Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Quality-by-design-engineered mitochondrial targeted nanoparticles for glioblastoma therapy†
Akanksha Dahifale‡
a,
Tejas Girish Agnihotri‡a,
Ankit Jain *b and
Aakanchha Jain*a
*b and
Aakanchha Jain*a
aDepartment of Pharmaceutics, National Institute of Pharmaceutical Education and Research (NIPER)-Ahmedabad, Palaj, Gandhinagar-382355, Gujarat, India. E-mail: jainaakanchha83@gmail.com; aakanchha.jain@niperahm.res.in
bDepartment of Pharmacy, Birla Institute of Technology & Science, Pilani-333031, Rajasthan, India. E-mail: ankit.j@pilani.bits-pilani.ac.in
First published on 28th October 2024
Abstract
Glioblastoma (GB, IDH-wildtype) constitutes the most aggressive primary malignant neoplasm with limited treatment modalities due to the blood–brain barrier (BBB) often restricting drug delivery. It also has an overall low survival rate with no curative solution, reinforcing the need for innovative formulation development for effective management of GB. This study explores a novel approach using triphenylphosphonium (TPP+)-conjugated chitosan nanoparticles for targeted mitochondrial delivery of temozolomide (TMZ) to GB cells. The conjugated nanoparticles were designed to leverage chitosan's biocompatibility and TPP's mitochondrial targeting ability. TMZ-loaded chitosan nanoparticles were systematically developed and optimized employing a Quality-by-Design (QbD) approach with a screening of factors (Taguchi design) followed by optimization (Box–Behnken design). The optimized nanoparticles had an average particle size of 138.1 ± 5 nm, PDI of 0.242 ± 0.04, and entrapment efficiency of 93.59 ± 3%. Further, a conjugate chitosan-TPP+ (CS-TPP+) was synthesized and validated, employing varied techniques such as NMR, FTIR, HPLC, zeta potential, and EDAX analysis. In vitro drug release in pH 5 phosphate buffer showed a sustained release for nanoparticulate formulations compared to the free drug solution further indicating that conjugation did not alter the release pattern of nanoparticles. With regards to intranasal delivery of the formulation, an ex vivo study carried out on goat nasal mucosa demonstrated greater retention of conjugated chitosan nanoparticles on nasal mucosa than free drug solution, and a mucin interaction study also corroborated this finding. In vitro cell line studies indicated nanoparticles' cytotoxic potential compared to TMZ solution. Overall, this study highlights the potential of TPP+-conjugated chitosan nanoparticles developed strategically for the targeted delivery of TMZ to mitochondria.
1. Introduction
According to WHO classification, 2021, glioblastoma, isocitrate dehydrogenase (IDH)-wildtype (GB), a type 4 astrocytic tumor, affects more than 60% of adults, primarily those in the 55–60 age range.1–3 Although it is categorized as a rare tumor, the condition is life-threatening and has a poor diagnosis, with a 14–15 months survival rate after a patient is diagnosed.4,5 GB, IDH-wildtype, is considered to be the most common primary malignant neoplasm of the brain, accounting for 54% of all types of gliomas. It is mostly located in the supratentorial region, i.e., frontal, temporal, parietal, and occipital lobes, with higher incidences in the frontal lobe followed by tumors that overlap multiple lobes and then temporal and parietal lobes.6 GB, IDH-wildtype is an aggressive and invasive disease; hence, to maximize patient outcomes from treatment, a multi-target approach must be used. Resection of the tumor, radiation therapy, and chemotherapy are now the main treatments for GB, IDH-wildtype.7 The standard chemotherapeutic drug authorized for the treatment of GB, IDH-wildtype is temozolomide (TMZ). However, the actual utilization of this compound is limited due to its comparatively short duration in the bloodstream, possible adverse effects on the whole body, and the invasive nature of the related treatment methods.8The presence of the blood–brain barrier (BBB) has been a formidable challenge for neurotherapeutic drug delivery, which makes it difficult for drugs to reach the brain effectively, underpinning the need for innovative ways to get around it and enhance drug delivery. The properties of colloidal-based particle systems referred to as nanocarriers and nanotechnology-based drug delivery systems, such as self-assembly, greater dissolution, delayed drug release, improved drug biological activity, and BBB penetrability, allow them to traverse the BBB.9,10 Numerous benefits, including better drug targeting and tumor cell penetrability, which boost therapeutic efficiency, are provided by nanoparticle-based drug delivery systems in the treatment of cancer.11,12 One approach involves the surface engineering of nanoparticles through biocompatible coatings. This strategy emphasizes on the optimization of surface ligands and development of stimuli-responsive delivery systems for targeted drug release within the tumor microenvironment.13,14 In this regard, targeted drug delivery to mitochondria holds great potential in the clinical intervention of cancer, primarily owing to the crucial involvement of mitochondria in cellular metabolic processes, energy generation, and programmed cell death. The selective delivery of medication to mitochondria can enhance the effectiveness of cancer therapy while minimizing negative effects on non-cancerous cells.15 The concept of “mitochondriotropics” often denotes small molecules with a strong innate affinity for mitochondria, as determined by their physicochemical features and reactivity. The most commonly used mitochondrial targeting agents are triphenylphosphonium cation (TPP+), dequalinium chloride, peptides, etc. One widely employed method for transporting these low-molecular-weight chemicals to mitochondria involves their attachment to mitochondriotropic lipophilic cations, such as TPP+. Lipophilic cations possess a delocalized positive charge, which enables them to traverse mitochondrial membranes and concentrate within or close to mitochondria because of the greater mitochondrial membrane potential, which is negative inside. As the driving force behind this mitochondrial-targeted drug delivery seems to be the passive mitochondrial membrane potential difference between normal and cancer cells alone, it is essential to consider associated sensing mechanisms that can detect changes in reactive oxygen species (ROS), pH, glutathione (GSH), the temperature of mitochondria to lessen undesirable effects on other organs. This becomes imperative in cases where drugs are unable to cross cellular membranes, let alone subcellular organelles like mitochondria.16 Furthermore, they are crucial to the process of apoptosis mediated by mitochondria.17,18
Another approach that circumvents the BBB and helps treat brain diseases is a noninvasive intranasal delivery system.19 Intranasal delivery bypasses BBB and provides better brain targeting efficiency, which has been supported by many preclinical findings.20–23 Numerous investigators presented preclinical and clinical data suggesting that nose-to-brain delivery is highly effective in treating various brain disorders, including GB, IDH-wildtype treatment approaches. Trigeminal and olfactory nerves are the pathways by which drugs delivered via intranasal administration enter the central nervous system.24 Drug delivery systems that can stick to the nasal mucosa and pass via the olfactory pathway are necessary for nasal delivery of drugs to the brain. A sustained and extended drug administration via the nasal route has been described using delivery systems such as mucoadhesive nanoparticles and nanogels which could be effectively targeted to the brain.25
In this study, biocompatible chitosan was selected as a mucoadhesive polymer for developing polymeric nanoparticles. The internalization of chitosan (CS) is aided by its positive charges, which are readily bonded with the negatively charged membrane, especially of the mitochondrial membrane. Because of their ability to synthesize adenosine triphosphate (ATP) and transport ions, mitochondria have a significantly greater potential than other organelles. As a result, triphenylphosphine (TPP+), a form of lipophilic cationic chemical, may be specifically targeted to mitochondria. A combination of CS, as well as TPP+, may smartly target the drug's delivery to the mitochondria, inducing mitochondrial depolarization that would cause a cell stress response, increase in ROS, and ultimately cause cytotoxicity in the tumor cells.26
Here, we aim to design triphenylphosphine (TPP+)-conjugated chitosan nanoparticles (TPP+-CS NPs) to transport TMZ specifically to the mitochondria and increase the antitumor efficacy. To target the mitochondria, grafting triphenyl phosphine onto the surface of (as-prepared) chitosan nanoparticles was done, and TMZ was encapsulated into NPs to study drug loading and the in vitro release mechanism. Additionally, in vitro cytotoxicity, and intracellular ROS levels were examined to assess the effectiveness of triphenyl phosphonium-linked chitosan nanoparticles for targeted mitochondrial delivery of TMZ.
