Open Access Article
Open Access ArticleAn NIR-driven biosensor based on the metal-enhanced fluorescence effect and a signal amplification strategy for miRNA detection†
Dabin Liuabf,
Wenzhang Zhuc,
Bin Qiu d and
Shiqian Zhang
d and
Shiqian Zhang *e
*e
aThe First Clinical College of Shandong University, Jinan, 250014, China
bDepartment of Gynecology, Fujian Maternity and Child Health Hospital, College of Clinical Medicine for Obstetrics & Gynecology and Pediatrics, Fujian Medical University, Fuzhou, 350001, China
cCollege of Materials Science and Engineering, Fuzhou University, Fuzhou, 350108, China
dCollege of Chemistry, Fuzhou University, Fuzhou, 350108, China
eDepartment of Gynecology, Qilu Hospital of Shandong University, Jinan, 250014, China. E-mail: Shiqianzhang@yeah.net
fDepartment of Gynecology, Fujian Provincial Governmental Hospital, Fuzhou, 350001, China
First published on 19th December 2024
Abstract
Research has shown that the expression level of microRNA-155 (miRNA-155) is positively correlated with clinical stage and depth of invasion in patients with cervical cancer and cervical intraepithelial neoplasia and tends to be highly expressed. Therefore, it is very important to develop sensitive miRNA-155 analysis methods for the early diagnosis, treatment, and prognostic evaluation of cervical cancer. In this study, a near-infrared light-driven fluorescent biosensor based on the metal-enhanced fluorescence effect of polydopamine-coated upconversion nanoparticle (UP/Au) and two toehold-mediated strand displacement (TMSD) steps was constructed for the detection of miRNA-155. The target miRNA-155 can undergo a TMSD1 reaction with single-stranded DNA1 (ssDNA1) modified on wrinkled silica (WSNs) to form the ssDNA1/miRNA-155 complex and expose toehold 2. Through the TMSD2 reaction, ssDNA2 adsorbed on the UP/Au surface reacts with ssDNA1 to form the ssDNA1/ssDNA2 complex, which replaces miRNA-155 and enables the recovery of the UP/Au fluorescence signal. The target miRNA-155 was reacted with the new ssDNA1 to amplify the detection signal, with a detection range of 0.5–20 pM and a detection limit of 19.76 fM for miRNA-155. In addition, the fluorescent biosensor has been applied for the analysis of miRNA-155 in serum samples, indicating its good practicality.
1 Introduction
MicroRNAs (miRNAs) are an important class of single-stranded, short-stranded, endogenous noncoding RNAs that play critical roles in various biological or pathological processes, such as the immune response, metabolism, hematopoietic differentiation, cell proliferation, differentiation, and apoptosis.1 More importantly, abnormal expression levels of miRNAs are associated with various diseases, including cancer, and can therefore serve as novel biomarkers for cancer classification, early disease diagnosis, and prognosis.2 Research has shown that the expression level of miRNA-155 is positively correlated with clinical stage and depth of invasion in patients with cervical cancer and cervical intraepithelial neoplasia and tends to be highly expressed.3 Therefore, developing sensitive miRNA-155 analysis methods not only helps in understanding the pathogenesis of cervical cancer but also improves the accuracy of its clinical diagnosis and prognosis.At present, the main methods for detecting miRNAs include northern blotting,4 microarray,5 and real-time quantitative polymerase chain reaction.6 Most of these methods have disadvantages such as low sensitivity, long time consumption, susceptibility to environmental influences, and high equipment requirements. In recent years, with the rapid development of biosensing technology, various new miRNA detection methods have been developed, such as electroanalytical biosensors,7,8 photoelectrochemical biosensors,9 and fluorescence biosensors,10 which have the advantages of low cost and high sensitivity. Among them, fluorescent biosensors have attracted attention because of their advantages, such as good repeatability, fast response speed, and simple equipment.11 However, traditional fluorescent biosensors often use fluorescent materials such as organic dyes,12 quantum dots,13 and noble metal nanoclusters14 as signal sources, which have problems such as poor luminescence stability and a low photobleaching threshold.15 In addition, most fluorescent biosensors and fluorescent materials use ultraviolet visible light as the excitation light source. However, ultraviolet visible light has high energy and can cause light damage to biological samples, and its penetration is poor, which limits its application in the field of biosensing.16
Upconversion nanoparticles (UCNPs), a special type of luminescent material, can emit high-energy ultraviolet visible light under low-energy near-infrared (NIR) excitation, resulting in anti-Stokes luminescence properties.17 Because the excitation light of UCNPs is NIR light (usually at 808 nm or 980 nm), it can not only effectively avoid light damage caused by ultraviolet visible excitation but also reduce interference from spontaneous background fluorescence.18 Therefore, UCNPs have broad application prospects in the field of fluorescence biosensing.19 However, high-quality UCNPs are usually prepared in organic solvents, and their surfaces are typically coated with oleic acid (OA), which is hydrophobic.20 The UCNPs used for biological utilization have high dispersivity and fixity in aqueous media, in addition to suitable surface functional groups for subsequent coupling of biomolecules or probe molecules.21 However, complex surface modifications often strongly affect the fluorescence properties of UCNPs. Therefore, improving the fluorescence properties of UCNPs after surface functionalization has become the focus of sensitive detection of low-abundance targets. Therefore, different methods need to be developed to functionalize the surface of hydrophobic UCNPs. Dopamine (DA) is prone to self-aggregation under alkaline aerobic conditions, resulting in the spontaneous formation of polydopamine (PDA) shells on various substrate surfaces.22 Owing to the presence of catechol groups, PDA has a strong chelating ability with metal cations, decreasing its ability to form metal nanoparticles (such as Ag NPs and Au NPs).23 On the basis of the metal-enhanced fluorescence effect, in situ deposited metal nanoparticles can use localized surface plasmon resonance (LSPR) to change irradiation and nonradiative attenuation, thereby increasing the quantum yield and enhancing fluorescence properties.24 The PDA shell can adsorb single-stranded DNA (ssDNA) through hydrogen bonding or π–π stacking between the aromatic groups of PDA and the nucleobases of ssDNA.25 In addition, the PDA shell can improve the water solubility of nanoparticles while also having good stability and biocompatibility, making it highly desirable for surface modification of UCNPs.
Owing to the low abundance of miRNAs, amplification techniques based on various signal amplifications have been widely used for miRNA detection in biological samples.26 Nucleic acid isothermal amplification technology has attracted much attention because of its high analysis efficiency, fast reaction speed, and good specificity.27 Nucleic acid isothermal amplification methods can be classified into two main categories: enzyme-assisted amplification methods28 and enzyme-free amplification methods.29 Among these methods, the enzyme-assisted amplification method relies on the activity of the enzyme, the reaction conditions are strictly limited, and there are disadvantages such as high cost and difficult storage, which limit its application.30 Therefore, enzyme-free amplification strategies such as catalytic hairpin amplification (CHA),31 hybridization chain reactions (HCR),32 and entropy-driven toe-mediated chain replacement reactions (TMSD)33 have been widely used for miRNA detection. TMSD achieves signal amplification through programmable hybrid reactions and isothermal cycles with a target.34 Compared with other signal amplification methods, it has the advantages of simple nucleic acid structure, high stability and fast reaction speed.35 In recent years, various new strategies for biological analysis and disease diagnosis have been developed by combining TMSD signal amplification technology with novel nanomaterials for the detection of in vivo and in vitro biomarkers.36 However, few fluorescent biosensors based on the metal-enhanced fluorescence effect of PDA-coated UCNPs and the TMSD signal amplification strategy for the measurement of miRNAs exist.
