Open Access Article
Open Access ArticleMechanistic insights into carbonic anhydrase IX inhibition by coumarins from Calendula officinalis: in vitro and in silico approaches†
Reem S. Alruhaimi‡
a,
Emadeldin M. Kamel‡ b,
Sulaiman M. Alnasserc,
Mohammed A. Alzoghaibid,
Al Mokhtar Lamsabhi
b,
Sulaiman M. Alnasserc,
Mohammed A. Alzoghaibid,
Al Mokhtar Lamsabhi ef and
Ayman M. Mahmoud
ef and
Ayman M. Mahmoud *gh
*gh
aDepartment of Biology, College of Science, Princess Nourah bint Abdulrahman University, Riyadh 11671, Saudi Arabia
bOrganic Chemistry Department, Faculty of Science, Beni-Suef University, Beni-Suef 62514, Egypt
cDepartment of Pharmacology and Toxicology, College of Pharmacy, Qassim University, Qassim 51452, Saudi Arabia
dPhysiology Department, College of Medicine, King Saud University, Riyadh, 11461, Saudi Arabia
eDepartamento de Química, Universidad Autónoma de Madrid, Campus de Excelencia UAM-CSIC Cantoblanco, Módulo 13, Madrid 28049, Spain
fInstitute for Advanced Research in Chemical Sciences (IAdChem), Universidad Autónoma de Madrid, Madrid 28049, Spain
gDepartment of Life Sciences, Faculty of Science and Engineering, Manchester Metropolitan University, Manchester M1 5GD, UK. E-mail: a.mahmoud@mmu.ac.uk
hMolecular Physiology Division, Zoology Department, Faculty of Science, Beni-Suef University, Beni-Suef 62514, Egypt. E-mail: ayman.mahmoud@science.bsu.edu.eg
First published on 23rd October 2024
Abstract
Given the critical role of carbonic anhydrase IX (CA IX) in various pathological conditions, there is a significant demand for new inhibitors to enhance patient outcomes and clinical management. In this study, we examined the inhibitory effectiveness of five coumarins derived from Calendula officinalis against CA IX using in vitro assays and computational modeling. Among the coumarins tested, xeroboside and isobaisseoside were identified as the most potent inhibitors. Kinetic studies indicated that xeroboside and isobaisseoside exhibit a mixed inhibition mode. Molecular docking analysis showed that the tested coumarins exhibit binding affinities and extensive polar interactions with CA IX. These coumarins demonstrated significant hydrophobic interactions and occupied the same binding site as acetazolamide (AAZ). Molecular dynamics (MD) indicated that xeroboside and isobaisseoside exhibited consistent trajectories and notable energy stabilization during their interaction with CA IX. MM/PBSA calculations showed that xeroboside displayed the lowest binding free energy (−27.26 ± 2.48 kJ mol−1). Potential Energy Landscape (PEL) analysis revealed distinct and stable conformational states for the CA IX–ligand complexes, with xeroboside exhibiting the most stable and lowest energy configuration. These computational findings are consistent with the experimental results, highlighting the potential efficacy of xeroboside and isobaisseoside as CA IX inhibitors. In conclusion, Calendula officinalis-derived coumarins are promising candidates as effective CA IX inhibitors.
1. Introduction
Carbonic anhydrases (CAs) are a family of zinc metalloenzymes that catalyze the reversible hydration of carbon dioxide to bicarbonate and a proton, a reaction crucial for maintaining acid–base balance in various tissues and organs.1 These enzymes are implicated in numerous physiological processes including respiration, acid-base homeostasis, and ion transport.1 Among the known isoforms in humans, CA IX is of particular interest due to its overexpression in several types of tumors, particularly under hypoxic conditions.2 CA IX plays a vital role in the regulation of intracellular and extracellular pH in cancer cells, facilitating their survival, proliferation, and invasion.3 The inhibition of CA IX has thus gained significant attention as a therapeutic strategy, aiming to disrupt the pH regulatory mechanism of cancer cells, thereby inhibiting tumor growth and metastasis.4 CA IX is remarkably expressed in cells with high growth and glycolysis rates and cells frequently exposed to acidic and hypoxic environments, such as cancer and endothelial cells (ECs).5 In the endothelium, the role of CA IX extends beyond pH regulation to aerobic glycolysis, EC migration and network formation.6 Targeting CA IX with specific inhibitors could lead to the development of novel anticancer therapies with reduced side effects compared to conventional treatments. In addition, CA IX inhibition might influence ECs and integrity of barriers in different tissues.Calendula officinalis (C. officinalis), commonly known as marigold, is a well-known medicinal plant with diverse phytochemical composition and broad spectrum of biological activities.7 The plant is rich in secondary metabolites, including flavonoids, triterpenoids, carotenoids, and essential oils, which contribute to its anti-inflammatory, antioxidant, antimicrobial, and anticancer properties.8–11 Among these compounds, coumarins are notable for their therapeutic potential.12 C. officinalis has been documented to exhibit different pharmacological effects,11 making its phytoconstituents, including coumarins prime candidates for further investigation in enzyme inhibition studies. The selection of coumarins from C. officinalis for studying CA IX inhibition is justified by the plant established use in traditional medicine,11 and previous studies indicating the effectiveness of coumarins in inhibiting various enzymes and pathways involved in cancer progression.13 Therefore, exploring the inhibitory effects of these natural compounds on CA IX could provide valuable insights into novel anticancer strategies and enhance the therapeutic applications of C. officinalis.
To comprehensively assess the inhibitory activity of coumarins on CA IX, we employed an integrated approach combining in vitro assays and in silico studies. The in vitro assays provide direct evidence of the potency of the isolated phytoconstituents. Complementing these experiments, molecular docking and molecular dynamics (MD) simulations offer a detailed understanding of the interaction mechanisms at the molecular level.14 Docking studies help predict the binding affinity and orientation of coumarins within the active site of CA IX, while MD simulations reveal the stability and dynamic behavior of the enzyme–inhibitor complexes over time. This integrated approach not only validates the inhibitory activity observed experimentally but also elucidates the underlying molecular interactions, paving the way for the development of more effective CA IX inhibitors.