2. Materials and methods
2.1 Materials
Temozolomide (TMZ), chitosan low molecular weight (<500 kDa), sodium tripolyphosphate (STPP), (3-carboxypropyl) triphenyl phosphonium bromide (TPP+), ROS fluorometric intracellular ROS kit (MAK145) and, dialysis tubing cellulose membrane (MWCO: 12–14 kDa) were obtained from Sigma-Aldrich (Mumbai, India). 1-Ethyl-3-(3-dimethyl aminopropyl) carbodiimide (EDC) and N-hydroxy succinimide (NHS) were received from Molychem, India. Minimum Essential Medium (MEM) and penicillin–streptomycin were obtained from Gibco Life Technologies (Grand Island, USA), while trypsin–EDTA solution and Alamar blue assay kit (DAL1100) were procured from Invitrogen (California, USA). All other chemicals employed were of analytical grade. Double distilled water, obtained from the Milli-Q Plus system, was utilized throughout the experiments.2.2 Cell culture
Human glioblastoma cells (U87MG) were obtained from the National Centre for Cell Science (NCCS), Pune, India. The cells were cultured in Minimum Essential Medium (MEM) supplemented with 10% v/v fetal bovine serum (FBS) and penicillin (100 IU mL−1)–streptomycin (100 μg mL−1) and incubated at 37 °C and 5% CO2.2.3 HPLC method development
A standard curve, also referred to as a calibration curve, is created by varying the concentrations of a standard drug, i.e., TMZ, within a linear range. This calibration curve is required to ascertain the analyte concentration in an unknown sample. It demonstrated linearity in two distinct solvents, Milli-Q, and PBS buffer (pH 5), using an established HPLC technique. One mg of TMZ was dissolved in one mL Milli-Q water to prepare the stock solution of 1000 μg mL−1. After serial dilutions, a concentration range of 1–250 μg mL−1 was obtained from this stock solution. After each concentration's injection, the peak area was calculated to plot the best-fitting line. The regression equation, y-intercept, slope, and correlation coefficient were computed. The linearity is considered acceptable if the regression coefficient is higher than 0.999.272.4 Fourier-transform infrared spectral analysis
Using an FTIR-spectrophotometer (Bruker, Germany), the spectra of individual TMZ, chitosan, sodium tripolyphosphate (STPP), and the physical mixture of TMZ, chitosan, and STPP were obtained to evaluate the compatibility between the drug and excipients. The spectra were scanned in the range of wavenumber 4000–400 cm−1. Equimolar quantities of TMZ, STPP, and chitosan were combined to form the physical combination, which was further homogenized by trituration with a mortar and pestle. The overlay of spectra then examined the drug's compatibility with the excipients.282.5 Formulation of TMZ-loaded chitosan nanoparticles (TMZ-loaded CSNPs) using QbD approach
Chitosan nanoparticles (CSNPs) were produced by the ionic gelation process, as reported by Pant et al.29 Sodium tripolyphosphate (STPP) and low molecular weight (LMW) chitosan were used in the synthesis. Briefly, a CS solution in 2% v/v acetic acid was prepared and then agitated for three hours at room temperature (RT) at 1000 rpm. 7 mL of the CS solution was then mixed dropwise with the prepared drug solution and STPP solution in 3 mL of distilled water. The mixture was continuously stirred for one hour using a magnetic stirrer set at 800 rpm. Further, for screening and optimization purpose, the Quality-by-Design approach was utilized.The Quality by Design (QbD) technique improves the final pharmaceutical product's value and quality. Creating a robust pharmaceutical product with the potential to achieve maximum therapeutic efficacy, quality attributes, and prolonged stability throughout its storage term is the main goal of employing QbD in the pharmaceutical industry. The Quality Target Product Profile (QTPP) and Critical Quality Attribute (CQA) criteria were determined to formulate nanoparticles. A QTPP was developed to direct the development of nanoparticles in line with the QbD approach for drug product development, aiming to improve their biopharmaceutical qualities and offer patients therapeutic advantages. Several patient-centered CQAs were found to meet QTPP requirements. The physical and formulation qualities of nanoparticles as the intended drug product are included in their quality characteristics.30 A subset of quality parameters that have a major influence on the product's performance was selected for this particular purpose. Particle size is crucial for the absorption of the nanoparticle. Entrapment efficiency is a measure of the developed delivery system's propensity to encapsulate particles, and zeta potential aids in determining the stability of the formulation. The CQAs of the nanoparticles are directly impacted by the choices made regarding the elements and parameters used as Critical Material Attributes (CMA) and Critical Process Parameters (CPP). The critical material attributes (CMAs), associated with the formulation's constituent parts, and the critical process parameters (CPPs), associated with the manufacturing techniques' instrumental parameters, were defined. This work aimed to optimize TPP+-conjugated TMZ-loaded CSNPs for positive surface charge transfer from the nose to the brain using the QbD technique. To create TPP+-conjugated TMZ-loaded CSNPs appropriate for nose-to-brain transport, the QTPP and the CQAs were determined before the trials. This formulation included the following characteristics: particle size, polydispersity index (PDI), zeta potential (ZP), and encapsulation efficiency (EE). To optimize the TPP+-conjugated TMZ-loaded CSNPs for nose-to-brain delivery through the QbD approach, the QTPP were identified and summarized in ESI Table 1 (Table S1†).
Design expert® software (Stat-Ease, Inc., Version 11, MN, USA) was used to screen and optimize the NPs.
| Name | Units | Levels | L[1] | L[2] |
|---|---|---|---|---|
| Chitosan conc. | mg mL−1 | 2 | 0.5 | 4 |
| TMZ conc. | mg mL−1 | 2 | 0.5 | 2 |
| STPP conc. | mg mL−1 | 2 | 0.5 | 2 |
| Stirring speed-1 | rpm | 2 | 200 | 1500 |
| Time-1 | h | 2 | 1 | 4 |
| Stirring speed-2 | rpm | 2 | 500 | 1500 |
| Time-2 | min | 2 | 15 | 60 |
| Std | Factor 1 | Factor 2 | Factor 3 | Factor 4 | Factor 5 | Factor 6 | Factor 7 |
|---|---|---|---|---|---|---|---|
| A: chitosan conc. (mg mL−1) | B: TMZ conc. (mg mL−1) | C: STPP conc. (mg mL−1) | D: stirring speed-1 (rpm) | E: time-1 (h) | F: stirring speed-2 (rpm) | G: time-2 (min) | |
| 1 | 0.5 | 0.5 | 0.5 | 200 | 1 | 500 | 15 |
| 2 | 0.5 | 0.5 | 0.5 | 1500 | 4 | 1500 | 60 |
| 3 | 0.5 | 2 | 2 | 200 | 1 | 1500 | 60 |
| 4 | 0.5 | 2 | 2 | 1500 | 4 | 500 | 15 |
| 5 | 4 | 0.5 | 2 | 200 | 4 | 500 | 60 |
| 6 | 4 | 0.5 | 2 | 1500 | 1 | 1500 | 15 |
| 7 | 4 | 2 | 0.5 | 200 | 4 | 1500 | 15 |
| 8 | 4 | 2 | 0.5 | 1500 | 1 | 500 | 60 |
| Factors (independent variables) | Levels of variables | ||
|---|---|---|---|
| Low (−1) | Medium (0) | High (+1) | |
| A: CS conc. (mg mL−1) | 0.5 | 1 | 1.5 |
| B: TMZ conc. (mg mL−1) | 0.5 | 0.75 | 1 |
| C: stirring speed-2 (rpm) | 800 | 1000 | 1200 |
| D: time-2 (min) | 20 | 40 | 60 |
| Run | Factor 1 | Factor 2 | Factor 3 | Factor 4 |
|---|---|---|---|---|
| A: chitosan conc. (mg mL−1) | B: TMZ conc. (mg mL−1) | C: stirring speed-2 (rpm) | D: time-2 (min) | |
| 1 | 1 | 0.5 | 1000 | 20 |
| 2 | 1.5 | 0.75 | 1200 | 40 |
| 3 | 1 | 0.75 | 1000 | 40 |
| 4 | 1 | 0.75 | 1200 | 20 |
| 5 | 1 | 0.75 | 1000 | 40 |
| 6 | 1.5 | 0.75 | 1000 | 20 |
| 7 | 1.5 | 0.75 | 1000 | 60 |
| 8 | 0.5 | 0.75 | 1000 | 20 |
| 9 | 1 | 1 | 1000 | 60 |
| 10 | 1.5 | 1 | 1000 | 40 |
| 11 | 1 | 0.75 | 1200 | 60 |
| 12 | 0.5 | 1 | 1000 | 40 |
| 13 | 0.5 | 0.5 | 1000 | 40 |
| 14 | 0.5 | 0.75 | 1000 | 60 |
| 15 | 1 | 0.5 | 1000 | 60 |
| 16 | 1 | 1 | 1200 | 40 |
| 17 | 1.5 | 1 | 1000 | 40 |
| 18 | 1 | 0.75 | 800 | 60 |
| 19 | 1 | 0.75 | 1000 | 40 |
| 20 | 1 | 1 | 800 | 60 |
| 21 | 1 | 0.5 | 1200 | 40 |
| 22 | 1 | 0.5 | 800 | 40 |
| 23 | 1.5 | 0.5 | 1000 | 40 |
| 24 | 1.5 | 0.75 | 800 | 40 |
| 25 | 0.5 | 0.75 | 1000 | 40 |
| 26 | 0.5 | 0.75 | 800 | 40 |
| 27 | 1 | 0.75 | 800 | 20 |
The two extreme levels of CS and TMZ conc. were chosen on performing preliminary and screening testing of the impact of factors above on PS, PDI, and % EE. Based on the preliminary study and screening study, broader ranges were selected. Software generated the medium levels of the enlisted factors based on the chosen levels. Using BBD, with 3 repetitions of the center point, 27 runs were obtained (Table 4) performed, and term significance was estimated by analysis of variance (ANOVA). Probability p-values (p < 0.05) denoted significance.32
2.6 Synthesis of triphenylphosphonium-chitosan (TPP+-CS) conjugate
NPs were conjugated by the method adopted by Huang et al. with minor modifications.33 A 100 mg quantity of low molecular weight-chitosan (LMWT-CS) was dissolved in 100 mL of acidified water (2% acetic acid solution; pH 2.5) under stirring until complete homogenization (3 h). Simultaneously, triphenylphosphonium (TPP+) was activated for 2 h using an aqueous solution of EDC/NHS. The activated TPP+ solution was then dropped into the CS solution and stirred overnight at low temperatures (CS![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) TPP+ in ratio 2
TPP+ in ratio 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1). Subsequently, the solution pH was adjusted to 9 using 1 N NaOH inducing precipitation of conjugated CS polymer. The precipitate was collected via centrifugation (10
1). Subsequently, the solution pH was adjusted to 9 using 1 N NaOH inducing precipitation of conjugated CS polymer. The precipitate was collected via centrifugation (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm, 20 min). The pellet subsequently was kept for dialysis (MWCO: 12–14 kDa) for 2 days to remove unconjugated materials. Then, the collected pellet was resuspended in water, frozen, and lyophilized.
000 rpm, 20 min). The pellet subsequently was kept for dialysis (MWCO: 12–14 kDa) for 2 days to remove unconjugated materials. Then, the collected pellet was resuspended in water, frozen, and lyophilized.
2.7 Structural validation of TPP+-CS conjugate
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm, 4 °C). Then, the supernatant was separated and injected into HPLC (HPLC 1260 Infinity, Agilent Technologies, CA, USA). Unconjugated TPP+ concentration was calculated by plotting the linearity of TPP+ with the developed HPLC method. This was further used to determine % conjugation efficiency.36
000 rpm, 4 °C). Then, the supernatant was separated and injected into HPLC (HPLC 1260 Infinity, Agilent Technologies, CA, USA). Unconjugated TPP+ concentration was calculated by plotting the linearity of TPP+ with the developed HPLC method. This was further used to determine % conjugation efficiency.36HPLC method was developed using HPLC (1260 Infinity, Agilent Technologies, CA, USA). Eclipse Plus C18 column (4.6 mm × 250 mm, 5 μm) was utilized for separation using a mobile phase consisting of ammonium formate buffer (10 mM) as an aqueous phase and acetonitrile (ACN) as an organic phase, respectively. Keeping the injection volume at 10 μL, analysis was done at 267 nm.
