Open Access Article
Open Access ArticleUsing waste to treat waste: facile synthesis of hollow carbon nanospheres from lignin for water decontamination†
Xiang
Liu
 *a,
Zixuan
Hao
a,
Chen
Fang
a,
Kun
Pang
a,
Jiaying
Yan
a,
Yingping
Huang
a,
Di
Huang
*a and
Didier
Astruc
*a,
Zixuan
Hao
a,
Chen
Fang
a,
Kun
Pang
a,
Jiaying
Yan
a,
Yingping
Huang
a,
Di
Huang
*a and
Didier
Astruc
 *ab
*ab
aEngineering Research Center of Eco-Environment in Three Gorges Reservoir Region of Ministry of Education, College of Materials and Chemical Engineering, China Three Gorges University, Yichang, Hubei 443002, China. E-mail: xiang.liu@ctgu.edu.cn; huangd94@iccas.ac.cn
bISM, UMR CNRS N°5255, Université de Bordeaux, 351 Cours de la Libération, 33405 Talence Cedex, France. E-mail: didier.astruc@u-bordeaux.fr
First published on 27th November 2023
Abstract
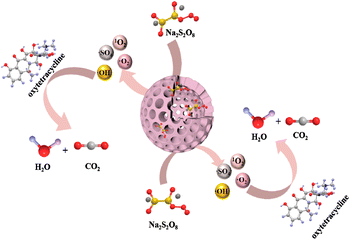
Lignin, the most abundant natural material, is considered as a low-value commercial biomass waste from paper mills and wineries. In an effort to turn biomass waste into a highly valuable material, herein, a new-type of hollow carbon nanospheres (HCNs) is designed and synthesized by pyrolysis of biomass dealkali lignin, as an efficient nanocatalyst for the elimination of antibiotics in complex water matrices. Detailed characterization shows that HCNs possess a hollow nanosphere structure, with abundant graphitic C/N and surface N and O-containing functional groups favorable for peroxydisulfate (PDS) activation. Among them, HCN-500 provides the maximum degradation rate (95.0%) and mineralization efficiency (74.4%) surpassing those of most metal-based advanced oxidation processes (AOPs) in the elimination of oxytetracycline (OTC). Density functional theory (DFT) calculations and high-resolution mass spectroscopy (HR-MS) were employed to reveal the possible degradation pathway of OTC elimination. In addition, the HCN-500/PDS system is also successfully applied to real antibiotics removal in complex water matrices (e.g. river water and tap water), with excellent catalytic performances.
1. Introduction
To date, sulfate radical (SO4˙−)-based advanced oxidation processes (AOPs) have aroused significant interest among scientists due to their excellent removal efficiency for multifarious organic contaminants in wastewater treatment, compared to the inadequate classical bio-degradation and physical adsorption methods.1 As a rule, sulfate radicals (SO4˙−) were generated by activation of persulfates, peroxydisulfate (PDS) and peroxymonosulfate (PMS).2 In contrast to PMS, PDS shows superior potential in wastewater treatment due to its cost effectiveness, lower activation energy and relative stability in water.3 Multifarious transition-metal catalysts, such as Fe,4 Co,5 Cu,6 Ni7 and Mn,8 have been developed for PDS activation in the generation of SO4˙−. However, these metal-based catalysts often pollute the water environment, due to metal leaching during activation.9 Therefore, it is a severe challenge to explore more cost-effective, eco-friendly and metal-free catalysts for PDS activation toward elimination of organic pollutants.10In order to solve the above problem, carbonaceous materials (including graphene,11 carbon nanosheets,12 carbon nanotubes,13 carbon nanodiamonds,14 carbon nanoribbons and biochars) have recently been proposed as the most promising non-metal PDS activators for efficient degradation of organic contaminants.15 In fact, our group has a long-term interest in the design and synthesis of metal-free carbonaceous nanomaterials and their applications in wastewater treatment.16 Turning biomass waste into a precious and useful material, herein, we first report hollow carbon nanospheres (HCNs) as an efficient nanocatalyst, upon pyrolysis of dealkali lignin, for the elimination of antibiotics in complex water matrices. Among them, lignin, one of the most abundant natural materials,17 is considered as a commercial low-value biomass waste from paper mills and wineries.18 First, the morphology and structure of HCNs are fully analyzed using various physical characterization techniques. The kinetic behavior, pH and anionic effect on the elimination of antibiotics over the HCNs/PDS system are investigated in detail. Finally, the degradation pathways, involvement of reactive oxygen species (ROS) and mechanism of oxytetracycline elimination over the HCNs/PDS system are studied by quenching tests, EPR, DFT and HR-MS.
