Open Access Article
Open Access ArticleSynthesis, in vitro biological evaluation and in silico studies of novel pyrrolidine derived thiosemicarbazones as dihydrofolate reductase inhibitors†
Hina Aftaba,
Saeed Ullahb,
Ajmal Khanbc,
Mariya al-Rashidad,
Talha Islamd,
Abdulrahman Alshammarie,
Norah A. Albekairie,
Parham Taslimi f,
Ahmed Al-Harrasi*b,
Zahid Shafiq
f,
Ahmed Al-Harrasi*b,
Zahid Shafiq *a and
Saeed Alghamdig
*a and
Saeed Alghamdig
aInstitute of Chemical Sciences, Bahauddin Zakariya University, Multan-60800, Pakistan. E-mail: zahidshafiq@bzu.edu.pk
bNatural and Medical Sciences Research Centre, University of Nizwa, P.O. Box 33, PC 616, Birkat Al Mauz, Nizwa, Sultanate of Oman. E-mail: aharrasi@unizwa.edu.om
cDepartment of Chemical and Biological Engineering, College of Engineering, Korea University, 145 Anam-ro, Seongbuk-gu, Seoul 02841, Republic of Korea
dDepartment of Chemistry, Forman Christian College (A Chartered University) Lahore, Pakistan
eDepartment of Pharmacology and Toxicology, College of Pharmacy, King Saud University, Post bezBox 2455, Riyadh, 11451, Saudi Arabia
fDepartment of Biotechnology, Faculty of Science, Bartin University, 74100 Bartin, Turkey
gDepartment of Pharmacy, Riyadh Security Forces Hospital, Ministry of Interior, Kingdom of Saudi Arabia
First published on 8th October 2024
Abstract
Dihydrofolate reductase (DHFR) is a crucial enzyme involved in folate metabolism and serves as a prime target for anticancer and antimicrobial therapies. In this study, a series of 4-pyrrolidine-based thiosemicarbazones were synthesized and evaluated for their DHFR inhibitory activity. The synthesis involved a multistep procedure starting from readily available starting materials, leading to the formation of diverse thiosemicarbazone 5(a–r) derivatives. These compounds were then subjected to in vitro assays to evaluate their inhibitory potential against DHFR enzyme. The synthesized compounds 5(a–r) exhibited potent inhibition with IC50 values in the range of 12.37 ± 0.48 μM to 54.10 ± 0.72 μM. Among all the derivatives 5d displayed highest inhibitory activity. Furthermore, molecular docking and ADME studies were performed to understand the binding interactions between the synthesized compounds and the active site of DHFR. The in vitro and in silico data were correlated to identify compounds with promising inhibitory activity and favorable binding modes. This comprehensive study provides insights into the structure–activity relationships of 4-pyrrolidine-based thiosemicarbazones as DHFR inhibitors, offering potential candidates for further optimization towards the development of novel therapeutic agents.
1. Introduction
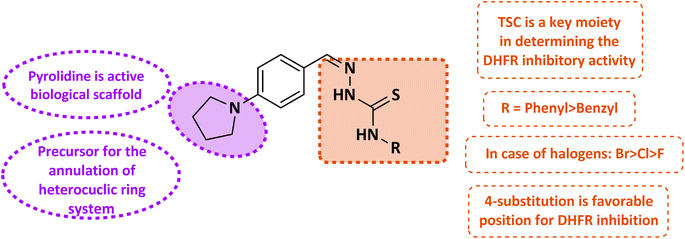
Folate metabolism has emerged as a promising and significant target for the creation of anti-cancer, anti-bacterial, and anti-parasitic therapeutic medicines in the last several years.1 Dihydrofolate reductase (DHFR) plays a crucial role in reducing 7,8-dihydrofolate (DHF) to 5,6,7,8-tetrahydrofolate (THF) in cooperation with NADPH.2 The production of thymidine (a precursor for DNA replication), purine nucleotides, glycine, methionine, serine, and N-formyl-methionyl tRNA all rely on this chemical as a precursor to the cofactors needed for these reactions.3 Cell development is restricted due to partial depletion of intracellular reduced folates caused by DHFR inhibition.4 Antibacterial DHFR inhibitors work by preventing the production of DNA, RNA, and proteins, which in turn stops the proliferation of cells. Consequently, DHFR becomes a prime target for anticancer and antibacterial drugs. A decrease in DHFR enzyme activity lowers the intracellular THF pool, which impacts the concentration of folate coenzymes and, by extension, the production of purines and pyrimidines.5,6 Since methyl-THF is required for homocysteine re-methylation to produce methionine, an essential component of S-adenosylmethionine (SAM) required for the majority of biological methylation reactions, this may also affect homocysteine levels and methylation processes. Higher eukaryotic cells must rely on exogenous folic acid because, unlike prokaryotes and lower eukaryotic cells, they are unable to produce folates de novo. So, to acquire medications to treat infectious disorders caused by protozoan parasites and bacteria, the folic acid production route is an easy target.7 Several DHFR inhibitors, as separate entities, have found clinical utility as antitumor agents.8–10 One of the most potent DHFR inhibitor methotrexate (MTX) is one the first chemotherapeutic agent discovered and is still a support in single agent and combination cancer chemotherapy.11One of the nitrogen heterocycles utilized extensively by medicinal chemists to derive molecules for the treatment of human diseases is the five-membered pyrrolidine ring. This saturated scaffold is of great interest for three reasons: (1) the ability to efficiently explore the pharmacophore space via sp3-hybridization; (2) the impact on the molecule's stereochemistry; and (3) the increased three-dimensional (3D) coverage caused by the ring's non-planarity; a phenomenon known as “pseudo rotation”.12 Additionally, the nitrogen atom of pyrrolidine ring has the capability to engage in hydrogen bonding interactions with amino acids residues present within active site of DHFR, thereby enhancing the compound's binding affinity. Its size and shape are well-suited for fitting into the active site pocket of DHFR. This facilitates favorable van der Waals interactions with nearby amino acids residues, which in turn enhances the stability of the enzyme–inhibitor complex.13 The incorporation of heteroatomic fragments into the molecules is a thoughtful decision, as they serve as valuable tools for adjusting physicochemical parameters and achieving optimal ADME outcomes for potential drug candidates.14,15
Thiosemicarbazones are a fascinating class of compounds. Numerous pharmacological and biological features have been discovered including their efficacy as anti-urease,16,17 chemo-sensing,18 antibacterial,19 antifungal,20 anticancer,21 antifolate,22,23 antidiabetic activity24 and antimicrobial activity.25 On the other hand, thiosemicarbazones also belong to one of the main families of DHFR (dihydrofolate reductase) inhibitors26,27 due to the formation of stable complexes with zinc ions present in the active side of DHFR.28,29 As thiosemicarbazones are known as chelate metal ions, this chelation helps in binding and inhibiting the enzyme. Furthermore, thiosemicarbazones possess a thioamide (–CSNH–) functional group, which may also participate in hydrogen bonding interactions with specific amino acids residues in the DHFR active site, these interactions helps to stabilize the inhibitor in the enzyme's binding pocket, enhancing its potency.28,30 The structural characteristics and redox activities of thiosemicarbazones contribute to their efficacy as anticancer agents, making them promising candidates for further exploration as DHFR inhibitors.31
Molecular hybridization involves merging different molecules to create new compounds. In our study, we synthesized novel 4-pyrrolidine based thiosemicarbazones by combining 4-pyrrolidinyl-benzaldehyde with thiosemicarbazide derivatives. These compounds were then evaluated for their effectiveness against the enzyme DHFR. Additionally, we conducted molecular docking, ADMET studies, and explored structure–activity relationships to understand the properties and potential of these compounds as DHFR inhibitors.