2.8 Preparation of TPP+-conjugated TMZ-loaded CSNPs
The conjugated TMZ-loaded CSNPs were made using a similar process to the one previously optimized for producing chitosan nanoparticles. The lyophilized conjugated CS-TPP+ polymer (1 mg mL−1) was dissolved in an aqueous solution of acetic acid using agitation [1000 rpm (stirring speed-1), 3 h (time-1)]. TPP (1.5 mg mL−1) and TMZ solutions (0.5 mg mL−1) were also prepared in Milli-Q water. Subsequently, the TMZ and TPP solutions were added dropwise to the CS-TPP+ solution while it was continuously stirred at 800 rpm (stirring speed-2) for 1 h (time-2), resulting in the synthesis of conjugated nanoparticles.2.9 Characterization of TMZ-loaded CSNPs and TPP+-conjugated TMZ-loaded CSNPs
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 30 min at RT.32,38 The quantity of free TMZ in the clear supernatant was estimated by the HPLC method at 330 nm. Free TMZ conc. was calculated by a previously developed TMZ calibration curve. Then, the %EE was calculated according to the following equation,
000 rpm for 30 min at RT.32,38 The quantity of free TMZ in the clear supernatant was estimated by the HPLC method at 330 nm. Free TMZ conc. was calculated by a previously developed TMZ calibration curve. Then, the %EE was calculated according to the following equation,
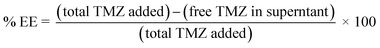 | (1) |
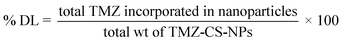 | (2) |
2.10 Morphological assessment of TMZ-loaded CSNPs and TPP+-conjugated TMZ-loaded CSNPs
The morphology of nanoparticle suspension was assessed using FE-SEM (SIGMA S300, Zeiss). During sample preparation, nanoparticles need to be diluted to get an image of discrete particles with better resolution and clarity. Nanoparticle formulation was diluted using Milli-Q water, and one drop of nanoparticles diluted solution was taken on a glass slide, air dried for 10 min, and again kept for drying overnight using the vacuum dryer. Then, samples were coated with conductive material (gold-coated). Because gold coating increases sample stability, protects samples, improves resolution and image quality, and enhances conductivity, it is essential to have accurate and dependable imaging of various materials in FE-SEM analysis. Images were captured under the accelerated electron voltage of 40 kV.422.11 Stability study
The TMZ-loaded CSNPs and TPP+-conjugated TMZ-loaded CSNPs were stored at 25 ± 1 °C and 4 ± 1 °C for 28 days. The aliquots of NPs were collected at different time intervals (0, 1, 3, 7, 14, and 28 days). The samples were subsequently characterized for particle size, PDI, and zeta potential. The results were reported as mean ± S. D. (n = 3).432.12 Ex vivo drug permeation study
Franz diffusion cells with a contact surface area of 2.26 cm2 and a receiver compartment capacity of 15 mL were used to conduct an ex vivo permeation study on goat nasal mucosa. It has been used for studies because goat nasal mucosa has similar physiological characteristics to human nasal mucosa. Fresh goat nasal mucosa was procured from a slaughterhouse. Adherent mucus and adipose tissues were meticulously dissected, and the tissue was trimmed to appropriate dimensions for subsequent mounting. The mounting orientation ensured the mucosal surface faced the donor compartment, while the serosal surface opposed the receiver compartment. To establish electrophysiological equilibrium, the tissue was pre-incubated for 15 min at 37 °C with saline solution filling both compartments. Subsequently, 1 mL of TMZ solution was introduced into the donor compartment. At predetermined intervals (5, 10, 15, 30, 60, 120, 240, 360, and 480 min), 1 mL aliquot was collected from the receiver compartment and immediately replaced with an equal volume of fresh media. The same procedure was performed for TPP+-conjugated TMZ-loaded CSNPs. HPLC method was developed to assess unknown sample concentration for drug penetration. The isocratic method was used, and the mobile phase composition was selected as a 90![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 ratio of phosphate buffer (pH 6.5) and acetonitrile at 330 nm and 1.0 mL min−1 flow rate with a run time of 7 min.
10 ratio of phosphate buffer (pH 6.5) and acetonitrile at 330 nm and 1.0 mL min−1 flow rate with a run time of 7 min.
2.13 Mucoadhesion study
A mucin binding assay was employed to assess the mucoadhesive potential of formulated nanoparticles based on alteration in their zeta potential. Deionized water was used to dissolve 1% w/v porcine mucin powder. The mucin solution was centrifuged at 5000 rpm for 10 min, and the resulting supernatant was utilized for mucoadhesion studies. An equal amount of formulation (TMZ solution and conjugated formulation) and 1% w/v mucin supernatant were mixed with a vortexer for one minute. The mixture was then left for four hours. Then, the zeta potential of the mucin solution was noted. Zeta potential measurement was performed on each combination with Zeta Sizer (Nano ZS, Malvern, United Kingdom). Mucoadhesion was evidenced by the change in zeta potential detected.44 The experiment was done in triplicate.2.14 Cytotoxicity study
Alamar blue assay (DAL1100, Invitrogen, USA) was performed to assess the cytotoxicity of the prepared formulation. U87MG cell line was used and cultured in Minimum Essential Medium (MEM) with 10% fetal bovine serum albumin (10% FBS). They were maintained in an incubator at 37 °C and 5% CO2. Cells were seeded in 96 well plates at a density of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per well. Then cells were treated with different concentrations of TMZ solution ranging from 1 to 5000 ppm for 24 h. The cells were also treated with TMZ-loaded CSNPs (concentration range: 0.1–100 ppm) and TPP+-conjugated TMZ-loaded CSNPs (concentration range: 1–100 ppm) diluted in a serum-free MEM medium. Control cells were treated with serum-free media without any treatment for 24 h. After 24 h of treatment, 10 μL of Alamar Blue Reagent (10 μg mL−1) was added to each well and kept in an incubator for 4 h. The fluorescence intensity of the solutions in each well was measured using a multimode UV microplate reader (Varioskan LUX, Thermo Fischer Scientific) at Ex/Em 560–590 nm. The cell viability was calculated by using formula,45
000 cells per well. Then cells were treated with different concentrations of TMZ solution ranging from 1 to 5000 ppm for 24 h. The cells were also treated with TMZ-loaded CSNPs (concentration range: 0.1–100 ppm) and TPP+-conjugated TMZ-loaded CSNPs (concentration range: 1–100 ppm) diluted in a serum-free MEM medium. Control cells were treated with serum-free media without any treatment for 24 h. After 24 h of treatment, 10 μL of Alamar Blue Reagent (10 μg mL−1) was added to each well and kept in an incubator for 4 h. The fluorescence intensity of the solutions in each well was measured using a multimode UV microplate reader (Varioskan LUX, Thermo Fischer Scientific) at Ex/Em 560–590 nm. The cell viability was calculated by using formula,45
 | (3) |
2.15 Intracellular reactive oxygen species (ROS) study
Intracellular ROS level was assessed by a fluorometric intracellular ROS kit (Sigma-Aldrich, MAK145). U87MG cell lines were seeded (MEM with 10% FBS) in a 96-well plate at a density of 1 × 104 cells per well. The cells were incubated at 37 °C and 5% CO2 overnight and then treated with master-mix solution (provided with the kit) for 1 h. The treatment (TMZ solution, TMZ-loaded CSNPs, and TPP+-conjugated TMZ-loaded CSNPs at their IC50 value) was then given for a specified period i.e., 3 h, 6 h, and 12 h. The next proceeding was done according to the manufacturer's protocol and the fluorescence intensity was measured with a multimode plate reader (Varioskan LUX Multimode, Thermo Fisher Scientific, USA) at excitation/emission wavelength at 520/605 nm. U87MG cells with media were considered as a control while hydrogen peroxide solution was taken as a positive control. The experiment was done in triplicate.2.16 Cellular uptake study
Cellular uptake study was done to track whether the prepared nanoparticles penetrate into the cells or not. To track the fate of TMZ, rhodamine B, a reporter dye (0.5% w/v) was integrated into nanoparticles (unconjugated as well as conjugated). It was then subjected to centrifugation to separate TMZ-loaded CSNPs from unloaded rhodamine B. Briefly, U87MG (50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per well) were seeded in 6-well plates that were covered with 18 mm coverslips. The were incubated overnight at 5% CO2 at 37 °C and then treated with TMZ solution, unconjugated TMZ-loaded CSNPs, and TPP+-conjugated TMZ-loaded CSNPs for 2 h and 4 h. Cells were fixed with 4% paraformaldehyde solution in PBS for 15 min and then stained with 10 μg of DAPI in PBS for 10 min followed by 2 times washing with PBS. Cellular uptake of nanoparticles was then observed by fluorescent microscopy.46
000 cells per well) were seeded in 6-well plates that were covered with 18 mm coverslips. The were incubated overnight at 5% CO2 at 37 °C and then treated with TMZ solution, unconjugated TMZ-loaded CSNPs, and TPP+-conjugated TMZ-loaded CSNPs for 2 h and 4 h. Cells were fixed with 4% paraformaldehyde solution in PBS for 15 min and then stained with 10 μg of DAPI in PBS for 10 min followed by 2 times washing with PBS. Cellular uptake of nanoparticles was then observed by fluorescent microscopy.46
2.17 Statistical analysis
All studies were performed in triplicate (mean ± S. D.) and graphs were made using Graph Pad Prism (Version 8.0.2). One-way Analysis of Variance (ANOVA), a parametric test was utilized for multigroup analysis with the Tukey–Kramer multiple comparison posthoc test (*p < 0.05, **p < 0.01, ***p < 0.001 represented statistically significant difference).3. Results and discussion
3.1 Spectrophotometric determination of TMZ by HPLC
HPLC method was employed to analyze the unknown concentration of TMZ in solution and formulation. The maximum absorption wavelength (λmax) of TMZ was found at 330 nm and the retention time was observed at 4.3 min. High correlation coefficients (R2 values of 0.9993 and 0.9998) indicate the linearity of the calibration curves for TMZ in both Milli-Q water and PBS buffer (pH 5), respectively. The HPLC chromatogram of TMZ is shown in ESI Fig. S1(a),† and the linear range of TMZ solution in Milli-Q water (1–250 μg mL−1) is shown in Fig. S1(b),† with regression equation as y = 27.227x + 20.119, with an R2 value of 0.9993. Fig. S1(c)† shows the linearity range of TMZ solution (1–100 μg mL−1) in PBS buffer (pH 5). The regression equation is y = 23.448x + 12.047, and its R2 value was found to be 0.9998.3.2 Fourier-transform infrared spectral analysis
Fourier transform infrared (FTIR) spectroscopy is a powerful analytical tool for characterizing drug molecules and excipients. By identifying their unique vibrational fingerprint regions in the infrared spectrum, FTIR spectroscopy allows for detecting potential interactions between these components. The spectra of drug and excipients, viz; CS, STPP, and TMZ, were obtained individually, and similarly, the spectrum of the physical mixture of drug and excipients was also taken. The overlay of spectra is represented in Fig. 1. The characteristic peaks of TMZ in the region of 3384.36 cm−1 (–NH stretching), 3038.32 cm−1 (–CH2), 1732.05 cm−1(–NH–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1678.72 cm−1 (amide bending), and 1451.67 cm−1 (–CH bending) were found to be retained in the spectrum of physical mixture with no addition or deletion of peaks indicating that there was no chemical interaction between drug and excipients thus, signifying the compatibility of TMZ with CS and STPP. A similar observation was seen for CS (characteristic peaks of 3358, 2924, 1655, 1575, 1378, 1156, 898 cm−1) and STPP (characteristic peaks of 2890, 1715, 1261, 1153, 1108 cm−1) where all significant peaks were retained in the spectrum of the physical mixture, further confirming the compatibility.
