Open Access Article
Open Access ArticleA DNAzymes-in-droplets assay for Burkholderia gladioli pathovar cocovenenans with single-bacterium sensitivity†
Xiaoqian
Li
,
Yangyang
Chang
,
Yunping
Wu
and
Meng
Liu
 *
*
School of Environmental Science and Technology, Key Laboratory of Industrial Ecology and Environmental Engineering (Ministry of Education), Dalian University of Technology, Dalian POCT Laboratory, Dalian, 116024, China. E-mail: mliu@dlut.edu.cn
First published on 17th January 2024
Abstract
Foodborne pathogens pose a serious risk to human health, and the simple and rapid detection of such bacteria in complex food matrices remains challenging. Herein, we present the selection and characterization of a novel RNA-cleaving fluorogenic DNAzyme, named RFD-BC1, with exceptional specificity for Burkholderia gladioli pv. cocovenenans (B. cocovenenans), a pathogen strongly associated with fatal food poisoning cases. RFD-BC1 was activated by a protein secreted specifically by whole viable B. cocovenenans and displayed an optimum pH distinct from the selection pH, with a rate constant of approximately 0.01 min−1 at pH 5.0. Leveraging this newly discovered DNAzyme, we developed a novel system, termed DNAzymes-in-droplets (DID), that integrates droplet microfluidics to achieve the rapid and selective detection of live B. cocovenenans with single-cell sensitivity. We believe that the approach described herein holds promise for combating specific bacterial pathogens in food samples, offering significant potential for broader applications in food safety and public health.
Introduction
Outbreaks linked to foodborne and medically important bacterial pathogens cause millions of deaths and hospitalizations per year.1 The environmental bacterium Burkholderia gladioli pv. cocovenenans (B. cocovenenans) was first discovered in 1933.2,3B. cocovenenans is pervasive in the natural environment and can cause fatal food poisoning by producing a heat-stable mitochondrial toxin called bongkrekic acid (BA), which has an acute toxicity of 1.41 mg kg−1 (LD50 by intravenous injection in mice).4–6 BA can efficiently inhibit the mitochondrial adenine nucleotide translocator, leading to human death.4–6 Outbreaks of BA food poisoning have been reported in Indonesia, Mozambique, and certain regions of China, with mortality rates reaching 40–60%.7–10 The most recent lethal food poisoning case was reported in Heilongjiang Province, China, in October 2020, resulting in nine deaths, with a fatality rate of 100%.10 Therefore, early and accurate detection of live pathogens is essential to prevent potential outbreaks and minimize the spread of epidemics.Although traditional microbiological culture is the gold standard for bacterial detection, it is time-consuming and can take days to weeks.11 Immunological and polymerase chain reaction (PCR) techniques offer faster and more sensitive detection but require costly instrumentation and trained personnel.12 Most importantly, they are unable to accurately distinguish live pathogens from dead ones, one of the greatest limitations in food safety and medical diagnostics. Therefore, there is a pressing need to develop rapid, accurate, inexpensive and sensitive methods for whole viable food-borne pathogen detection.
Various catalytically active DNA molecules, also known as deoxyribozymes or DNA enzymes, have been identified through in vitro selection from random sequence DNA pools.13–17 RNA-cleaving DNAzymes (RCDs) represent a widely studied class of deoxyribozymes18–22 and have been successfully used as fluorescent biosensors for various live bacterial pathogens, including Escherichia coli, Clostridium difficile, Helicobacter pylori, Klebsiella pneumoniae, Legionella pneumophila, Salmonella typhimurium, Staphylococcus aureus, and Fusobacterium nucleatum.23–31 A major advantage of RCD-based assays is that RCDs are activated by a specific protein target left behind by metabolically active cells, making them ideal for live pathogen detection. In light of these successes, this study aimed to explore the feasibility of utilizing RCDs as effective molecular recognition elements for live B. cocovenenans.