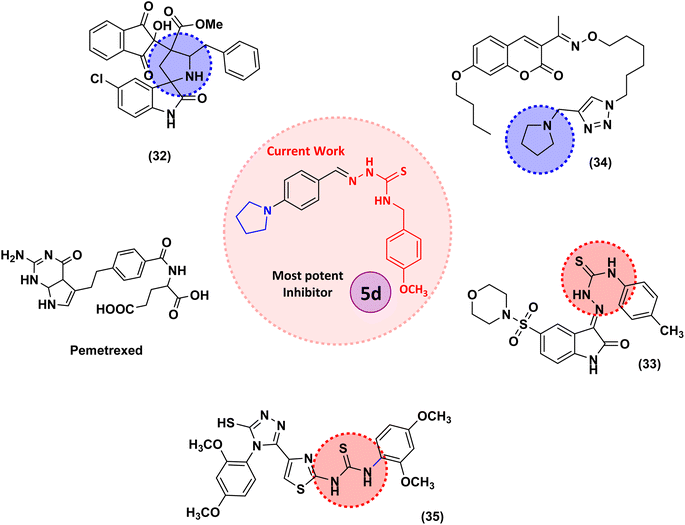
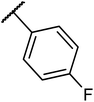
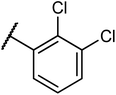
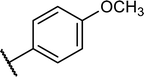
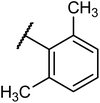
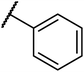
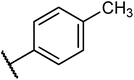
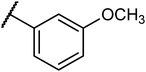
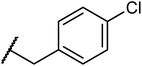
Structures of some pyrrolidine and thiosemicarbazones based dihydrofolate reductase (DHFR) inhibitors reported in literature are shown in (Fig. 1).32–35
2. Results and discussion
2.1 Chemistry
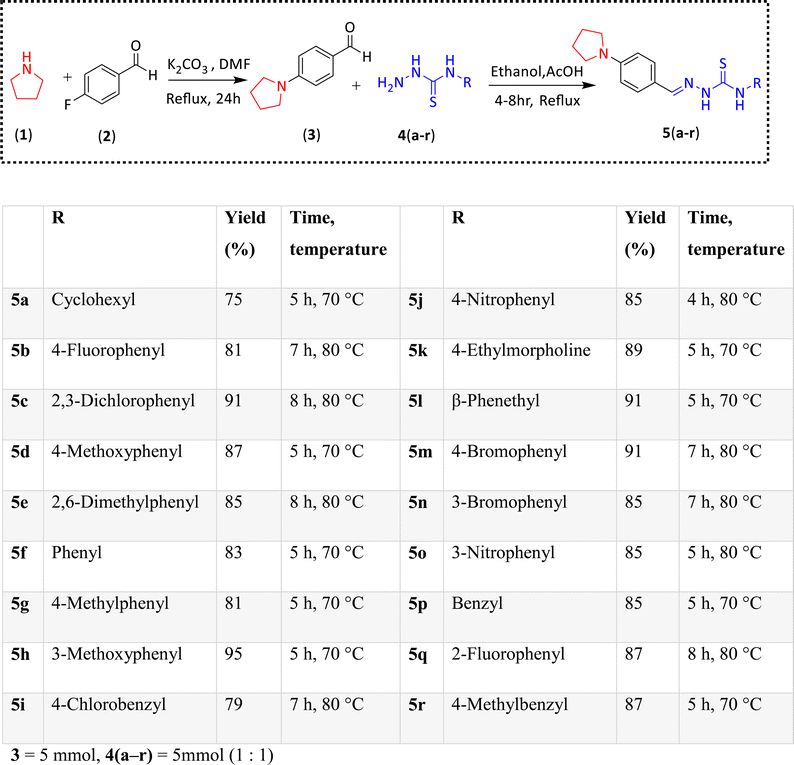
Pyrrolidine and thiosemicarbazone moieties have a wide range of biological and pharmacological effects. Therefore, it is reasonable to integrate these two components into a unified molecular framework to create more potent biologically active compounds, such as those depicted in Fig. 1. The desired compounds were synthesized by modifying pyrrolidine moiety through the introduction of 4-fluorobenzaldehyde. This modification primarily involves replacing the NH group of pyrrolidines with 4-fluorobenzaldehyde instead of the hydrogen atom at the N–H position of pyrrolidine, and forming thiosemicarbazones by reacting with thiosemicarbazides, thus enhancing their biological activity. The targeted compounds 5(a–r) were prepared by the substitution at N–H position which was done by the reaction of pyrrolidine (1) with 4-fluorobenzaldehyde (2) using anhydrous K2CO3 and DMF at 90 °C for 12 h, giving 4-(pyrrolidin-1-yl)benzaldehyde (3). The synthesis of thiosemicarbazones was achieved by reaction of compound (3) with thiosemicarbazides 4(a–r). The synthetic route in synthesis of 4-pyrrolidine based thiosemicarbazones 5(a–r) was displayed in Scheme 1.The structure of synthesized compounds was confirmed using various advanced spectroscopic techniques. In the 1H-NMR spectra of the pyrrolidine-based thiosemicarbazones, a prominent single peak appeared within the chemical shift range of δ 11.9–10.1 ppm, revealing of NH-protons presence. Furthermore, the protons of N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH were prominent between δ 7.95 and 9.13 ppm. Notably, C–H protons in the benzene moiety exhibited distinct signals within the range of δ 7.0–8.0 ppm, while those in the pyrrolidine group were identified within δ 1.0–5.0 ppm. Furthermore, negative signals observed in 13C-NMR spectra suggested the presence of CH2 groups in the compounds. Other characteristic signals were detected at δ 23.5, 33.9, 175.8 ppm, corresponding to CH3 and CH, as well as C
CH were prominent between δ 7.95 and 9.13 ppm. Notably, C–H protons in the benzene moiety exhibited distinct signals within the range of δ 7.0–8.0 ppm, while those in the pyrrolidine group were identified within δ 1.0–5.0 ppm. Furthermore, negative signals observed in 13C-NMR spectra suggested the presence of CH2 groups in the compounds. Other characteristic signals were detected at δ 23.5, 33.9, 175.8 ppm, corresponding to CH3 and CH, as well as C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) S functionalities. In order to determine the purity of compounds, HPLC analysis was carried out using CH3CN
S functionalities. In order to determine the purity of compounds, HPLC analysis was carried out using CH3CN![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O = 80
H2O = 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 eluent system with 263 nm wavelength. All the compounds exhibited great than 95% purity.