2. Results and discussion
2.1. Characterization of HCNs
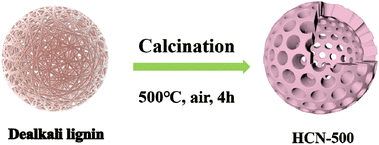
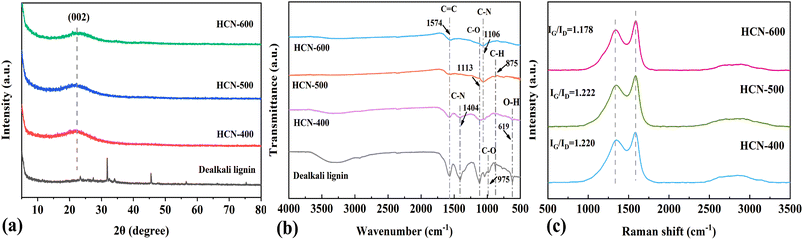
As shown in Scheme 1, dealkali lignin was pyrolyzed at 400 °C, 500 °C and 600 °C under air for 4 h to obtain the hollow carbon nanospheres HCN-400, HCN-500 and HCN-600, respectively. As recorded in Fig. 1a, the nature of the crystalline phase of HCN samples and dealkali lignin was evaluated by XRD. The characteristic diffraction peaks of dealkali lignin disappeared after high-temperature calcination, while a broad peak of 22°, corresponding to the graphene (002) lattice plane clearly appeared in all of HCN-400, HCN-500 and HCN-600, illustrating the formation of graphitic carbon in these HCNs.19 The surface functional groups of HCNs and dealkali lignin were also characterized by FT-IR. Fig. 1b shows that the characteristic absorption peaks of 1574, 1404, 1113, 1106 and 875 cm−1, assigned to the stretching of –C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–, –C–N, C–O, C–N, and C–H, were recorded in all of HCN-400, HCN-500 and HCN-600.20
C–, –C–N, C–O, C–N, and C–H, were recorded in all of HCN-400, HCN-500 and HCN-600.20
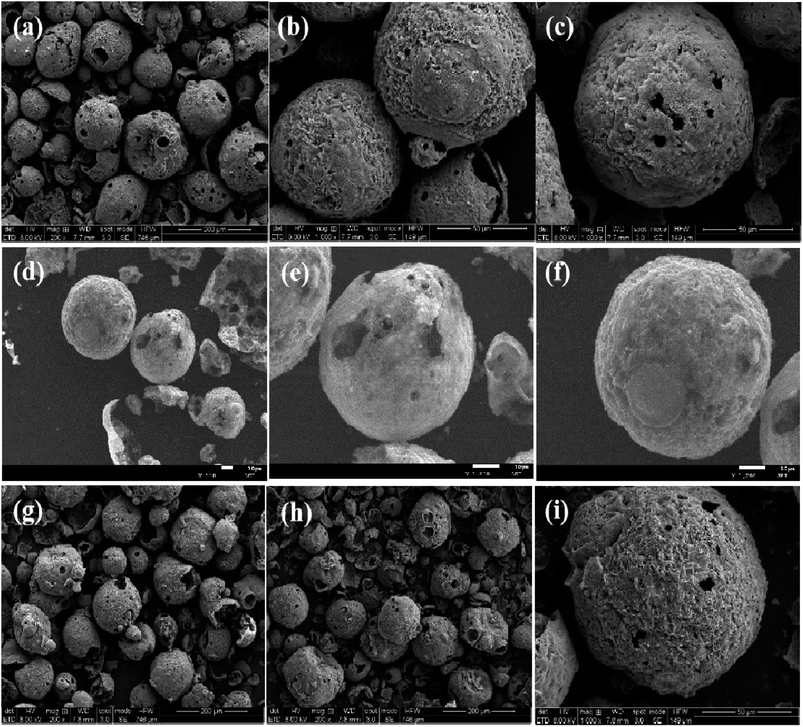
The characteristic absorption peaks of dealkali lignin were totally suppressed, suggesting that dealkali lignin was fully transformed into hollow carbon nanospheres. As shown in Fig. 1c, two broad peaks at 1587 cm−1 and 1335 cm−1 were recorded in HCNs and assigned to the G band and D band. Then, the IG/ID value of HCN-500 (1.222) was slightly larger than those of HCN-400 (1.220) and HCN-600 (1.178). This suggested a relatively higher content of graphitic carbon in HCN- 500, favorable for PDS activation.21 In Fig. S1† and Table 1, BET analysis was employed to check the textures of HCNs. The results showed that the surface area, pore volume and pore size of HCNs increased together with the enhancement of the calcination temperature. HCN-600 possessed a much larger BET surface area (533.76 m2 g−1), pore volume (0.56 cm3 g−1) and pore size (4.2 nm) than those of HCN-400 (331.49 m2 g−1, 0.27 cm3 g−1 & 3.33 nm) and HCN-500 (390.11 m2 g−1, 0.35 cm3 g−1 & 3.64 nm). This implied that HCN-600 with large surface areas may be favorable for adsorption, rather than catalysis. The morphologies of HCNs were also measured by scanning electron microscopy (SEM). Fig. S2† shows that dealkali lignin presents a uniform hollow nanosphere structure with a mean size of 108.4 μm. After high-temperature carbonization, the uniform hollow nanosphere structure was preserved in HCN-400 (105.1 μm, Fig. 2a–c and S3†), HCN-500 (100.1 μm, Fig. 2d–f and S4†) and HCN-600 (95.2 μm, Fig. 2g–i and S5†), and the size of the HCNs decreased with the increase of calcination temperature.