In this study, we report the in vitro selection and characterization of RNA-cleaving fluorogenic DNAzymes (RFDs) that selectively recognize B. cocovenenans. Detailed features, including fluorescence signaling, kinetics, metal-ion specificities, and pH dependences, were thoroughly investigated. Notably, we found that even though the in vitro selection was conducted at pH 7.0, the RFDs exhibited high catalytic activity at pH 5.0, showing a significant ∼3-fold rate enhancement. To our knowledge, this is the first instance of bacteria-responsive RCDs that function optimally at low pH. This feature enhances the inherent chemical stability of RNA phosphodiester bonds, making RCDs more suitable for practical applications. Furthermore, we found that a trans-acting RFD variant also exhibited catalytic activity, with an apparent rate constant of 0.01 min−1. Finally, we present a biosensing platform termed DNAzymes-in-droplets (DID), which enables the selective detection of viable B. cocovenenans at single-cell sensitivity in culture- and amplification-free reactions within 2 h.
Results and discussion
In vitro selection of fluorogenic DNAzymes activated by B. cocovenenans
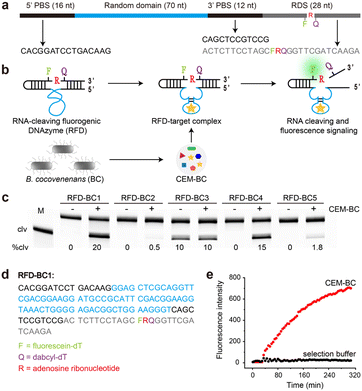
The detailed procedure for the in vitro selection is provided in the ESI.† As depicted in Fig. 1a, the DNA library consisted of a 5′ primer-binding site (5′ PBS), a 70-nucleotide (nt) random region (N70), a 3′ primer-binding site (3′ PBS), and an RNA-containing DNA substrate (RDS). The RDS contained one ribonucleotide, rA, flanked by a pair of modified deoxyribothymidines, one labelled with fluorescein (F) and the other with DABCYL (Q). In the absence of B. cocovenenans, the DNAzyme remained inactivated (Fig. 1b). In contrast, in the presence of live B. cocovenenans, the DNAzyme cleaved the single RNA moiety in the RDS, causing fluorescence enhancement. This RNA-cleaving fluorogenic DNAzyme for B. cocovenenans was termed RFD-BC.Before the selection process, a crude extracellular mixture (CEM) from viable B. cocovenenans (named CEM-BC) was prepared by removing cells grown overnight in Luria Bertani medium (LB). Our in vitro selection strategy is schematically summarized in Fig. S1.† Initially, a pool of approximately 1014 DNA sequences was incubated in the selection buffer (1× SB; 50 mM HEPES, 150 mM NaCl, 15 mM MgCl2, and 0.01% Tween 20, pH 7.0) for 5 h at room temperature. This negative selection step served to eliminate any self-cleaving DNAzymes. The uncleaved DNA molecules were then purified by 10% denaturing polyacrylamide gel electrophoresis (dPAGE) and subsequently incubated with the positive selection target, CEM-BC, in 1× SB for 2 h. The cleaved DNA products were purified by dPAGE, amplified by PCR, and used for the next selection cycle.
A total of 14 selection iterations were conducted. The round-14 DNA pool underwent deep sequencing, and the top 5 sequences were chemically synthesized (Table S1†). We then tested the RNA-cleaving activities of these sequences in the presence of CEM-BC. As shown in Fig. 1c, a DNAzyme named RFD-BC1 exhibited the highest cleavage activity (20%). The sequence of RFD-BC1, provided in Fig. 1d, was selected for further investigation. We also tested the real-time fluorescence signalling capability of RFD-BC1 (Fig. 1e). RFD-BC1 demonstrated a significant fluorescence signal in the presence of CEM-BC, while no fluorescence signal enhancement was observed in 1× SB alone.