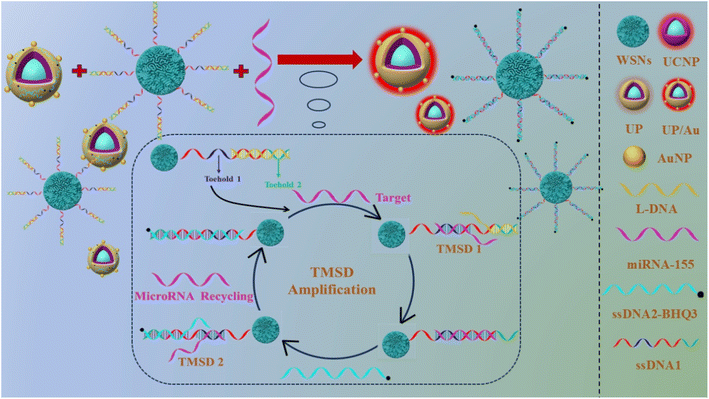
In this work, an NIR-excited signal-amplified fluorescence biosensor was constructed for the sensitive detection of miRNA-155 (as shown in Scheme 1). The capture probe WSNs–ssDNA1 was prepared via an amide reaction between single-stranded DNA1 (ssDNA1) and wrinkled silica (WSNs). The preparation process of the signal probe was as follows: UCNPs were prepared via a high-temperature coprecipitation assay. In the water in the oil microlotion system, NaIO4 induces fast oxidative polymerization of DA on UCNP to obtain water-soluble PDA-coated UCNP composites (UPs). On the one hand, when PDA was used for the in situ reduction of HAuCl4, Au NPs were coated on the surface of UP to obtain UP/Au nanocomposites, which can effectively enhance the UCL. On the other hand, the UP/Au-ssDNA2 signal probe was prepared on the basis of the strong ability of PDA to adsorb ssDNA. Owing to the presence of the quencher BHQ3 in ssDNA2, the fluorescence of UP/Au can be effectively quenched. When the target miRNA-155 is present, it can undergo a toehold region-mediated chain substitution reaction with ssDNA1 modified on the WSNs surface. ssDNA1 exposes a new toehold region and reacts with ssDNA2 modified on the UCNP surface, forming the ssDNA1/ssDNA2 complex. At the same time, miRNA-155 is replaced and reacts with the new ssDNA1, achieving cyclic amplification of the signal. This work proposes a near-infrared light-excited signal amplification fluorescence biosensor, which has the advantages of high sensitivity, simple operation, and fast reaction speed, providing a new method for miRNA detection.
2 Experimental
2.1 Materials
ErCl3·6H2O, TmCl3·6H2O, YCl3·6H2O, GdCl3·6H2O, YbCl3·6H2O, oleic acid (OA), octadecene (ODE), ammonium fluoride (NH4F), sodium hydroxide (NaOH), dopamine hydrochloride (DA·HCl), ammonium hydroxide solution (NH3·H2O, 30 wt%), sodium periodate (NaIO4), chloroauric acid (HAuCl4), cetylpyridinium bromide (CPB), urea, tetraethyl orthosilicate (TEOS), succinic anhydride, (3-aminopropyl)-trimethoxysilane (APTMS), calcium chloride (CaCl2), 1-ethyl-3-(3-dimethyllaminopropyl)carbodiimide hydrochloride (EDC), and N-hydroxy succinimide (NHS) were obtained from Aladdin Reagent Co., Ltd (Shanghai, China). Methanol (CH3OH), ethanol (C2H5OH), cyclohexane (C6H12), N,N-dimethylformamide (DMF) and isopropanol (C3H8O) were obtained from Shanghai Chemical Reagent Company (Shanghai, China). In addition, polyoxyethylene (5) nonylphenyl ether (Lgepal CO-520, branched, Mw = 441) was obtained from Shanghai Macklin Biochemical Technology. Phosphate-buffered saline (PBS) (0.01 M, pH 7.5) and HEPES buffer solution (0.02 M, pH 8.5) were purchased from Sangon, Inc. (Shanghai, China). The oligonucleotides were synthesized and purified via HPLC by Sangon Biotech Co. The base sequences from left to right (from 5–3) of the miRNAs are listed below:ssDNA1: NH2-AAAAAAAAAAACCCCTATCACGATTAGCATTAAAATCGTCT
ssDNA2: AGACGATTTTAATGCTAATCGTGATAGGH-BHQ3
L-DNA: TCACGATT TTAATGCTAATCGTGA
miRNA-155: UUAAUGCUAAUCGUGAUAGGGGU
miRNA-222: AGCUACAUCUGGCUACUGGGU
miRNA-214: UGCCUGUCUACACUUGCUGCUGC
miRNA-21: UAGCUUAUCAGACUGAUGUUGA.
2.2 Instruments
UV-vis absorption spectra were acquired with a LAMBDA 950-visible spectrophotometer (PerkinElmer, USA). The fluorescence spectra were recorded via a FluoroMax-4 (Horiba Jobin yvon, France). Fourier-transform infrared (FTIR) analysis was implemented with a Nicolet 5700 instrument (Thermal Power Company, USA). X-ray diffraction (XRD) patterns were recorded via a Miniflex600 instrument (Rigaku Corporation, Japan). Zeta potentials were measured on a Nano-ZS90 instrument (Malvern Instruments, UK). X-ray photoelectron spectroscopy (XPS) was performed with a Miniflex600 (Thermo Fisher Technologies, America). The results of the specific surface area and aperture analysis were obtained with a 3Flex instrument (Mack Instruments, America). Transmission electron microscopy (TEM), high-resolution transmission electron microscopy (HRTEM) and scanning transmission electron microscopy-high-angle annular dark-field (STEM-HAADF) were performed via a Talos F200i (Thermo Fisher Technologies, America).2.3 Synthesis of naked core UCNPs
In accordance with the literature37 with slight modifications, NaErF4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.5%Tm was synthesized via a high-temperature coprecipitation method. ErCl3·6H2O (760 mg), TmCl3·6H2O (3.84 mg), and 42.0 mL of OA or ODE (volume ratio 2
0.5%Tm was synthesized via a high-temperature coprecipitation method. ErCl3·6H2O (760 mg), TmCl3·6H2O (3.84 mg), and 42.0 mL of OA or ODE (volume ratio 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5) were mixed and heated to 160 °C in argon gas for 30 min until the raw materials were completely dissolved, after which the mixture was cooled to 40 °C. A 10 mL methanol mixture containing 296.4 mg of NH4F and 200 mg of NaOH was added to the above mixture, and after reacting at 40 °C for 30 min, the mixture was heated to 110 °C and maintained for 20 min to remove water and methanol. The mixed liquid was subsequently heated to 300 °C and allowed to react for 1.5 h to form nanocrystals. After the reaction was complete, the mixture was cooled and collected, and the product (NaErF4
5) were mixed and heated to 160 °C in argon gas for 30 min until the raw materials were completely dissolved, after which the mixture was cooled to 40 °C. A 10 mL methanol mixture containing 296.4 mg of NH4F and 200 mg of NaOH was added to the above mixture, and after reacting at 40 °C for 30 min, the mixture was heated to 110 °C and maintained for 20 min to remove water and methanol. The mixed liquid was subsequently heated to 300 °C and allowed to react for 1.5 h to form nanocrystals. After the reaction was complete, the mixture was cooled and collected, and the product (NaErF4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.5%Tm) was washed with ultrapure water and anhydrous ethanol.