2. Materials and methods
2.1. Phytochemical investigation
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.51
1.51![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) followed by the DCM-EtOAc (8
1) followed by the DCM-EtOAc (8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2), and MeOH to produce 56 fractions. These fractions were collected and combined based on their TLC profile into six main fractions (A1–A6). Fraction A3 was further chromatographed over a silica gel column eluted with dioxane-hexane (4.5
2), and MeOH to produce 56 fractions. These fractions were collected and combined based on their TLC profile into six main fractions (A1–A6). Fraction A3 was further chromatographed over a silica gel column eluted with dioxane-hexane (4.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5.5) to afford five main subfractions after TLC comparison and combinations (A3.1–A3.5). Subfraction A3.1 underwent spontaneous crystallization to afford the pure form of compound 2 (26 mg). Subfraction A3.3 was subjected to chromatographic purification over Sephadex LH-20 column eluted with MeOH to produce the purified compounds 4 (31 mg) and 5 (19 mg). The fraction A4 revealed an interesting TLC profile and was subjected to fractionation using a silica gel column using chloroform–MeOH solvent (90
5.5) to afford five main subfractions after TLC comparison and combinations (A3.1–A3.5). Subfraction A3.1 underwent spontaneous crystallization to afford the pure form of compound 2 (26 mg). Subfraction A3.3 was subjected to chromatographic purification over Sephadex LH-20 column eluted with MeOH to produce the purified compounds 4 (31 mg) and 5 (19 mg). The fraction A4 revealed an interesting TLC profile and was subjected to fractionation using a silica gel column using chloroform–MeOH solvent (90![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10) of increasing polarity. A total of six main subfractions were obtained after TLC investigation and combination (A4.1–A4.6). Subfractions A4.2 and A4.3 were purified over the Sephadex LH-20 column eluted with MeOH to afford compound 1 (28 mg) and compound 3 (32 mg), respectively. The purity of the isolated compounds was confirmed by spectroscopic analysis.
10) of increasing polarity. A total of six main subfractions were obtained after TLC investigation and combination (A4.1–A4.6). Subfractions A4.2 and A4.3 were purified over the Sephadex LH-20 column eluted with MeOH to afford compound 1 (28 mg) and compound 3 (32 mg), respectively. The purity of the isolated compounds was confirmed by spectroscopic analysis.2.2. In vitro CA IX inhibition assay
The effect of coumarins on CA IX activity conducted as previously described with minor modification,15,16 using 4-nitrophenyl acetate (4-NPA) as a substrate. A buffer solution containing HEPES and Tris–HCl (20 mM) was used as the reaction medium. Acetazolamide (AAZ), a clinically used CA inhibitor, was selected as the standard drug for this study. Compounds 1–5 and AAZ were prepared in DMSO, diluted in the assay buffer to different concentrations, and then utilized in the experiments. The reaction mixture consisted of 150 μL of the prepared reaction medium, 20 μL of the aqueous CA IX solution (0.1 mg mL−1 in deionized water), 20 μL of 4-NPA (0.7 mM in ethanol), and 20 μL of the tested compounds. The production of the yellow-colored 4-nitrophenolate was monitored at 400 nm and the experiment was performed in triplicate. The kinetic analysis of the CA IX inhibition was performed using varying concentrations of the tested compounds and 4-NPA. Following substrate addition, the change in absorbance was monitored every minute for 30 minutes at 25 °C. To elucidate the inhibition mechanism, a Lineweaver–Burk plot was constructed by plotting the reciprocal of the substrate concentration (1/[S]) against the reciprocal of the reaction velocity (1/Vmax). The inhibition constant (Ki) was determined by creating a double reciprocal plot of the inhibitor concentration ([I]) against the slope of the Lineweaver–Burk plot.2.3. Statistical analysis
The results are presented as the mean ± standard deviation (SD), with each data point reflecting the average of three independent experiments. Comparison of the IC50 was conducted using one-way ANOVA followed by Tukey's test on GraphPad Prism 8.0. A P value <0.05 was considered significant.2.4. In silico investigations
3. Results and discussion
3.1. Phytochemical study
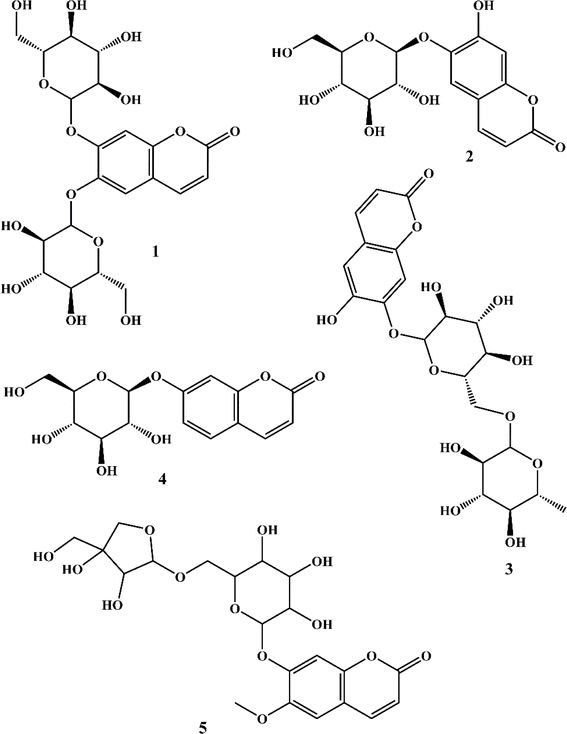
The phytochemical investigation of the aerial parts of C. officinalis led to the isolation and identification of five coumarins. The chemical structures of these isolated compounds (Fig. 1) were elucidated through spectroscopic analysis, comparison of TLC profiles with known standards, and referencing existing scientific literature. Using these methods, the coumarins were identified as 6,7-di-O-glucopyranosyl esculetin (1),34 aesculin (2),35 isobaisseoside (3),36 skimmin (4),37 and xeroboside (5).383.2. Inhibitory activity of C. officinalis-derived coumarins on CA IX
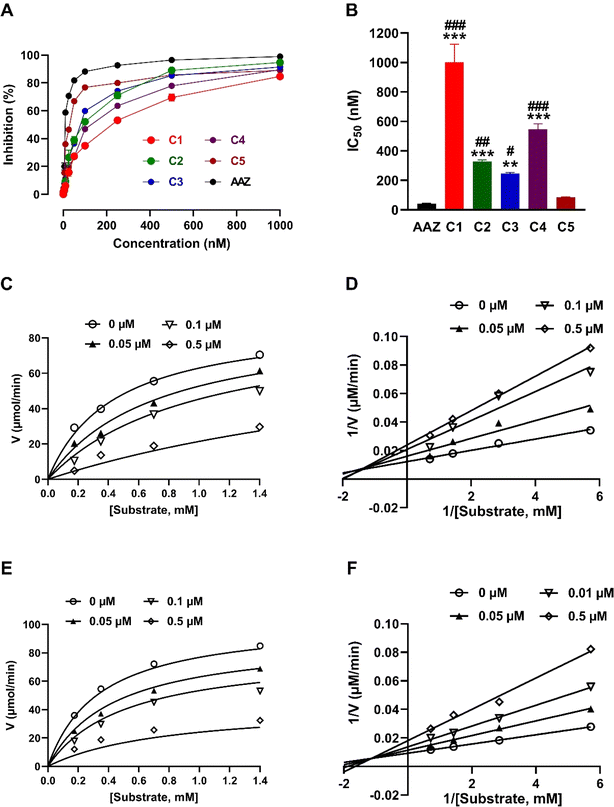
Given the involvement of CA IX in different physiological and pathological processes, including its overexpression in hypoxic tumor environments and its contribution to tumor growth and survival,1,2 there is a pressing need to identify effective inhibitors to modulate its activity. The dose–response curves and IC50 values for the isolated coumarins and AAZ on CA IX activity are presented in Fig. 2. As shown in Fig. 2A, the compounds and AAZ showed a concentration-dependent inhibition of CA IX activity. The IC50 values (Fig. 2B), indicating the concentration at which 50% inhibition of CA IX activity is achieved, varied among the compounds. Xeroboside demonstrated the most potent inhibition with an IC50 value of 85.34 ± 3.01 nM, following AAZ (IC50 = 42.87 ± 1.39 nM). Isobaisseoside also showed potent inhibition with an IC50 of 247.61 ± 7.27 nM, and aesculin and skimmin displayed moderate inhibition with IC50 values of 327.05 ± 11.46 nM and 547.93 ± 35.66 nM, respectively. The least potent inhibitor was 6,7-di-O-glucopyranosyl esculetin, with an IC50 value of 1.01 ± 0.12 μM. Statistical analysis revealed significant differences between the IC50 value of compounds 1–4 when compared with either AAZ or compound 5. Also, a non-significant difference was observed between IC50 value AAZ and compound 5 (Fig. 2B).To further characterize the inhibitory effects of isobaisseoside and xeroboside on CA IX, kinetic analyses were performed, and the Michaelis–Menten (Fig. 2C and E) and Lineweaver–Burk (Fig. 2D and F) plots were constructed. The Michaelis–Menten plot for isobaisseoside (Fig. 2E) showed a decrease in the maximum reaction velocity (Vmax) with increasing concentration (Ki = 126 nM), suggesting a mixed-type inhibition. The Lineweaver–Burk plot (Fig. 2F) corroborated this, as the lines intersect to the left of the y-axis, indicating changes in both Vmax and Km. Similar to isobaisseoside, xeroboside exhibited a mixed-type inhibition pattern (Fig. 2C and D). The decrease in Vmax with increasing concentration was evident from the Michaelis–Menten plot (Ki = 77.2 nM). The Lineweaver–Burk plot showed intersecting lines, confirming the mixed inhibition. The reference inhibitor AAZ demonstrated non-competitive inhibition.