| Catalyst | Surface area (m2 g−1) | Pore volume (cm3 g−1) | Pore size (nm) |
|---|---|---|---|
| HCN-400 | 331.49 | 0.27 | 3.33 |
| HCN-500 | 390.11 | 0.35 | 3.64 |
| HCN-600 | 533.76 | 0.56 | 4.2 |
2.2. Catalytic performances of HCNs
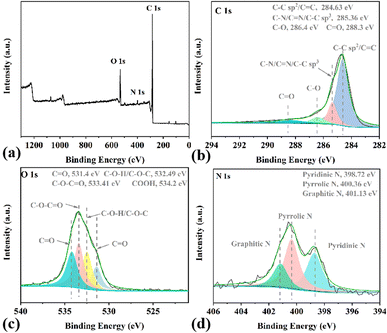
First, the compared catalytic performances of HCN-400, HCN-500 and HCN-600 in OTC degradation were investigated (Fig. S6a†). OTC degradation tests were conducted with OTC (20 mg L−1), PDS (0.1 g L−1) and the catalyst (0.5 g L−1) in 50 mL H2O at 30 °C. The results show that OTC was totally degraded by HCN-400, HCN-500 and HCN-600 via PDS activation in 60 min, 40 min and 3 min, respectively. Moreover, single PDS only provided a 10% degradation efficiency in OTC degradation, and the physical adsorption of OTC by HCNs was then performed without PDS (Fig. S6b†), demonstrating that HCN-600 exhibited a 95% adsorption efficiency for OTC in 15 min, due to its largest BET surface area and pore volume (Table 1). However, HCN-400 and HCN-500 had only 32% and 48% adsorption efficiency for OTC in 40 min. Therefore, HCN-500 was chosen as the optimal nanocatalyst for further study.In order to probe why HCN-500 was highly efficient in OTC degradation via PDS activation, EDX elemental mapping was employed to investigate the accurate localization of the C, N, S and O elements in HCN-500. As described in Fig. 3a, HCN-500 presented a uniform hollow nanosphere structure with a mean size of 100.1 μm. In Fig. 3b–e, it is clear that HCN-500 mainly consisted of C and O elements, while the content of N and S elements was at a trace level, demonstrating that few N and S atoms were co-doped into hollow carbon nanospheres. In Fig. 3f, the sum spectrum also exhibits the co-existence of the C, O, N and S elements in HCN-500. In addition, the surface elemental composition and chemical valence of HCN-500 were further measured by XPS. In Fig. 4a, C, N and O elements are identified with the sum XPS spectrum of HCN-500, verifying the presence of the C, N and O elements. In the C 1s spectrum, four typical peaks are observed at 284.63, 285.36, 286.40 and 288.30 eV corresponding to C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C/C–C, C
C/C–C, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N/C–N, C–O and C
N/C–N, C–O and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O in Fig. 4b, respectively. As recorded in Fig. 4c, the O 1s is fitted into four peaks with C
O in Fig. 4b, respectively. As recorded in Fig. 4c, the O 1s is fitted into four peaks with C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O (531.40 eV), C–O–C (532.49 eV), C–O/C
O (531.40 eV), C–O–C (532.49 eV), C–O/C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O (533.41 eV), and C–OOH (534.20 eV), illustrating the existence of N and O-containing functional groups at the HCN-500 surface. In Fig. 4d, the N 1s spectrum is deconvoluted into three typical peaks at pyridinic-N (398.72 eV), pyrrolic-N (400.36 eV), and graphitic-N (401.13 eV). This confirms the presence of graphitic N, which is favorable for PDS activation.22
O (533.41 eV), and C–OOH (534.20 eV), illustrating the existence of N and O-containing functional groups at the HCN-500 surface. In Fig. 4d, the N 1s spectrum is deconvoluted into three typical peaks at pyridinic-N (398.72 eV), pyrrolic-N (400.36 eV), and graphitic-N (401.13 eV). This confirms the presence of graphitic N, which is favorable for PDS activation.22
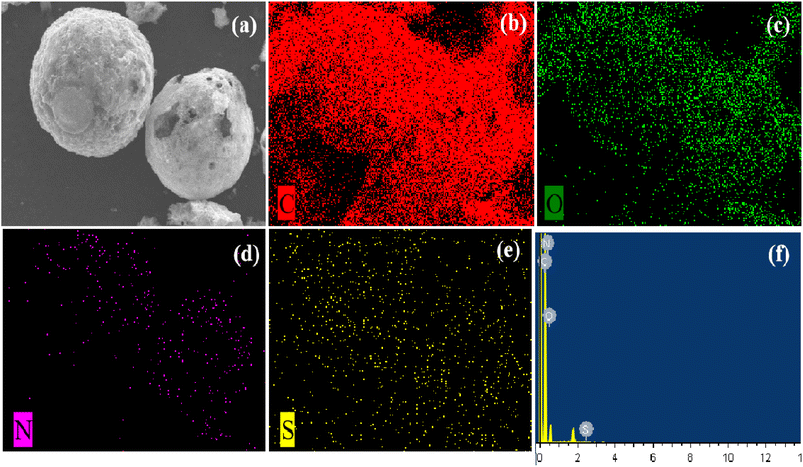 | ||
| Fig. 3 (a) HADDF-STEM, (b) C, (c) O, (d) N, (e) S compositional mapping and (f) sum spectrum of HCN-500. | ||
In summary, these results confirmed that dealkali lignin was successfully transformed into N and S-doped hollow carbon nanospheres at 500 °C calcination, with abundant graphitic C/N and surface N and O-containing functional groups favorable for PDS activation.