O), 1678.72 cm−1 (amide bending), and 1451.67 cm−1 (–CH bending) were found to be retained in the spectrum of physical mixture with no addition or deletion of peaks indicating that there was no chemical interaction between drug and excipients thus, signifying the compatibility of TMZ with CS and STPP. A similar observation was seen for CS (characteristic peaks of 3358, 2924, 1655, 1575, 1378, 1156, 898 cm−1) and STPP (characteristic peaks of 2890, 1715, 1261, 1153, 1108 cm−1) where all significant peaks were retained in the spectrum of the physical mixture, further confirming the compatibility.
 | ||
| Fig. 1 (a) Overlay of FTIR spectra of TMZ, STPP, CS, and their physical mixture as a pre-formulation study indicating compatibility between TMZ and excipients. | ||
3.3 Formulation of TMZ-loaded CSNPs
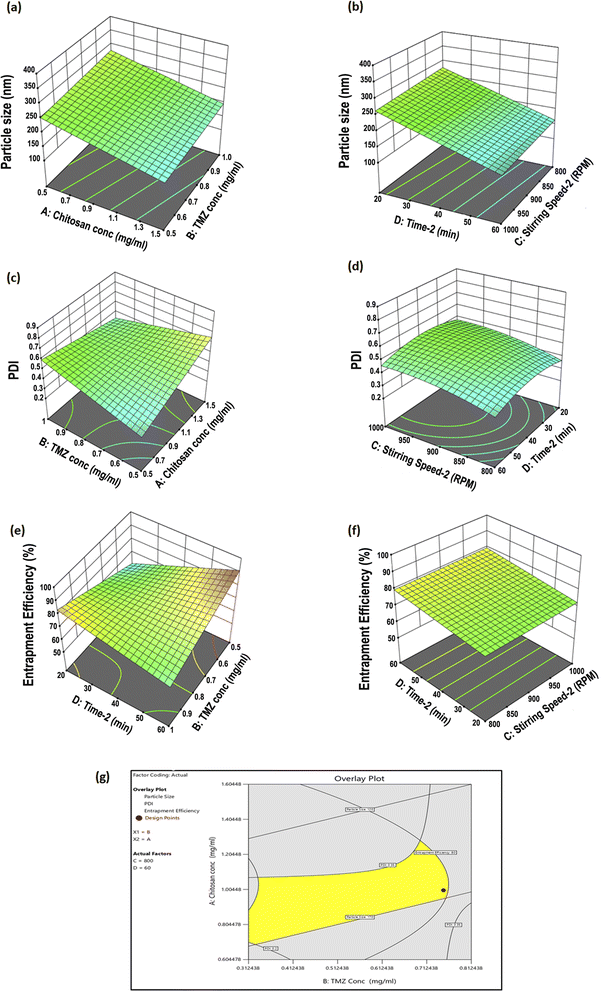
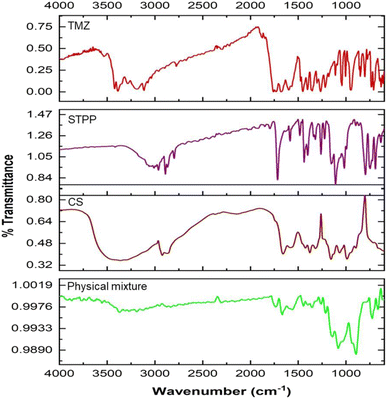
CSNPs were formulated using biocompatible and biodegradable polymer like CS and STPP as crosslinking agent. By creating crosslinks with the CS polymer, the STPP helped to entrap TMZ in the polymer matrix and promote the formulation of CSNPs. The resultant mixture was continuously stirred for 1 h using the ionic gelation process to formulate TMZ-loaded CSNPs.3.3.1.1 Particle size. The particle size of TMZ-loaded CSNPs was distributed in a range of 318–3515 nm. The particle size of NPs suspensions was analyzed using Zetasizer (Nano ZS, Malvern, United Kingdom). Table 5 shows the particle size of the TMZ-loaded CSNPs for the formulations batch 1 to 8. Batch 1–3 and 5–8 had larger particle sizes than batch 4. From these results, the level of factors has been selected for further optimization purposes. Out of all selected factors, TMZ conc. (F value of 19.60) and stirring speed-2 (F value of 11.96) showed a positive effect on particle size demonstrated by the Pareto chart and ANOVA Table of Taguchi design. Both the factors crossed the t-value limit in the Pareto chart, signifying the possible impact on particle size. Further p values of both factors were less than 0.05, again highlighting their significant impact on particle size. (Fig. 2(a) and ESI Table S2(a)†).
| Run | Factor 1 | Factor 2 | Factor 3 | Factor 4 | Factor 5 | Factor 6 | Factor 7 | Response 1 | Response 2 | Response 3 |
|---|---|---|---|---|---|---|---|---|---|---|
| A: chitosan conc. (mg mL−1) | B: TMZ conc. (mg mL−1) | C: STPP conc. (mg mL−1) | D: stirring speed-1 (rpm) | E: time-1 (h) | F: stirring speed 2 (rpm) | G: time-2 (min) | Particle size (nm) | PDI | Entrapment efficiency (%) | |
| 1 | 4 | 0.5 | 2 | 1500 | 1 | 1500 | 15 | 748.7 | 0.267 | 59.2787 |
| 2 | 4 | 2 | 0.5 | 1500 | 1 | 500 | 60 | 1594 | 0.683 | 14.1182 |
| 3 | 4 | 2 | 0.5 | 200 | 4 | 1500 | 15 | 3515 | 0.293 | 28.0109 |
| 4 | 0.5 | 0.5 | 0.5 | 200 | 1 | 500 | 15 | 318.9 | 0.489 | 56.5438 |
| 5 | 0.5 | 2 | 2 | 200 | 1 | 1500 | 60 | 3132 | 0.857 | 38.3676 |
| 6 | 0.5 | 2 | 2 | 1500 | 4 | 500 | 15 | 1336 | 1 | 46.4461 |
| 7 | 4 | 0.5 | 2 | 200 | 4 | 500 | 60 | 726.8 | 0.692 | 98.3426 |
| 8 | 0.5 | 0.5 | 0.5 | 1500 | 4 | 1500 | 60 | 1493 | 0.832 | 63.5691 |
 | ||
| Fig. 2 Pareto chart analysis during screening by Taguchi design (a) particle size, (b) PDI, (c) % entrapment efficiency. | ||
3.3.1.2 PDI. The PDI of TMZ-loaded CSNPs was distributed in a wide range from 0.26 to 0.85. The PDI of NPs was analyzed using Zetasizer. Table 5 shows the PDI of the CSNPs for the formulation batch 1–8. The PDI obtained in batches 2 and 5–8 was larger than those obtained in batches 1 and 3–4. The Pareto chart and ANOVA table (Fig. 2(b) and Table S2(b)†) indicated that the time-2 (F value of 20.15), chitosan conc. (F value of 17.99), and stirring speed-2 (F value of 10.60) showed a significant impact on PDI. Time-2 showed a negative impact, meaning as time increased, there was a decrease in the PDI, but chitosan conc. and stirring speed-2 showed positive effects, meaning as CS conc. or stirring speed-2 increased, there was an increase in PDI.
3.3.1.3 % entrapment efficiency (% EE). The % EE of TMZ-loaded CSNPs was obtained in a wide range from 14.11% to 98.34%. The entrapment of TMZ in CSNPs suspensions was analyzed using the HPLC method. Table 5 shows the entrapment efficiency of the TMZ-loaded CSNPs for the formulation batches 1–8. The entrapment obtained in batches 1, 4, 7, and 8 was larger than those obtained in batches 2, 3, 5, and 6. By analyzing the Pareto chart and ANOVA Table (Fig. 2(c) and Table S2(c)†), it was found that the TMZ conc. (F value 8.52) showed a significant impact on entrapment efficiency. The % EE decreases with increasing TMZ concentration. This negative relationship between TMZ conc. and % EE suggests that % EE falls with increasing TMZ concentration.
Design Expert® software (version 13) was employed to evaluate model fit based on adjusted R2 and prediction R2. For PS, a linear model provided the optimal fir. On the other hand, quadratic models were chosen for both PDI and % EE. These selections were made based on the models with the highest adjusted and predicted R2 values. After model reduction, the final equations for the responses related to different factors and interactions in terms of coded variables were as follows,
| PS = + 215.80 − 40.73 (CS conc.) + 12.62 (TMZ conc.) − 7.26 (stirring speed-2) − 56.98 (time-2) | (4) |
| PDI = +5059 + 0.0400 (CS conc.) + 0.0121 (TMZ conc.) + 0.0610 (stirring speed-2) − 0.0092 (time-2) − 0.1197 (CS conc. × TMZ conc.) + 0.0462 (CS conc. × stirring speed-2) − 0.0345 (stirring speed-2)2 − 0.0595 (Time-2)2 | (5) |
| % EE = +76.93 + 1.62 (CS conc.) − 0.3585 (TMZ conc.) + 3.56 (time-2) − 11.36 (TMZ conc. × time-2) − 11.99 (CS conc.)2 | (6) |
3.3.2.1 Influence of investigated factors on particle size. ANOVA analysis showed that CS conc. (A), and time-2 (D) were the significant factors affecting average PS (p < 0.05), while TMZ conc. (B) and stirring speed (C) had no significant effect on particle size (Table S3(a)†). As seen in the 3D surface plot illustrated in Fig. 3(a and b), both factors (CS and time-2) were inversely correlated with particle size. The results demonstrated that particle size was significantly affected by CS conc. and time-2. The optimum formula with the smallest particle size was obtained when the CS conc. was 1 mg mL−1, and the particle size was found to increase below and above this conc. Below this concentration, the concentration of CS was not sufficient to form effective cross-linkage with STPP. With a CS–STPP ratio of 1
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.5, the particle size decreased due to the enhanced density of cross-linkage between CS and STPP. However, increasing the TMZ conc. above a certain limit could also lead to particle size enlargement due to the accumulation of excess TMZ on the surface of the NPs. Also, time-2 helps in reducing the size of the particles, but if it falls below a specific point, aggregation forms and the size of the particles grows. Optimum size nanoparticles (<200 nm) were obtained at 60 min stirring.