The efficacy of the tested coumarins against CA IX demonstrates the potential of these natural products as therapeutic agents. Xeroboside and isobaisseoside exhibited the highest inhibition activity among the tested coumarins and the superior activity of xeroboside can be attributed to its lower IC50 and higher inhibition rate compared to other coumarins. The structural features of xeroboside, including the presence of specific functional groups, may contribute to its enhanced binding affinity and inhibitory potency. The kinetic analysis revealed that isobaisseoside and xeroboside exhibited mixed-type inhibition, which suggests that they can bind to both the enzyme active site and an allosteric site, altering both Vmax and Km. This mode of inhibition may provide a therapeutic advantage by offering multiple binding interactions that stabilize the inhibitor–enzyme complex. However, it is noteworthy to highlight that the two-site model of mixed inhibition might not be mechanistically relevant as reported by Pesaresi.39 Using a statistical approach and by combining statistical analysis of enzyme inhibition cases with a theoretical investigation of inhibition models, Pesaresi concluded that mixed inhibitors bind exclusively to the active site, ruling out allosteric involvement.39 Consistently, we propose that our inhibitors interact primarily with the active site, causing a conformational change that affects substrate binding and enzyme activity. This would explain the mixed inhibition observed in our kinetic data without necessarily invoking an allosteric site. While the inhibitors do not completely block substrate binding as in pure competitive inhibition, they still influence the enzyme activity, likely through modulation of the active site conformation. The outcomes of the computational studies conducted in this work support the idea that the inhibitors bind within or near the catalytic region of CA IX, potentially altering the active site structure and leading to mixed inhibition behavior. AAZ, a well-known CA inhibitor, displayed non-competitive inhibition, consistent with its mechanism of action that involves binding to an allosteric site, leading to a decrease in Vmax without affecting Km. The comparison with AAZ highlights the efficacy of the natural coumarins, particularly xeroboside and isobaisseoside, as potent CA IX inhibitors.
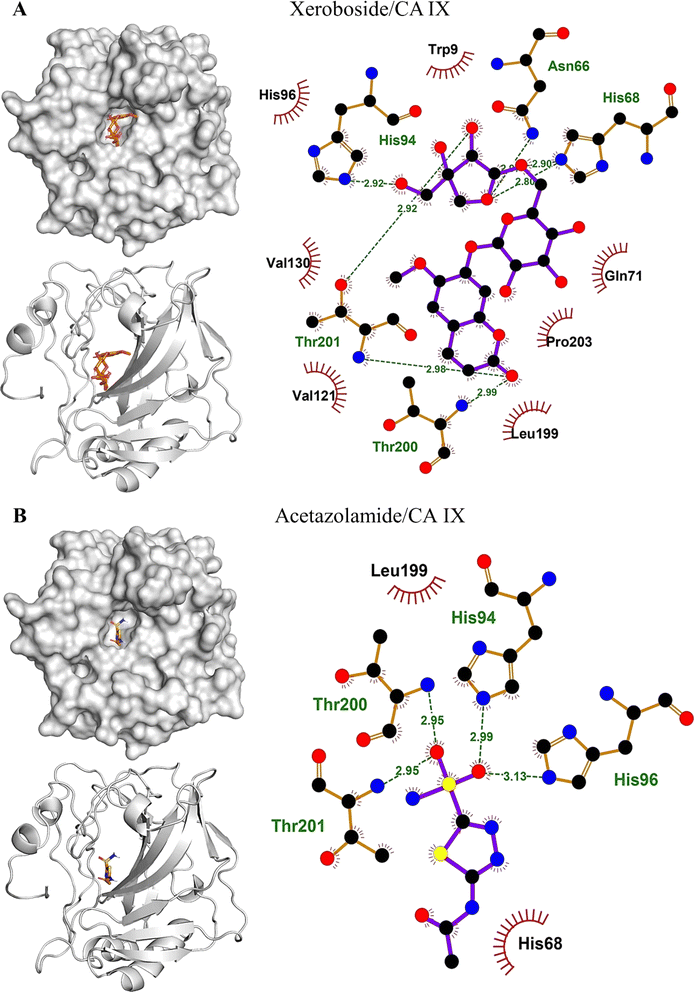
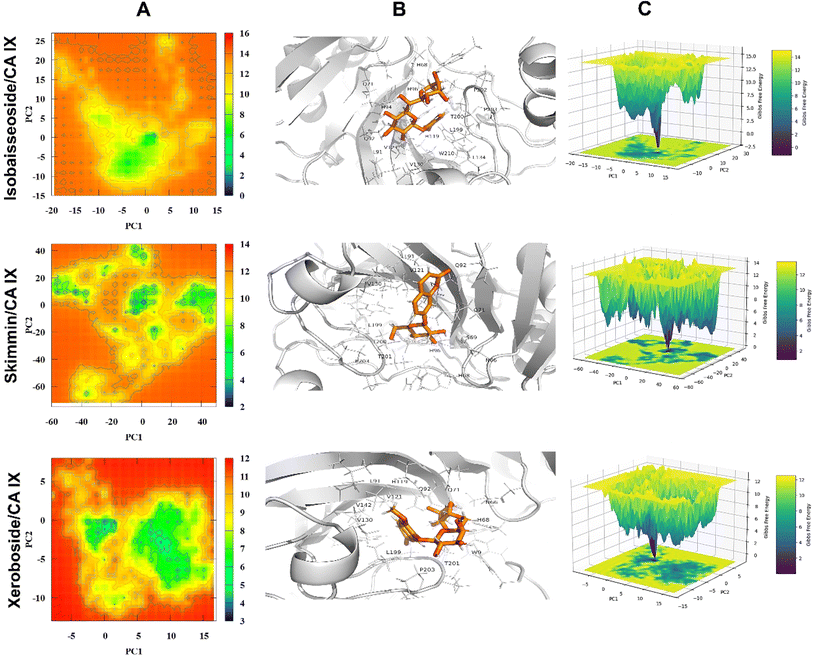
3.3. Molecular docking analysis
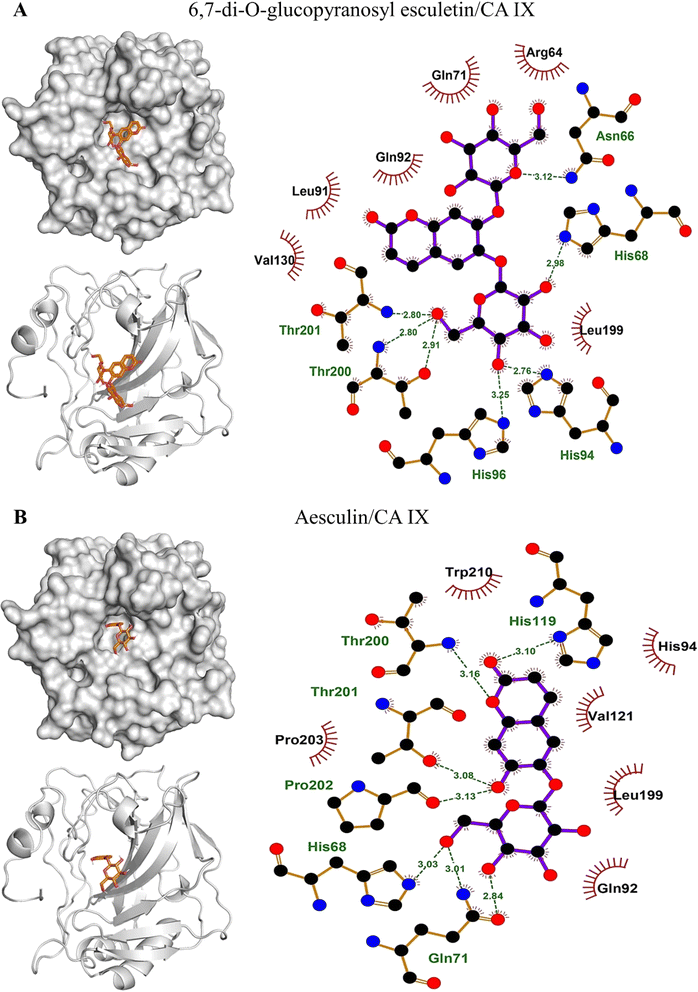