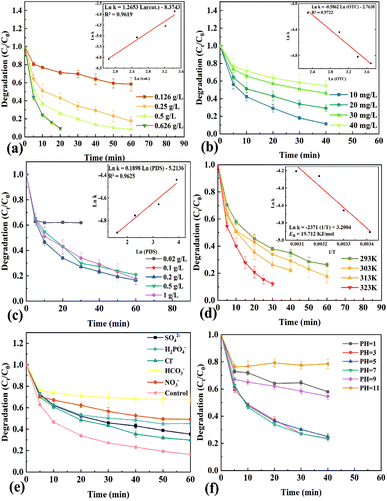
2.3. Kinetic study
The kinetic parameters of OTC degradation (such as the catalyst dosage, initial OTC concentration, PDS amount and degradation temperature) were systematically studied for further actual wastewater treatment. First, OTC degradation tests were performed with different concentrations of HCN-500 catalyst in the range of 0.126 g L−1 to 0.626 g L−1. Fig. 5a shows that the degradation rate was dependent on HCN-500 catalyst concentration, suggesting pseudo-first-kinetics in terms of catalyst concentration. Then, experiments were carried out to observe OTC degradation with different initial concentrations of OTC (10 to 40 mg L−1). As shown in Fig. 5b, the OTC degradation rate decreased together with the increase of the initial OTC concentration. OTC degradation tests were conducted with various PDS dosages from 0.02 g L−1 to 1 g L−1 (Fig. 5c). The OTC degradation efficiency reached its maximum at 0.1 g L−1 of PDS, whereas the OTC degradation efficiency was suppressed with excess PDS. Finally, the effect of degradation temperatures on OTC degradation was recorded (Fig. 5d). The OTC degradation rate heightened with the increase of temperature from 293 to 323 K. Based on the Arrhenius law, the Ea of OTC degradation over HCN-500/PDS is calculated to be 19.71 kJ mol−1. As shown in Fig. S7,† 3-aminophenol, sulfamethoxazole, tetracycline, phenol and Rhodamine B were degraded by the HCN-500/PDS system in 40 min.2.4. Anion effect
Recently, a variety of inorganic anions (including SO42−, H2PO4−, Cl−, HCO3− and NO3−), which significantly affected OTC degradation, were frequently found in both surface water and groundwater. Therefore, the effects of SO42−, H2PO4−, Cl−, HCO3− and NO3− on OTC degradation over the HCN-500/PDS system were further investigated (Fig. 5e). SO42−, H2PO4−, Cl− and NO3− exhibited a slight influence on OTC degradation over the HCN-500/PDS system, whereas HCO3− presented suppression effect on OTC degradation due to its efficient quenching ability for ˙OH. These results imply that the HCN-500/PDS system exhibited an acceptable anti-interference ability for common inorganic anions in OTC degradation.2.5. pH effect
The pH is of great significance in the generation of reactive oxygen species (ROS) and elimination of environmental pollutants.23 The OTC degradation over the HCN-500/PDS system was carried out at various initial pH values of the reaction medium from 1 to 11. HCN-500 showed a superior catalytic performance in OTC degradation via PDS activation from pH = 3 to pH = 7, whereas the OTC degradation rate was significantly suppressed under too acidic (pH ≤ 1) or alkaline (pH ≥ 9) conditions, demonstrating that the HCN-500/PDS system exhibited a good catalytic performance in OTC degradation under pH = 3–7 conditions.2.6. Stability of HCN-500
In general, the stability of the heterogeneous catalyst plays a key role in OTC degradation for further industrial applications.24 When OTC degradation was over, the HCN-500 nanocatalyst was isolated and recycled by simple filtration. Then, another mixture of OTC and PDS was put into the reaction medium for the next degradation reaction. As the number of HCN-500 catalyst cycles increased, the OTC degradation efficiency gradually decreased (Fig. S8†). The 3rd reused HCN-500 only provided 55% degradation efficiency. Gratifyingly, however, the OTC degradation efficiency recovered to 95% again after the reactivation of 3rd reused HCN-500 at 500 °C for 2 h. Then, the reactivated HCN-500 was further tested by SEM. As displayed in Fig. S9,† the hollow nanospheres-shaped morphology of reactivated HCN-500 remained almost the same as that of the fresh one, demonstrating that HCN-500 was an efficient metal-free, heterogeneous and recyclable nanocatalyst in antibiotics degradation via PDS activation.2.7. Mechanism insight
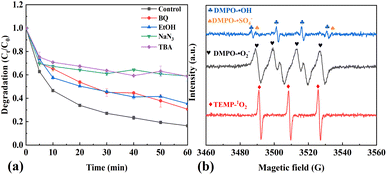
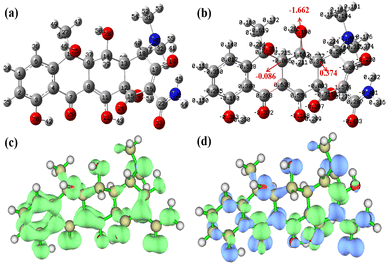
In order to reveal the mechanism of OTC degradation over the HCN-500/PDS system, the reactive oxygen species in the HCN-500/PDS system were determined by quenching tests and EPR analysis. First, quenching tests were carried out with different scavengers. Explicitly, benzoquinone (BQ), EtOH, NaN3 and tert-butyl alcohol (TBA) were applied as effective inhibitors for O2˙−, ˙OH, 1O2 (reactive oxygen species, ROS) and SO4˙−, respectively.25 As illustrated in Fig. 6a, BQ and EtOH exhibited a slight influence on OTC degradation, whereas TBA and NaN3 played a significant inhibiting effect on OTC degradation, revealing that 1O2 and SO4˙− were the major reactive species in the HCN-500/PDS system. Then, the ROS in HCN-500/PDS were further identified by EPR. As depicted Fig. 6b, indeed the signals of O2˙−, ˙OH, 1O2 and SO4˙− were recorded in the HCN-500/PDS system, confirming that 1O2 and SO4˙− were the major ROS in the HCN-500/PDS system.In the present work, the atomic charge, Fukui function and dual descriptor of OTC were also calculated to predict electrophilic reaction active sites during the degradation process (Fig. 7).26 First, OTC was optimized using density functional theory (DFT) calculations with the Gaussian 16 program at the B3LYP/6-311+(d, p) level.27 Water was selected as the solvent based on the polarizable continuum model using the integral equation formalism variant (IEFPCM). The Mulliken charge is shown in Fig. 7a & b, the charge value on C14 is the lowest (−1.662), and the charges of C9 and C13 are −0.086 and 0.374, respectively. Hence, the electron density on the C14–C9 bond is much higher than that on the C14–C13 bond, suggesting that the C14–C9 bond shows a great tendency to be attacked by reactive species. The Fukui function and dual descriptor of OTC were calculated using the Multiwfn software (Fig. 7c & d).28 The green color represents a positive charge, whereas the blue color represents a negative charge. Electrons are uniformly distributed on the OTC molecule as revealed by Fukui function analysis, and the active sites could not be pointed out.