1.5, the particle size decreased due to the enhanced density of cross-linkage between CS and STPP. However, increasing the TMZ conc. above a certain limit could also lead to particle size enlargement due to the accumulation of excess TMZ on the surface of the NPs. Also, time-2 helps in reducing the size of the particles, but if it falls below a specific point, aggregation forms and the size of the particles grows. Optimum size nanoparticles (<200 nm) were obtained at 60 min stirring.
3.3.2.2 Influence of investigated factors on PDI. The PDI of TMZ-loaded CSNPs varied from 0.25 to 0.69, as shown in Table 6. ANOVA analysis indicated that all factors, A, B, C, and D, significantly affected the PDI of the prepared NPs (p < 0.05) respectively (Table S3(b)†). The main effects of A, B, and C on PDI are represented by the 3D surface plots in Fig. 3(c and d). PDI significantly increased by increasing the CS concentration and stirring speed-2. A decrease in the TMZ concentration and an increase in stirring time-2 were associated with a significant decrease in PDI.
| Runs | Factor 1 | Factor 2 | Factor 3 | Factor 4 | Response 1 | Response 2 | Response 3 |
|---|---|---|---|---|---|---|---|
| A: chitosan conc. (mg mL−1) | B: TMZ conc. (mg mL−1) | C: stirring speed-2 (rpm) | D: time-2 (min) | Particle size (nm) | PDI | EE (%) | |
| 1 | 1 | 0.5 | 1000 | 20 | 349.23 | 0.49 | 57.12 |
| 2 | 1.5 | 0.75 | 1200 | 40 | 150.6 | 0.6 | 78.91 |
| 3 | 1 | 0.75 | 1000 | 40 | 106.9 | 0.43 | 75.54 |
| 4 | 1 | 0.75 | 1200 | 20 | 303.1 | 0.38 | 83.94 |
| 5 | 1 | 0.75 | 1000 | 40 | 370 | 0.69 | 69.54 |
| 6 | 1.5 | 0.75 | 1000 | 20 | 169.4 | 0.48 | 54.62 |
| 7 | 1.5 | 0.75 | 1000 | 60 | 205.2 | 0.49 | 74.08 |
| 8 | 0.5 | 0.75 | 1000 | 20 | 212.8 | 0.39 | 57.48 |
| 9 | 1 | 1 | 1000 | 60 | 109.3 | 0.52 | 66.64 |
| 10 | 1.5 | 1 | 1000 | 40 | 135.3 | 0.56 | 64.01 |
| 11 | 1 | 0.75 | 1200 | 60 | 102.6 | 0.25 | 81 |
| 12 | 0.5 | 1 | 1000 | 40 | 329.3 | 0.56 | 63.22 |
| 13 | 0.5 | 0.5 | 1000 | 40 | 209.4 | 0.36 | 64.41 |
| 14 | 0.5 | 0.75 | 1000 | 60 | 188.2 | 0.4 | 67.36 |
| 15 | 1 | 0.5 | 1000 | 60 | 114.2 | 0.43 | 89.55 |
| 16 | 1 | 1 | 1200 | 40 | 239.9 | 0.38 | 59.79 |
| 17 | 1.5 | 1 | 1000 | 40 | 214.8 | 0.47 | 69.1 |
| 18 | 1 | 0.75 | 800 | 60 | 117.5 | 0.38 | 81.29 |
| 19 | 1 | 0.75 | 1000 | 40 | 104.5 | 0.4 | 78.19 |
| 20 | 1 | 1 | 800 | 60 | 201.9 | 0.36 | 73.49 |
| 21 | 1 | 0.5 | 1200 | 40 | 138.3 | 0.33 | 83.61 |
| 22 | 1 | 0.5 | 800 | 40 | 274.8 | 0.35 | 77.65 |
| 23 | 1.5 | 0.5 | 1000 | 40 | 147.8 | 0.8 | 54.17 |
| 24 | 1.5 | 0.75 | 800 | 40 | 178.9 | 0.41 | 70.68 |
| 25 | 0.5 | 0.75 | 1000 | 40 | 303.1 | 0.43 | 67.83 |
| 26 | 0.5 | 0.75 | 800 | 40 | 282.1 | 0.38 | 59.65 |
| 27 | 1 | 0.75 | 800 | 20 | 231.3 | 0.36 | 84.4 |
3.3.2.3 Influence of investigated factors on % EE. The % EE of TMZ-loaded CS NPs ranged from 54.62% to 89.55% as shown in Table 6, indicating that the NPs highly entrapped TMZ. As shown by the 3D surface plot in Fig. 3(e and f), TMZ conc. and chitosan conc. significantly affected % EE. The results of the ANOVA showed that the TMZ conc. and time-2 had a significant impact on % EE (p < 0.05). Additionally, the chitosan conc. demonstrated a significant effect on % EE (p < 0.05) (Table S3(c)†). Increasing the CS conc. led to a decreased % EE. This could be due to the increased viscosity of CS solution with higher CS conc., which restricted loading the TMZ cargo into the CS matrix.
| Responses | R2 | Adjusted R2 | Predicted R2 | Constrains | Predicted results | Observed results |
|---|---|---|---|---|---|---|
| Particle size (nm) | 0.3538 | 0.2364 | 0.1222 | Minimize | 148.77 | 143.96 |
| PDI | 0.7384 | 0.6222 | 0.4970 | Minimize | 0.347 | 0.34 |
| % EE | 0.5639 | 0.4601 | 0.2665 | Maximize | 90.28 | 93.29 |
3.4 Conjugation of TPP+-CS conjugate
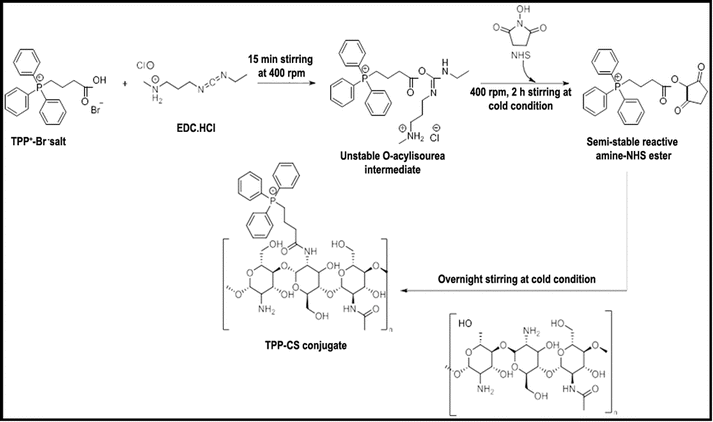
The protocol described in Scheme 1 of the chemical synthesis technique was followed in the synthesis of the TPP+-CS conjugate. TPP+ carboxylic group activation was first accomplished by interaction with coupling chemicals, specifically EDC·HCl and NHS. Following a series of steps, an amine-reactive O-acyl-isourea intermediate was formed, which later transformed into the more stable amine-reactive NHS ester. After activation, the required conjugate (TPP+-CS) was formed and linked by a stable amide bond when the NHS ester was added to an acidified aqueous solution of CS-polymer. The grafted polymer was isolated by centrifugation and subsequently subjected to lyophilization, resulting in a brownish-yellow powder.3.5 Structural validation of TPP+-CS conjugate
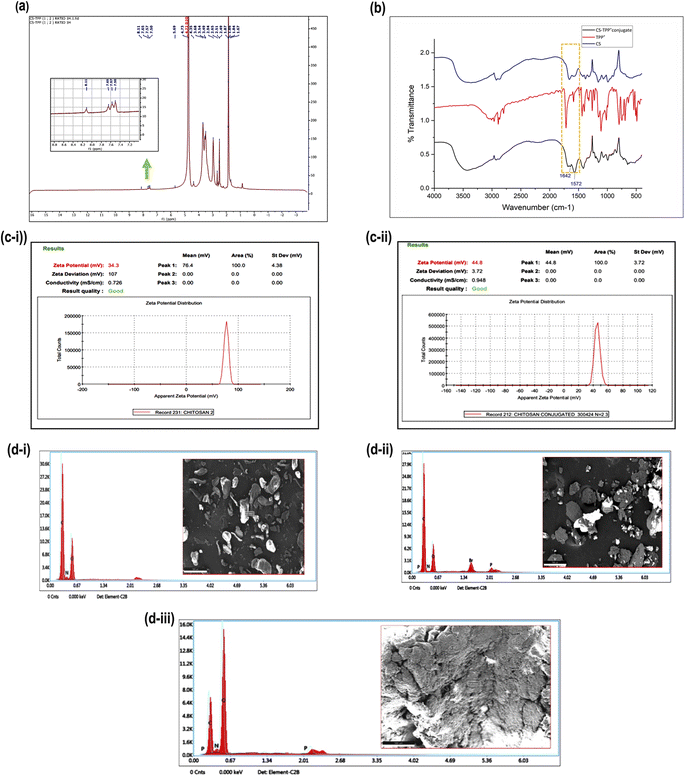
Fig. 4(a) displays the 1H NMR spectrum of TPP+-CS. The existence of signals at the aromatic area of the spectrum between 7.80 and 8.57 ppm, which are relevant to the three phenyl rings of the added triphenylphosphonium groups, confirmed conjugation. However, because of their relatively low intensity and overlap with the CS peaks at the aliphatic region of the spectrum, the methylene protons' signals in TPP+ could not be differentiated.Fig. 4(b) shows the FTIR spectra of TPP+-CS, TPP+, and CS, which range from 400 to 4000 cm−1. A prominent new absorption peak in the TPP+-CS spectra was seen at 1572 cm−1. This peak corresponds to the absorption bands that are linked to the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C bonds present in the phenyl rings that are typical of the TPP+ moiety. Additionally, the TPP+-CS spectra showed extra peaks at 1642 cm−1 compared to the CS spectrum, suggesting the effective creation of an amide bond between the amino groups of CS and the carboxylic group of TPP+.