RFD-BC1 with metal ion specificities and pH dependences
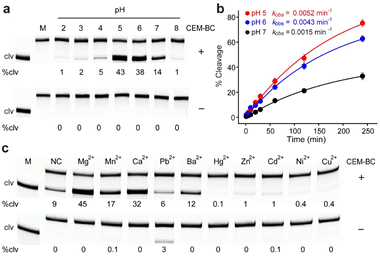
We examined the effect of pH on the catalytic efficiency of RFD-BC1 (Fig. S2†). As shown in Fig. 2a, RFD-BC1 demonstrated catalytic activity across a range of pH values (2.0 to 8.0) in the presence of CEM-BC. Notably, the highest efficiency was observed at pH 5.0, with 43% cleavage. For comparison, we determined the observed rate constants (kobs) for RFD-BC1 at pH 5.0, 6.0 and 7.0 to be 0.52 × 10−2, 0.43 × 10−2, and 0.15 × 10−2 min−1, respectively (Fig. 2b). To our knowledge, most bacteria-responsive DNAzymes were derived to perform catalysis under neutral conditions.23–31 However, several deoxyribozymes and ribozymes that can catalyze chemical reactions at acidic pH have also been reported.32–35 The existence of this RFD-BC1 further indicates that neither the molecular recognition nor catalytic properties of nucleic acids are lost in acidic environments. At low pH, nucleic acid protonation can favor the structural folding of DNAzyme or participate in the cleavage reaction. We also tested the cleavage activity of RFD-BC1 at different temperatures (Fig. S3†). Robust cleavage activity was observed at 25 °C.Considering the importance of metal ions in achieving high catalytic efficiency for various DNAzymes,36,37 the effect of monovalent metal ions (Na+, Li+, K+, Cs+, and NH4+) on RFD-BC1 activity was studied (Fig. S4†). Interestingly, RFD-BC1 displayed catalytic activity in the presence of CEM-BC, even in the absence of monovalent metal ions, resulting in 11% cleavage. Li+, Cs+, and NH4+ induced strong cleavage (20–26%) in the presence of CEM-BC, while Na+ and K+ led to reduced activity (3–5%). The effects of divalent metal ions (Mg2+, Mn2+, Ca2+, Pb2+, Ba2+, Hg2+, Zn2+, Cd2+, Ni2+, and Cu2+) on RFD-BC1 activity were also investigated (Fig. 2c). RFD-BC1 displayed non-metal-selective behaviour, exhibiting high catalytic activity with Mg2+, Mn2+, Ca2+ and Ba2+ (causing 12–45% of cleavage), and reduced activity with Zn2+, Cd2+, Ni2+, and Cu2+ (causing 0.1–1% of cleavage), while remaining inactive in the presence of Hg2+. Notably, Pb2+ induced weak cleavage (3%) in the absence of CEM-BC. Taken together, these results suggest that DNAzymes have successfully evolved to be activated by live B. cocovenenans, aligning with the objective of the SELEX experiment. All subsequent experiments were carried out using a reaction buffer (1× RB) composed of 50 mM acetate (pH 5.0 at 25 °C), 15 mM MgCl2, and 0.01% Tween 20.
RFD-BC1 is likely activated by a secreted protein in live B. cocovenenans
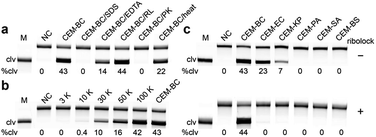
We next investigated the cleavage reactions of RFD-BC1 with CEM-BC and the crude intracellular mixture produced by B. cocovenenans (CIM-BC, see the SI for details). The results revealed that RFD-BC1 exhibited much stronger cleavage with CEM-BC than with CIM-BC (46% vs. 8%, Fig. S5†), suggesting that CEM-BC indeed contained a much higher amount of the target. To determine the nature of the targets in CEM-BC (i.e., proteins, peptides or small-molecule metabolites), CEM-BC was treated with sodium dodecyl sulfate (SDS) and proteinase K (PK). As shown in Fig. 3a, no cleavage activity was observed, suggesting that the target is likely a protein. We then investigated whether the protein target could be a ribonuclease present in CEM-BC. However, adding an RNase inhibitor to CEM-BC did not reduce the activity of RFD-BC1 (44% cleavage). Moreover, when CEM-BC was denatured at 90 °C for 10 min, it still effectively activated RFD-BC1 (22% cleavage), suggesting the thermal stability of the protein target. In addition, RFD-BC1 exhibited reduced activity with CEM-BC containing 15 mM EDTA (14% cleavage). To estimate the molecular weight of the target protein, we conducted a molecular sizing experiment, revealing that the protein fell within the range of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 to 30
000 to 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 Daltons (Fig. 3b). This conclusion was drawn because the 10 kDa filtrate of CEM-EC did not cause cleavage of RFD-BC1.
000 Daltons (Fig. 3b). This conclusion was drawn because the 10 kDa filtrate of CEM-EC did not cause cleavage of RFD-BC1.
