Open Access Article
Open Access ArticleSol–gel derived ceramic nanocomposite CNFs anchored with a nanostructured CeO2 modified graphite electrode for monitoring the interaction of a selective tyrosine kinase inhibitor capmatinib with dsDNA†
Ahmed Z. Alanazi a,
Khalid Alhazzani
a,
Khalid Alhazzani a,
Mohamed M. El-Wekil
a,
Mohamed M. El-Wekil b,
Al-Montaser Bellah H. Ali
b,
Al-Montaser Bellah H. Ali b,
Mahmoud Darweeshc and
Hossieny Ibrahim
b,
Mahmoud Darweeshc and
Hossieny Ibrahim *de
*de
aDepartment of Pharmacology and Toxicology, College of Pharmacy, King Saud University, Riyadh, Saudi Arabia
bDepartment of Pharmaceutical Analytical Chemistry, Faculty of Pharmacy, Assiut University, Assiut, Egypt
cDepartment of Medical Biochemistry and Microbiology, Uppsala University, Uppsala, Sweden
dDepartment of Chemistry, Faculty of Science, Assiut University, Assiut 71516, Egypt. E-mail: Hossieny.Ibrahim@aun.edu.eg
eSchool of Biotechnology, Badr University in Assiut, Assiut 2014101, Egypt
First published on 29th October 2024
Abstract
In the current study, the potential interaction mechanisms between capmatinib (CAP), a selective tyrosine kinase inhibitor, and calf thymus double-stranded DNA (ds-DNA) were evaluated. In this research, we construct an amplified electrochemical platform based on a disposable pencil graphite electrode (PGE) modified with nanostructured CeO2 decorated carbon nanofiber ceramic film (CeNPs@CNF-CF) for monitoring CAP–dsDNA interaction at physiological pH. The morphology and structure of the obtained CeNPs@CNF nanocomposite were characterized. The CeNPs@CNF-CF/PGE was characterized by scanning electron microscopy (SEM). The CAP–dsDNA interaction was examined using cyclic voltammetry (CV) and square wave voltammetry (SWV) techniques. Voltammetric experiments were conducted using CeNPs@CNF-CF/PGE. The interaction of CAP with dsDNA was investigated after applying different incubation times. The addition of dsDNA to the CAP solution decreased the peak currents of the latter and led to a negative shift in peak potentials, suggesting that the electrostatic type of interaction is the most likely to occur. SWV was employed to quantify dsDNA, demonstrating excellent sensitivity (LOD = 5 × 10−8 M). The binding constant (Kb) of CAP and dsDNA was calculated to be 4.54 ± 0.18 × 104 M−1 using SW voltammetric data.
1. Introduction
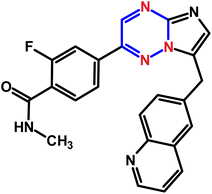
Tyrosine kinase inhibitors (TKIs) represent a class of targeted therapeutic agents that have revolutionized the treatment of various cancers by interfering with the signaling pathways essential for tumor growth and survival. Capmatinib (CAP, Fig. 1), a selective tyrosine kinase inhibitor (TKI) targeting the c-MET receptor, has demonstrated significant potential in treating various cancers, particularly non-small cell lung cancer (NSCLC).1DNA (deoxyribonucleic acid) is the molecule that carries genetic information in living organisms. DNA's structure and its ability to store and transmit genetic information make it a critical target for many drugs. Understanding the interaction between drugs and DNA is pivotal in the fields of pharmacology, medicinal chemistry, and molecular biology.2,3 Drug–DNA interactions form the foundation of numerous therapeutic strategies, particularly in the treatment of cancers, genetic disorders, and infectious diseases.4–8 Cancer remains one of the leading causes of mortality worldwide, necessitating the continuous development of effective therapeutic strategies. By elucidating the mechanisms of anticancer drug–DNA interactions, researchers can design more selective and potent anti-cancer agents, ultimately improving therapeutic outcomes and reducing side effects.
The interaction between a drug and dsDNA is dependent on the specific characteristics of the molecule. There are three ways that small compounds, including many drugs, can bind to dsDNA, including electrostatic interaction, intercalation, and groove binding.9,10 A literature survey reveals that drug–DNA interactions can be studied by a variety of analytical techniques, such as spectroscopy11 chromatography,12,13 colorimetric14,15 and electrochemical methods.16–18 Among the techniques mentioned, voltammetry is the simplest, combining the sensitivity of the measurements with their speed, and is currently one of the cheapest techniques available for studying drug–DNA interactions.19–23 In electrochemical research, numerous efforts have been made to develop modified electrodes based on various nanomaterials, including metal nanoparticles,24,25 carbon nanomaterials,26,27 and metal oxides nanostructured.28,29
Among several types of carbon nanomaterials, carbon nanofibers (CNF) offer distinct surface morphology, excellent electrical conductivity, outstanding chemical stability, and a high surface area. Another interesting kind of nanomaterial is cerium oxide nanoparticles (CeNPs), owing to their large oxygen storage capacity, excellent electrocatalytic activity, environmentally benign nature, and low cost.30 Moreover, CeNPs play a very important role in improving the electrochemical sensing detection performance.31–33 Nevertheless, the low conductivity and aggregation of CeNPs limit their application in electrochemical analysis. Therefore, the integration of CeNPs with highly conductive CNF is necessary to synthesize an electrocatalyst with synergistic features that enhance the electrical conductivity, large electrochemically active surface area (EASA), and electrocatalytic activity.