0.5%Tm) was washed with ultrapure water and anhydrous ethanol.
2.4 Preparation of UCNPs with a core–shell structure
The synthesis of core–shell UCNPs was carried out as follows: 446 mg of GdCl3·6H2O, 310 mg of YbCl3·6H2O, and 21.0 mL of OA and ODE mixture (volume ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) were mixed, heated to 160 °C for 30 min in argon gas, and then cooled to 80 °C. A total of 5.0 mL of the prepared NaErF4
1) were mixed, heated to 160 °C for 30 min in argon gas, and then cooled to 80 °C. A total of 5.0 mL of the prepared NaErF4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.5%Tm dispersed in cyclohexane was added to the above mixture, which was subsequently heated to 110 °C to remove the cyclohexane and then cooled to 40 °C. A total of 10 mL of methanol solution containing 296.4 mg of NH4F and 200 mg of NaOH was added, and the mixture was heated to 110 °C and maintained for 40 min to remove water and methanol. The temperature was subsequently increased to 300 °C and maintained for 1.5 h. After the reaction was complete, the mixture was cooled to room temperature, centrifuged to collect the product (core shell UCNPs covered with oleic acid, OA-UCNPs), and rinsed with ultrapure water and anhydrous ethanol.
0.5%Tm dispersed in cyclohexane was added to the above mixture, which was subsequently heated to 110 °C to remove the cyclohexane and then cooled to 40 °C. A total of 10 mL of methanol solution containing 296.4 mg of NH4F and 200 mg of NaOH was added, and the mixture was heated to 110 °C and maintained for 40 min to remove water and methanol. The temperature was subsequently increased to 300 °C and maintained for 1.5 h. After the reaction was complete, the mixture was cooled to room temperature, centrifuged to collect the product (core shell UCNPs covered with oleic acid, OA-UCNPs), and rinsed with ultrapure water and anhydrous ethanol.
2.5 Preparation of the UP/Au composite
The UCNP@PDA composite (UP) was synthesized via a modified water-in-oil microemulsion assay.38 First, 3 mL of Lgepal CO-520 was added to 10 mL of a cyclohexane solution of OA-UCNPs (5 mg mL−1) and agitated for 20 min. Next, 0.35 mL of ammonia solution (28%) was added to the mixture, followed by ultrasonic treatment for 15 min and stirring for 30 min. Finally, 0.5 mL of DA·HCl solution (10 mg mL−1) was added. After agitating for 30 min, 0.05 mL of NaIO4 solution (12.5 mg mL−1) was added, and the mixture was agitated for 10 min. The products were precipitated with ethanol and rinsed with ethanol/water. HAuCl4 (0.3 wt%) at different volumes (0.05, 0.15, 0.30, 0.45, 0.50, and 0.75 mL) was added to 200 μL of 100 mg mL−1 UP aqueous solution. Subsequently, different volumes of HEPES buffer (0.02 M, pH 8.5) were used to make the total volume of the reaction mixture 2 mL, and the mixture was agitated for 1.5 h. Finally, the obtained product (UP/Au) was collected by centrifugation, dispersed in 200 μL of ultrapure water and stored at 4 °C.2.6 Synthesis of wrinkled silica
First, 1 g of cetylpyridinium bromide (CPB) and 0.6 g of 20.0 mM urea were dissolved in 30 mL of ultrapure water. Subsequently, 15 mL of cyclohexane and 0.98 mL of isopropanol were cooled and agitated at room temperature for 30 min. Then, 2.5 g of tetraethyl silicate (TEOS) was added to the mixture, which was agitated at room temperature for 30 min and then reacted at 70 °C for 1 h. Next, the mixture was incubated at 10 °C for 2 h, after which it was allowed to react at 70 °C for 12 h. After washing with ethanol, the product (WSNs) was dried at 70 °C and calcined in air at 550 °C for 6 h.2.7 Construction of fluorescent biosensors
2.8 Fluorescence detection of miRNA-155
To ensure the binding of lock DNA (L-DNA) to ssDNA1, toehold region 2 is locked. 200 μL of WSNs–ssDNA1 and 40 μL of 10 μM L-DNA were mixed and incubated at 95 °C for 5 min, followed by natural cooling to room temperature for no more than 3 h. After centrifugation, the product (labelled WSNs–ssDNA1/L) was resuspended in 200 μL of PBS buffer solution (0.01 M, pH 7.4). Then, 20 μL of WSNs–ssDNA1/L, 20 μL of UP/Au-ssDNA2, 110 μL of PBS buffer solution (0.01 M, pH 7.4), and different volumes of 1 μM miRNA-155 were mixed, and the total volume was 400 μL with ultrapure water. The above mixture was incubated at 37 °C for 70 min, after which the fluorescence at 655 nm was recorded under excitation light at 980 nm.2.9 Actual sample analysis
Before analysis, human serum samples were diluted 20 times in 20 mM Tris-HCl (pH 7.4), and miRNA-155 in the human serum samples was detected via the standard addition approach. The design method was subsequently used for analysis, and the specific steps were the same as those for miRNA-155 detection described in Section 2.8. Finally, recovery experiments were carried out by adding various contents of miRNA-155. The labelled solution was tested separately with commercial detection kits according to the above methods.3 Results and discussion
3.1 Characterization of WSNs–ssDNA1
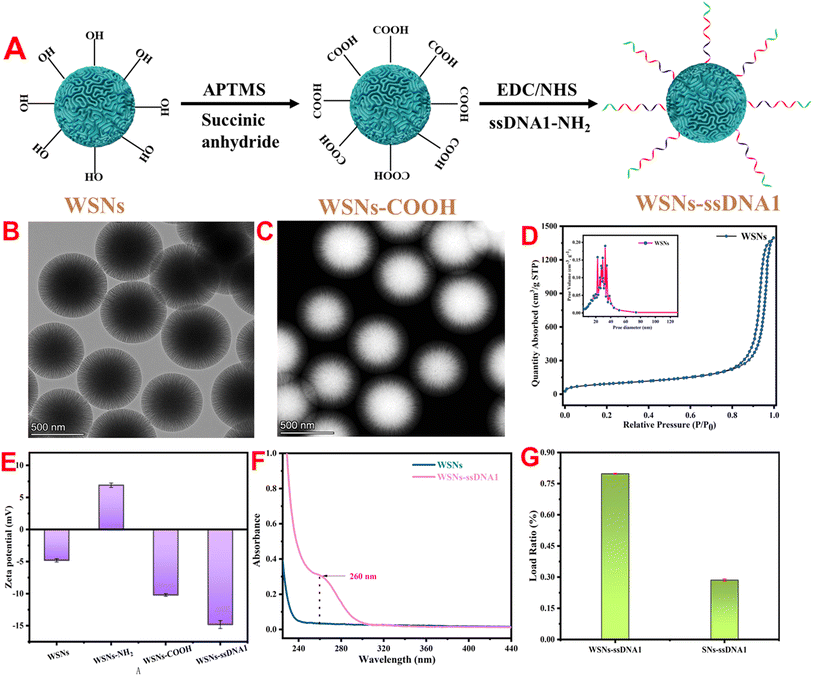