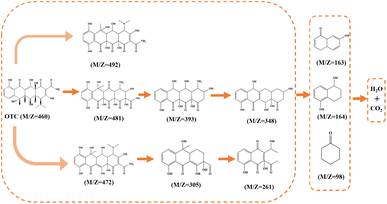
However, the C14–C9 bond has the richest electron density distribution as revealed by dual descriptor analysis (Fig. 7d), which is consistent with the inference of Mulliken charge analysis. In order to verify the OTC degradation pathways over the HCN-500/PDS system, the totally degraded OTC solution was further tested by the total content of organic carbon (TOC) analysis and high-resolution mass spectroscopy (HR-MS) to determine the degradation intermediates. The TOC analysis indicated a 74.4% mineralization efficiency for OTC, which is much higher than those of most metal-based AOPs,29 and complete mineralization into CO2 and H2O. As recorded in Fig. S10,† the OTC peak of m/z = 460 disappeared in HR-MS, demonstrating that OTC molecules were totally decomposed into small molecules of CO2 and H2O. Ten kinds of organic intermediates in OTC degradation were inferred from the mass library (Fig. 8). According to the quenching tests, TOC results, EPR analysis, DFT calculation, HR-MS analysis, and relevant literature reports,30 the plausible mechanism of OTC degradation via the HCN-500/PDS system is proposed in Fig. 9. First, OTC molecules and PDS molecules are adsorbed and attached at the surface of HCN-500. Then, the PDS molecule is immediately decomposed into O2˙−, ˙OH, 1O2 and SO4˙− by HCN-500. Finally, the OTC molecules react with O2˙−, ˙OH, 1O2 and SO4˙−, and are disintegrated into intermediates that are further mineralized into CO2 and H2O.
2.8. Antibiotics degradation in complex water matrices
In order to evaluate the practical application potential of the HCN-500/PDS system in wastewater treatment, ultrapure water was displaced to use in OTC degradation via the HCN-500/PDS system with some typical water bodies, such as river water (Yangtze River) and tap water (laboratory). As displayed in Fig. 10, the OTC degradation efficiency is slightly promoted in both river water and tap water, suggesting significant potential applications of the HCN-500/PDS system in real antibiotics removal.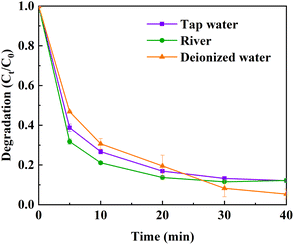 | ||
| Fig. 10 OTC degradation catalyzed by the HCN-500/PDS system in river water and tap water. Conditions: 20 mg L−1 of OTC, 0.5 g L−1 of HCN-500, 0.1 g L−1 of PDS at 30 °C. | ||
2.9. Comparison
The comparison of HCN-500/PDS with other catalytic systems in OTC degradation is summarized and recorded in Table S1.†31 It shows that the HCN-500/PDS system provided a 95.0% degradation rate and a 74.4% mineralization efficiency in 40 min, which exceed those of most metal-based AOPs in the elimination of oxytetracycline, demonstrating that the HCN-500/PDS system is a superior catalytic system for efficient elimination of antibiotics.3. Conclusion
In summary, a new-type of hollow carbon nanospheres (HCNs) has been designed and synthesized as an efficient nanocatalyst by pyrolysis of dealkali lignin for efficient elimination of antibiotics in complex water matrices. Detailed characterization confirmed that dealkali lignin was successfully transformed into N- and S-doped hollow carbon nanospheres upon 500 °C calcination, with abundant graphitic C/N and surface N- and O-containing functional groups favorable for PDS activation. Among them, HCN-500 provided the maximum degradation rate (95.0%) and mineralization efficiency (74.4%), which surpassed those of most metal-based AOPs in OTC elimination. The quenching experiments and EPR results suggest that SO4˙− and 1O2 are the major reactive species in the HCN-500/PDS system. Note that the ROS inorganic radicals involved are much more efficient than transition metal radicals,32 which, in addition, are polluting, contrary to the ROS. The DFT calculations and HR-MS revealed the most probable degradation pathway of OTC. The HCN-500/PDS system has also been successfully applied to real antibiotics removal in complex water matrices (e.g. river water and tap water), with excellent catalytic performance. In an effort to turn biomass waste into a valuable material, the discovery of HCN-500 from lignin provides new eco-friendly and economically opportunities in both biomass reclamation and wastewater treatment.Data availability
Data will be made available on request.Author contributions
X. Liu, D. Huang and D. Astruc: conceptualization, supervision and funding acquisition. X. Liu, Z. Hao, C. Fang, K. Pang, J. Yan, Y. Huang: experiment and data curation. X. Liu: writing – original draft, writing – review & editing.Conflicts of interest
The authors declare no competing financial interest.Acknowledgements
Financial support from the NSFC (No. 21805166, 21972073, 22136003 and 22206188), the 111 Project of China (No. D20015), the Natural Science Foundation of Hubei Province, China (No. 2022CFB275), University of Bordeaux and CNRS is gratefully acknowledged, and the authors thank eceshi (https://www.eceshi.com) for the FT-IR and Raman tests and Yuan Zhou from Shiyanjia Lab (https://www.shiyanjia.com) for the EPR test.References
- (a) Y. Shang, X. Xu, B. Gao, S. Wang and X. Duan, Chem. Soc. Rev., 2021, 50, 5281–5322 RSC; (b) J. Wang and R. Zhuan, Sci. Total Environ., 2020, 701, 135023 CrossRef CAS PubMed; (c) J. Ma, X. Yang, X. Jiang, J. Wen, J. Li, Y. Zhong, L. Chi and Y. Wang, Chem. Eng. J., 2020, 389, 123422 CrossRef CAS; (d) L. Peng, Y. Shang, B. Gao and X. Xu, Appl. Catal., B, 2021, 282, 119484 CrossRef CAS; (e) Y. Peng, M. Cui, Z. Zhang, S. Shu, X. Shi, J. T. Brosnahan, C. Liu, Yu. Zhang, P. Godbold, X. Zhang, F. Dong, G. Jiang and S. Zhang, ACS Catal., 2019, 9, 10803–10811 CrossRef CAS.