C bonds present in the phenyl rings that are typical of the TPP+ moiety. Additionally, the TPP+-CS spectra showed extra peaks at 1642 cm−1 compared to the CS spectrum, suggesting the effective creation of an amide bond between the amino groups of CS and the carboxylic group of TPP+.
Fig. 4(c-i and ii) indicates the zeta potential of CS and CS-TPP+-conjugate is 34.3 mV and 44.8 mV, respectively. A significant interaction wherein the positively charged triphenyl phosphonium ion efficiently conjugates with chitosan is implied by the observed difference in zeta potentials between chitosan (CS) and its conjugate with triphenyl phosphonium ion (TPP+-CS).47
TPP method was developed and R2 was observed to be 0.999 and the regression equation was found to be 3.0109x + 57.32.
Once the conjugation was done, a reliable method for measuring unconjugated TPP was established using HPLC. The standard TPP+ chromatogram profile is shown in Fig. S2(a),† and the TPP linearity plot is given in Table S4,† which helps identify TPP concentrations that are not conjugated with line equation displayed in Fig. S2(b).† This mathematical knowledge enables precise conjugation efficiency calculations as a percentage, which is crucial for evaluating the efficacy of the conjugation process.
This study evaluated the percentage conjugation efficiency of TPP+ using HPLC methodology. The amount of unconjugated TPP+ in the reaction mixture was measured so that the percentage conjugation could be calculated by deducting it from the amount of TPP+ that was initially added. The resultant % conjugation for TPP+-CS was 45.04 ± 4% (n = 3, mean ± SD).
The elemental composition of TPP+, CS, and their conjugate (TPP+-CS) were analyzed using energy dispersive X-ray spectroscopy (EDAX) elemental mapping in conjunction with scanning electron microscopy (SEM) images, as shown in Fig. 4(d) The EDAX analysis verified that each structure included the expected components. In particular, the elemental analysis showed that the CS structure contains carbon, nitrogen, and oxygen (i). As predicted, TPP+ (ii) revealed the presence of carbon, nitrogen, oxygen, phosphorus, and bromine. The EDAX findings for the (iii) CS-TPP+ conjugate showed that carbon, oxygen, nitrogen, and phosphorus were present.
3.6 Preparation of TPP+-conjugated TMZ-loaded CSNPs
Conjugated nanoparticles were made utilizing the same procedure that was used to optimize the CSNPs production process by BBD. Conjugated chitosan was prepared utilizing a TPP+-CS conjugate polymer, then TMZ was added. Sodium tripolyphosphate (STPP) was utilized as the crosslinking agent. These conjugated nanoparticles were made using the same ionic gelation procedure that is utilized to prepare CSNPs.3.7 Characterization of CSNPs and conjugated TPP+-conjugated TMZ-loaded CSNPs
Prepared TMZ-loaded CSNPs and TPP+-conjugated TMZ-loaded CSNPs were evaluated for particle size, polydispersity index, zeta potential, % entrapment, and % drug loading.3.8 In vitro drug release study
TMZ solution showed almost 95% drug release (pH 5 PBS) in 1 h while TMZ-loaded CSNPs and TPP+-conjugated TMZ-loaded CSNPs showed 70% and 54% drug release after 1 h, respectively, and elicited sustained release up to 24 h. Both the NPs formulations were tested for drug release kinetics at different time intervals in pH 5 PBS. Different kinetic models such as first order, zero order, Higuchi, Korsmeyer–Peppas, Hixson–Crowell, and Weibull were used to analyze the drug release data for the optimized TMZ-loaded CSNPs and conjugated CSNPs. Both the formulations' release profiles (Fig. 6), most closely followed the Weibull model, as demonstrated by the largest correlation coefficients (R2) in Tables 8 and 9. The release of a medication from a pharmaceutical dosage form is described mathematically by the Weibull release kinetics model. In situations when conventional models, such as zero-order or first-order kinetics, would not match well, this model is very helpful in characterizing drug release patterns. By allowing for parameter flexibility, the Weibull model can describe intricate release behaviors.48 Due to their capacity to control drug release through diffusion, degradation, and swelling processes, chitosan nanoparticles display Weibull release kinetics. This implies that the drug is delivered in two stages; first, rapid release (by diffusion), and then gradual and controlled release (probably via matrix disintegration).49 For deciding the best-fit model, 2 parameters, viz; AIC and MSC were taken into consideration. Akaike Information Criterion, or AIC, is a metric for assessing the relative merits of statistical models for a particular collection of data. It offers a method for choosing models by weighing the model's complexity and goodness of fit. Since a lower AIC value suggests a balance between goodness-of-fit and model complexity, it denotes a better-fitting model. A statistic called the Model Selection Criterion (MSC) evaluates a model's goodness-of-fit relative to other models. It is beneficial in pharmacokinetics and other areas where choosing the best model is essential. A greater MSC score denotes a better model-to-data fit.50,51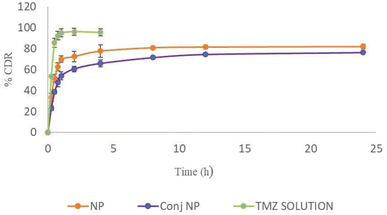 | ||
| Fig. 6 In vitro drug release profile of TMZ from NPs, conjugated NPs, and TMZ solution in pH 5 PBS at 200 rpm at 37 ± 1 °C (n = 3). | ||
| Weibull | Korsmeyer–Peppas | Higuchi | Hixson–Crowell | First order | Zero order | |
|---|---|---|---|---|---|---|
| R2 | 0.9833 | 0.8991 | −1.0499 | −1.3792 | 0.6554 | −3.103 |
| AIC | 63.4273 | 82.9821 | 117.1162 | 118.9038 | 95.7197 | 125.4434 |
| MSC | 2.6949 | 1.0653 | −1.7792 | −1.9281 | 0.0039 | −2.4731 |
| Weibull | Korsemeyer–Peppas | Higuchi | Hixson–Crowell | First order | Zero order | |
|---|---|---|---|---|---|---|
| R2 | 0.9915 | 0.928 | 0.0948 | −0.1905 | 0.4242 | −2.1551 |
| AIC | 44.3555 | 63.7604 | 87.0809 | 89.8211 | 82.5571 | 42.0267 |
| MSC | 3.5188 | 1.5783 | −0.7537 | −1.0278 | −0.3014 | −1.6370 |
3.9 Morphological assessment of NPs
Scanning electron microscopy (SEM) image of the optimized TMZ-loaded CSNPs & TPP+-conjugated TMZ-loaded CSNPs is shown in Fig. 7(a and b). The figure depicted that the particles have remarkably homogenous core structures and are spherical in shape. These nanoparticles' measured sizes, which fall between 100 and 200 nm, agree with the Z-average readings found using Zetasizer analysis.52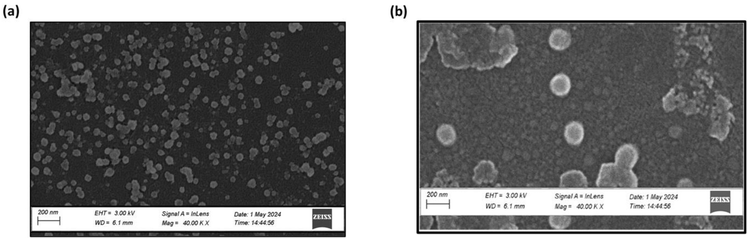 | ||
| Fig. 7 Surface morphology of (a) TMZ-loaded CSNPs, (b) TPP+-conjugated TMZ-loaded CSNPs showing uniform and spherical morphology. | ||
3.10 Stability study
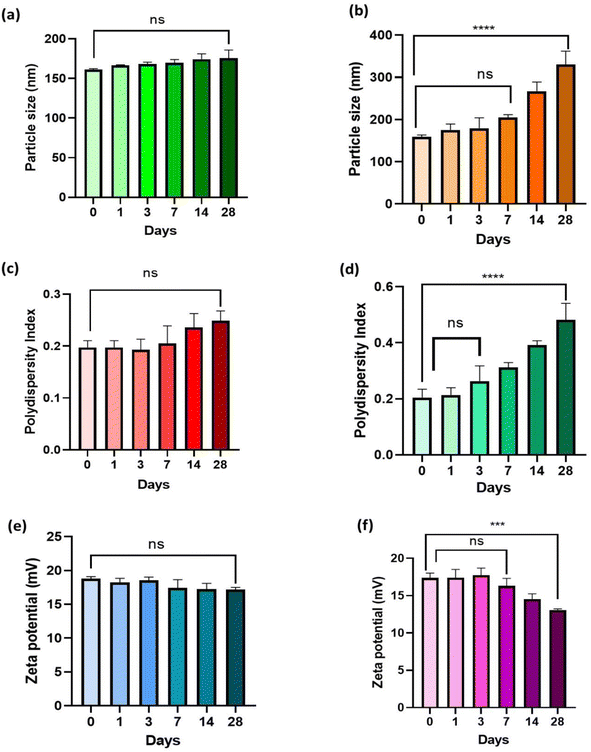
As seen in Fig. 8, the results of the stability study were evaluated in terms of zeta potential (mV), polydispersity index (PDI), and particle size (nm). The findings demonstrated that TPP+-conjugated TMZ-loaded CSNPs showed no significant difference at p > 0.05 in terms of particle size, PDI, and zeta potential at 4 ± 1 °C even after 28 days. However, at 25 ± 1 °C, particle size and PDI of TMZ-loaded CSNPs tend to increase over a period of time and a statistically significant difference was observed (p < 0.0001) from 0 to 28 days (Fig. 8(a–d)) while zeta potential decreased over 28 days and a significant difference was observed (p < 0.001) from day 0 to day 28 (Fig. 8(e and f)). The same trend was observed in the case of TMZ-loaded CSNPs stored at 25 ± 1 °C and found to be stable at 4 ± 1 °C for 28 days (Fig. S3†) (n = 3, mean ± SD).3.11 Ex vivo study
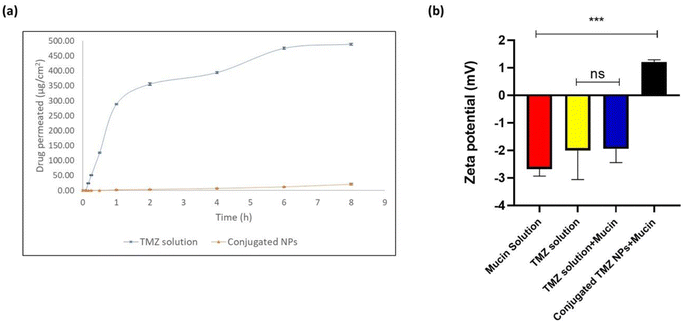
Permeation profile was obtained for TMZ solution and optimized TPP+-conjugated TMZ-loaded CSNPs on an isolated nasal mucosa. The high permeation of the drug shown by TMZ solution indicated the burst release of the surface drug. The cumulative amount of TMZ permeated from TMZ solution and optimized TPP+-conjugated TMZ-loaded CSNPs were 383.7 ± 3.5 μg and 47.54 ± 5.2 μg, respectively, after 8 h. The TMZ solution showed enhanced drug permeation of 488.84 ± 3.64 μg cm−2 while conjugated NPs showed 21.04 ± 2.33 μg cm−2 after 8 h. Consequently, the steady-state flux for TMZ solution at 8 h was also found to be almost 30 times more when compared to conjugated CSNPs (61.10 ± 0.56 μg cm−2 h−1 for TMZ solution vs. 2.62 ± 0.29 μg cm−2 h−1 for conjugated CSNPs through the nasal mucosa). The results are shown in Fig. 9(a). Chitosan nanoparticles have a high mucoadhesive property, which can cause them to adhere to the nasal mucosa, potentially slowing their permeation. Nanoparticles may be surface-modified to enhance their mucoadhesive properties and interaction with the olfactory mucosa. This modification can prolong their residence time in the nasal cavity, increasing the chances of interaction with olfactory neurons.533.12 Mucoadhesion study