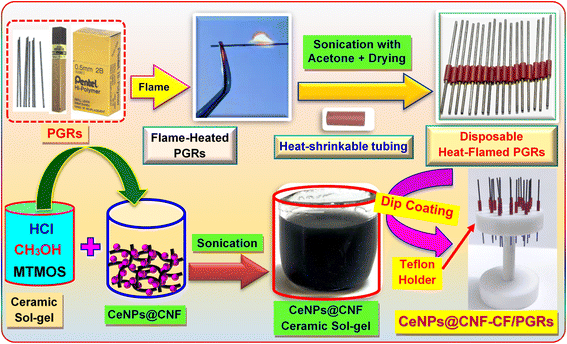
In electroanalytical studies, different types of carbon-based electrochemical sensors required extensive cleaning operations because their surfaces were rapidly deactivated. Among the carbon-based electrodes, the commercially available pencil graphite electrode (PGE) is the most desirable owing to its disposability, low technology, low cost, good mechanical rigidity, and electronic conductivity. Interestingly, the convenient and rapid mechanical surface renewal of the PGE obviates the need for an additional regeneration step.34–38 Since pencil graphite rod (PGR) is a composite material with different hardness due to the different ratios of graphite, clay, and binder, using the PGRs as the working electrode directly may inevitably result in the appearance of some unexpected electrochemical signals. Consequently, we sought to cover the PGRs with a conductive ceramic layer composed of a CeNPs@CNF nanocomposite. Thus, we aimed to encapsulate the PGRs with a conductive ceramic film consisting of a CeNPs@CNF nanocomposite.
The sol–gel method is a straightforward procedure for obtaining carbon ceramic electrodes (CCE), which are a very popular type of carbon-based electrodes.39,40 Even though CCEs provide excellent sensitivity and precision, their use involves a labor-intensive process, requiring detailed preparation, electrode polishing, and surface cleaning before each electrochemical test, which can be quite time-consuming. Consequently, this study introduces a new ceramic nanocomposite film coated on PGE (CeNPs@CNF-CF/PGE), designed as a single-use electrode for detection of double-strand DNA (ds-DNA) and monitoring CAP–dsDNA interaction. As far as we know, there is no study in the literature based on voltammetric monitoring of the interaction between CAP and dsDNA. This research represents the first exploration of the mechanism by which CAP binds to dsDNA, employing CV and SWV as investigative tools. Moreover, the binding constant of the CAP–dsDNA interaction was obtained by the titration of CAP with dsDNA at physiological pH using CeNPs@CNF-CF/PGE.
2. Experimental
2.1. Reagents and instruments
ESI describe the details.†2.2. Synthesis of CeNPs@CNF
The CNF powder underwent an impurity removal process by being treated with a HNO3/H2SO4 mixture (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3) and stirred continuously for 8 hours at 50 °C.41 Following this, the black mixture was centrifuged to extract the cleaned CNF, which was finally dried in an oven at 60 °C for 10 hours. According to a prior study,42 the CeNPs@CNF nanocomposite was successfully synthesized. In short, 2.5 g of cerium nitrate was combined with 40 mL of deionized water and stirred for 30 minutes. Concurrently, 1.0 g of CNF was added to isopropanol. After mixing the two solutions, around 5 mL of 0.5 M NaOH was gradually added. Then, the mixture was placed in a Teflon-lined autoclave at 75 °C for about 5 h. In the next step, the black residue was washed after filtration. The resulting CeNPs@CNF nanocomposite was placed in an oven and dried at 80 °C for 10 hours. For comparison, CeNPs was obtained by using the similar method, except that CNF was not added (Fig. S2, ESI†).
3) and stirred continuously for 8 hours at 50 °C.41 Following this, the black mixture was centrifuged to extract the cleaned CNF, which was finally dried in an oven at 60 °C for 10 hours. According to a prior study,42 the CeNPs@CNF nanocomposite was successfully synthesized. In short, 2.5 g of cerium nitrate was combined with 40 mL of deionized water and stirred for 30 minutes. Concurrently, 1.0 g of CNF was added to isopropanol. After mixing the two solutions, around 5 mL of 0.5 M NaOH was gradually added. Then, the mixture was placed in a Teflon-lined autoclave at 75 °C for about 5 h. In the next step, the black residue was washed after filtration. The resulting CeNPs@CNF nanocomposite was placed in an oven and dried at 80 °C for 10 hours. For comparison, CeNPs was obtained by using the similar method, except that CNF was not added (Fig. S2, ESI†).
