
| Fig. 7 The system's linearity test and fitting curve of (a) on-chip qPCR instrument; (b) commercial qPCR instrument. | ||
| On-chip qPCR instrument | Commercial qPCR instrument | |||||||
|---|---|---|---|---|---|---|---|---|
| Copy numbers (copies per μL) | 2.128 | 2.128 × 101 | 2.128 × 102 | 2.128 × 103 | 2.128 | 2.128 × 101 | 2.128 × 102 | 2.128 × 103 |
| Ct value | 32.436 | 29.531 | 27.765 | 22.195 | 34.732 | 31.033 | 27.283 | 24.442 |
| k | −3.249 | −3.462 | ||||||
| E% | 103.14 | 94.46 | ||||||
The potential reason for the lower R2 value of on-chip qPCR results than commercial qPCR instruments may be related to the tiny bubbles inside the channel. The on-chip qPCR requires less sample volume, and the unignored bubbles generated by the injection or the sample itself will affect the thermal uniformity of the chip and the dynamics of the reaction liquid system. Although we have carried out hydrophilic treatment on the inside of the channel to reduce the generation of bubbles, it still needs more efforts to completely avoid the phenomenon by improving the manufacturing technique. In addition, due to the high surface-to-volume (SVR) of on-chip qPCR, the potential inhibition of qPCR may also affect R2.26–28
Additionally, the efficiencies of both on-chip qPCR and commercial qPCR devices were summarized in the Table 5. The amplification efficiencies of both on-chip PCR instrument and commercial PCR instrument are all within the reasonable range of 90–110%. It is worth noting that the amplification efficiency of on-chip PCR instrument is different from that of commercial PCR instrument. This could be affected by factors such as inhibitors in samples, primers adsorption during amplification, or temperature accuracy of the device.29 The difference in amplification efficiency can be minimized in subsequent studies by optimizing the amplification solution of on-chip qPCR.
4 Conclusions
In this work, we present a rapid microfluidic SNPs detection device based on on-chip qPCR. Compared to conventional devices, our approach enables expedited qPCR amplification, facilitating efficient CYP2C19 gene typing. Utilizing the same probe-based assay kit, our method exhibits heightened specificity, significantly reducing the detection time from 80 minutes to a mere 15 minutes (40 PCR cycles). Compared with commercial instruments, on-chip qPCR has the same judgment results, similar system linearity, and higher amplification efficiency. Additionally, compared with other silicon-based qPCR chips, this chip is constructed by using a convenient two-step method and offers low manufacturing costs, which potentially reduces single-test costs to an acceptable level. This makes our chip promising for POCT. In future investigations, we will explore additional applications of qPCR methods, and enhance the overall versatility of our device.Data availability
Data are available upon request from the authors.Author contributions
Haobo Wang (first author): conceptualization, methodology, investigation, analysis, writing – original draft; Chi Yan: visualization, resources; validation, writing – review & editing; Hua Tong: methodology, data curation; Xiaojun Ye: software, visualization; Xiao Yuan: project administration, conceptualization, funding acquisition, resources; Cui Liu (corresponding author): supervision, project administration, resources; Hongbo Li (corresponding author): supervision.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors extend their gratitude to Shiyanjia Lab (http://www.shiyanjia.com) for providing invaluable assistance with the morphology and XPS analysis. The authors extend their gratitude to Mr Jianwei Zhu from the United Gene Group for providing invaluable assistance with the SNPs analysis.References
- X. M. Hou, W. Z. Han, Q. Gan, Y. Liu and W. Y. Fang, J. Clin. Lab. Anal., 2018, 32, e22369 CrossRef
.
- S. Matetzky, B. Shenkman, V. Guetta, M. Schechter, R. Bienart, I. Goldenberg, I. Novikov, H. Pres, N. Savion, D. Varon and H. Hod, Circulation, 2004, 109, 3171–3175 CrossRef CAS PubMed
.
- C. R. Lee, J. A. Luzum, K. Sangkuhl, R. S. Gammal, M. S. Sabatine, C. M. Stein, D. F. Kisor, N. A. Limdi, Y. M. Lee, S. A. Scott, J. S. Hulot, D. M. Roden, A. Gaedigk, K. E. Caudle, T. E. Klein, J. A. Johnson and A. R. Shuldiner, Clin. Pharmacol. Ther., 2022, 112, 959–967 CrossRef CAS
.
- X. H. Yan, S. N. Zhang, J. P. Liang, Y. Cai, J. L. Zhu, C. Zhang, D. S. Li, K. Hua, Y. L. Cui and W. L. Hui, Artif. Cells, Nanomed., Biotechnol., 2019, 47, 636–643 CrossRef CAS
.
- S. M. Naushad, K. K. Vattam, Y. K. D. Devi, T. Hussain, S. Alrokayan and V. K. Kutala, Gene, 2021, 784, 145592 CrossRef CAS
.