The comparative zeta potential analysis of mucin solution, TMZ solution, TMZ solution + mucin, and conjugated TMZ NPs + mucin is shown in Fig. 9(b), and is suggested to reflect the interaction between mucin, TMZ solution, and conjugated NPs.The zeta potential of mucin solution, TMZ solution, TMZ solution + mucin, and conjugated TMZ NPs + mucin were found to be −2.67 ± 0.210 mV, −2.005 ± 0.858 mV, −1.94 ± 0.409 mV, and +1.21 ± 0.066 mV, respectively. The negative zeta potential of temozolomide in aqueous solutions is primarily due to its chemical structure, which includes ionizable functional groups like an amide group (–CONH), and a methyl group (–CH3) that can carry negative charges under physiological conditions. These charges affect the electric potential at the surface of the molecule or its aggregates in solution, resulting in a negative zeta potential. Further mucin solution was also found to have negative zeta potential due to sialic acid ionization. From Fig. 9(b), it was seen that there was no significant difference between TMZ solution and TMZ solution mixed with mucin implying no interaction between mucin and drug solution. On the other hand, positive zeta potential observed in the case of conjugated TMZ NPs might be due to ammonium ions (NH3+) present in chitosan coupled with positively charged mitochondriotropic moiety, i.e., TPP+ which contribute towards overall positive zeta potential.54 There was a statistically significant difference observed between mucin solution and conjugated TMZ NPs at ***p < 0.001 implying that there might be a possibility that linked conjugated chitosan nanoparticles and mucin will have a significant electrostatic interaction decreasing the overall zeta potential of mucin.55
3.13 Cytotoxicity study
IC50 value, a half maximal inhibitory concentration that kills 50% of the cell, can be used to measure the potency, safety, and efficacy of the drug, though the IC50 value is not the sole parameter to decide the potency and safety of the compound. TMZ solution, TMZ-loaded CSNPs, and TPP+-conjugated TMZ-loaded CSNPs were subjected to Alamar Blue assay for cytotoxicity. Data was processed in GraphPad Prism 8.0.2, and the IC50 value was determined using non-linear regression analysis. It was observed that the IC50 value of TMZ solution was found to be 1646 ppm after 24 h of treatment. For TMZ-loaded CSNPs formulation, the IC50 value was observed at 17.06 ppm while for TPP+-conjugated TMZ-loaded CSNPs, it was 16.05 ppm after 24 h of treatment. The overall results indicated that conjugated NPs were more cytotoxic (around 100 times) than plain TMZ solution signifying their potency in killing GB cells.3.14 ROS study
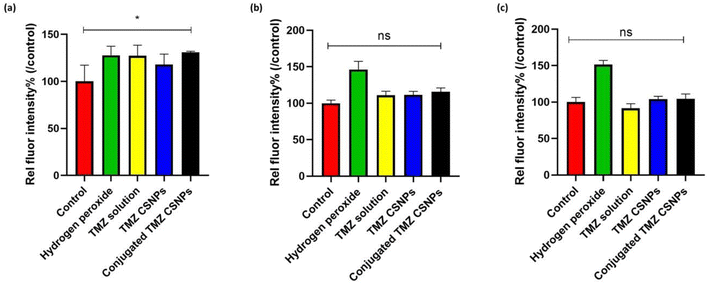
Intracellular redox homeostasis significantly influences the regulation of signaling cascades that govern apoptosis and proliferation in malignant cells. Several established chemotherapeutic agents, including TMZ, possess the capacity to induce ROS, thereby causing DNA damage.56 Our findings also demonstrated that there was a significant increase in intracellular ROS production in TPP+-conjugated TMZ-loaded CSNPs compared to a control group, after 3 h of treatment (Fig. 10(a)). However, over a period, the extent of the increase in fluorescence intensity, which is directly proportional to ROS production, decreased. (Fig. 10(b) and (c)). It is important to understand that both normal and cancer cells rely on Nrf2 transcription network to counteract oxidative stress. Nrf2 regulates a suit of antioxidant genes, ensuring redox balance. Due to its protective function, Nrf2 activity is upregulated in GB cells after TMZ exposure.57 TMZ is also known to modulate cell viability by influencing autophagy, and hypoxia-inducible factor-1α activity. Various reports suggested that chaperone-mediated autophagy (CMA) is responsible for effective TMZ treatment through the promotion of apoptosis. CMA activity is initiated by a transient surge of ROS within cytoplasm, originating from mitochondria subsequent to endoplasmic reticulum stress.58 This might be the possible reason behind the transient increase of ROS levels up to 3 h.3.15 Cellular uptake study
Qualitative drug internalization into glioblastoma cell lines, i.e., U87MG cells, was performed with rhodamine B-labeled formulation. The experiment was done after 2 h and 4 h of treatment with formulation and it was seen that fluorescence intensity of rhodamine B was increased from 2 h to 4 h of treatment (Fig. S4†). It was also observed that compared to the TMZ solution, TPP-conjugated TMZ-loaded CSNPs showed higher rhodamine B intensity in both instances, viz., 2 h and 4 h, indicating greater cellular uptake of conjugated NPs. A similar finding was reported by Mohammadian et al.464. Conclusion
Mitochondria play a crucial role in cell apoptosis, and their dysfunction is associated with numerous diseases, including cancer. This has led to the development of mitochondria-targeted drug delivery systems. While most early efforts focused on attaching targeting moieties to therapeutic or sensing molecules, nanoparticle-based systems have gained significant attention in recent years. This study reports the development and optimization of chitosan nanoparticles with temozolomide chosen as a chemotherapeutic drug. A QbD strategy was utilized with Taguchi design for screening variables followed by BBD design for optimization purposes, leading to improved entrapment efficiency, decreased particle size, and PDI. We then successfully synthesized mitochondria-targeted chitosan-TPP+ conjugated nanoparticles with carbodiimide reaction and validated them with varied techniques like FTIR, EDAX, NMR, HPLC, and zeta potential measurements. The in vitro release profile of conjugated nanoparticles in acidic pH was comparable to that of unconjugated nanoparticles suggesting a negligible impact of conjugation procedure. Stability study indicated that conjugated nanoparticles were stable at 4 °C for 28 days. Ex vivo study on goat nasal mucosa proved that conjugated CSNPs exhibited increased retention on mucosa, thereby reducing the drug permeation by virtue of stronger electrostatic interaction between negatively charged mucin and positively charged TPP and CS in conjugated nanoparticles. This finding was also supported by a mucin interaction study and evidenced by the alteration of zeta potential. In vitro cell line studies showed greater cytotoxicity potential for conjugated nanoparticles than free TMZ solution on U87MG cells. Further conjugated nanoparticles showed increased intracellular ROS generation compared to control cells, thereby proving to cause damage to malignant cells. In conclusion, this study successfully developed and optimized mitochondria-targeted CSNPs for temozolomide delivery, demonstrating enhanced nasal mucosa retention and improved physicochemical properties, stability, and cytotoxicity against glioblastoma cells offering a promising approach for glioblastoma therapy.Data availability
The data supporting this article have been included as part of the ESI.†Author contributions
TGA and Aakanchha J. conceptualized and designed this study. AD and TGA were involved in all the experimental studies and manuscript writing/editing, leading to the completion of the manuscript. TGA performed analyzed and interpreted the results. Ankit J. reviewed and edited the manuscript. Aakanchha J. reviewed the manuscript and supervised the study.Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
The authors are grateful to the Department of Pharmaceuticals, Ministry of Chemical and Fertilizers, and the Director, National Institute of Pharmaceutical Education and Research (NIPER)-Ahmedabad, for providing the necessary facilities and chemicals. Ankit Jain expresses gratitude to the Birla Institute of Technology and Science, Pilani for generous financial support in publishing this article as open access.References
- S. Agnihotri, K. E. Burrell, A. Wolf, S. Jalali, C. Hawkins, J. T. Rutka and G. Zadeh, Arch. Immunol. Ther. Exp., 2013, 61, 25–41 CrossRef CAS.
- H. Ohgaki and P. Kleihues, Acta Neuropathol., 2005, 109, 93–108 CrossRef.
- F. Gaillard, B. Mazini and C. Kearns, from https://radiopaedia.org/articles/glioblastoma-idh-wildtype, accessed on 23/10/24.
- J. Zhang, X. Xiao, J. Zhu, Z. Gao, X. Lai, X. Zhu and G. Mao, Int. J. Nanomed., 2018, 13, 3039–3051 CrossRef CAS.