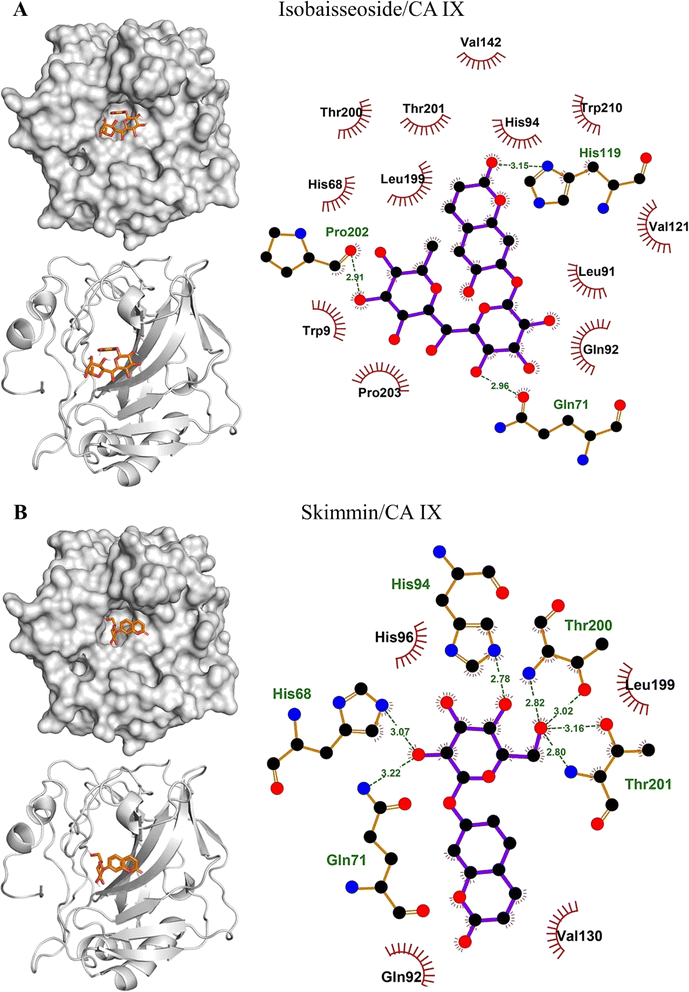
The binding affinities and interacting residues of the isolated coumarins with CA IX were evaluated using molecular docking. Fig. 3, 4, and 5 represent the results of docking simulations, depicting the positioning of different ligands in the binding site of the target enzyme and showcasing residues involved in polar and hydrophobic interactions. Among tested coumarins, 6,7-di-O-glucopyranosyl esculetin and xeroboside demonstrated the lowest binding energies (−8.1 and −8.3 kcal mol−1), respectively (Table 1). However, the remaining coumarins displayed comparable binding energies ranging from −7.3 to −7.7 kcal mol−1, and AAZ had a binding energy of −6.5 kcal mol−1 (Table 1). These findings highlight the potential of these coumarins as effective CA IX inhibitors. The binding site of CA IX displayed a dense network of hydrophobic interactions for all the tested coumarins. These hydrophobic interactions were crucial in ligand–enzyme complexes stability, thereby enhancing the binding affinity and inhibitory potential of the tested compounds. The poses revealed docking of all compounds into the binding site of CA IX, which is also occupied by the reference drug AAZ. This consistent docking pattern suggested that the coumarins and the reference drug share a common binding site, highlighting the potential of these coumarins as effective CA IX inhibitors. In addition, isolated compounds showed a high extent of polar interactions, significantly contributing to the stability and efficacy of these inhibitors.| Lowest binding energy (kcal mol−1) | Polar interacting residues | Hydrophobic interacting residues | |
|---|---|---|---|
| 6,7-di-O-glucopyranosyl esculetin | −8.1 | Thr201, Thr200, His96, His94, His68, and Asn66 | Val130, Leu91, Gln92, Gln71, Arg64, and Leu199 |
| Aesculin | −7.7 | Thr200, Thr201, Pro202, His86, Gln71, and His119 | Trp210, His94, Val121, Leu199, Gln92, and Pro203 |
| Isobaisseoside | −7.3 | Pro202, His119, and Gln71 | Thr200, His68, Thr201, Leu199, His94, Val142, Trp210, Val121, Leu91, Gln91, Trp9, and Pro203 |
| Skimmin | −7.4 | His68, Gln71, Thr201, His94, and Thr200 | His96, Leu199, Val130, and Gln92 |
| Xeroboside | −8.3 | His94, Asn66, Hi68, Thr201, and Thr200 | His96, Trp9, Gln71, Pro203, Leu199, Val130, and Val121 |
| AAZ | −6.5 | His94, His96, Thr200, and Thr201 | His68 and Leu199 |
3.4. Molecular dynamics (MD) simulations
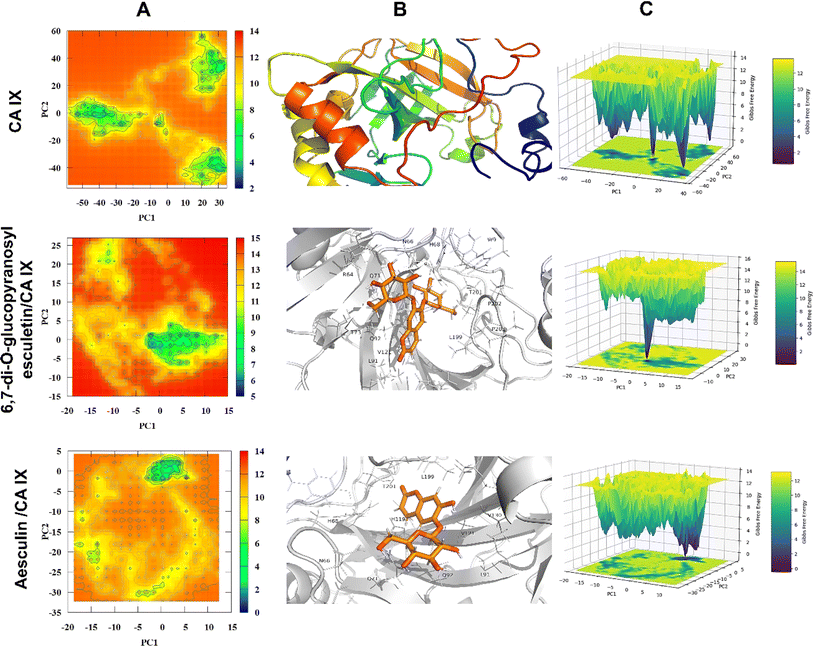