20 eluent system with 263 nm wavelength. All the compounds exhibited great than 95% purity.
2.2 Pharmacology
Compound 5j was the least potent member of the series with IC50 value of 54.10 ± 0.72 μM.
2.3 Structure–activity relationship
Thiosemicarbazones exhibit the remarkable ability to hinder the activity of the DHFR enzyme, thereby disrupting folate metabolism pathways crucial for DNA synthesis and cellular proliferation. This exceptional characteristic renders thiosemicarbazones promising candidates for anti-cancer and anti-microbial therapies. A novel series of thiosemicarbazones, based on 4-pyrrolidinyl-benzaldehyde, has been synthesized and assessed for their efficacy against the dihydrofolate reductase enzyme. The IC50 values and percentage inhibition of these synthesized compounds were determined and is presented in Table 1. Thiosemicarbazone and 4-pyrrolidinylbeznaldehyde functioned as significant structural frameworks, and the phenyl or benzyl group bonded to the N4 nitrogen of thiosemicarbazone was crucial in establishing an indisputable structure–activity link. The inhibitory potential of thiosemicarbazone is explored by changing the R group on the thiosemicarbazides moiety. The R group is varied with aromatic, non-aromatic and aliphatic groups. The compounds 5f and 5p with phenyl and benzyl rings respectively, by comparison, phenyl derivative has more inhibition potential than benzyl derivative. In case of compound 5l and 5d with phenethyl and 4-methoxyphenyl moieties showed maximum inhibition potencies. Compounds 5j and 5o with para and meta-nitro group on the phenyl ring, among them meta-substitution is slightly active (5o, IC50 = 45.28 ± 0.63 μM) as compared to para substitution (5j, IC50 = 54.10 ± 0.72 μM). Compounds 5d and 5h containing methoxy substitution at the para and meta positions of the phenyl ring respectively, 5d showed much better inhibitory potency with IC50 value 12.37 ± 0.48 μM of than 5h with IC50 value of 23.11 ± 0.35 μM. Electron donating substituents, like alkyl groups, have the ability to increase the electron density around the benzene ring through resonance effects. This amplified electron density raises stronger interactions between the inhibitor molecule and the enzymes active site, potentially leading to more potent inhibition. Moreover, the positive inductive effect (+I) exerted by alkyl groups on the benzene rings further intensifies its electron density. This phenomenon enhances the inhibitor's capacity to engage with the enzyme's active site. Therefore, with increase in alkyl substitution inhibition potential is increased as we see in compound 5e when compared with 5g and 5r. In case of methyl substitutions at para position of phenyl and benzyl rings of compound 5g and 5r with IC50 = 14.37 ± 0.29 μM and IC50 = 42.16 ± 0.63 μM respectively, 5g is more potent as compared to 5r. The compound 5e with dimethyl substitution (IC50 = 15.30 ± 0.26 μM) slightly increases inhibition potency than one methyl substitution. In case of halogens, with the increase in number of substitutions, potency is somewhat decreased, possibly due to electron-withdrawing effect. This effect makes any carbon center even more electron-deficient than before also causes negative inductive effect (−I). Their activity order is Br > Cl > F. Compounds 5m and 5n with bromo substitution at para and meta positions of phenyl ring respectively, among them 5m (IC50 = 16.27 ± 0.26 μM) is more potent than 5n (IC50 = 22.06 ± 0.37 μM). Compounds 5c, and 5i with 2,3-dichlorophenyl and 4-chlorobenzyl exhibited moderate inhibition with IC50 = 36.20 ± 0.69, IC50 = 27.46 ± 0.50 respectively. While in case of cyclohexyl containing compound 5a moderate activity against enzyme was observed (IC50 = 29.11 ± 0.38 μM).The study reveals a clear correlation between the chemical structure of the compounds and their inhibitory potency, as showed in Fig. 2. Notably, compounds featuring para substitution of the phenyl ring exhibit heightened potency in the series. Among these, compounds 5d and 5l, distinguished by 4-methoxyphenyl and phenethyl substitution, emerge as most potent in the series. Conversely, compound 5j, with 4-nitro phenyl substitution, displays diminished activity. It becomes evident that the introduction of electron-withdrawing substituents on the phenyl ring leads to decreased inhibitory potential, whereas electron-donating substitutions enhance activity. This highlights the crucial role of understanding the electronic characteristics of substituents in modulating biological activity, thereby facilitating the rational design of more effective compounds.
2.4 Molecular docking studies
The crystal structure of human dihydrofolate reductase (DHFR) in complex with the NADP and inhibitor SRI-9439 was downloaded from the PDB (PDB id: 1kms, 1.09 Å). To validate the docking protocol, the co-crystallized inhibitor was re-docked, the docking method was able to successfully reproduce the experimentally observed conformation for the ligand. The NADPH co-factor was retained for the docking studies in accordance with previous studies.36 All compounds (5a–r) were docked and were found to bind in the same area of the active site as the co-crystallized inhibitor (Fig. 3). For reference, molecular docking of methotrexate was also carried out to map out important interactions with the binding site amino acids. Details of binding site interactions are given in Table 2.| Code | Hydrogen bond | Distance (Å) | Hydrophobic | Other | ||