- H. H. Zhou, Int. Congr. Ser., 2002, 1244, 51–61 CrossRef CAS
.
- Y. Choi, Y. Song, Y. T. Kim, H. Kim, Y. M. Park, S. J. Lee, S. G. Im and K. G. Lee, Sens. Actuators, B, 2021, 344, 130307 CrossRef CAS
.
- X. Zhao, X. Li, W. Yang, J. Peng, J. Huang and S. Mi, Analyst, 2021, 146, 5102–5114 RSC
.
- Y. Shang, J. Sun, Y. Ye, J. Zhang, Y. Zhang and X. Sun, Crit. Rev. Food Sci. Nutr., 2020, 60, 201–224 CrossRef CAS PubMed
.
- J. Guo, S. Chen, J. Guo and X. Ma, J. Mater. Sci. Technol., 2021, 60, 90–104 CrossRef CAS
.
- B. Ma, J. Li, K. Chen, X. Yu, C. Sun and M. Zhang, Foods, 2020, 9, 278 CrossRef CAS PubMed
.
- J. Guo, S. Chen, S. Tian, K. Liu, J. Ni, M. Zhao, Y. Kang, X. Ma and J. Guo, Biosens. Bioelectron., 2021, 181, 113160 CrossRef CAS PubMed
.
- J. Wang, H. Jiang, L. Pan, X. Gu, C. Xiao, P. Liu, Y. Tang, J. Fang, X. Li and C. Lu, Front. bioeng. biotechnol., 2023, 11, 1020430 CrossRef
.
- S. S. Abdullaev, R. H. Althomali, A. R. Khan, H. S. Jabbar, M. Abosoda, A. Ihsan, S. Aggarwal, Y. F. Mustafa, I. H. Khlewee and A. m. Jabbar, Talanta, 2024, 273, 125896 CrossRef PubMed
.
- X. Wu, Y. Zhao, C. Guo, C. Liu, Q. Zhang, Y. Chen, Y. Liu and X. Zhang, Biosens. Bioelectron., 2023, 241, 115676 CrossRef CAS PubMed
.
- Y. K. Jung, J. Kim and R. A. Mathies, Biosens. Bioelectron., 2016, 79, 371–378 CrossRef CAS PubMed
.
- C. Wang, M. Liu, Z. Wang, S. Li, Y. Deng and N. He, Nano Today, 2021, 37, 101092 CrossRef CAS
.
- B. Majeed, B. Jones, D. S. Tezcan, N. Tutunjyan, L. Haspeslagh, S. Peeters, P. Fiorini, M. Op de Beeck, C. Van Hoof, M. Hiraoka, H. Tanaka and I. Yamashita, Jpn. J. Appl. Phys., 2012, 51, 04DL01 CrossRef
.
- L. Zhang, Q. Cai, R. S. Wiederkehr, M. Fauvart, P. Fiorini, B. Majeed, M. Tsukuda, T. Matsuno and T. Stakenborg, Lab Chip, 2016, 16, 4012–4019 RSC
.
- N. Aggarwal, Y. Liang, J. L. Foo, H. Ling, I. Y. Hwang and M. W. Chang, Biosens. Bioelectron., 2023, 222, 115002 CrossRef CAS
.
- S. J. Mun, W. Jang, H.-S. Park, Y. J. Lim, T.-J. Yang and K. W. Bong, Biosens. Bioelectron., 2023, 241, 115670 CrossRef CAS PubMed
.
- H. Wang, H. Tong, C. Liu, X. Yuan, X. Ye, S. Xiong, J. Xu and H. Li, Mater. Sci. Semicond. Process., 2024, 174, 108208 CrossRef CAS
.
- D. Podbiel, R. Zengerle and J. Hoffmann, Microfluid. Nanofluid., 2020, 24, 16 CrossRef CAS
.
- Z. Cao, Y. Ye, G. Li, R. Zhang, S. Dong and Y. Liu, Biosens. Bioelectron., 2022, 213, 114499 CrossRef CAS
.
- U. Perwez, I. Aziz, F. Ahmed and M. R. Khan, Processes, 2019, 7, 919 CrossRef CAS
.
- N. Ramalingam, M. E. Warkiani and T. H.-Q. Gong, Biomicrofluidics, 2017, 11, 034110 CrossRef PubMed
.
- J. J. Chen and X. C. Qiu, Microsyst. Technol., 2024 DOI:10.1007/s00542-024-05675-2
.
- M. A. Shoffner, J. Cheng, G. E. Hvichia, L. J. Kricka and P. Wilding, Nucleic Acids Res., 1996, 24, 375–379 CrossRef CAS PubMed
.
- X. Zhu, J. Zhao, A. Hu, J. Pan, G. Deng, C. Hua, C. Zhu, Y. Liu, K. Yang and L. Zhu, Micromachines, 2020, 11, 186 CrossRef PubMed
.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4ra03913k |
| This journal is © The Royal Society of Chemistry 2024 |