The objective of MD simulations was to elucidate the long-term stability and interactions within the coumarins–CA IX complexes, thereby illuminating the inhibition mechanisms and various binding modes. By carefully examining the simulation trajectories, we aimed to gain comprehensive insights into how the structural dynamics of the compounds influence their CA IX inhibitory potential. Understanding these molecular mechanisms is critical for the strategic design of novel CA IX-targeting therapeutics. We focused on complexes identified through initial docking analyses that exhibited the most favorable binding affinities and conducted an in-depth evaluation using 100 ns MD simulations. This rigorous analysis included key parameters such as interaction energies, MM/PBSA binding free energies, root mean square deviations (RMSD), radius of gyration (Rg), potential energy landscape (PEL), root mean square fluctuations (RMSF), solvent accessible surface area (SASA), and hydrogen bonding profile. Our examination covered both CA IX and its complexes with coumarins, aiming at gaining understanding of their interaction characteristics and dynamic behavior. These insights are essential for elucidating the inhibitory effects of the compounds and guiding the development of effective CA IX inhibitors.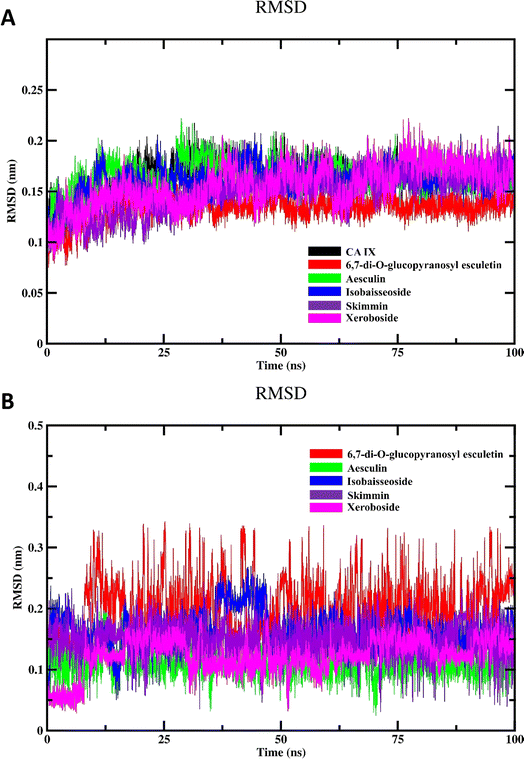 | ||
| Fig. 6 MD simulation of CA IX and its complexes with C. officinalis-derived coumarins; (A) backbone RMSD of the unbound CA IX and CA IX–coumarin complexes and (B) RMSD of isolated compounds. | ||
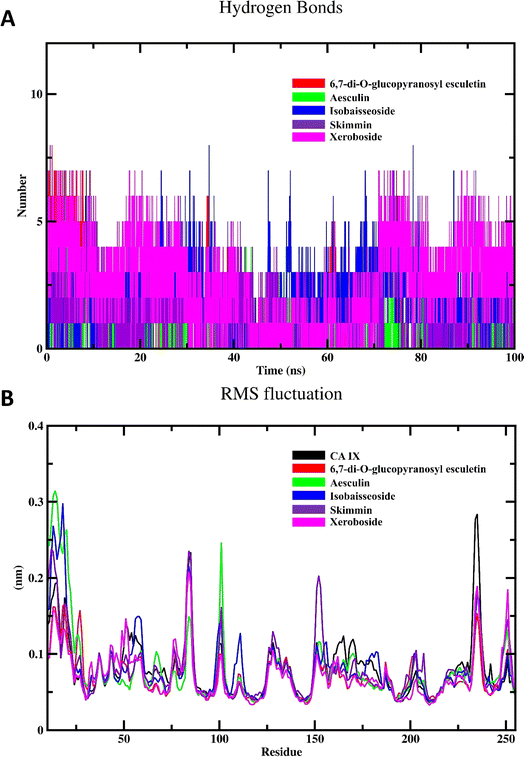
To investigate the inherent movement and conformational changes of the coumarin molecules independent of their interaction with CA IX, we investigated their RMSD values (Fig. 6B). The RMSD values were obtained by aligning the trajectory frames to the initial structure of each drug and calculating the deviation of the coordinates. This method enables an assessment of how much each drug deviates from its initial conformation over the 100 ns simulation period. Such an analysis is crucial in drug design research to evaluate the stability and dynamic behavior of drug molecules. As shown in Fig. 6B, the RMSD analysis of different coumarins revealed notable variations in their RMSD profiles. The compounds exhibited fluctuation patterns in their RMSD values indicating that they maintained relatively stable conformations throughout the simulation. This stability suggests the presence of strong and consistent interactions between isolated coumarins and CA IX.