- (a) N. P. Shetti, S. J. Malode, R. S. Malladi, S. L. Nargund, S. S. Shukla and T. M. Aminabhavi, Microchem. J., 2019, 146, 387–392 CrossRef CAS; (b) M. Moradi, B. Kakavandi, A. Bahadoran, S. Giannakis and E. Dehghanifard, Sep. Purif. Technol., 2022, 285, 120313 CrossRef CAS; (c) H. Zhang, Y. Mei, F. Zhu, F. Yu, S. Komarneni and J. Ma, Chemosphere, 2022, 306, 135635 CrossRef CAS; (d) W. D. Oh and T. T. Lim, Chem. Eng. J., 2019, 358, 110–133 CrossRef CAS; (e) S. Cai, Q. Zhang, Z. Wang, S. Hua, D. Ding, T. Cai and R. Zhang, Appl. Catal., B, 2021, 291, 120093 CrossRef CAS; (f) A. M. Pai, M. M. Shanbhag, T. Maiyalagan, S. A. Alqarni and N. P. Shetti, Diamond Relat. Mater., 2023, 140, 110561 CrossRef CAS; (g) G. Velvizhi, K. Balakumar, N. P. Shetti, E. Ahmad, K. K. Pant and T. M. Aminabhavi, Bioresour. Technol., 2022, 343, 126151 CrossRef CAS; (h) G. Velvizhi, C. Goswami, N. P. Shetti, E. Ahmad, K. K. Pant and T. M. Aminabhavi, Fuel, 2022, 313, 122678 CrossRef CAS; (i) G. Velvizhi, P. J. Jacqueline, N. P. Shetti, K. Latha, G. Mohanakrishna and T. M. Aminabhavi, J. Environ. Manage., 2023, 345, 118527 CrossRef CAS PubMed.
- (a) R. Yin, W. Guo, H. Wang, J. Du, Q. Wu, J. Chang and N. Ren, Chem. Eng. J., 2019, 357, 589–599 CrossRef CAS; (b) X. Duan, H. Sun and S. Wang, Acc. Chem. Res., 2018, 51, 678–687 CrossRef CAS PubMed; (c) W. Ren, G. Nie, P. Zhou, H. Zhang, X. Duan and S. Wang, Environ. Sci. Technol., 2020, 54, 6438–6447 CrossRef CAS PubMed; (d) Y. Bu, H. Li, W. Yu, Y. Pan, L. Li, Y. Wang, L. Pu, J. Ding, G. Gao and B. Pan, Environ. Sci. Technol., 2021, 55, 2110–2120 CrossRef CAS; (e) X. Huang, X. Zhou, Q. Su, J. Zhou, G. Qian, N. Gao and J. Liu, Chem. Eng. J., 2019, 363, 133–140 CrossRef CAS.
- M. Wang, Y. Wang, Y. Li, C. Wang, S. Kuang, P. Ren and B. Xie, Chem. Eng. J., 2023, 464, 142558 CrossRef CAS.
- C. Fang, Z. Hao, Y. Wang, Y. Huang, D. Huang and X. Liu, J. Cleaner Prod., 2023, 405, 136912 CrossRef CAS.
- Y. Shi, D. Feng, S. Ahmad, L. Liu and J. Tang, Chem. Eng. J., 2023, 454, 140244 CrossRef CAS.
- F. Xu, Y. Wang, C. Wang, W. Huang and X. Liu, Fuel, 2023, 332, 126116 CrossRef CAS.
- Z. Hao, Y. Huang, Y. Wang, X. Meng, X. Wang and X. Liu, Chemosphere, 2022, 308, 136521 CrossRef CAS PubMed.
- (a) X. Liu, W. Hou, Yu. Huang, H. Zhao, Z. Song and Y. Huang, Chem. Eng. J., 2022, 433, 133822 CrossRef CAS; (b) S. Zhu, X. Li, J. Kang, X. Duan and S. Wang, Environ. Sci. Technol., 2019, 53, 307–315 CrossRef CAS PubMed; (c) Y. Qi, J. Li, Y. Zhang, Q. Cao, Y. Si, Z. Wu, M. Akram and X. Xu, Appl. Catal., B, 2021, 286, 119910 CrossRef CAS; (d) J. Chen, X. Zhou, Y. Zhu, Y. Zhang and C. Huang, Water Res., 2020, 186, 116371 CrossRef CAS PubMed; (e) R. Yin, B. Jing, S. He, J. Hu, G. Lu, Z. Ao, C. Wang and M. Zhu, Water Res., 2021, 190, 116720 CrossRef CAS; (f) T. Lian, Y. Wang, B. Wu, F. Yang and N. V. Tarakina, J. Hazard. Mater., 2023, 442, 130070 CrossRef CAS PubMed.
- B. Wang, C. Cheng, M. Jin, J. He, H. Zhang, W. Ren, J. Li, D. Wang and Y. Li, Angew. Chem., Int. Ed., 2022, 61, e202207268 CrossRef CAS.
- X. Huang, Q. Su, S. Han, J. Zhou, G. Qian and N. Gao, J. Hazard. Mater., 2020, 389, 122051 CrossRef CAS.
- S. She, Y. Wang, R. Chen, F. Yi, C. Sun, J. Hu, Z. Li, G. Lu and M. Zhu, Chemosphere, 2021, 266, 128929 CrossRef CAS.
- (a) W. Ren, L. Xiong, X. Yuan, Z. Yu, H. Zhang, X. Duan and S. Wang, Sci. Technol., 2019, 53, 14595–14603 CrossRef CAS; (b) W. Ren, L. Xiong, G. Nie, H. Zhang, X. Duan and S. Wang, Environ. Sci. Technol., 2020, 54, 1267–1275 CrossRef CAS.
- P. Shao, J. Tian, F. Yang, X. Duan, S. Gao, W. Shi, X. Luo, F. Cui, S. Luo and S. Wang, Adv. Funct. Mater., 2018, 28, 1705295 CrossRef.