To achieve the coupling of WSNs with ssDNA1, surface modification of WSNs is necessary. The specific modification process is shown in Fig. 1A. The structure and morphology of the WSNs were characterized via TEM and STEM-HAADF. TEM images revealed a particle size of 620 nm, with good dispersion and a spherical shape with a central radial dendrite structure (Fig. 1B). STEM-HAADF revealed that the WSNs had a folded structure (Fig. 1C). Compared with traditional silica (SN), WSNs have a larger specific surface area and more active modification sites because of their unique wrinkled structure. First, the specific surface area and pore size distribution of SN and WSNs were determined by nitrogen adsorption desorption isotherms. Fig. 1D shows that the specific surface area of WSNs is 320.18 m2 g−1, and the average pore size is 39.15 nm, which is 3.51 times and 2.1 times greater than that of SN (Fig. S1†), respectively. To improve the biocompatibility of WSNs and achieve coupling with ssDNA1, WSNs were modified in two steps. First, surface amination of WSNs (WSNs–NH2) was achieved through APTMS. As shown in Fig. S2† (blue line), the FTIR spectrum shows a wide absorption band belonging to the stretching vibration of –NH2 at 3440 cm−1, and the absorption peak at 1641 cm−1 is related to the deformation vibration of –NH2.39 Second, carboxyl-functionalized WSNs (WSNs–COOH) were obtained by reacting WSNs–NH2 with succinic anhydride. Fig. S2† (green line) shows symmetric and asymmetric stretching vibrations corresponding to COO− at 1730 cm−1 and 1409 cm−1.40WSNs–ssDNA1 were prepared via an amide reaction between ssDNA1 and WSNs. The surface modification of WSNs and coupling with ssDNA1 are accompanied by changes in the potential and UV-vis spectrum. Fig. 1E shows that the zeta potentials of WSNs, WSNs–NH2, WSNs–COOH, and WSNs–ssDNA1 are −4.78, 6.92, −10.19, and −14.8 mV, respectively. In addition, UV-vis spectroscopy (Fig. 1F) revealed that after coupling with ssDNA1 through an amide reaction, a characteristic absorption peak belonging to ssDNA1 appeared at 260 nm. These results indicate the successful modification of WSNs with ssDNA1. To achieve sensitive detection of target miRNAs through TMSD, it is necessary to couple ssDNA1 as much as possible to improve detection sensitivity. Fig. S1† shows that the specific surface area of WSNs is 3.51 times greater than that of SNs, indicating that WSNs can be coupled with more ssDNA1. Fig. 1G shows that the loading rates of ssDNA1 on WSNs and SNs are 79.8% and 28.2%, respectively, verifying that WSNs have better ssDNA1 loading capacity than SNs do.
3.2 Characterization of UP/Au-ssDNA2
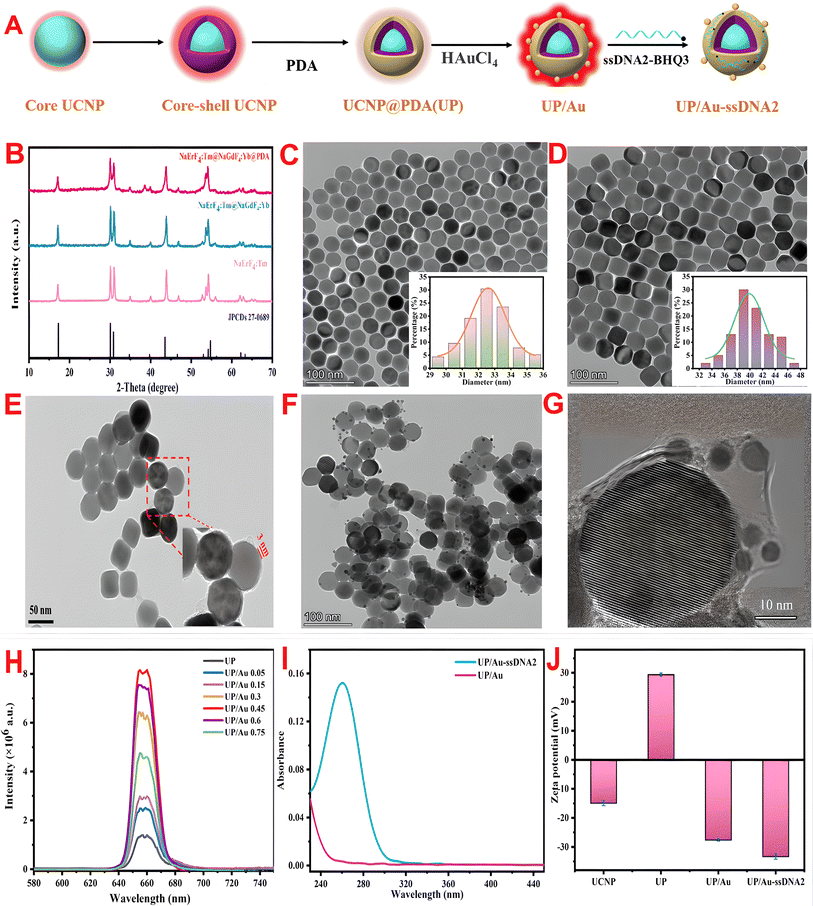
The preparation process of UP/Au-ssDNA2 is shown in Fig. 2A. The products obtained at each step were characterized via TEM, HRTEM, XRD, UV-vis spectroscopy, zeta potential, and FTIR. First, the crystal structure of the prepared material was characterized via XRD. As shown in Fig. 2B, the diffraction peak positions of the core UPCP, core–shell UPCP, and PDA-coated UCNP are consistent with the diffraction peak positions of the standard hexagonal NaErF4 (JCPDS 27-00689) at 17.1°, 30.0°, 30.8°, 43.5°, 53.2°, and 53.7°, indicating that the synthesized UCNP has a hexagonal crystal structure and that the PDA coating has no effect on its crystal phase structure. This phenomenon was also verified in the TEM images (Fig. 2C–E). In addition, after the shell was coated, the particle size increased from 35.5 nm to 40.26 nm, suggesting the formation of a core–shell structure. To enhance the biocompatibility of the UCNPs, a PDA shell with a thickness of approximately 3 nm was formed on their surface via the water-in-oil microemulsion method (Fig. 2E). In the infrared spectrum of UCNP (Fig. S3,† red line), the absorption bands at 1561 cm−1 and 1462 cm−1 belong to the stretching vibration of COO− in oleic acid molecules, whereas the absorption bands at 2930 cm−1 and 2852 cm−1 correspond to the stretching vibration of the –CH2 bond in oleic acid molecules. After coating with PDA, the absorption band (Fig. S3,† cyan line) corresponding to the COO− group disappeared, and new absorption bands appeared at 1606 cm−1 (aromatic ring stretching vibration and N–H bending vibration) and 1295 cm−1 (phenolic C–O stretching vibration), confirming the successful coating of PDA.Owing to the presence of catechol groups, PDA has a strong chelating ability with metal cations and is spontaneously reduced to Au NPs in situ, effectively enhancing UCL. As shown in Fig. 2F and G, the TEM and HRTEM images confirmed that the Au NPs were deposited on the surface of the UP composite, which was verified by EDS (Fig. S4†) and XPS (Fig. S5†). In addition, after the deposition of AuNPs, an ultraviolet absorption peak attributed to AuNPs appeared at 526 nm, and a redshift occurred relative to the typical UV absorption peak at 520 nm for AuNPs (Fig. S6†). This may be due to the surface plasmon resonance frequency (LSPR) of the AuNPs shifting to a higher wavelength. To verify the fluorescence enhancement effect of Au NPs on UP, the influence of distinct amounts of HAuCl4 on the fluorescence intensity of UP/Au was investigated. Fig. 2H shows that the UP composite has bright red fluorescence, and its luminescence mechanism is described in the ESI (Fig. S7†). With increasing HAuCl4 dosage, UP/Au first tends to increase but then decreases. When the volume of HAuCl4 was 0.45 mL, the fluorescence enhancement factor of UCNP at 655 nm reached 5.67. This may be due to the LSPR of AuNPs matching the UCL wavelength of the UCNPs, leading to LSPR coupling and local electric field enhancement, improving the emission efficiency and enhancing the fluorescence intensity.41 Therefore, a dose of 0.45 mL of HAuCl4 was chosen to construct the fluorescent biosensor.