The results of the Rg calculations for the unbound CA IX and its complexes with various coumarins are shown in Fig. 7A. The Rg profile provides insights into structural stability and compactness of the enzyme and/or its complexes over the course of the simulation.40 Throughout the majority of the simulation time, the Rg profiles of the different coumarin–CA IX complexes demonstrated a consistent and stabilized behavior, comparable to that observed for the unbound enzyme. This indicates that the binding of coumarins does not disrupt the overall compactness of the enzyme structure. Interestingly, among all the complexes studied, xeroboside (the compound with the lowest IC50 value) exhibited the lowest average Rg value. This suggested that the xeroboside–CA IX complex is more compact compared to the other coumarin–CA IX complexes. A lower Rg value often correlates with a more tightly bound and stable complex, which could contribute to the enhanced inhibitory potency of xeroboside. The compact nature of the xeroboside–CA IX complex might facilitate stronger interaction with the active site, thus explaining its superior inhibitory activity.
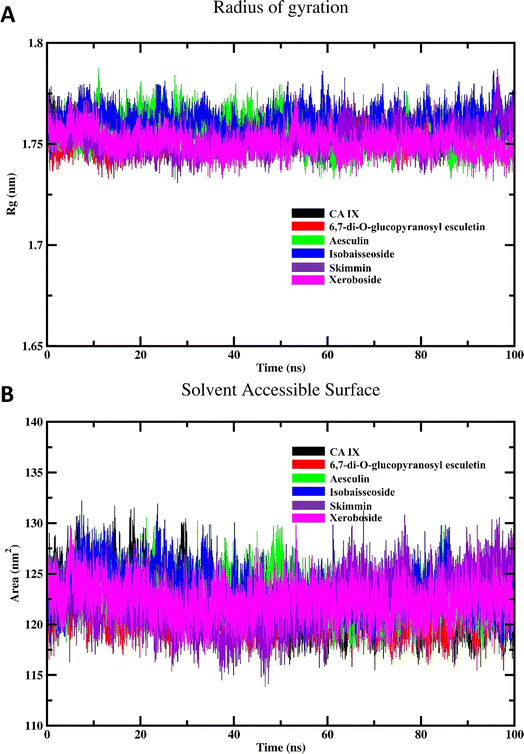 | ||
| Fig. 7 MD simulation of CA IX and its complexes with C. officinalis-derived coumarins; (A) radius of gyration of CA IX and CA IX–coumarin complexes and (B) SASA of CA IX and various complexes. | ||
The results of the SASA calculations for the free CA IX enzyme and its complexes with various coumarins are depicted in Fig. 7B. The SASA profile provides insights into the surface exposure of the enzyme and its complexes to the solvent, which is a critical factor in understanding the interaction and stability of the complexes.14 Throughout the majority of the simulation span, the SASA profiles of different coumarin–CA IX complexes demonstrated an equilibrated behavior similar to that observed for the unbound enzyme. This indicates that the binding of coumarins does not significantly alter the overall solvent exposure of the enzyme structure. During the simulation, a noticeable decrease in SASA values for all systems was observed. This reduction in SASA values suggested that the enzyme and its complexes become more compact over time, with less surface area exposed to the solvent. Such a decrease in SASA is indicative of a stable binding interaction where the enzyme–inhibitor complex achieves a more energetically favorable conformation by minimizing its solvent-exposed surface area. The equilibrated and reduced SASA profiles across the different complexes suggested that the coumarins contribute to the stabilization of the enzyme by promoting a more compact structure. This stabilization is crucial for the inhibitory activity, as it indicates that the enzyme–inhibitor complex achieves a conformation that is less prone to destabilizing interactions with the solvent.
The SASA findings are consistent with the Rg results, reinforcing the notion that the binding of coumarins leads to a stabilized and compact enzyme–inhibitor complex. The observed decrease in SASA values further supports the idea that these complexes are energetically favorable and structurally stable. This stability is essential for the effective inhibition of CA IX by the coumarins, as a stable complex is less likely to dissociate and more likely to maintain its inhibitory function.
The RMSF profile of the free CA IX and its complexes with different coumarins is depicted in Fig. 8B. RMSF analysis provides insights into the flexibility of individual residues within the protein structure over the course of the MD simulation. High RMSF values indicate regions of the protein that exhibit significant flexibility, while low RMSF values denote more rigid regions. The RMSF profiles of the CA IX–coumarin complexes were found to be similar to that of the free enzyme. This observation suggested that the binding of coumarins with CA IX does not induce significant conformational changes or alter the flexibility of the enzyme residues in a substantial manner. Despite the similarity in RMSF profiles, subtle differences were observed in specific regions, which may reflect localized interactions between the enzyme and coumarins. These localized interactions can stabilize certain regions of the enzyme, potentially enhancing the inhibitory effects of the coumarins without causing widespread conformational changes.