|---|---|---|---|---|---|---|
| Alkyl | Pi-alkyl | Pi–pi stacked | ||||
| 5a | SER59 | 1.74 | ALA9 | PHE34 | ||
| ASP21 | 2.1 | NDP202 | LEU22 | |||
| SER59 | 2.1 | ILE60 | ||||
| NDP202 | ||||||
| 5b | VAL115 | 1.9 | PRO61 | LEU22 | PHE31 | VAL8 halogen (fluorine) |
| ASP21 | 2.67 | PRO61 | PHE34 | GLU30 halogen (fluorine) | ||
| ILE7 | SER59 pi-lone pair | |||||
| ALA9 | ||||||
| 5c | PHE31 | 2.74 | ILE60 | PHE34 | PHE31 | |
| PHE31 | 2.91 | VAL115 | LEU22 | |||
| ILE60 | ||||||
| 5d | SER59 | 2.12 | VAL115 | PHE34 | PHE31 | |
| ILE7 | 3.06 | NDP202 | LEU22 | PHE34 | ||
| VAL115 | 2.71 | NDP202 | ||||
| NDP202 | 2.97 | |||||
| 5e | LEU22 | 2.43 | PRO26 | ILE60 | PHE31 | |
| SER59 | 1.86 | PRO61 | PRO61 | |||
| ASP21 | 1.74 | PRO61 | ||||
| ASN64 | 3.03 | |||||
| 5f | SER59 | 2.14 | LEU22 | PHE31 | ||
| PHE31 | 2.58 | NDP202 | ||||
| 5g | SER59 | 2.15 | ARG32 | PHE34 | PHE31 | |
| NDP202 | LEU22 | PHE34 | ||||
| 5h | VAL115 | 2.1 | PRO26 | PHE34 | PHE31 | |
| PRO61 | LEU22 | |||||
| ILE7 | ILE60 | |||||
| PRO61 | ||||||
| ALA9 | ||||||
| NDP202 | ||||||
| 5i | VAL115 | 2.03 | ALA9 | PHE31 | ||
| ASN64 | 2.08 | LEU22 | TRP24 | PHE34 (pi–pi T-shaped) | ||
| ASN64 | 1.79 | TRP24 | ||||
| PHE31 | ||||||
| PHE31 | ||||||
| ALA9 | ||||||
| LEU22 | ||||||
| NDP202 | ||||||
| 5j | THR136 | 2.31 | PRO61 | LEU22 | PHE34 | GLU30 attractive charge |
| lig:H33 – VAL115:O | 2.16 | PRO61 | SER59 pi-lone pair | |||
| PHE34:HA:O25 | 2.96 | ILE7 | ||||
| ALA9 | ||||||
| 5k | ASP21 | 1.99 | LEU67 | ILE60 | ASP21 salt bridge; attractive charge | |
| SER59 | 2.17 | |||||
| ASP21 | 3.01 | |||||
| SER59 | 1.94 | |||||
| ASP21 | 2.37 | |||||
| 5l | SER59 | 1.66 | ILE60 | PHE31 | ||
| ASP21 | 1.64 | PRO61 | ||||
| PRO26 | ||||||
| PRO61 | ||||||
| 5m | VAL115 | 1.9 | ALA9 | LEU22 | PHE31 | SER59 pi-lone pair |
| ASP21 | 2.6 | PRO61 | PRO61 | PHE34 | ||
| ILE7 | ILE7 | |||||
| ALA9 | ||||||
| 5n | SER59 | 1.76 | LEU22 | PHE31:Br24 | PHE31 | |
| PHE31 | 2.33 | ILE60 | PHE34 (pi–pi T-shaped) | |||
| LEU22 | ||||||
| NDP202 | ||||||
| 5o | ASP21 | 1.94 | VAL115 | PHE34 | ||
| VAL115 | 2.68 | NDP202 | LEU22 | |||
| LEU22 | 3.17 | ILE60 | ||||
| NDP202 | ||||||
| LEU22 | ||||||
| PRO26 | ||||||
| 5p | ASP21 | 1.94 | VAL115 | PHE34 | ||
| VAL115 | 2.68 | NDP202 | LEU22 | |||
| LEU22 | 3.17 | ILE60 | ||||
| NDP202 | ||||||
| LEU22 | ||||||
| PRO26 | ||||||
| 5q | SER59 | 1.73 | ILE60 | PHE31 | THR56 halogen (fluorine) | |
| ILE60 | 1.92 | LEU22 | ||||
| PHE31 | 2.85 | ILE60 | ||||
| NDP202 | ||||||
| 5r | PRO61 | 2.72 | LEU67 | PHE34 | ||
| ILE60 | ILE60 | |||||
| LEU22 | ||||||
| ILE60 | ||||||
| NDP202 | ||||||
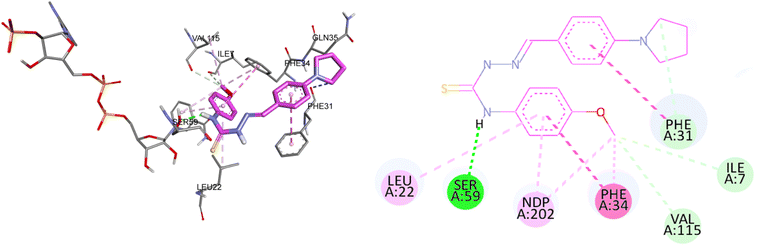
Compounds 5d, the most active inhibitor of hDHFR was selected for in depth analysis of binding site interactions (Fig. 4). One of the NH groups was making a hydrogen bond with Ser59. Two carbon hydrogen bonds were observed between methoxy carbon and Val115 and Ile7, the same methoxy carbon was also making alkyl interactions with Val115 and NADP, and pi alkyl interaction with Phe34. The methoxy substituted phenyl ring was making pi alkyl contacts with the NADP and Leu22 and a pi–pi stacked contact with Leu34. While the other phenyl ring was making a pi–pi stacked interaction with Phe31.
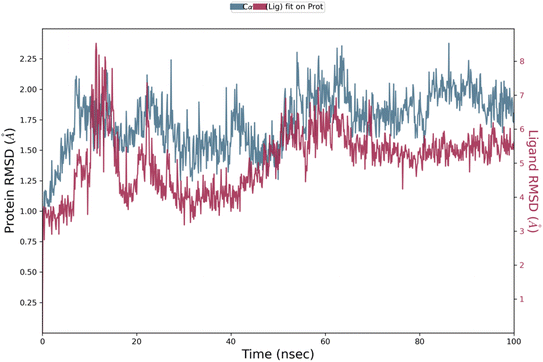
2.5 Molecular dynamics simulation studies
Analysis of protein–ligand RMSD show that the complex remained stable throughout the 100 ns simulation time period. The Cα protein RMSD value remained between 1.0 Å to 2.3 Å during the whole simulation mostly around 2.0 Å during the last 50 ns of simulation (Fig. 5). In ligand fit on protein RMSD, the value varied during the first 25 ns up-to 8.0 Å, then remained stable between 5.0 Å to 6.0 Å and stabilized around 5.5 Å towards the end of simulation. In protein RMSF, Cα protein all residues showed value less than 3.0 Å (Fig. 6). Protein secondary structure was % total SSE at 43.78, while α-helix was 14.58 and β-strand at 29.21. Ligand RMSF values of fit ligand on protein showed all atoms have RMSF values between 2–3 Å where pyrrolidine moiety and methyl of anisole moiety showed relatively higher RMSF values as compared to other atoms in the ligand (ESI Fig. 1†). Protein–ligand contacts showed GLY 20 showed mostly hydrogen bonds and water bridges interactions with the highest interaction fraction around 0.6. Ligand–protein contacts showed nitrogen from thiosemicarbazide moiety formed interactions with GLY 20 for 55% of simulation time (ESI Fig. 2†). Ligand torsion profile showed most fluctuation and movement in the bonds of pyrrolidine and anisole moieties in the compound while thiosemicarbazide moiety remained relatively stable in bonds torsion (ESI Fig. 3†).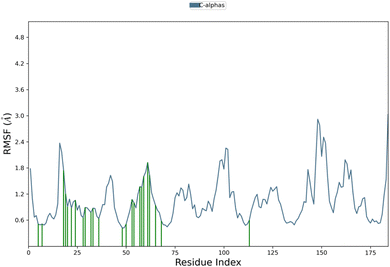 | ||
| Fig. 6 Protein RMSF graph of 5d complexed with hDHFR. Residues that interact with the ligand are shown in green vertical bars. | ||
2.6 ADME studies