- G. Iacob, E. B. Dinca and J. Med, Life, 2009, 2, 386–393 Search PubMed.
- A. F. Tamimi and M. Juweid, Exon Publ., 2017, pp. 143–153 Search PubMed.
- F. Hanif, K. Muzaffar, K. Perveen, S. M. Malhi and S. U. Simjee, Asian Pac. J. Cancer Prev., 2017, 18, 3–9 Search PubMed.
- L. Chu, A. Wanga, L. Ni, X. Yan, Y. Song, M. Zhao, K. Sun, H. Mu, S. Liu, Z. Wu and C. Zhang, Drug Delivery, 2018, 25, 1634–1641 CrossRef CAS PubMed.
- M. Zhao, D. van Straten, M. L. D. Broekman, V. Préat and R. M. Schiffelers, Theranostics, 2020, 10, 1355–1372 CrossRef CAS.
- W. Liao, S. Fan, Y. Zheng, S. Liao, Y. Xiong, Y. Li and J. Liu, Curr. Med. Chem., 2019, 26, 5862–5874 CrossRef CAS.
- T. G. Agnihotri, S. S. Gomte and A. Jain, Drug Dev. Ind. Pharm., 2022, 48, 585–601 CrossRef CAS PubMed.
- C. Janrao, S. Khopade, A. Bavaskar, S. S. Gomte, T. G. Agnihotri and A. Jain, J. Biomater. Sci., Polym. Ed., 2022, 1–62 Search PubMed.
- Y. Li, G. Deng, X. Hu, C. Li, X. Wang, Q. Zhu, K. Zheng, W. Xiong and H. Wu, Nanomedicine, 2022, 17, 1253–1279 CrossRef CAS.
- V. Peddinti, B. Rout, T. G. Agnihotri, S. S. Gomte and A. Jain, J. Liposome Res., 2023, 1–19 Search PubMed.
- J. Hou, X. Yu, Y. Shen, Y. Shi, C. Su and L. Zhao, Nanoscale Res. Lett., 2017, 12, 1–9 CrossRef CAS PubMed.
- H. Cho, Y. Y. Cho, M. S. Shim, J. Y. Lee, H. S. Lee and H. C. Kang, Biochim. Biophys. Acta, Mol. Basis Dis., 2020, 1866, 165808 CrossRef CAS PubMed.
- G. van Loo, X. Saelens, M. van Gurp, M. MacFarlane, S. J. Martin and P. Vandenabeele, Cell Death Differ., 2002, 9, 1031–1042 CrossRef CAS.
- R. W. Horobin, S. Trapp and V. Weissig, J. Controlled Release, 2007, 121, 125–136 CrossRef CAS.
- N. A. Emad, B. Ahmed, A. Alhalmi, N. Alzobaidi and S. S. Al-Kubati, J. Drug Delivery Sci. Technol., 2021, 64, 102642 CrossRef CAS.
- P. Shah, P. Dubey, B. Vyas, A. Kaul, A. K. Mishra, D. Chopra and P. Patel, Artif. Cells, Nanomed., Biotechnol., 2021, 49, 511–522 CrossRef CAS.
- O. Esim, A. Savaser, C. K. Ozkan, A. Oztuna, B. A. Goksel, M. Ozler, C. Tas and Y. Ozkan, J. Drug Delivery Sci. Technol., 2020, 59, 101919 CrossRef CAS.
- K. M. Noorulla, M. Yasir, F. Muzaffar, R. S, M. M. Ghoneim, A. S. Almurshedi, A. J. Tura, S. Alshehri, T. Gebissa, S. Mekit, M. M. Ahmed and A. Zafar, J. Drug Delivery Sci. Technol., 2022, 67, 102939 CrossRef CAS.
- M. Z. Ahmad, A. H. Bin Sabri, Q. K. Anjani, J. Domínguez-Robles, N. Abdul Latip and K. A. Hamid, Pharmaceuticals, 2022, 15, 370 CrossRef CAS PubMed.
- T. G. Agnihotri, G. S. Jadhav, B. Sahu and A. Jain, Drug Delivery Transl. Res., 2022, 12, 3104–3120 CrossRef PubMed.
- A. R. Khan, M. Liu, M. W. Khan and G. Zhai, J. Controlled Release, 2017, 268, 364–389 CrossRef CAS PubMed.
- J. Hou, X. Yu, Y. Shen, Y. Shi, C. Su and L. Zhao, Nanoscale Res. Lett., 2017, 1–9 Search PubMed.
- B. Gürten, E. Yenigül, A. D. Sezer and S. Malta, Braz. J. Pharm. Sci., 2018, 54, e17513 Search PubMed.
- M. Gitte, S. Sudhakar Gomte, T. Girish Agnihotri, N. Singh and A. Jain, J. Mol. Liq., 2023, 374, 121281 CrossRef CAS.
- A. Pant and J. S. Negi, Eur. J. Pharm. Sci., 2018, 112, 180–185 CrossRef CAS.
- T. Shinde, T. G. Agnihotri, S. S. Gomte, N. Sharma and A. Jain, Bionanoscience, 2024, 1–14 Search PubMed.
- N. Sreeharsha, K. Rajpoot, M. Tekade, D. Kalyane, A. B. Nair, K. N. Venugopala and R. K. Tekade, Pharmaceutics, 2020, 12, 920 CrossRef CAS PubMed.
- W. K. Delan, M. Zakaria, B. Elsaadany, A. N. ElMeshad, W. Mamdouh and A. R. Fares, Int. J. Pharm., 2020, 577, 119038 CrossRef CAS PubMed.
- H. Huang, H. Wu, Y. Huang, S. Zhang, Y. Lam and N. Ao, J. Mater. Res., 2018, 33, 2586–2597 CrossRef CAS.
- K. K. Arafa, M. A. Hamzawy, S. A. Mousa and I. M. El-Sherbiny, RSC Adv., 2022, 12, 21690–21703 RSC.
- H. Huang, H. Wu, Y. Huang, S. Zhang, Y. Lam and N. Ao, J. Mater. Res., 2018, 33, 2586–2597 CrossRef CAS.
- S. Salave, S. D. Shinde, D. Rana, B. Sahu, H. Kumar, R. Patel, D. Benival and N. Kommineni, Pharmaceutics, 2023, 15, 608 CrossRef CAS PubMed.
- T. G. Agnihotri, D. Badgujar, N. Sharma and A. Jain, Chromatographia, 2023, 86, 295–307 CrossRef CAS.
- S. Ahmad, I. Khan, J. Pandit, N. A. Emad, S. Bano, K. I. Dar, M. M. A. Rizvi, M. D. Ansari, M. Aqil and Y. Sultana, Int. J. Biol. Macromol., 2022, 221, 435–445 CrossRef CAS PubMed.
- R. Fernández-Urrusuno, P. Calvo, C. Remuñán-López, J. L. Vila-Jato and M. José Alonso, Pharm. Res., 1999, 16, 1576–1581 CrossRef.
- M. Gitte, S. Sudhakar Gomte, T. Girish Agnihotri, N. Singh and A. Jain, J. Mol. Liq., 2023, 374, 121281 CrossRef CAS.
- S. Zameer, J. Ali, D. Vohora, A. K. Najmi and M. Akhtar, J. Drug Targeting, 2021, 29, 199–216 CrossRef CAS.
- T. Banerjee, S. Mitra, A. Kumar Singh, R. Kumar Sharma and A. Maitra, Int. J. Pharm., 2002, 243, 93–105 CrossRef CAS PubMed.
- International Council for Harmonisation, ICH Harmon. Tripart. Guidel., 2003, vol. 4, p. 24 Search PubMed.
- G. S. El-Feky and G. M. Zayed, Int. J. Appl. Pharm., 2019, 11, 106–112 CrossRef CAS.
- A. M. Silva, H. L. Alvarado, G. Abrego, C. Martins-Gomes, M. L. Garduño-Ramirez, M. L. García, A. C. Calpena and E. B. Souto, Pharmaceutics, 2019, 11, 1–11 Search PubMed.
- J. Mohammadian, S. Mahmoudi, P. Pourmohammad and M. Pirouzpanah, Naunyn-Schmiedebergs Arch. Pharmakol., 2020, 393, 2315–2323 CrossRef CAS.
- N. A. Edwards, G. Fisher, G. H. Harris and N. Kellichan, J. Chromatogr. A, 2014, 1323, 49–56 CrossRef CAS.
- C. Corsaro, G. Neri, A. M. Mezzasalma and E. Fazio, Polymers, 2021, 13, 2897 CrossRef CAS.
- S. Dash, P. N. Murthy, L. Nath and P. Chowdhury, Acta Pol. Pharm., 2010, 67, 217–223 CAS.
- H. Akaike, J. Econ., 1981, 16, 3–14 Search PubMed.
- I. Lopes Alves, D. Vállez García, A. Parente, J. Doorduin, R. Dierckx, A. M. Marques da Silva, M. Koole, A. Willemsen and R. Boellaard, EJNMMI Res., 2017, 7, 17 CrossRef PubMed.
- S. Soleymanfallah, Z. Khoshkhoo, S. E. Hosseini and M. Azizi, Food Sci. Nutr., 2022, 10, 3272–3281 CrossRef CAS PubMed.
- L. Illum, Eur. J. Pharm. Sci., 2000, 1, 1–18 CrossRef.
- D. Vllasaliu, R. Exposito-Harris, A. Heras, L. Casettari, M. Garnett, L. Illum and S. Stolnik, Int. J. Pharm., 2010, 400, 183–193 CrossRef CAS PubMed.
- E. M. A. Hejjaji, A. M. Smith and G. A. Morris, Int. J. Biol. Macromol., 2018, 120, 1610–1617 CrossRef CAS PubMed.
- M. Seyfrid, V. Marschall and S. Fulda, Anticancer Drugs, 2016, 27, 953–959 CrossRef CAS PubMed.
- J. A. Campos-Sandoval, M. C. Gómez-García, J. de los Santos-Jiménez, J. M. Matés, F. J. Alonso and J. Márquez, Neurochem. Int., 2021, 149, 105136 CrossRef CAS PubMed.
- A. Lo Dico, D. Salvatore, C. Martelli, D. Ronchi, C. Diceglie, G. Lucignani and L. Ottobrini, Cells, 2019, 8, 1–17 CrossRef.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4ra04748f |
| ‡ These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2024 |