To verify the strong adsorption of ssDNA2 on the surface of UP/Au via PDA for ssDNA, the ultraviolet visible absorption spectra of UP/Au before and after ssDNA2 adsorption were measured. As shown in Fig. 2I (blue line), UP/Au adsorbed with ssDNA2 exhibited characteristic DNA absorption at 260 nm. In addition, the potential of UP/Au before and after ssDNA2 was also measured. Fig. 2J shows that the potential potentials of UP/Au and UP/Au-ssDNA2 are −27.6 and −33.3 mV, respectively, which may be due to the presence of many phosphate groups in DNA. The results indicated that ssDNA2 was successfully adsorbed on the surface of UP/Au, and the adsorption efficiency of UP/Au on ssDNA2 was calculated to be 66.9%.
3.3 Feasibility of the detection method
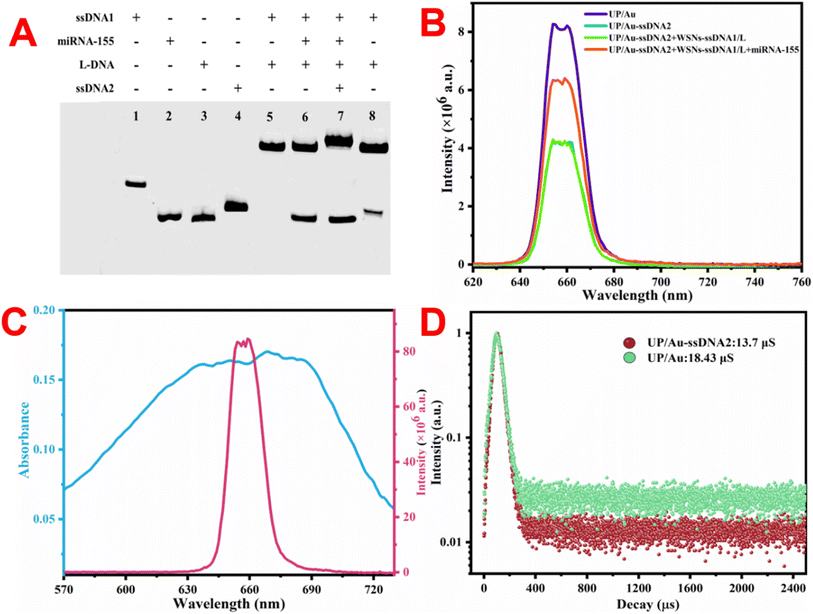
This work proposes an NIR-excited signal-amplified fluorescence biosensor for the sensitive measurement of miRNA-155. To verify the feasibility of the proposed method, the characteristic TMSD was verified via polyacrylamide gel-electrophoresis (PAGE). As shown in Fig. 3A, lane 5 demonstrated that miRNA-155 can successfully bind to ssDNA1 and replace L-DNA with toehold-1. Lane 7 shows that after miRNA-155 was bound to ssDNA1, ssDNA2 could use toehold-2 to bind to ssDNA1 with the exposure of toehold-2, while miRNA-155 was replaced. Lane 8 indicates that ssDNA2 cannot react with ssDNA1 in the absence of miRNA-155. The PAGE results revealed that the designed nucleic acid sequence can be used to detect miRNA-155 via two TMSD reactions. Next, we verified whether the constructed fluorescent biosensor could detect miRNA-155. As shown in Fig. 3B, after ssDNA2 adsorbs on the surface of UP/Au through π–π packing and hydrogen bonding,42 the fluorescence of UP/Au is effectively quenched. This is because the UV-vis light absorption peak of BHQ3 carried by SSDNA2 can fully cover the fluorescence emission peak of UP/Au (Fig. 3C), resulting in a FRET process between BHQ3 and UP/Au. Fig. 3D shows that the fluorescence lifetime of UP/Au decreased after ssDNA2 adsorption, which confirmed the occurrence of the FRET process. In addition, when WSNs–ssDNA1 were added, the fluorescence of the detection system did not change. Only when WSNs–ssDNA1 and miRNA-155 are added at the same time can the fluorescence signal of the detection system be effectively recovered (Fig. 3B). This is due to the simultaneous addition of WSNs–ssDNA1 and miRNA-155, and two TMSD reactions are triggered sequentially. During this process, ssDNA2, which was originally adsorbed on UP/Au, left the surface of UP/Au and bound to ssDNA1 connected to WSNs, forming a DNA double-stranded structure, leading to the fluorescence recovery of UP/Au; the recovery efficiency was 51.88%. Owing to the dependence of weak interactions such as π–π stacking and hydrogen bonding on the adsorption of ssDNA2 by UP/Au, it is necessary to investigate the stability of UP/Au-ssDNA2. As displayed in Fig. S8,† the quenching efficiency of UP/Au-ssDNA2 remained almost unchanged within 30 days, indicating that UP/Au-ssDNA2 has good anti-desorption ability and can maintain stability in physiological environments, making it feasible for practical sample analysis.3.4 Optimization of testing conditions
To achieve optimal detection performance via the TMSD signal amplification strategy, key parameters such as the ssDNA2 concentration, reaction time, ssDNA1 concentration, and reaction temperature were optimized during the detection process. The fluorescence recovery efficiency (F1 − F)/F was used to evaluate the effect of each parameter on the detection performance, where F and F1 represent the fluorescence intensity of the reaction system before and after the addition of the target miRNA, respectively. The detection of the proposed method is based on the fluorescence changes caused by BHQ3 carried on ssDNA2 moving away from the UP/Au surface. Therefore, the influence of different ssDNA2 concentrations on the detection performance of the constructed sensor was first investigated. As displayed in Fig. 4A, the fluorescence recovery efficiency gradually increased with increasing concentrations of ssDNA2 in the range of 50–300 nM. When the content exceeded 300 nM, the recovery efficiency reached a plateau. The effects of the reaction time and ssDNA1 concentration on the detection system were also investigated. The fluorescence recovery efficiencies of the detection system are both highest when the reaction time is 70 min (Fig. 4B) and the concentration of ssDNA1 is 250 nM (Fig. 4C). Finally, the influence of the reaction temperature on the detection performance was studied, and Fig. 4D shows that the optimal reaction temperature was 40 °C.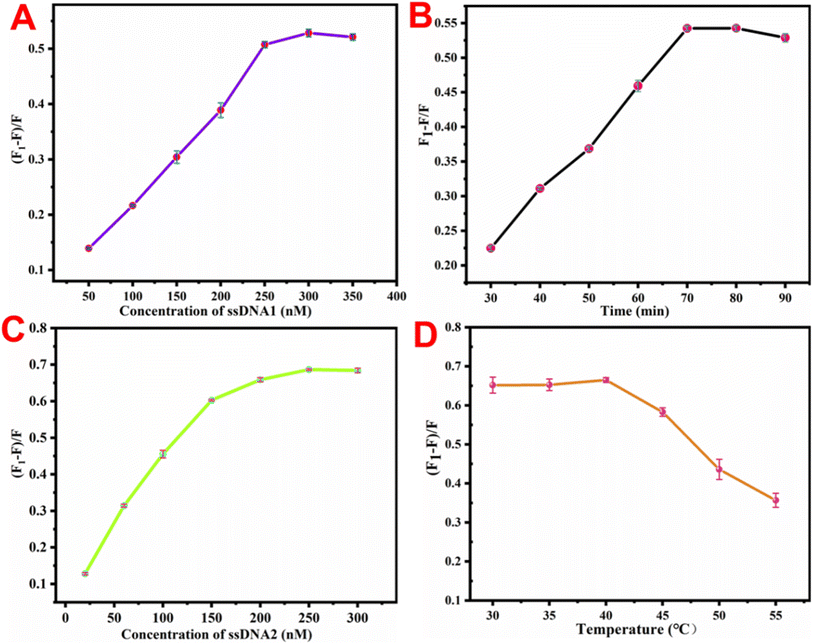 | ||
| Fig. 4 Influence of (A) ssDNA2 concentration, (B) reaction time, (C) ssDNA1 concentration and (D) temperature on the detection system. | ||
3.5 Detection performance of the proposed method
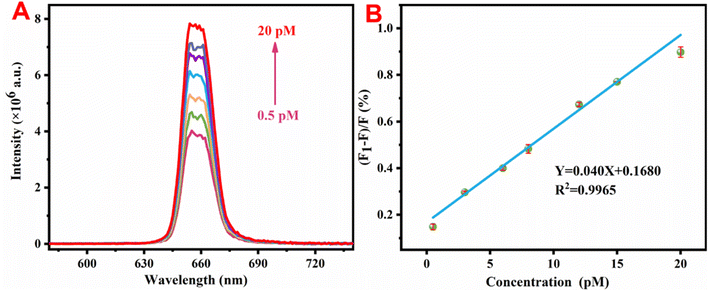
Under optimal experimental conditions, the detection performance of the constructed sensor was studied. Fig. 5A shows that as the content of miRNA-155 increased, the fluorescence intensity of UP/Au progressively increased. Fig. 5B shows a good linear relationship between the luminescence intensity and the miRNA-155 content within the range of 0.5–20 pM, corresponding to a calibration equation of Y = 0.040X + 0.1680 (R2 = 0.9965), where Y represents the fluorescence recovery efficiency of UP/Au and X represents the concentration of miRNA-155 added. A higher R2 reflects the excellent linear response of the constructed fluorescent biosensor to miRNA-155. The limit of detection (LOD) is calculated according to the following formula: LOD = 3 × δ/S, where δ represents the noise level, which is the standard deviation from 10 blank experiments, and S is the slope of the linear regression equation in the sensitivity test. Therefore, the calculated LOD was 19.76 fM, which is superior to those of the other methods reported in Table 1 for determining miRNA-155.| Methods | Linear range (pM) | Detection limit (fM) | Reference |