In silico ADME evaluation of all compounds 5a–r was carried out using Swiss ADME. Overall, all compounds indicated favorable ADME profile as indicated by several physicochemical parameters given in (Table 3). Oral bioavailability of a compound is represented in the form of a bioavailability radar diagram based on parameters such lipophilicity, size, polarity, solubility, saturation rate and molecular flexibility. All compounds were predicted to have good oral bioavailability, as a selected example radar diagram of most active compound 5d is shown in (Fig. 7). All compounds were predicted to have good to moderate water solubility, and with high gastrointestinal (GI) absorption. Interestingly most of the compounds were predicted to cross the blood–brain barrier (BBB). This ability to cross BBB, coupled with potent DHFR inhibition activity can be tailored for the design of novel anticancer drugs targeting brain tumors.| Code | MW | RB | HBA | HBD | TPSA | log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P P |
log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) S S |
GI | BBB | B score |
|---|---|---|---|---|---|---|---|---|---|---|
| 5a | 330.49 | 6 | 1 | 2 | 71.75 | 3.53 | −4.60 | High | Yes | 0.55 |
| 5b | 343.43 | 6 | 2 | 2 | 71.75 | 3.70 | −5.95 | High | No | 0.55 |
| 5c | 393.33 | 6 | 1 | 2 | 71.75 | 4.45 | −6.87 | High | Yes | 0.55 |
| 5d | 354.47 | 7 | 2 | 2 | 80.98 | 3.32 | −5.79 | High | No | 0.55 |
| 5e | 352.50 | 6 | 1 | 2 | 71.75 | 4.01 | −5.73 | High | Yes | 0.55 |
| 5f | 324.44 | 6 | 1 | 2 | 71.75 | 3.38 | −5.68 | High | Yes | 0.55 |
| 5g | 338.47 | 6 | 1 | 2 | 71.75 | 3.72 | −6.06 | High | Yes | 0.55 |
| 5h | 354.47 | 7 | 2 | 2 | 80.98 | 3.38 | −5.79 | High | No | 0.55 |
| 5i | 372.91 | 7 | 1 | 2 | 71.75 | 3.95 | −6.67 | High | Yes | 0.55 |
| 5j | 369.44 | 7 | 3 | 2 | 117.57 | 2.98 | −5.51 | High | No | 0.55 |
| 5k | 361.50 | 8 | 3 | 2 | 84.22 | 1.99 | −4.14 | High | No | 0.55 |
| 5l | 352.50 | 8 | 1 | 2 | 71.75 | 3.70 | −6.48 | High | Yes | 0.55 |
| 5m | 403.34 | 6 | 1 | 2 | 71.75 | 4.02 | −6.48 | High | Yes | 0.55 |
| 5n | 403.34 | 6 | 1 | 2 | 111.66 | 4.01 | −6.48 | High | Yes | 0.55 |
| 5o | 369.44 | 7 | 3 | 2 | 117.57 | 2.98 | −5.51 | High | No | 0.55 |
| 5p | 338.47 | 7 | 1 | 2 | 71.75 | 3.42 | −6.08 | High | Yes | 0.55 |
| 5q | 342.43 | 6 | 2 | 2 | 71.75 | 3.73 | −7.59 | High | Yes | 0.55 |
| 5r | 352.50 | 7 | 1 | 2 | 71.75 | 3.74 | −6.46 | High | Yes | 0.55 |
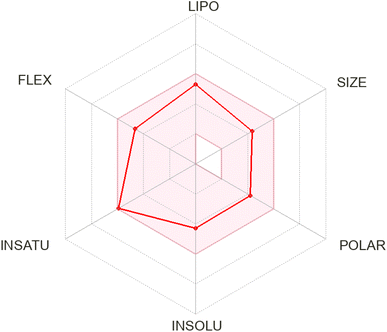 | ||
| Fig. 7 Bioavailability radar diagram for compound 5d showing favorable oral bioavailability profile. | ||
3. Experimental
3.1 Chemistry
All of the chemicals needed to synthesize 4-(pyrrolidin-1-yl)benzaldehyde-based thiosemicarbazones were purchased from Sigma-Aldrich. Petroleum ether, ethyl acetate, glacial acetic acid, ethanol, and methanol were among the chemicals and solvents purchased from Merck and utilized in their original forms. Aluminum-backed silica gel plates were used to monitor the start and finish of the reaction. 1H and 13C-NMR spectra of were obtained at 25 °C using a Bruker Ascend 600 MHz NMR spectrometer in deuterated solvent such as DMSO-d6 (600 MHz for 1H and 151 MHz for 13C). Chemical shifts (ppm) were used to portray NMR spectra, and coupling constants (J) were shown in hertz (Hz) to show signal multiplicity HPLC was carried out on Agilent, Germany (liquid chromatographic column 150 mm × 4.6 mm (id) packed with 5-micron C18; 263 nm).3.2 General method for the synthesis of 4-(pyrrolidin-1-yl)benzaldehyde (3)
Compound (3) was synthesized using a method that had been previously reported.37 To summarize, pyrrolidine (1 mmol) was dissolved in 4.0 ml of DMF. After that, the solution was heated to 80 °C, while being stirred, anhydrous K2CO3 (2 mmol) was added. After 30 minutes, 4-fluorobenzaldehyde (1 mmol) was added in reaction mixture. The heating was continued for 12 hours. Once the reaction was finished, the mixture was cooled to room temperature and added to the ice water drop by drop. The product was precipitated out, filtered and dried. The formed product was pure and used without additional purification.3.3 General method for the synthesis and characterization of compounds 5(a–r)
A previously reported38 one-step condensation method was employed to synthesize thiosemicarbazones, with some modifications to the solvent, in a tropical setting. First, thiosemicarbazides (4a–r) (5 mmol) and compound (3) (0.1 g, 5 mmol) were mixed in a 20 ml ethanol solution in an oven-dried, single-neck round bottom flask. As a catalyst, a small amount (three to four drops) of glacial acetic acid was added. After that, the mixture was refluxed for four to eight hours while thin layer chromatography (TLC) was used for monitoring the reaction's development. The resultant solid product was filtered and dried once the reaction was finished. The residue was then crystallized using ethanol to obtain the required thiosemicarbazones 5(a–r). Characterization data of each synthesized compound is given below.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O = 80
H2O = 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20; tR: 2.164 min, purity: 98.7%, C18H26N4S, QTOF MS ES+ (m/z): [M + H]+, calcd: 331.1956, found: 331.1956.