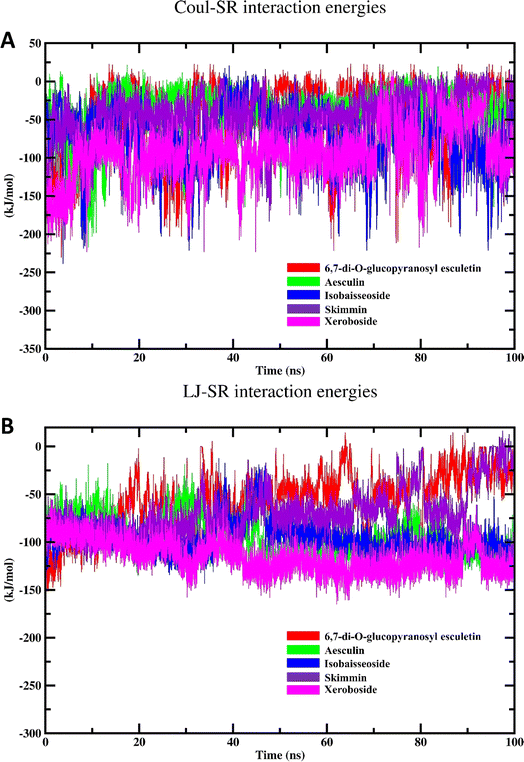
| Coul-SR interaction energy | LJ-SR interaction energy | |||
|---|---|---|---|---|
| Average (kJ mol−1) | RMSD (nm) | Average (kJ mol−1) | RMSD (nm) | |
| 6,7-di-O-glucopyranosyl esculetin | −46.27 ± 4.7 | 45.69 | −58.32 ± 3.1 | 37.69 |
| Aesculin | −37.90 ± 3.0 | 7.80 | −95.40 ± 1.9 | 17.74 |
| Isobaisseoside | −72.47 ± 3.8 | 36.79 | −96.67 ± 1.73 | 12.66 |
| Skimmin | −42.43 ± 3.1 | 32.14 | −68.74 ± 2.5 | 11.46 |
| Xeroboside | −94.09 ± 3.6 | 32.45 | −115.83 ± 1.7 | 11.20 |
| ΔEvdw | ΔEele | ΔGsolv | ΔGgas | ΔGtotal | |
|---|---|---|---|---|---|
| 6,7-di-O-glucopyranosyl esculetin | −11.71 ± 0.79 | −13.42 ± 1.93 | 19.46 ± 1.31 | −25.13 ± 2.11 | −5.67 ± 2.35 |
| Aesculin | −25.08 ± 0.64 | −7.94 ± 1.64 | 20.86 ± 1.11 | −33.02 ± 1.77 | −12.16 ± 2.06 |
| Isobaisseoside | −25.13 ± 1.76 | −15.16 ± 1.38 | 26.30 ± 1.30 | −40.29 ± 2.20 | −14.00 ± 2.67 |
| Skimmin | −17.96 ± 1.65 | −11.66 ± 1.53 | 18.76 ± 3.19 | −29.62 ± 2.25 | −10.86 ± 3.72 |
| Xeroboside | −34.62 ± 1.78 | −31.58 ± 1.96 | 38.94 ± 0.19 | −66.20 ± 2.63 | −27.26 ± 2.48 |
4. Conclusion
This study examined the inhibitory capabilities of C. officinalis-derived coumarins against CA IX using an integrated approach, including in vitro and in silico investigations. The results identified xeroboside as the most effective CA IX inhibitor followed by isobaisseoside and both compounds displayed a mixed inhibition mode. Molecular docking revealed polar and hydrophobic interactions between the isolated coumarins and CA IX. MD simulations offered insights into the dynamic behavior of the coumarins within the CA IX binding site. Xeroboside and isobaisseoside exhibited consistent trajectories, with lower Coul-SR and LJ-SR interaction energy values, indicating favorable electrostatic and van der Waals interactions. Xeroboside showed extensive hydrogen bonding with CA IX, affirming its high inhibitory potential. Additionally, xeroboside exhibited lower average Rg and SASA values, suggesting a more compact structure and reduced solvent exposure of the xeroboside–CA IX complex. The MM/PBSA analysis indicated that xeroboside had the most favorable binding free energies when interacting with CA IX. Furthermore, the PEL analysis showed distinct and stable conformational states for the CA IX–ligand complexes, with xeroboside presenting the most stable and lowest energy configuration. These computational results are consistent with our experimental findings, highlighting the potential of xeroboside and isobaisseoside as potent CA IX inhibitors. Therefore, C. officinalis-derived coumarins are promising CA IX inhibitors, pending in vitro, in vivo, and clinical investigations to determine their exact mechanism(s) of action.Data availability
The manuscript and ESI† contain all data supporting the reported results.Conflicts of interest
All authors declare no conflicts of interest in relation to the manuscript.Acknowledgements
Princess Nourah bint Abdulrahman University Researchers Supporting Project Number (PNURSP2024R381), Princess Nourah bint Abdulrahman University, Riyadh, Saudi Arabia. This work was carried out with the support from the project PID2023-150717NB-I00 from Ministerio de Ciencia, Innovacion y Universidades in Spain and the PRIES-CM project Ref: Y2020/EMT-6290 from the Comunidad Autónoma de Madrid. The authors express their gratitude to the Centro de Computación Científica of the UAM (CCC-UAM) for providing the computing time.References
- M. Imtaiyaz Hassan, B. Shajee, A. Waheed, F. Ahmad and W. S. Sly, Bioorg. Med. Chem., 2013, 21, 1570–1582 Search PubMed.
- A. Queen, H. N. Bhutto, M. Yousuf, M. A. Syed and M. I. Hassan, Semin. Cancer Biol., 2022, 86, 899–913 CrossRef CAS PubMed.
- S.-H. Lee and J. R. Griffiths, Cancers, 2020, 12, 1616 CrossRef CAS PubMed.
- M. Logozzi, C. Capasso, R. Di Raimo, S. Del Prete, D. Mizzoni, M. Falchi, C. T. Supuran and S. Fais, J. Enzyme Inhib. Med. Chem., 2019, 34, 272–278 CrossRef CAS PubMed.
- J. K. Rasmussen and E. Boedtkjer, J. Cereb. Blood Flow Metab., 2018, 38, 492–505 CrossRef CAS PubMed.
- J. Y. Lee, M. Onanyan, I. Garrison, R. White, M. Crook, M. F. Alexeyev, N. Kozhukhar, V. Pastukh, E. R. Swenson, C. T. Supuran and T. Stevens, Am. J. Physiol.: Lung Cell. Mol. Physiol., 2019, 317, L188–L201 CrossRef CAS PubMed.
- D. N. Olennikov and N. I. Kashchenko, Molecules, 2022, 27, 8626 CrossRef CAS PubMed.
- N. F. Komissarenko, V. T. Chernobai and A. I. Derkach, Chem. Nat. Compd., 1988, 24, 675–680 CrossRef.
- A. Raal and K. Kirsipuu, Nat. Prod. Res., 2011, 25, 658–662 CrossRef CAS PubMed.
- H. Neukirch, M. D'Ambrosio, S. Sosa, G. Altinier, R. Della Loggia and A. Guerriero, Chem. Biodiversity, 2005, 2, 657–671 CrossRef CAS PubMed.
- K. Shahane, M. Kshirsagar, S. Tambe, D. Jain, S. Rout, M. K. M. Ferreira, S. Mali, P. Amin, P. P. Srivastav, J. Cruz and R. R. Lima, Pharmaceuticals, 2023, 16, 611 CrossRef CAS PubMed.
- E. H. M. Hassanein, A. M. Sayed, O. E. Hussein and A. M. Mahmoud, Oxid. Med. Cell. Longevity, 2020, 2020, 1675957 Search PubMed.
- D. Pal and S. Saha, in Plant-derived Bioactives: Chemistry and Mode of Action, ed. M. K. Swamy, Springer Singapore, Singapore, 2020, pp. 205–222, DOI:10.1007/978-981-15-2361-8_9.
- R. S. Alruhaimi, A. M. Mahmoud, I. Elbagory, A. F. Ahmeda, A. A. El-Bassuony, A. M. Lamsabhi and E. M. Kamel, Bioorg. Chem., 2024, 147, 107397 CrossRef CAS PubMed.
- J. A. Verpoorte, S. Mehta and J. T. Edsall, J. Biol. Chem., 1967, 242, 4221–4229 CrossRef CAS PubMed.