|---|---|---|---|
| Chemiluminescent | 10–100 | 400 | 43 |
| Electrochemiluminescence | 5–200 | 32 | 44 |
| Electrochemiluminescence | 0.05–5 | 50 | 45 |
| SERS | 10–450 | 100 | 46 |
| SERS | 0.1–500 | 36.7 | 47 |
| Fluorescence | 1–1000 | 50 | 48 |
| Fluorescence | 0.1–100 | 33 | 49 |
| Fluorescence | 0.1–100 | 54.14 | 50 |
| Fluorescence | 0.5–20 | 19.76 | This work |
3.6 Specificity and stability of the proposed method
To prevent false positive signals from miRNAs of similar length and sequence to the target miRNA-155 from interfering with the accuracy of the experimental results, several different miRNAs were selected as interference factors to explore the specificity of the fluorescent sensors for the target. To better demonstrate the application of sensor construction in actual samples, the concentration of interfering miRNAs during detection was increased to 10 times that of miRNA-155. As shown in Fig. 6A, only the experimental group with miRNA-155 added produced significant fluorescence signal recovery, while the fluorescence recovery efficiency of the experimental group with interfering miRNA added was negligible compared with that of the experimental group with miRNA-155 added, indicating that the sensing platform has excellent specificity for detecting miRNA-155. In addition, the stability of the constructed sensing platform was also studied. Fig. 6B shows that the fluorescence response signal to the target miRNA-155 at a concentration of 3 pM remained almost unchanged within one month. The above results indicate that the constructed sensing platform has excellent specificity and stability for the detection of the target miRNA-155.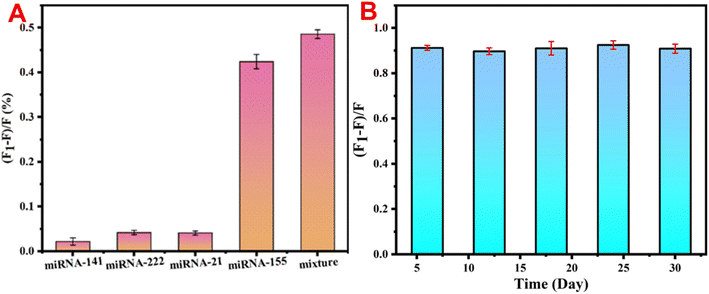 | ||
| Fig. 6 (A) Effects of different miRNAs on the fluorescence intensity of the detection system and (B) stability of the detection system. | ||
3.7 Actual sample analysis
To validate the accuracy and reliability of the constructed sensor in serum sample detection, the standard addition method was used to detect miRNA-155 in diluted human serum samples. Table 2 indicates that the spiked recovery rate in actual samples ranges from 97.57% to 104.1%, with a relative standard deviation (RSD, n = 3) of less than 5%. The small standard deviation and good recovery rate confirmed that the developed method can be used to detect miRNA-155 in complex biological samples.| Samples | Added (pM) | Found (pM) | Recovery (%) | RSD (%) |
|---|---|---|---|---|
| Serum1 | 2.5 | 2.439 | 97.57 | 4.9 |
| 7.0 | 7.280 | 104.0 | 4.2 | |
| 8.5 | 8.647 | 101.7 | 3.5 | |
| Serum 2 | 4.0 | 3.921 | 98.03 | 1.4 |
| 6.0 | 6.245 | 104.1 | 3.6 | |
| 14 | 14.53 | 103.8 | 3.0 |
4 Conclusion
In this work, a fluorescence biosensor was constructed on the basis of the fluorescence enhancement effect caused by in situ deposition of AuNPs and the target cycle amplification strategy induced by TMSD. Compared with other similar detection platforms, it has the following advantages: first, it uses a single red light UCNP excited by near-infrared light as the fluorescence emission source. Compared with other fluorescent materials, UCNPs have less background radiation, reduce the occurrence of false positive signals, and have excellent and stable luminescence performance. Second, the PDA wrapped around the UCNP shell enhances the biocompatibility of the UCNPs. Third, the introduction of a metal-enhanced fluorescence effect greatly improved the fluorescence performance of the UCNPs coated with PDA. Fourth, the entire detection process does not require the involvement of enzymes, avoiding detection errors caused by enzyme inactivation. Finally, WSNs have a rich surface structure, providing many reaction sites for subsequent modifications. The above analysis indicates that the fluorescent biosensor has great potential for future biosensing and clinical diagnosis and treatment.Ethical statement
Human serum samples were attained from Fujian Maternity and Child Health Hospital. Before the laboratory study, written informed consent was signed by all volunteers. All procedures were performed under the guidelines approved by Fujian Maternity and Child Health Hospital. The experimental procedure was approved by the Medical Ethics Committee of Fujian Maternity and Child Health Hospital.Data availability
The authors confirm that the data supporting the findings of this study are available within the article.Author contributions
Dabin Liu, Wenzhang Zhu and Shiqian Zhang designed research. Dabin Liu and Wenzhang Zhu performed the experiments. Dabin Liu, Wenzhang Zhu and Bin Qiu analyzed data. All author wrote and revised the manuscript.Conflicts of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.Acknowledgements
This work was supported by Fujian Provincial Natural Science Foundation of China (2021J01422) and Joint Funds for the innovation of science and Technology, Fujian province (2021Y9176).References
- O. Kurataka, H. Nishiyama, D. Kuriki and T. Ochiya, et al., Connecting the dots in the associations between diet, obesity, cancer, and microRNAs, Semin. Cancer Biol., 2023, 93, 52–69, DOI:10.1016/j.semcancer.2023.05.001.