20; tR: 2.164 min, purity: 98.7%, C18H26N4S, QTOF MS ES+ (m/z): [M + H]+, calcd: 331.1956, found: 331.1956.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O = 80
H2O = 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20; tR: 2.088 min, purity: 98.2%, C18H19FN4S, QTOF MS ES+ (m/z): [M + H]+, calcd: 343.1392, found: 343.1392.
20; tR: 2.088 min, purity: 98.2%, C18H19FN4S, QTOF MS ES+ (m/z): [M + H]+, calcd: 343.1392, found: 343.1392.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O = 80
H2O = 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20; tR: 3.956 min, purity: 98.3%, C18H18Cl2N4S, QTOF MS ES+ (m/z): [M + H]+, calcd: 393.0707, found: 393.0706.
20; tR: 3.956 min, purity: 98.3%, C18H18Cl2N4S, QTOF MS ES+ (m/z): [M + H]+, calcd: 393.0707, found: 393.0706.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O = 80
H2O = 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20; tR: 1.977 min, purity: 98.3%, C19H22N4OS, QTOF MS ES+ (m/z): [M + H]+, calcd: 355.1592, found: 355.1582.
20; tR: 1.977 min, purity: 98.3%, C19H22N4OS, QTOF MS ES+ (m/z): [M + H]+, calcd: 355.1592, found: 355.1582.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O = 80
H2O = 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20; tR: 2.300 min, purity: 97.9%, C20H24N4S, QTOF MS ES+ (m/z): [M + H]+, calcd: 353.1799, found: 353.1789.
20; tR: 2.300 min, purity: 97.9%, C20H24N4S, QTOF MS ES+ (m/z): [M + H]+, calcd: 353.1799, found: 353.1789.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O = 80
H2O = 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20; tR: 2.150 min, purity: 97.5%, C18H20N4S, QTOF MS ES+ (m/z): [M + H]+, calcd: 325.1486, found: 325.1463.
20; tR: 2.150 min, purity: 97.5%, C18H20N4S, QTOF MS ES+ (m/z): [M + H]+, calcd: 325.1486, found: 325.1463.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O = 80
H2O = 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20; tR: 2.459 min, purity: 97.7%, C19H22N4S, QTOF MS ES+ (m/z): [M + H]+, calcd: 339.1643, found: 339.1630.
20; tR: 2.459 min, purity: 97.7%, C19H22N4S, QTOF MS ES+ (m/z): [M + H]+, calcd: 339.1643, found: 339.1630.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O = 80
H2O = 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20; tR: 2.166 min, purity: 97.1%, C19H22N4OS, QTOF MS ES+ (m/z): [M + H]+, calcd: 355.1592, found: 355.1582.
20; tR: 2.166 min, purity: 97.1%, C19H22N4OS, QTOF MS ES+ (m/z): [M + H]+, calcd: 355.1592, found: 355.1582.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O = 80
H2O = 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20; tR: 2.558 min, purity: 96.3%, C19H21ClN4S, QTOF MS ES+ (m/z): [M + H]+, calcd: 373.1253, found: 373.1253.
20; tR: 2.558 min, purity: 96.3%, C19H21ClN4S, QTOF MS ES+ (m/z): [M + H]+, calcd: 373.1253, found: 373.1253.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O = 80
H2O = 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20; tR: 2.344 min, purity: 98.8%, C18H19N5O2S, QTOF MS ES+ (m/z): [M + H]+, calcd: 370.1337, found: 370.1337.
20; tR: 2.344 min, purity: 98.8%, C18H19N5O2S, QTOF MS ES+ (m/z): [M + H]+, calcd: 370.1337, found: 370.1337.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O = 80
H2O = 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20; tR: 1.610 min, purity: 98.9%, C18H27N5OS, QTOF MS ES+ (m/z): [M + H]+, calcd: 362.2014, found: 362.2014.
20; tR: 1.610 min, purity: 98.9%, C18H27N5OS, QTOF MS ES+ (m/z): [M + H]+, calcd: 362.2014, found: 362.2014.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O = 80
H2O = 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20; tR: 2.608 min, purity: 98.4%,C20H24N4S, QTOF MS ES+ (m/z): [M + H]+, calcd: 353.1799, found: 353.1799.
20; tR: 2.608 min, purity: 98.4%,C20H24N4S, QTOF MS ES+ (m/z): [M + H]+, calcd: 353.1799, found: 353.1799.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O = 80
H2O = 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20; tR: 3.025 min, purity: 97.4%, C18H19BrN4S, QTOF MS ES+ (m/z): [M + H]+, calcd: 403.0592, found: 403.0503.
20; tR: 3.025 min, purity: 97.4%, C18H19BrN4S, QTOF MS ES+ (m/z): [M + H]+, calcd: 403.0592, found: 403.0503.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O = 80
H2O = 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20; tR: 3.022 min, purity: 97.2%, C18H19BrN4S, QTOF MS ES+ (m/z): [M + H]+, calcd: 403.0592, found: 403.0522.
20; tR: 3.022 min, purity: 97.2%, C18H19BrN4S, QTOF MS ES+ (m/z): [M + H]+, calcd: 403.0592, found: 403.0522.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O = 80
H2O = 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20; tR: 2.207 min, purity: 97.9%, C18H19N5O2S, QTOF MS ES+ (m/z): [M + H]+, calcd: 370.1337, found: 370.1337.
20; tR: 2.207 min, purity: 97.9%, C18H19N5O2S, QTOF MS ES+ (m/z): [M + H]+, calcd: 370.1337, found: 370.1337.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O = 80
H2O = 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20; tR: 2.202 min, purity: 98.1%, C19H22N4S, QTOF MS ES+ (m/z): [M + H]+, calcd: 339.1643, found: 339.1643.
20; tR: 2.202 min, purity: 98.1%, C19H22N4S, QTOF MS ES+ (m/z): [M + H]+, calcd: 339.1643, found: 339.1643.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O = 80
H2O = 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20; tR: 2.164 min, purity: 98.6%, C18H19FN4S, QTOF MS ES+ (m/z): [M + H]+, calcd: 343.1392, found: 343.1395.
20; tR: 2.164 min, purity: 98.6%, C18H19FN4S, QTOF MS ES+ (m/z): [M + H]+, calcd: 343.1392, found: 343.1395.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O = 80
H2O = 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20; tR: 2.603 min, purity: 98.3%, C20H24N4S, QTOF MS ES+ (m/z): [M + H]+, calcd: 353.1799, found: 353.1799.