- (a) T. Zhang, Y. Chen, Y. Wang, J. Le Roux, Y. Yang and J. P. Croue, Environ. Sci. Technol., 2014, 48, 5868–5875 CrossRef CAS PubMed; (b) R. Salih, Z. Velickovic, M. Milosevic and V. P. Pavlovic, J. Environ. Manage., 2023, 326, 116838 CrossRef CAS PubMed; (c) P. J. M. Carrott and M. R. Carrott, Bioresour. Technol., 2007, 98, 2301–2312 CrossRef; (d) Y. Sun, T. Wang, C. Han, X. Lv, L. Bai, X. Sun and P. Zhang, Bioresour. Technol., 2022, 344, 126186 CrossRef CAS; (e) C. M. Hung, C. W. Chen, C. P. Huang, S. S. Lam, Y. Y. Yang and C. D. Dong, Technol, 2022, 354, 127166 CrossRef CAS; (f) X. Zhang, C. M. Navarathna, W. Leng, T. Karunaratne, R. V. Thirumalai, Y. Kim, C. U. Pittman Jr, T. Mlsna, Z. Cai and J. Zhang, Chem. Eng. J., 2021, 417, 129199 CrossRef CAS; (g) N. Srisasiwimon, S. Chuangchote, N. Laosiripojana and T. Sagawa, ACS Sustainable Chem. Eng., 2018, 6, 13968–13976 CrossRef CAS; (h) H. Wang, W. Guo, B. Liu, Q. Wu, H. Luo, Q. Zhao, Q. Si and F. Sseguya, Water Res., 2019, 160, 405–414 CrossRef CAS PubMed.
- (a) X. Liu, Y. Liu, H. Qin, Z. Ye, X. Wei, W. Miao, D. Yang and S. Mao, Environ. Sci. Technol., 2022, 56, 2665–2676 CrossRef CAS PubMed; (b) J. Tan, J. Wang, Z. Tan, M. Yu, Z. Yang, Z. Ren, Y. Li, Y. Zhang and X. Lin, Chem. Eng. J., 2023, 451, 138504 CrossRef CAS; (c) Z. Hao, W. Hou, C. Fang, Y. Huang and X. Liu, J. Hazard. Mater., 2022, 439, 129618 CrossRef CAS PubMed; (d) F. Xu, J. Yan, Y. Wang and X. Liu, iScience, 2023, 27, 106504 CrossRef.
- (a) M. Martin-Martinez, M. F. F. Barreiro, A. M. T. Silva, J. L. Figueiredo, J. L. Faria and H. T. Gomes, Appl. Catal., B, 2017, 219, 372–378 CrossRef CAS; (b) N. Supanchaiyamat, K. Jetsrisuparb, J. T. N. Knijnenburg, D. C. W. Tsang and A. J. Hunt, Bioresource. Technol., 2019, 272, 570–581 CrossRef CAS; (c) H. Zhou, X. Li, H. Jin and D. She, Bioresour. Technol., 2022, 346, 126652 CrossRef CAS; (d) C. Dong, J. Cheng, C. Chen, C. Huang and C. Hung, Bioresour. Technol., 2023, 374, 128768 CrossRef CAS.
- (a) M. Cao, Y. Hu, W. Cheng, S. Huan, T. Bai, Z. Niu, Y. Zhao, G. Yue, Y. Zhao and G. Han, Chem. Eng. J., 2022, 436, 135233 CrossRef CAS; (b) Q. Wu, X. Ye, Y. Lv, R. Pei, M. Wu and M. Liu, Chemosphere, 2020, 258, 127276 CrossRef CAS; (c) D. S. Bajwa, G. Pourhashem, A. H. Ullah and S. G. Bajwa, Ind. Crops Prod., 2019, 139, 111526 CrossRef CAS.
- (a) B. Kong, J. Tang, Y. Zhang, T. Jiang, X. Gong, C. Peng, J. Wei, J. Yang, Y. Wang, X. Wang, G. Zheng, C. Selomulya and D. Zhao, Nat. Chem., 2016, 8, 171–178 CrossRef CAS; (b) C. Kütahya, Y. Zhai, S. Li, S. Liu and J. Li, Angew. Chem., Int. Ed., 2021, 60, 10983–10991 CrossRef; (c) X. Xu, J. Gao, Q. Tian, X. Zhai and Y. Liu, Appl. Surf. Sci., 2017, 411, 170–176 CrossRef CAS.
- H. Xu, J. Yan, Y. Xu, Y. Song, H. Li, J. Xia, C. Huang and H. Wan, Appl. Catal., B, 2013, 129, 182–193 CrossRef CAS.
- M. Shen, X. Zhang, S. Zhao, Y. Cai and S. Wang, Chemosphere, 2023, 314, 137728 CrossRef CAS PubMed.
- (a) H. Wang, W. Guo, B. Liu, Q. Si, H. Luo, Q. Zhao and N. Ren, Appl. Catal., B, 2020, 279, 119361 CrossRef CAS; (b) R. Yin, W. Guo, H. Wang, J. Du, Q. Wu, J. Chang and N. Ren, Chem. Eng. J., 2019, 357, 589–599 CrossRef CAS.
- Z. Wang, J. Jiang, S. Pang, Y. Zhou, C. Guan, Y. Gao, J. Li, Y. Yang, W. Qiu and C. Jiang, Environ. Sci. Technol., 2018, 52, 11276–11284 CrossRef CAS.
- N. Thomas, D. D. Dionysiou and S. C. Pillai, J. Hazard. Mater., 2021, 404, 124082 CrossRef CAS.
- (a) T. Zeng, X. Zhang, S. Wang, H. Niu and Y. Cai, Environ. Sci. Technol., 2015, 49, 2350–2357 CrossRef CAS; (b) Y. Wang, D. Cao and X. Zhao, Chem. Eng. J., 2017, 328, 1112–1121 CrossRef CAS.