- S. Iqbal, M. Saleem, M. K. Azim, M. Taha, U. Salar, K. M. Khan, S. Perveen and M. I. Choudhary, Bioorg. Chem., 2017, 72, 89–101 CrossRef CAS PubMed.
- N. Guex and M. C. Peitsch, Electrophoresis, 1997, 18, 2714–2723 CrossRef CAS PubMed.
- E. F. Pettersen, T. D. Goddard, C. C. Huang, G. S. Couch, D. M. Greenblatt, E. C. Meng and T. E. Ferrin, J. Comput. Chem., 2004, 25, 1605–1612 CrossRef CAS PubMed.
- C. Lee, W. Yang and R. G. Parr, Phys. Rev. B: Condens. Matter Mater. Phys., 1988, 37, 785 CrossRef CAS PubMed.
- A. D. Becke, Phys. Rev. A: At., Mol., Opt. Phys., 1988, 38, 3098 CrossRef CAS PubMed.
- W. J. Hehre, L. Radom, P. V. R. Schleyer and J. A. Pople, Ab Initio Molecular Orbital Theory, Wiley New York, 1986 Search PubMed.
- M. J. Frisch, G. W. Trucks, H. B. Schlegel, G. E. Scuseria, M. A. Robb, J. R. Cheeseman, G. Scalmani, V. Barone, G. A. Petersson, H. Nakatsuji, X. Li, M. Caricato, A. V. Marenich, J. Bloino, B. G. Janesko, R. Gomperts, B. Mennucci, H. P. Hratchian, J. V. Ortiz, A. F. Izmaylov, J. L. Sonnenberg, D. Williams-Young, F. Ding, F. Lipparini, F. Egidi, J. Goings, B. Peng, A. Petrone, T. Henderson, D. Ranasinghe, V. G. Zakrzewski, J. Gao, N. Rega, G. Zheng, W. Liang, M. Hada, M. Ehara, K. Toyota, R. Fukuda, J. Hasegawa, M. Ishida, T. Nakajima, Y. Honda, O. Kitao, H. Nakai, T. Vreven, K. Throssell, J. A. Montgomery Jr, J. E. Peralta, F. Ogliaro, M. J. Bearpark, J. J. Heyd, E. N. Brothers, K. N. Kudin, V. N. Staroverov, T. A. Keith, R. Kobayashi, J. Normand, K. Raghavachari, A. P. Rendell, J. C. Burant, S. S. Iyengar, J. Tomasi, M. Cossi, J. M. Millam, M. Klene, C. Adamo, R. Cammi, J. W. Ochterski, R. L. Martin, K. Morokuma, O. Farkas, J. B. Foresman and D. J. Fox, GaussView 5.0. Wallingford, E.U.A., Gaussian 16, Revision B.01, Gaussian, Inc., Wallingford CT, 2016 Search PubMed.
- O. Trott and A. J. Olson, J. Comput. Chem., 2010, 31, 455–461 CrossRef CAS PubMed.
- S. A. Ramadan, E. M. Kamel, M. A. Ewais, A. A. Khowailed, E. H. M. Hassanein and A. M. Mahmoud, Environ. Sci. Pollut. Res., 2023, 30, 49197–49214 CrossRef CAS PubMed.
- P. Bauer, B. Hess and E. Lindahl, November, 2022, 16, 2022 Search PubMed.
- M. J. Abraham, T. Murtola, R. Schulz, S. Páll, J. C. Smith, B. Hess and E. Lindahl, SoftwareX, 2015, 1–2, 19–25 CrossRef.
- J. Huang, S. Rauscher, G. Nawrocki, T. Ran, M. Feig, B. L. de Groot, H. Grubmüller and A. D. MacKerell Jr, Nat. Methods, 2017, 14, 71–73 CrossRef CAS PubMed.
- A. D. MacKerell Jr, D. Bashford, M. Bellott, R. L. Dunbrack Jr, J. D. Evanseck, M. J. Field, S. Fischer, J. Gao, H. Guo, S. Ha, D. Joseph-McCarthy, L. Kuchnir, K. Kuczera, F. T. K. Lau, C. Mattos, S. Michnick, T. Ngo, D. T. Nguyen, B. Prodhom, W. E. Reiher, B. Roux, M. Schlenkrich, J. C. Smith, R. Stote, J. Straub, M. Watanabe, J. Wiórkiewicz-Kuczera, D. Yin and M. Karplus, J. Phys. Chem. B, 1998, 102, 3586–3616 CrossRef PubMed.
- B. Hess, C. Kutzner, D. Van Der Spoel and E. Lindahl, J. Chem. Theory Comput., 2008, 4, 435–447 CrossRef CAS PubMed.
- M. Parrinello and A. Rahman, J. Appl. Phys., 1981, 52, 7182–7190 CrossRef CAS.
- S. Genheden and U. Ryde, Expert Opin. Drug Discovery, 2015, 10, 449–461 CrossRef CAS PubMed.
- E. Wang, H. Sun, J. Wang, Z. Wang, H. Liu, J. Z. H. Zhang and T. Hou, Chem. Rev., 2019, 119, 9478–9508 CrossRef CAS PubMed.
- M. S. Valdés-Tresanco, M. E. Valdés-Tresanco, P. A. Valiente and E. Moreno, J. Chem. Theory Comput., 2021, 17, 6281–6291 CrossRef PubMed.
- S. Lin, M. Liu, S. Wang, S. Li, Y. Yang and J. Shi, Chinese Journal of Traditional Chinese Medicine, 2008, 33, 1708–1710 CAS.
- R. Liu, Q. Sun, A. Sun and J. Cui, J. Chromatogr. A, 2005, 1072, 195–199 CrossRef CAS PubMed.
- S. D. Sarker, P. G. Waterman and J. A. Armstrong, J. Nat. Prod., 1995, 58, 1109–1115 CrossRef CAS.
- T. S. Wu, Phytochemistry, 1987, 26, 873–875 CrossRef CAS.
- S. Sibanda, B. Ndengu, G. Multari, V. Pompi and C. Galeffi, Phytochemistry, 1989, 28, 1550–1552 CrossRef CAS.
- A. Pesaresi, J. Enzyme Inhib. Med. Chem., 2023, 38, 2245168 CrossRef PubMed.
- E. M. Kamel, H. A. Alqhtani, M. Bin-Jumah, H. A. Rudayni, A. A. El-Bassuony and A. Mokhtar Lamsabhi, Bioorg. Chem., 2024, 150, 107609 CrossRef CAS PubMed.
- E. M. Kamel, A. Bin-Ammar, A. A. El-Bassuony, M. M. Alanazi, A. Altharawi, A. F. Ahmeda, A. S. Alanazi, A. M. Lamsabhi and A. M. Mahmoud, RSC Adv., 2023, 13, 12361–12374 RSC.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4ra05984k |
| ‡ Contributed as first author. |
| This journal is © The Royal Society of Chemistry 2024 |