2.3. Modification of PGE
The construction of CeNPs@CNF-CF modified PGE (CeNPs@CNF-CF/PGE) is shown in Scheme 1. The detailed description is presented in ESI†.3. Results and discussion
3.1. Morphological studies
Material characterizations were carried out to validate the presented structure. Accordingly, the obtained CeNPs@CNF was characterized via XRD (Fig. S3, ESI†). The scanning electron microscopy (SEM) images presented in Fig. 2A show CeNPs deposited on the surface of CNF (CeNPs@CNF) and a smooth network structure of CNF (Fig. 2A, inset). In addition, transmission electron microscopy (TEM) micrograph of CeNPs@CNF nanocomposite (Fig. 2B) demonstrated that the uniform decoration of nanostructured CeO2 on the surface of CNF was successfully accomplished. According to the high-resolution TEM (HRTEM) image of the CeNPs@CNF (Fig. 2C), the observed lattice fringe spacings 0.29 nm and 0.34 nm are associated with the CeNPs crystal planes (200) and (111), respectively. Moreover, EDX study offers clear evidence for the elemental composition of CeNPs@CNF nanocomposite (Fig. S4A†). As shown by the spectrum, Ce, O, and C are components for the nanocomposite. Fig. S4B† exhibits the EDX mapping diagram of the nanocomposite, which authenticates that CNF was successfully decorated with CeNPs. SEM analysis was used to study PGE surface morphologies in the absence and presence of CeNPs@CNF ceramic film. As indicated in Fig. 2E and F, the PGE is uniformly covered with CeNPs@CNF-CF and has a different morphology than the bare PGE (Fig. 2D).3.2. Electrochemical analysis of CeNPs@CNF-CF/PGE surface by CV
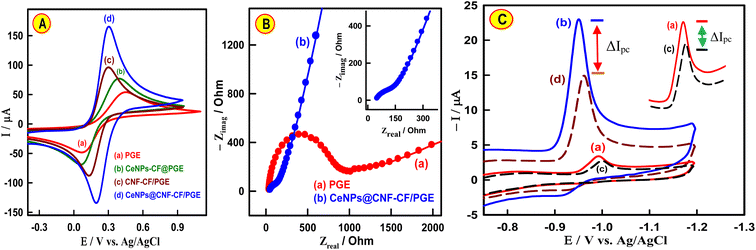
The redox peak current of [Fe(CN)6]3−/4− is extensively used as an electrochemical probe. Fig. 3A represents the CV of a 5 × 10−3 M [Fe(CN)6]3−/4− system containing KCl solution (0.1 M) for evaluating the properties of the bare PGE, CeNPs-CF/PGE, CNF-CF/PGE and CeNPs@CNF-CF/PGE. From the comparison of redox peak currents, it is distinctly observed that CeNPs@CNF-CF/PGE (curve d) displays outstanding electrocatalytic signal towards the [Fe(CN)6]3−/4− compared to PGE (curve a), CeNPs-CF/PGE (curve b), and CNF-CF/PGE (curve c). In addition, a redox peak was seen with a separation peak (ΔEP) of 0.35 V for bare PGE, while the ΔEP values for CeNPs-CF/PGE, CNF-CF/PGE and CeNPs@CNF-CF/PGE are 0.32 V, 0.17 V and 0.10 V, respectively. Interestingly, the ΔEP value for CeNPs@CNF-CF/PGE is smaller than those of bare PGE, CeNPs-CF/PGE and CNF-CF/PGE, indicating the rate of electron transfer and redox peak current for CeNPs@CNF-CF/PGE is significantly increased which is a result of the excellent electrical conductivity of the combination of CeNPs and CNF ceramic film.In order to verify CV results, the electron transfer behavior of CeNPs@CNF-CF modified PGE was investigated by electrochemical impedance spectroscopy (EIS). Fig. 3B shows the Nyquist plots of EIS for PGE and CeNPs@CNF-CF/PGE in 5.0 mM [Fe (CN)6]3−/4− redox. The semicircle diameters of the EIS are proportional to the electron transfer resistance (RCT). From the graphs, it is observed that a small semicircle with an RCT of about 110 Ω recorded for CeNPs@CNF-CF/PGE (curve b) while the curve of bare PGE (curve a) revealed a greater diameter of semicircles (RCT = 980 Ω). The significant change of RCT as a result of modification of PGE indicated that CeNPs@CNF-CF has been successfully attached to PGE surface.
The EASA of the working electrodes was estimated using the Randles–Sevcik equation.43 It can be estimated from the slope of the plot of IPa vs. ν1/2 (Fig. S5†). Based on the calculation, the EASA values were 0.14 ± 0.005 cm2 and 0.62 ± 0.017 cm2 for unmodified PGE and CeNPs@CNF-CF/PGE, respectively. Therefore, the EASA increased approximately 23% after PGE was wrapped with the proposed CeNPs@CNF nanocomposite ceramic film.