20; tR: 2.603 min, purity: 98.3%, C20H24N4S, QTOF MS ES+ (m/z): [M + H]+, calcd: 353.1799, found: 353.1799.4. Conclusion
In conclusion, the comprehensive study into the potential of 4-pyrrolidine-based thiosemicarbazones 5(a–r) as DHFR inhibitors through in vitro and in silico studies sheds light on their promising therapeutic applications. The synthesized compounds 5(a–r) demonstrated notable inhibitory activity against DHFR with IC50 value in the range of 12.37 ± 0.48 μM to 54.10 ± 0.72 μM, showcasing their potential as effective agents against various diseases, including cancer and microbial infections. Amongst all derivatives, 5d is more potent towards DHFR enzyme. Furthermore, the molecular docking simulations provided valuable insights into the binding interactions between the compounds 5(a–r) and the target enzyme, elucidating their mechanism of action at the molecular level. These findings underscore the significance of further exploration and optimization of 4-pyrrolidine-based thiosemicarbazones for the development of novel DHFR inhibitors with enhanced efficacy and reduced toxicity, thereby offering new avenues for drug discovery and therapeutic invention.Data availability
The data supporting this article have been included as part of the ESI.†Conflicts of interest
The authors have declared no conflict of interest.Acknowledgements
Authors are thankful to the Researchers Supporting Project number (RSP2024R491), King Saud University, Riyadh, Saudi Arabia. Z. S. is thankful to the Alexander von Humboldt Foundation for the award of the Georg Forster Research Fellowship for Experienced Researchers.References
- A. Aggarwal, O. Ginsburg and T. Fojo, Cancer economics, policy and politics: what informs the debate? Perspectives from the EU, Canada and US, J. Cancer Policy, 2014, 2(1), 1–11 CrossRef CAS.
- P. Singh, M. Kaur and S. Sachdeva, Mechanism inspired development of rationally designed dihydrofolate reductase inhibitors as anticancer agents, J. Med. Chem., 2012, 55(14), 6381–6390 CrossRef CAS PubMed.
- A. Wróbel, M. Baradyn, A. Ratkiewicz and D. Drozdowska, Synthesis, biological activity, and molecular dynamics study of novel series of a trimethoprim analogs as multi-targeted compounds: Dihydrofolate reductase (DHFR) inhibitors and DNA-binding agents, Int. J. Mol. Sci., 2021, 22(7), 3685 CrossRef PubMed.
- C. Capasso and C. T. Supuran, Sulfa and trimethoprim-like drugs–antimetabolites acting as carbonic anhydrase, dihydropteroate synthase and dihydrofolate reductase inhibitors, J. Enzyme Inhib. Med. Chem., 2014, 29(3), 379–387 CrossRef CAS.
- A. Rao and S. Tapale, A study on dihydrofolate reductase and its inhibitors: A review, Int. J. Pharm. Sci. Res., 2013, 4(7), 2535 Search PubMed.
- E. O. Osman, S. H. Emam, A. Sonousi, M. M. Kandil, A. M. Abdou and R. A. Hassan, Design, synthesis, anticancer, and antibacterial evaluation of some quinazolinone-based derivatives as DHFR inhibitors, Drug Dev. Res., 2023, 84(5), 888–906 CrossRef CAS.
- G. Tcherkez, E. Boex-Fontvieille, A. Mahé and M. Hodges, Respiratory carbon fluxes in leaves, Curr. Opin. Plant Biol., 2012, 15(3), 308–314 CrossRef.
- M. V. Raimondi, O. Randazzo, M. La Franca, G. Barone, E. Vignoni, D. Rossi and S. Collina, DHFR inhibitors: reading the past for discovering novel anticancer agents, Molecules, 2019, 24(6), 1140 CrossRef.
- H. Li, F. Fang, Y. Liu, L. Xue, M. Wang, Y. Guo, X. Wang, C. Tian, J. Liu and Z. Zhang, Inhibitors of dihydrofolate reductase as antitumor agents: design, synthesis and biological evaluation of a series of novel nonclassical 6-substituted pyrido [3, 2-d] pyrimidines with a three-to five-carbon bridge, Bioorg. Med. Chem., 2018, 26(9), 2674–2685 CrossRef PubMed.
- W. D. Alrohily, M. E. Habib, S. M. El-Messery, A. Alqurshi, H. El-Subbagh and E.-S. E. Habib, Antibacterial, antibiofilm and molecular modeling study of some antitumor thiazole based chalcones as a new class of DHFR inhibitors, Microb. Pathog., 2019, 136, 103674 CrossRef.
- Y.-C. Hsieh, N. E. Skacel, N. Bansal, K. W. Scotto, D. Banerjee, J. R. Bertino and E. E. Abali, Species-specific differences in translational regulation of dihydrofolate reductase, Mol. Pharmacol., 2009, 76(4), 723–733 CrossRef PubMed.
- G. Li Petri, M. V. Raimondi, V. Spanò, R. Holl, P. Barraja and A. Montalbano, Pyrrolidine in drug discovery: a versatile scaffold for novel biologically active compounds, Top. Curr. Chem., 2021, 379, 1–46 CrossRef PubMed.
- A. A. Bhat, I. Singh, N. Tandon and R. Tandon, Structure activity relationship (SAR) and anticancer activity of pyrrolidine derivatives: Recent developments and future prospects (A review), Eur. J. Med. Chem., 2023, 246, 114954 CrossRef CAS PubMed.
- M. Tasleem, J. Pelletier, J. Sévigny, Z. Hussain, A. Khan, A. Al-Harrasi, A. F. El-Kott, P. Taslimi, S. Negm, Z. Shafiq and J. Iqbal, Synthesis, in vitro, and in silico studies of morpholine-based thiosemicarbazones as ectonucleotide pyrophosphatase/phosphodiesterase-1 and-3 inhibitors, Int. J. Biol. Macromol., 2024, 266, 131068 Search PubMed.
- A. Gomtsyan, Heterocycles in drugs and drug discovery, Chem. Heterocycl. Compd., 2012, 48, 7–10 CrossRef CAS.
- A. Hamad, M. A. Khan, K. M. Rahman, I. Ahmad, Z. Ul-Haq, S. Khan and Z. Shafiq, Development of sulfonamide-based Schiff bases targeting urease inhibition: Synthesis, characterization, inhibitory activity assessment, molecular docking and ADME studies, Bioorg. Chem., 2020, 102, 104057 Search PubMed.
- M. T. Shehzad, A. Khan, M. Islam, A. Hameed, M. Khiat, S. A. Halim, M. U. Anwar, S. R. Shah, J. Hussain and R. Csuk, Synthesis and urease inhibitory activity of 1, 4-benzodioxane-based thiosemicarbazones: Biochemical and computational approach, J. Mol. Struct., 2020, 1209, 127922 CrossRef CAS.
- M. Arooj, M. Zahra, M. Islam, N. Ahmed, A. Waseem and Z. Shafiq, Coumarin based thiosemicarbazones as effective chemosensors for fluoride ion detection, Spectrochim. Acta, Part A, 2021, 261, 120011 CrossRef CAS PubMed.
- M. D. Altıntop, Z. A. Kaplancıklı, G. A. Çiftçi and R. Demirel, Synthesis and biological evaluation of thiazoline derivatives as new antimicrobial and anticancer agents, Eur. J. Med. Chem., 2014, 74, 264–277 CrossRef.