- X. Liu, W. Liu, S. Xin, S. Gao, S. Huo, W. Fu, M. Gao and H. Xie, J. Environ. Chem. Eng., 2023, 11, 110576 CrossRef CAS.
- M. Priyadarshini, I. Das, M. M. Ghangrekar and L. Blaney, J. Environ. Manage., 2022, 316, 115295 CrossRef CAS PubMed.
- M. J. Frisch, G. W. Trucks, H. B. Schlegel, G. E. Scuseria, M. A. Robb, J. R. Cheeseman, G. Scalmani, V. Barone, G. A. Petersson, H. Nakatsuji, X. Li, M. Caricato, A. V. Marenich, J. Bloino, B. G. Janesko, R. Gomperts, B. Mennucci, H. P. Hratchian, J. V. Ortiz, A. F. Izmaylov, J. L. Sonnenberg, F. Williams, F. Ding, F. Lipparini, F. Egidi, J. Goings, B. Peng, A. Petrone, T. Henderson, D. Ranasinghe, V. G. Zakrzewski, J. Gao, N. Rega, G. Zheng, W. Liang, M. Hada, M. Ehara, K. Toyota, R. Fukuda, J. Hasegawa, M. Ishida, T. Nakajima, Y. Honda, O. Kitao, H. Nakai, T. Vreven, K. Throssell, J. A. Montgomery Jr, J. E. Peralta, F. Ogliaro, M. J. Bearpark, J. J. Heyd, E. N. Brothers, K. N. Kudin, V. N. Staroverov, T. A. Keith, R. Kobayashi, J. Normand, K. Raghavachari, A. P. Rendell, J. C. Burant, S. S. Iyengar, J. Tomasi, M. Cossi, J. M. Millam, M. Klene, C. Adamo, R. Cammi, J. W. Ochterski, R. L. Martin, K. Morokuma, O. Farkas, J. B. Foresman and D. J. Fox, Gaussian 16 Rev. C.01, 2016 Search PubMed.
- (a) T. Lu and F. Chen, J. Comput. Chem., 2021, 33, 580–592 CrossRef PubMed; (b) K. Tian, L. Hu, L. Li, Q. Zheng, Y. Xin and G. Zhang, Chin. Chem. Lett., 2022, 33, 4461–4477 CrossRef CAS; (c) L. Chen, J. Duan, P. Du, W. Sun, B. Lai and W. Liu, Water Res., 2022, 221, 118747 CrossRef CAS.
- (a) C. H. Han, H. D. Park, S. B. Kim, V. Yargeau, J. W. Choi, S. H. Lee and J. A. Park, Water Res., 2020, 172, 115514 CrossRef CAS; (b) D. Liu, M. Li, X. Li, F. Ren, P. Sun and L. Zhou, Chem. Eng. J., 2020, 387, 124008 CrossRef CAS.
- (a) S. Gao, Z. Wang, H. Wang, Y. Jia, N. Xu, X. Wang, J. Wang, C. Zhang, T. Tian and W. Shen, Appl. Surf. Sci., 2022, 599, 153917 CrossRef CAS; (b) T. T. Nguyen, D. G. Kim and S. O. Ko, Chemosphere, 2022, 307, 135870 CrossRef CAS PubMed; (c) Y. Yang, G. Zeng, D. Huang, C. Zhang, D. He, C. Zhou, W. Wang, W. Xiong, X. Li, B. Li, W. Dong and Y. Zhou, Appl. Catal., B, 2020, 272, 118970 CrossRef CAS; (d) N. An, M. Zhao, X. Zheng, Q. Wang, X. Huang, B. Sun, Y. Shen, J. Wang, B. Chen and R. Liu, J. Hazard. Mater., 2022, 424, 127444 CrossRef CAS; (e) C. Zhang, Z. Ouyang, Y. Yang, X. Long, L. Qin, W. Wang, Y. Zhou, F. Qin and C. Lai, Chem. Eng. J., 2022, 448, 137370 CrossRef CAS; (f) Y. Chen, R. Yin, L. Zeng, W. Guo and M. Zhu, J. Hazard. Mater., 2021, 412, 125256 CrossRef CAS; (g) N. M. Viet, B. L. Giang, N. L. M. Tri, P. Thao, T. H. Pham, F. Z. Kamand and T. M. Al Tahtamouni, J. Water Process Eng., 2019, 32, 100954 CrossRef; (h) X. Li, K. Cui, Z. Guo, T. Yang, Y. Cao, Y. Xiang, h. Chen and M. Xi, Chem. Eng. J., 2020, 379, 122324 CrossRef CAS; (i) Y. Yang, J. Zhu, Q. Zeng, X. Zeng, G. Zhang and Y. Niu, J. Taiwan Inst. Chem. Eng., 2023, 145, 104775 CrossRef CAS; (j) G. Pan, J. Wei, M. Xu, J. Li, L. Wang, Y. Li, N. Cui, J. Li and Z. Wang, J. Hazard. Mater., 2023, 445, 130479 CrossRef CAS PubMed; (k) J. Cao, Z. Yang, W. Xiong, Y. Zhou, Y. Wu, M. Jia, S. Sun, C. Zhou, Y. Zhang and R. Zhong, Sep. Purif. Technol., 2020, 250, 117237 CrossRef CAS; (l) L. Wei, J. Li, C. Zhou, B. Song, F. Qin, W. Wang, H. Luo, D. Qin, C. Huang, C. Zhang and Y. Yang, Chin. Chem. Lett., 2023, 34, 107893 CrossRef CAS; (m) L. Wang, X. Ran, B. Xiao, L. Lei, J. Zhu, X. Xi, G. Feng, R. Li and J. Feng, J. Environ. Chem. Eng., 2022, 10, 108330 CrossRef CAS.
- D. Astruc, Acc. Chem. Res., 1991, 24, 36–42 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3sc05275c |
| This journal is © The Royal Society of Chemistry 2024 |