3.3. Voltammetric behavior of CAP at CeNPs@CNF-CF/PGE
Cyclic voltammetric (CV) responses of CAP were recorded on CeNPs@CNF-CF/PGE in phosphate buffer solution (PBS, pH 7.4) and compared with bare PGE (Fig. 3C). In general, CAP shows a reduction peak in the cathodic scan, but no peak was observed in the reverse anodic scan, suggesting that CAP reduction is an irreversible process.38 The PGE responded with a small peak signal at −994 mV (Fig. 3C, curve a). More acceptably, CeNPs@CNF-CF/PGE presented a higher current response (23 ± 0.58 μA) than bare PGE (3.5 ± 0.20 μA) toward electrochemical sensing of CAP (Fig. 3C, curve b). In addition, a shift in cathodic peak potential (EPc) to −950 mV was observed, which could be attributed to large EASA and enhanced electrocatalytic activity. Therefore, the sensing platform based on CeNPs@CNF-CF/PGE could have high sensitivity towards the monitoring of CAP–dsDNA interaction. To confirm that the difference in cathodic currents (ΔI) of the CAP in the absence and presence of dsDNA was studied at PGE and CeNPs@CNF-CF/PGE (Fig. 3C, curves c and d). The low ΔI value of −0.8 μA was observed when using bare PGE (Fig. 3C, curve c). After modifying the PGE with CeNPs@CNF-CF (Fig. 3C, curve d), the cathodic current of CAP significantly decreased in the presence of dsDNA (ΔI = −8.2 μA).The interaction between dsDNA and drugs is influenced by a myriad of factors, one of the most critical being the pH of the supporting electrolyte. Thus, the cathodic current of CAP on the CeNPs@CNF-CF/PGE was initially studied in the broad pH range offered by PBS (3.0–8.0). From Fig. S6,† it is evident that the reduction current of CAP increased gradually from 3.0 to 7.0, and it subsequently attained its maximum current value at pH 7.0 with a favorable peak shape. Obviously, the slope of the EPc − pH dependence was 60 mV pH−1, which implies equal protons and electrons in the reduction of –N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–C
C–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N– group in CAP at CeNPs@CNF-CF/PGE.38 Furthermore, SWV was employed to optimize various experimental conditions to have the best sensing response of CAP on the sensing electrode. These parameters include adsorption time and deposition potential (Fig. S7†). Next, CV measurements were obtained at various scan rates (ν) ranging from 0.05 to 0.40 V s−1 in pH 7.0 PBS to investigate how scan rate impacts CAP behaviour (Fig. S8A†). Moreover, the adsorption-controlled nature of the process at the CeNPs@CNF-CF/PGE is supported by the slope of log
N– group in CAP at CeNPs@CNF-CF/PGE.38 Furthermore, SWV was employed to optimize various experimental conditions to have the best sensing response of CAP on the sensing electrode. These parameters include adsorption time and deposition potential (Fig. S7†). Next, CV measurements were obtained at various scan rates (ν) ranging from 0.05 to 0.40 V s−1 in pH 7.0 PBS to investigate how scan rate impacts CAP behaviour (Fig. S8A†). Moreover, the adsorption-controlled nature of the process at the CeNPs@CNF-CF/PGE is supported by the slope of log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Ipc versus log
Ipc versus log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ν, as shown in Fig. S8B† and expressed as: log
ν, as shown in Fig. S8B† and expressed as: log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Ipc (μA) = 0.75
Ipc (μA) = 0.75![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) log
log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ν (mV s−1) − 0.26 (R2 = 0.997). The slope value (0.75) confirms the adsorption-controlled nature of the process.
ν (mV s−1) − 0.26 (R2 = 0.997). The slope value (0.75) confirms the adsorption-controlled nature of the process.
3.4. Reproducibility, repeatability and storage stability studies