- Y. Qin, R. Xing, S. Liu, K. Li, X. Meng, R. Li, J. Cui, B. Li and P. Li, Novel thiosemicarbazone chitosan derivatives: Preparation, characterization, and antifungal activity, Carbohydr. Polym., 2012, 87(4), 2664–2670 CrossRef CAS.
- A. Pyrih, M. Jaskolski, A. K. Gzella and R. Lesyk, Synthesis, structure and evaluation of anticancer activity of 4-amino-1, 3-thiazolinone/pyrazoline hybrids, J. Mol. Struct., 2021, 1224, 129059 CrossRef CAS.
- J. F. Machado, F. Marques, T. Pinheiro, M. J. Villa de Brito, G. Scalese, L. Pérez-Díaz, L. Otero, J. P. António, D. Gambino and T. S. Morais, Copper (I)-Thiosemicarbazone Complexes with Dual Anticancer and Antiparasitic Activity, ChemMedChem, 2023, 18(14), e202300074 CrossRef CAS PubMed.
- M. Scaccaglia, S. Pinelli, L. Manini, B. Ghezzi, M. Nicastro, J. Heinrich, N. Kulak, P. Mozzoni, G. Pelosi and F. Bisceglie, Gold (III) complexes with thiosemicarbazone ligands: insights into their cytotoxic effects on lung cancer cells, J. Inorg. Biochem., 2024, 251, 112438 CrossRef CAS PubMed.
- M. T. Shehzad, A. Hameed, M. Al-Rashida, A. Imran, M. Uroos, A. Asari, H. Mohamad, M. Islam, S. Iftikhar and Z. Shafiq, Exploring antidiabetic potential of adamantyl-thiosemicarbazones via aldose reductase (ALR2) inhibition, Bioorg. Chem., 2019, 92, 103244 CrossRef CAS PubMed.
- S. Umamatheswari, B. Balaji, M. Ramanathan and S. Kabilan, Synthesis, stereochemistry, antimicrobial evaluation and QSAR studies of 2, 6-diaryltetrahydropyran-4-one thiosemicarbazones, Eur. J. Med. Chem., 2011, 46(4), 1415–1424 CrossRef CAS PubMed.
- M. Ertas, Z. Sahin, E. F. Bulbul, C. Bender, S. N. Biltekin, B. Berk, L. Yurttas, A. M. Nalbur, H. Celik and Ş. Demirayak, Potent ribonucleotide reductase inhibitors: Thiazole-containing thiosemicarbazone derivatives, Arch. Pharm., 2019, 352(11), 1900033 CrossRef CAS PubMed.
- A. Imran, M. T. Shehzad, S. J. A. Shah, T. Al Adhami, M. Laws, K. M. Rahman, R. D. Alharthy, I. A. Khan, Z. Shafiq and J. Iqbal, Development and exploration of novel substituted thiosemicarbazones as inhibitors of aldose reductase via in vitro analysis and computational study, Sci. Rep., 2022, 12(1), 5734 CrossRef CAS.
- X. Jiang, L. A. Fielding, H. Davis, W. Carroll, E. C. Lisic and J. E. Deweese, Inhibition of topoisomerases by metal thiosemicarbazone complexes, Int. J. Mol. Sci., 2023, 24(15), 12010 CrossRef CAS PubMed.
- R. H. Dhabale, S. Shah, N. Tiwari and P. Patani, Review of Semicarbazone, Thiosemicarbazone, and their transition metal complexes, and their biological activities, J. Pharm. Negat. Results, 2022, 2416–2424 Search PubMed.
- D. S. Kalinowski, P. Quach and D. R. Richardson, Thiosemicarbazones: the new wave in cancer treatment, Future Med. Chem., 2009, 1(6), 1143–1151 Search PubMed.
- Y. Yu, D. S. Kalinowski, Z. Kovacevic, A. R. Siafakas, P. J. Jansson, C. Stefani, D. B. Lovejoy, P. C. Sharpe, P. V. Bernhardt and D. R. Richardson, Thiosemicarbazones from the old to new: iron chelators that are more than just ribonucleotide reductase inhibitors, J. Med. Chem., 2009, 52(17), 5271–5294 CrossRef CAS.
- N. Arumugam, A. I. Almansour, R. S. Kumar, V. S. Krishna, D. Sriram and N. Dege, Stereoselective synthesis and discovery of novel spirooxindolopyrrolidine engrafted indandione heterocyclic hybrids as antimycobacterial agents, Bioorg. Chem., 2021, 110, 104798 Search PubMed.
- A. Ragab, Y. A. Ammar, A. Ezzat, A. M. Mahmoud, M. B. I. Mohamed, S. Abdou and R. S. Farag, Synthesis, characterization, thermal properties, antimicrobial evaluation, ADMET study, and molecular docking simulation of new mono Cu (II) and Zn (II) complexes with 2-oxoindole derivatives, Comput. Biol. Med., 2022, 145, 105473 CrossRef CAS PubMed.
- N. W. Hassan, A. Sabt, M. A. El-Attar, M. Ora, A. E.-D. A. Bekhit, K. Amagase, A. A. Bekhit, A. S. Belal and P. A. Elzahhar, Modulating leishmanial pteridine metabolism machinery via some new coumarin-1, 2, 3-triazoles: Design, synthesis and computational studies, Eur. J. Med. Chem., 2023, 253, 115333 Search PubMed.
- G. S. Hassan, S. M. El-Messery, F. A. Al-Omary, S. T. Al-Rashood, M. I. Shabayek, Y. S. Abulfadl, E.-S. E. Habib, S. M. El-Hallouty, W. Fayad and K. M. Mohamed, Nonclassical antifolates, part 4. 5-(2-Aminothiazol-4-yl)-4-phenyl-4H-1, 2, 4-triazole-3-thiols as a new class of DHFR inhibitors: Synthesis, biological evaluation and molecular modeling study, Eur. J. Med. Chem., 2013, 66, 135–145 CrossRef CAS PubMed.
- M. B. Tufail, M. A. Javed, M. Ikram, M. H. Mahnashi, B. A. Alyami, Y. S. Alqahtani, A. Sadiq and U. Rashid, Synthesis, pharmacological evaluation and Molecular modelling studies of pregnenolone derivatives as inhibitors of human dihydrofolate reductase, Steroids, 2021, 168, 108801 Search PubMed.
- R. A. Markandewar, H. M. Zia and M. Baseer, Impact of reaction dynamics on synthesis of novel nitrogen containing aldehydes, Orient. J. Chem., 2013, 29(4), 1531–1534 Search PubMed.
- D. Taşdemir, A. Karaküçük-İyidoğan, M. Ulaşli, T. Taşkin-Tok, E. E. Oruç-Emre and H. Bayram, Synthesis, molecular modeling, and biological evaluation of novel chiral thiosemicarbazone derivatives as potent anticancer agents, Chirality, 2015, 27(2), 177–188 CrossRef.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4ra05071a |
| This journal is © The Royal Society of Chemistry 2024 |