- K. Taewan and C. M. Croce, MicroRNA: trends in clinical trials of cancer diagnosis and therapy strategies, Exp. Mol. Med., 2023, 55, 1314–21, DOI:10.1038/s12276-023-01050-9.
- M. Joanna, A. Mielczarek-Palacz and J. M. Gola, et al., MicroRNAs as potential biomarkers in gynecological cancers, Biomedicines, 2023, 11(6), 1704, DOI:10.3390/biomedicines11061704.
- G. S. Pall, C. Codony-Servat and A. Hamilton, et al., Carbodiimide-mediated cross-linking of RNA to nylon membranes improves the detection of siRNA, miRNA and piRNA by northern blot, Nucleic Acids Res., 2007, 35, e60, DOI:10.1093/nar/gkm112.
- D. J. Xu, K. L. Di and Z. Y. Li, et al., MicroRNAs in extracellular vesicles: sorting mechanisms, diagnostic value, isolation, and detection technology, Biosens. Bioelectron., 2022, 10, 948959, DOI:10.3389/fbioe.2022.948959.
- N. A. Schultz, C. Dehlendorff and J. S. Johansen, et al., MicroRNA biomarkers in whole blood for detection of pancreatic cancer, JAMA, J. Am. Med. Assoc., 2014, 311, 392–404, DOI:10.1001/jama.2013.284664.
- Y. D. Sun, Y. X. Han and K. Zhang, et al., An ultra-sensitive biosensor for circulating microRNA detection with Fe single-atom enhanced cathodic luminol-O2 electrochemiluminescence, Nano Res., 2024, 17, 7926–7934, DOI:10.1007/s12274-024-6767-2.
- Y. Jiang, J. M. Huang and S. Eda, et al., A rapid, sensitive, and simple-to-use biosensor for on-site detection of attomolar level microRNA biomarkers from serum extracellular vesicles, Sens. Actuators, B, 2022, 369, 132314, DOI:10.1016/j.snb.2022.132314.
- Y. Yu, F. J. Mo and Y. Z. Fu, et al., Ultrasensitive microRNA photoelectric assay based on a mimosa-like CdS-NiS/Au Schottky junction, Anal. Chem., 2023, 95, 12097–103, DOI:10.1021/acs.analchem.3c02153.
- Y. Xu, X. M. Li and P. Liao, et al., Janus wireframe DNA cube-based 3D nanomachine for rapid and stable fluorescence detection of exosomal microRNA, Biosens. Bioelectron., 2022, 212, 114405, DOI:10.1016/j.bios.2022.114405.
- M. Y. He, N. Shang and X. B. Hu, et al., Ultrasensitive fluorescence detection of microRNA through DNA-induced assembly of carbon dots on gold nanoparticles with no signal amplification strategy, Microchim. Acta, 2022, 189, 217, DOI:10.1007/s00604-022-05309-2.
- A. Karami and M. Hasani, Freezing-assisted nano-sweeping of short oligonucleotides using spherical nucleic acids: a fluorometric platform for quantification of microRNAs, Sens. Actuators, B, 2022, 365, 131899, DOI:10.1016/j.snb.2022.131899.
- Y. T. Yang, J. L. Liu and Y. Q. Chai, et al., Highly efficient electrochemiluminescence of MnS:CdS@ZnS core shell quantum dots for ultrasensitive detection of microRNA, Anal. Chem., 2022, 94(18), 6874–81, DOI:10.1021/acs.analchem.2c00970.
- M. Li, X. Xu and Q. Y. Cai, et al., Label-free detection of microRNA: two-stage signal enhancement with hairpin assisted cascade isothermal amplification and light-up DNA-silver nanoclusters, Microchim. Acta, 2020, 187(2), 141, DOI:10.1007/s00604-019-4094-1.
- X. Zhu, J. Zhang and Y. Zhang, et al., Recent progress of rare-earth doped upconversion nanoparticles: synthesis, optimization, and applications, Adv. Sci., 2019, 6(22), 1901358, DOI:10.1002/advs.201901358.
- S. Zha, H. L. Li and A. H. All, et al., Sensitive and responsive upconversion nanoprobes for fluorescence turn-on detection of glucose concentration, Mater. Des., 2023, 227, 111800, DOI:10.1016/j.matdes.2023.11180.
- C. Chen, H. Lei and H. Yan, et al., An aptasensor for ampicillin detection in milk by fluorescence resonance energy transfer between upconversion nanoparticles and Au nanoparticles, Food Chem.:X, 2022, 15, 100439, DOI:10.1016/j.fochx.2022.100439.
- R. X. Wang, J. J. Cheng and H. Q. Chen, et al., Construction of an upconversion luminescence composite nanoprobe for ratiometric single particle imaging detection of hydrogen peroxide in food, Food Chem., 2024, 461, 140928, DOI:10.1016/j.foodchem.2024.140928.
- J. Kim, J. H. Kwon and J. Lee, et al., Rapid and background-free detection of avian influenza virus in opaque sample using NIR-to-NIR upconversion nanoparticle-based lateral flow immunoassay platform, Biosens. Bioelectron., 2018, 112, 209–215, DOI:10.1016/j.bios.2018.04.047.
- H. Ling, D. M. Guan and Q. Liu, et al., Effect of surface modification on the luminescence of individual upconversion nanoparticles, Small, 2024, 20, 2309035, DOI:10.1002/smll.202309035.
- F. Jia, G. L. Li and H. L. Cong, et al., Investigation of rare earth upconversion fluorescent nanoparticles in biomedical field, Nanotechnol. Rev., 2019, 8, 1–17, DOI:10.1515/ntrev-2019-0001.
- T. Chen, Y. H. Xu and J. Q. Liu, et al., A signal amplification system constructed by bi-enzymes and bi-nanospheres for sensitive detection of norepinephrine and miRNA, Biosens. Bioelectron., 2019, 124–125, 224–232, DOI:10.1016/j.bios.2018.10.030.
- Y. Li, L. Xu and X. Sui, et al., Cellulose sponge supported palladium nanoparticles as recyclable cross-coupling catalysts, ACS Appl. Mater. Interfaces, 2017, 9, 17155–17162, DOI:10.1021/acsami.7b03600.
- A. Campu, I. Muresan and M. Focsan, et al., Innovative, flexible, and miniaturized microfluidic paper-based plasmonic chip for efficient near-infrared metal enhanced fluorescence biosensing and imaging, ACS Appl. Mater. Interfaces, 2023, 15, 55925–55937, DOI:10.1021/acsami.3c08658.