The proposed electrode has been constructed in the seven diverse electrodes (n = 7) with the SWV method for 85 nM CAP recorded consecutively to examine the reproducibility (Fig. S9A†). The RSD% value obtained was lower than 2.7%, indicating that the disposable sensing electrode has excellent reproducibility. Additionally, the repeatability of CeNPs@CNF-CF/PGE was analyzed using the same electrode ten times, as shown in Fig. S9B,† in which the obtained RSD value was 1.8%. As a result, a high repeatability of the CAP sensing platform was obtained due to its simplicity of construction. Furthermore, the storage stability CeNPs@CNF-CF/PGE was evaluated by keeping the constructed electrode at room temperature and using it for measurement on different days after preparation (Fig. S9C†). Remarkably, the current responses of the sensor only decrease by 2.5% after 40 days of continuous measurements, which indicates that the CeNPs@CNF ceramic film was not affected by environmental temperature, humidity, or air oxidation.3.5. Voltammetric studies of CAP–dsDNA interaction
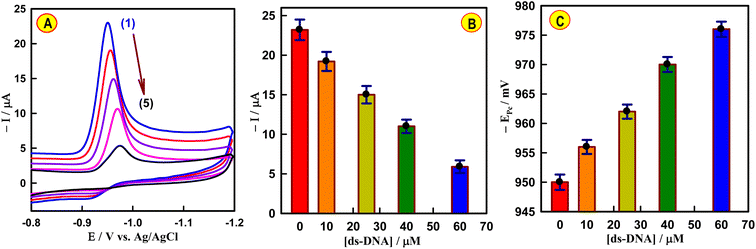
Interactions between drugs and dsDNA have often been conducted at the physiological pH (PBS, pH 7.4).44 Square wave voltammetric (SWV) curves of the interaction were initially performed by incubating a CAP solution (4.5 × 10−7 M) with dsDNA (6.5 × 10−6 M) in PBS of pH 7.4 at the disposable CeNPs@CNF-CF/PGE. The SWV curves were recorded after different incubation times ranging from 0.5 to 10 min. As depicted in Fig. S10† the current CAP responses were observed to decrease significantly up to 6 min, after which they remained constant; thus, the optimized incubation time was 6 min.Then, CV curves of CAP were measured after the addition of variable amounts of dsDNA at CeNPs@CNF-CF/PGE to examine the mode of binding (Fig. 4A). When calf thymus ds-DNA was injected into CAP solution, a drop in the cathodic signal was observed, suggesting the existence of an interaction between CAP and dsDNA (Fig. 4B). The change in peak potential provides valuable information about the mode of interaction between CAP and dsDNA. As seen in Fig. 4C, as more dsDNA was added, the peak potential of CAP shifted to greater negative values, indicating that the electrostatic binding mode was more likely to occur.9,45,46
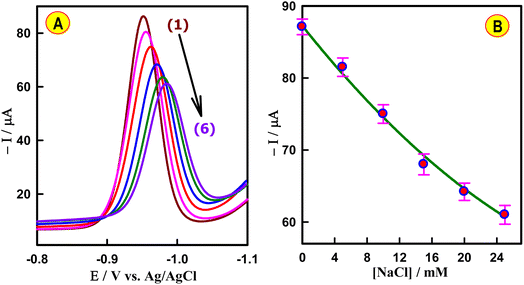
To reveal the nature of the CAP–dsDNA interaction, the effect of ionic strength was evaluated using the SWV technique. As illustrated in Fig. 5A, increasing the NaCl concentration caused the peak potential of CAP to shift in a more negative direction. In addition, the current signal of 4.5 × 10−7 M CAP after injection of 6.5 × 10−6 M dsDNA was gradually decreased with the increasing concentration (0–25 mM) of NaCl (Fig. 5B). These results imply that the concentration of NaCl has a significant influence on the reaction process, demonstrating that the electrostatic interaction between CAP and dsDNA is dominant.
3.6. Calculation of the binding constant
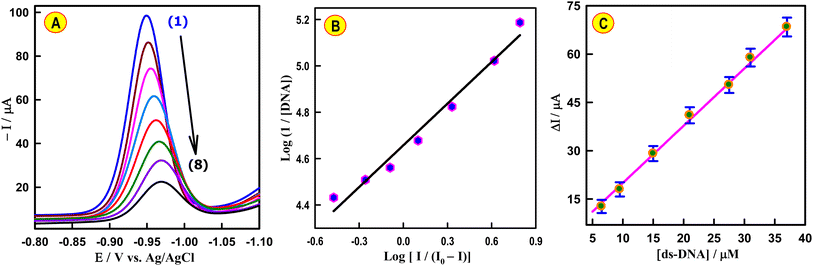
SWV was employed to conduct current titration, with the concentration of CAP remaining constant while the concentration of dsDNA was varied in PBS (pH 7.4) at CeNPs@CNF-CF/PGE (Fig. 6A). The changes in the observed reduction peak current of CAP upon the addition of an increasing amount of dsDNA can be used for quantifying the binding constant (Kb) from a straight line plot (Fig. 6B) according to eqn (1).47
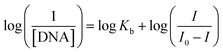 | (1) |
ΔG0 = −RT![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Ln Ln![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Kb Kb
| (2) |
The negative value of ΔG0 (−45.4 ± 0.65 kJ mol−1) proves the CAP and dsDNA interaction and indicates that binding occurred spontaneously.
3.7. Analytical feature of CAP–dsDNA interaction
The decline in the cathodic current of the anticancer drug CAP, observed with each injection of dsDNA, can be utilized to monitor the dsDNA concentration at a physiological pH of 7.4 (Fig. 6A). The cathodic current of CAP decreased upon the addition of dsDNA concentration. The decrease in the sensing response was proportional to the dsDNA concentration (6.5 to 37 μM). The calibration curve was plotted between the ΔI and dsDNA concentrations, as described in Fig. 6C. The regression equation was found to be ΔI (μA) = 1.8 CdsDNA (μM) + 2.2 (R2 = 0.996). Based on S/N = 3, the LOD value was calculated to be 5.0 × 10−8 M. To apply the proposed sensor for analytical application, the effects of some common interfering substances such as inorganic ions (Fe3+, Ca2+, Mg2+, Zn2+ and Cl−) and alanin, EDTA, glutaric acid, and ascorbic in the determination of ds-DNA was examined by SWV under the optimized conditions. The recovery values were from 96.5 to 103.0% with the relative standard deviation (RSD) below 2.3%. The results offer an opportunity to use CAP drug as a new indicator for monitoring ds-DNA concentration.4. Conclusions
The interaction of the anticancer drug CAP with dsDNA at the physiological pH value of 7.4 was investigated for the first time. In this investigation, an amplified sensor based on a disposable PGE modified with ceramic film (CeNPs@CNF-CF) for monitoring CAP–dsDNA interaction was constructed. From the comparison of redox peak currents, it is distinctly observed that CeNPs@CNF-CF/PGE displays outstanding electrocatalytic activity towards the [Fe(CN)6]3−/4− redox couple compared to PGE. Interestingly, the EASA increased approximately 23% after PGE was wrapped with the proposed CeNPs@CNF nanocomposite ceramic film. The CAP–dsDNA interaction was examined using CV and SWV techniques. As more dsDNA was added, the peak potential of CAP shifted to greater negative values, indicating that the electrostatic binding mode was more likely to occur. The binding of CAP to double-stranded DNA was examined following various incubation times. The binding constant of the CAP–dsDNA was estimated as 4.54 ± 0.18 × 104 M−1, which indicates that the most stable complex is formed at physiological pH.Data availability
The data that support the findings of this study are available on request from the corresponding author.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors extended their appreciation to Distinguished Scientist Fellowship Program at King Saud University, Riyadh, Saudi Arabia for funding this work through Research Supporting Project Number (RSPD2024R563).References
- Y. L. Wu, L. Zhang, D. W. Kim, X. Liu, D. H. Lee, J. C. H. Yang, M. J. Ahn, J. F. Vansteenkiste, W. C. Su, E. Felip, V. Chia, S. Glaser, P. Pultar, S. Zhao, B. Peng, M. Akimov and D. S. W. Tan, Phase Ib/II study of capmatinib (INC280) plus gefitinib after failure of epidermal growth factor receptor (EGFR) inhibitor therapy in patients with EGFR-mutated, MET factor-dysregulated non-small-cell lung cancer, J. Clin. Oncol., 2018, 36, 3101–3109 CrossRef CAS.
- C. B. Chen, J. Chen, J. Wang, Y. Y. Zhu and J. H. Shi, Combined spectroscopic and molecular docking approach to probing binding interactions between lovastatin and calf thymus DNA, Luminescence, 2015, 30, 1004–1010 CrossRef CAS PubMed.
- K. Y. Chen, K. L. Zhou, Y. Y. Lou and J. H. Shi, Exploring the binding interaction of calf thymus DNA with lapatinib, a tyrosine kinase inhibitor: Multi-spectroscopic techniques combined with molecular docking, J. Biomol. Struct. Dyn., 2019, 37, 576–583 CrossRef CAS PubMed.
- M. Hasanzadeh and N. Shadjou, Pharmacogenomic study using bio- and nanobioelectrochemistry: Drug–DNA interaction, Mater. Sci. Eng. C, 2016, 61, 1002–1017 CrossRef CAS PubMed.
- M. Sirajuddin, S. Ali and A. Badshah, Drug–DNA interactions and their study by UV–visible, fluorescence spectroscopies and cyclic voltammetry, J. Photochem. Photobiol., B, 2013, 124, 1–19 CrossRef CAS PubMed.
- S. U. Rehman, T. Sarwar, M. A. Husain, H. M. Ishqi and M. Tabish, Studying non-covalent drug–DNA interactions, Arch. Biochem. Biophys., 2015, 576, 49–60 CrossRef PubMed.
- J. Devi and S. Pachwania, Recent advancements in DNA interaction studies of organotin(IV) complexes, Inorg. Chem. Commun., 2018, 91, 44–62 CrossRef CAS.
- L. S. Foteeva, M. Matczuk and A. R. Timerbaev, Analytical methodology for determination of interactions between metallodrugs and DNA: A critical examination, TrAC, Trends Anal. Chem., 2017, 90, 107–113 CrossRef CAS.
- K. Morawska, T. Popławski, W. Ciesielski and S. Smarzewska, Interactions of lamotrigine with single- and double-stranded DNA under physiological conditions, Bioelectrochemistry, 2020, 136, 107630 CrossRef CAS PubMed.
- Z. Krejcova-Sirlova, J. Barek and V. Vyskocil, Voltammetric studies of the interaction of genotoxic 2-nitrofluorene with DNA, Bioelectrochemistry, 2023, 149, 108326 CrossRef CAS PubMed.
- C. Z. Huang, Y. F. Li and P. Feng, A spectrophotometric study on the interaction of neutral red with double-stranded DNA in large excess, Talanta, 2001, 55, 321–328 CrossRef CAS PubMed.
- D. G. Sar, M. Montes-Bayón, E. B. González and A. Sanz-Medel, Speciation studies of cisplatin adducts with DNA nucleotides via elemental specific detection (P and Pt) using liquid chromatography-inductively coupled plasma-mass spectrometry and structural characterization by electrospray mass spectrometry, J. Anal. At. Spectrom., 2006, 21, 861–868 RSC.
- C. Caramelo-Nunes, T. Tente, P. Almeida, J. C. Marcos and C. Tomaz, Specific berenil DNA interactions: An approach for separation of plasmid isoforms by pseudo-affinity chromatography, Anal. Biochem., 2011, 412(2), 153–158 CrossRef CAS.