- Y. Cen, W. J. Deng and X. Chu, et al., Core–shell–shell multifunctional nanoplatform for intracellular tumor-related mRNAs imaging and near-infrared light tTriggered Photodynamic-photothermal synergistic therapy, Anal. Chem., 2017, 89(19), 10321–10328, DOI:10.1021/acs.analchem.7b02081.
- S. Egloff, N. Melnychuk and A. S. Klymchenko, et al., Enzyme-free amplified detection of cellular microRNA by light-harvesting fluorescent nanoparticle probes, Biosens. Bioelectron., 2021, 179, 113084, DOI:10.1016/j.bios.2021.113084.
- R. J. Deng, K. X. Zhang and J. H. Li, et al., Isothermal amplification for microRNA detection: from the test tube to the cell, Acc. Chem. Res., 2017, 50, 1059–1068, DOI:10.1021/acs.accounts.7b00040.
- F. Ma, M. Liu and C. Y. Zhang, et al., Sensitive quantification of microRNAs by isothermal helicase-dependent amplification, Anal. Chem., 2017, 89, 6182–6187, DOI:10.1021/acs.analchem.7b01113.
- S. Y. Li, F. Zhang and C. Q. Cai, et al., Efficient dual-amplification system for G-quadruplex-based non-enzymatic fluorescence detection of microRNA, Sens. Actuators, B, 2018, 263, 87–93, DOI:10.1016/j.snb.2018.02.009.
- F. Hu, W. C. Zhang and Y. Q. Gu, et al., An electrochemical biosensor for sensitive detection of microRNAs based on target-recycled non-enzymatic amplification, Sens. Actuators, B, 2018, 271, 15–23, DOI:10.1016/j.snb.2018.05.081.
- L. Huang, Y. Zhou and Q. Tang, et al., Accurate and sensitive dual-response fluorescence detection of microRNAs based on an upconversion nanoamplicon with red emission, RSC Adv., 2024, 14(45), 32911–32921, 10.1039/d4ra05061d.
- J. Wei, H. M. Wang and F. Wang, et al., A smart, autocatalytic, DNAzyme biocircuit for in vivo, amplified, microRNA imaging, Angew. Chem., Int. Ed., 2020, 59(15), 5965–5971, DOI:10.1002/anie.201911712.
- Y. L. Yang, J. Z. Yang and Z. He, et al., CRISPR/Cas13a catalyzed self-assembly of quantum dot-DNA hydrogel for microRNA assay, Sens. Actuators, B, 2023, 393, 134249, DOI:10.1016/j.snb.2023.134249.
- P. Miao and Y. G. Tang, Cascade toehold-mediated strand displacement reaction for ultrasensitive detection of exosomal microRNA, CCS Chem., 2021, 3, 2331–2339, DOI:10.31635/ccschem.020.202000458.
- X. M. Zhou, H. Y. Geng and S. Bi, et al., NIR-driven photoelectrochemical-fluorescent dual-mode biosensor based on bipedal DNA walker for ultrasensitive detection of microRNA, Biosens. Bioelectron., 2024, 247, 115916, DOI:10.1016/j.bios.2023.115916.
- X. L. Song, Q. Ding and S. Q. Liu, et al., Application of the dimeric g-quadruplex and toehold-mediated strand displacement reaction for fluorescence biosensing of ochratoxin A, Biosens. Bioelectron., 2021, 192, 113537, DOI:10.1016/j.bios.2021.113537.
- G. Y. Hu, D. P. Yue and H. X. Lyu, et al., Dual-mode upconversion sensors for detecting differently charged biotargets based on the oxidase-mimicking activity of Ce4+ and electrostatic control, Talanta, 2024, 277, 126392, DOI:10.1016/j.talanta.2024.126392.
- W. Zhang, Z. F. Liao and X. Q. Meng, et al., Fast coating of hydrophobic upconversion nanoparticles by NaIO4-induced polymerization of dopamine: positively charged surfaces and in situ deposition of Au nanoparticles, Appl. Surf. Sci., 2020, 527, 146821, DOI:10.1016/j.applsci.2020.146821.
- H. Y. Yang, Q. Shen and Q. Ding, et al., Liposome fusion assisted delivery of silica nanoquenchers for rapid detection of exosomal MicroRNAs, Sens. Actuators, B, 2024, 415, 135978, DOI:10.1016/j.snb.2024.135978.
- L. Chen, T. Gao and Y. Li, et al., Polycarboxylate functionalized magnetic nanoparticles Fe3O4@SiO2@CS-COOH: preparation, characterization, and immobilization of bovine serum albumin, Int. J. Biol. Macromol., 2024, 260, 129617, DOI:10.1016/j.ijbiomac.2024.129617.
- C. Joyce, S. M. Fothergill and F. Xie, et al., Recent advances in gold-based metal enhanced fluorescence platforms for diagnosis and imaging in the near-infrared, Mater. Today Adv., 2020, 7, 100073, DOI:10.1016/j.mtadv.2020.100073.
- Y. Y. Gao, X. R. Xue and K. Wei, et al., A target-triggered strand displacement-assisted target recycling based on carbon dots-based fluorescent probe and MSNs@PDA nanoparticles for miRNA amplified detection and fluorescence imaging, Microchim. Acta, 2024, 191, 351, DOI:10.1007/s00604-024-05883-2.
- O. L. Bodulev, M. D. Zakharov and I. Y. Sakharov, et al., Chemiluminescent plate assay of microRNA-155 coupled with catalytic hairpin assembly with oligonucleotide release (CHAOR), Talanta, 2024, 270, 125525, DOI:10.1016/j.talanta.2023.125525.
- H. K. Li, Q. Q. Cai and G. T. Jie, et al., Ratiometric electrochemical biosensor based on silver nanoparticles coupled with walker amplification for sensitive detection of microRNA, Sens. Actuators, B, 2022, 353, 131115, DOI:10.1021/acssensors.2c02574.
- Y. J. Zheng, C. Li and S. H. Weng, et al., Dual-mode biosensor for femtomolar miRNA-155 detection by electrochemiluminescence and adsorptive stripping voltammetry, Microchem. J., 2021, 165, 106091, DOI:10.1016/j.microc.2021.106091.
- M. Mohammadniaei, J. Yoon and J. W. Choi, et al., Spectro electrochemical detection of microRNA-155 based on functional RNA immobilization onto ITO/GNP nanopattern, J. Biotechnol., 2018, 274, 40–46, DOI:10.1016/j.jbiotec.2018.01.010.
- H. Liang, L. L. Jiang and X. Yang, et al., DNA-guided one-dimensional plasmonic nanostructures for the SERS bioassay, ACS Sens., 2023, 8, 1192–1199, DOI:10.1021/acssensors.2c02574.
- W. C. He, S. T. Li and W. T. Xu, et al., AuNPs-DNAzyme molecular motor biosensor mediated by neighborhood click chemistry reactions for the ultrasensitive detection of microRNA-155, Sens. Actuators, B, 2019, 290, 503–511, DOI:10.1016/j.snb.2019.04.012.
- X. M. Miao, Z. Y. Cheng and P. Wang, et al., Label-free platform for microRNA detection based on the fluorescence quenching of positively charged gold nanoparticles to silver nanoclusters, Anal. Chem., 2018, 90, 1098–1103, DOI:10.1021/acs.analchem.7b01991.
- D. Zhu, Y. Q. Wei and L. H. Wang, et al., Encoding DNA frameworks for amplified multiplexed imaging of intracellular microRNAs, Anal. Chem., 2021, 93, 2226–2234, DOI:10.1021/acs.analchem.0c04092.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4ra07080a |
| This journal is © The Royal Society of Chemistry 2024 |