- M. S. Han, A. K. Lytton-Jean, B. K. Oh, J. Heo and C. A. Mirkin, Colorimetric screening of DNA-binding molecules with gold nanoparticle probes, Angew Chem. Int. Ed. Engl., 2006, 45, 1807–1810 CrossRef CAS PubMed.
- S. Das and G. S. Kumar, Molecular aspects on the interaction of phenosafranine to deoxyribonucleic acid: Model for intercalative drug–DNA binding, J. Mol. Struct., 2008, 872(1), 56–63 CrossRef CAS.
- F. Tahernejad-Javazmi, M. Shabani-Nooshabadi and H. Karimi-Maleh, Gold nanoparticles and reduced graphene oxide-amplified label-free DNA biosensor for dasatinib detection, New J. Chem., 2018, 42(19), 16378–16383 RSC.
- Y. M. Temerk, M. S. Ibrahim, M. Kotb and W. Schuhmann, Interaction of antitumor flavonoids with dsDNA in the absence and presence of Cu(II), Anal. Bioanal. Chem., 2013, 405, 3839–3846 CrossRef CAS PubMed.
- Y. M. Temerk, M. S. Ibrahim and M. Kotb, Voltammetric and spectroscopic studies on binding of antitumor morin, morin-Cu complex and morin-b-cyclodextrin with DNA, Spectrochim. Acta, Part A, 2009, 71, 1830–1836 CrossRef CAS PubMed.
- Y. M. Temerk, M. S. Ibrahim, H. Ibrahim and W. Schuhmann, Comparative studies on the interaction of anticancer drug irinotecan with dsDNA and ssDNA, RSC Adv., 2018, 8, 25387–25395 RSC.
- Y. M. Temerk, M. S. Ibrahim and M. Kotb, Interactions of an anticancer drug formestane with single and double stranded DNA at physiological conditions, J. Photochem. Photobiol., B, 2015, 149, 27–36 CrossRef CAS PubMed.
- Y. M. Temerk and M. S. Ibrahim, Electrochemical studies and spectroscopic investigations on the interaction of an anticancer drug flutamide with DNA and its analytical applications, J. Electroanal. Chem., 2015, 736, 1–7 CrossRef CAS.
- Y. M. Temerk, M. S. Ibrahim and M. Kotb, Interactions of an anticancer drug lomustine with single and double stranded DNA at physiological conditions analyzed by electrochemical and spectroscopic methods, J. Electroanal. Chem., 2016, 769, 62–71 CrossRef CAS.
- Y. M. Temerk and M. S. Ibrahim, Binding mode and thermodynamic studies on the interaction of the anticancer drug dacarbazine and dacarbazine–Cu(II) complex with single and double stranded DNA, J. Pharm. Biomed. Anal., 2014, 95, 26–33 CrossRef CAS PubMed.
- M. S. Ibrahim and Y. M. Temerk, A novel electrochemical sensor based on gold nanoparticles decorated functionalized carbon nanofibers for selective determination of xanthine oxidase inhibitor febuxostat in plasma of patients with gout, Sens. Actuators, B, 2021, 347, 130626 CrossRef.
- H. M. Ali, I. A. Alhagri and M. S. Ibrahim, Fabrication of an electrochemical sensor based on gold nanoparticle functionalized nanocarbon black hybrid nanocomposite for sensitive detection of anti-cancer drug formestane in biological and pharmaceutical samples, J. Electroanal. Chem., 2022, 907, 116067 CrossRef CAS.
- M. S. Ibrahim and Y. M. Temerk, Surface decoration of functionalized carbon black nanoparticles with nanosized gold particles for electrochemical sensing of diuretic spironolactone in patient plasma, Microchem. J., 2022, 178, 107425 CrossRef.
- M. S. Ibrahim and Y. M. Temerk, Gold nanoparticles anchored graphitized carbon nanofibers ionic liquid electrode for ultrasensitive and selective electrochemical sensing of anticancer drug irinotecan, Microchim. Acta, 2020, 187, 579 CrossRef PubMed.
- Y. M. Temerk and M. S. Ibrahim, Fabrication of a novel electrochemical sensor based on Zn–In2O3 nanorods coated glassy carbon microspheres paste electrode for square wave voltammetric determination of neuroprotective hibifolin in biological fluids and in the flowers of hibiscus vitifolius, J. Electroanal. Chem., 2016, 782, 9–18 CrossRef CAS.
- M. S. Ibrahim, Y. M. Temerk and H. Ibrahim, A novel megestrol acetate electrochemical sensor based on conducting functionalized acetylene black–CeO2NPs nanohybrids decorated glassy carbon microspheres, Talanta, 2019, 200, 324–332 CrossRef.
- D. Manoj, R. Manigandan, S. Rajendran and L. C. Ponce, Self-assembled dendrite-like 3D-CeO2 nanostructures for non-enzymatic vitamin B2 sensor, Mater. Lett., 2021, 295, 129834 CrossRef CAS.
- M. S. Ibrahim and Y. M. Temerk, A novel electrochemical sensor based on functionalized glassy carbon microparticles@CeO2 core–shell for ultrasensitive detection of breast anticancer drug exemestane in patient plasma and pharmaceutical dosage form, Microchem. J., 2021, 167, 106264 CrossRef.
- R. M. K. Mohamed, S. H. Mohamed, A. M. Asran, I. H. Alsohaimi, H. M. Hassan and M. S. Ibrahim, Carbon microspheres uniformly decorated with ceria nanoparticles as an ultrasensitive platform for electrochemical sensing of antihypertensive drug lacidipine in patient plasma and pharmaceutical formulation, Microchem. J., 2023, 187, 108422 CrossRef CAS.
- M. S. Ibrahim, Y. M. Temerk and H. Ibrahim, Fabrication of a new biosensor based on a Sn doped ceria nanoparticle modified glassy carbon paste electrode for the selective determination of the anticancer drug dacarbazine in pharmaceuticals, RSC Adv., 2017, 7, 32357–32366 RSC.
- E. M. Hussien, M. S. Rizk, A. M. Daoud and R. T. El-Eryan, An eco-friendly pencil graphite sensor for voltammetric analysis of the antidepressant vilazodone hydrochloride, Electroanalysis, 2022, 34, 1402–1410 CrossRef CAS.
- Y. Altunkaynak, O. Yavuz and A. Levent, Firstly electrochemical examination of vildagliptin at disposable graphite sensor: Sensitive determination in drugs and human urine by square-wave voltammetry, Microchem. J., 2021, 170, 106653 CrossRef CAS.
- R. T. Hussain, A. K. M. S. Islam, M. Khairuddean and F. B. M. Suah, A polypyrrole/GO/ZnO nanocomposite modified pencil graphite electrode for the determination of andrographolide in aqueous samples, Alexandria Eng. J., 2022, 61, 4209–4218 CrossRef.
- H. Ibrahim and Y. Temerk, A novel disposable electrochemical sensor based on modifying graphite pencil lead electrode surface with nanoacetylene black for simultaneous determination of antiandrogens flutamide and cyproterone acetate, J. Electroanal. Chem., 2020, 859, 113836 CrossRef CAS.
- A. Z. Alanazi, K. Alhazzani, M. M. El-Wekil, A. M. B. Ali, M. Darweesh and H. Ibrahim, A novel disposable ultrasensitive sensor based on nanosized ceria uniformly loaded carbon nanofiber nanoceramic film wrapped on pencil graphite rods for electrocatalytic monitoring of a tyrosine kinase inhibitor capmatinib, Talanta, 2024, 279, 126610 CrossRef CAS PubMed.
- J. Węgiel, B. Burnat and S. Skrzypek, A graphene oxide modified carbon ceramic electrode for voltammetric determination of gallic acid, Diamond Relat. Mater., 2018, 88, 137–143 CrossRef.
- C. Ferrag, M. Noroozifar and K. Kerman, Thiol functionalized carbon ceramic electrode modified with multi-walled carbon nanotubes and gold nanoparticles for simultaneous determination of purine derivatives, Mater. Sci. Eng. C, 2020, 110, 110568 CrossRef CAS PubMed.
- A. G. Osorio, I. C. L. Silveira, V. L. Bueno and C. P. Bergmann, H2SO4/HNO3/HCl- Functionalization and its effect on dispersion of carbon nanotubes in aqueous media, Appl. Surf. Sci., 2008, 255, 2485–2489 CrossRef CAS.
- H. Guo, T. Liu, J. Li, G. Chen and P. Jia, Synthesis and characterization of cerium oxide quantum dots loaded biodegradable dextran matrix for effective pain management, J. Cluster Sci., 2020, 31, 831–837 CrossRef CAS.
- A. J. Bard and L. R. Faulkner. Electrochemical Methods, Fundamentals and Applications. New York: Wiley; 1980 Search PubMed.
- N. Arshad, U. Yunus, S. Razzque, M. Khan, S. Saleem, B. Mirza and N. Rashid, Electrochemical and spectroscopic investigations of isoniazid and its analogs with dsDNA at physiological pH: Evaluation of biological activities, Eur. J. Med. Chem., 2012, 47, 452–461 CrossRef CAS.
- S. Shahzad, B. Dogan-Topal, L. Karadurmus, M. G. Caglayan, T. Taskin Tok, B. Uslu, A. Shah and S. A. Ozkan, Electrochemical, spectroscopic and molecular docking studies on the interaction of calcium channel blockers with dsDNA, Bioelectrochemistry, 2019, 127, 12–20 CrossRef CAS PubMed.
- E. Jabeen, N. K. Janjua, S. Ahmed, I. Tahiri, M. Kashif and A. Javed, DNA binding interaction studies of flavonoid complexes of Cu(II) and Fe(II) and determination of their chemotherapeutic potential, Inorg. Chim. Acta, 2019, 496, 119048 CrossRef CAS.
- R. Hajian, Z. Tayebi and N. Shams, Fabrication of an electrochemical sensor for determination of doxorubicin in human plasma and its interaction with DNA, J. Pharm. Anal., 2017, 7, 27–33 CrossRef.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4ra06158f |
| This journal is © The Royal Society of Chemistry 2024 |