We examined the response of RFD-BC1 to CEM samples produced by six viable bacterial cells. As shown in Fig. 3c, Escherichia coli BL21 (EC) and Klebsiella pneumoniae (KP) were also capable of activating RFD-BC1, leading to nonspecific cleavage products. However, the addition of an RNase inhibitor (i.e., ribolock) completely inhibited the nonspecific cleavage activity of RFD-BC1. Therefore, we hypothesized that the observed nonspecific cleavage was most likely due to the presence of various RNases expressed by bacteria. The addition of the RNase inhibitor will significantly suppress the activity of RNases, thus improving the specificity.28 Previously, it was demonstrated that removal of the internal F and Q modifications can efficiently improve the activity but reduce the specificity of the DNAzyme.28 However, we found that the presence of the F and Q modifications had no effect on the catalytic activity (with a kobs of 0.006 min−1) and specificity of RFD-BC1 (Fig. S6†). These experiments indicate that this DNAzyme is highly specific to the protein target secreted in B. cocovenenans.
The fluorescence response of RFD-BC1 in the presence of different concentrations of live B. cocovenenans was also examined (Fig. S7†). Remarkably, RFD-BC1 was able to detect B. cocovenenans at concentrations as low as 103 colony-forming units (CFU) per mL without requiring cell culture. This detection limit is comparable to those reported for other bacteria-responsive DNAzymes, such as those for Escherichia coli (104 CFU mL−1),23,24Helicobacter pylori (104 CFU mL−1),27 and Staphylococcus aureus (103 CFU mL−1).26
Sequence optimization by nucleotide truncation
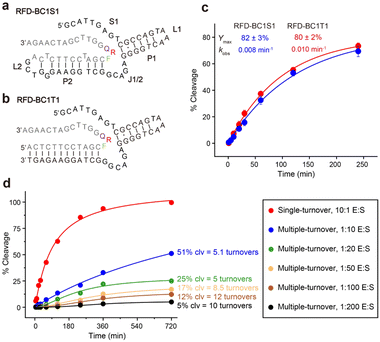
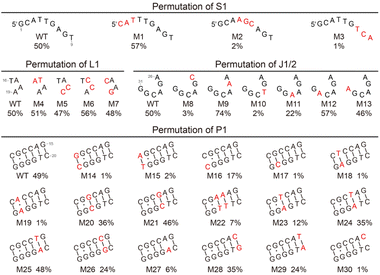
We then investigated the possibility of shortening the sequence of RFD-BC1. We found that at least 45 nucleotides could be deleted from the 5′ end without compromising its functionality. The resulting shortened version, named RFD-BC1S1 (Fig. 4a), consists of a single-stranded region (S1), two short duplexes (P1 and P2), two hairpin loops (L1 and L2), and one interhelical unpaired element (J1/2). Note that it is possible that S1 is directly involved in some tertiary interactions that are essential for structure folding and catalytic function. The existence of P2 was further confirmed by using an engineered trans-acting DNAzyme system, denoted RFD-BC1T1, which efficiently cleaved the matching external RDS (Fig. 4b). Fig. 4c shows the cleavage of RDS by RFD-BC1S1 and RFD-BC1T1 in the presence of CEM-BC, with the kobs of 0.008 min−1 and 0.01 min−1, and the final cleavage yield of 82% and 80% at 25 °C, respectively (Fig. S8†). We also tested whether RFD-BC1T1 could perform multiple-turnover cleavage reactions (Fig. S9†). It was indicated that approximately 12 turnovers were obtained after a 12 h incubation wherein a 100-fold excess of substrate was present (Fig. 4d). These data indicate that RFD-BC1T1 is a very efficient DNAzyme.To assess the essentiality of each nucleotide within RFD-BC1T1, 30 mutant constructs of RFD-BC1T1 were examined (Fig. 5 and S10†). (1) For S1, the remaining nucleotides from T4 to T9 are important as mutation to each of them significantly reduced %clv (M2 and M3). In contrast, G1, C2, and A3 can be mutated (M1). (2) For L1, T16, A17, A18, and A19 can tolerate mutations (M4, M5, M6, and M7). (3) For J1/2, A26, A28, and C29 are important nucleotides because mutating any of these nucleotides caused a noticeable reduction in %clv (M8, M10, and M11). However, G27, G30, and G31 can be mutated (M9, M12, and M13); interestingly, mutation of G27 to A27 could significantly enhance the %clv from 50% to 74%. (4) For P1, mutating the C10–G25 pair into the G–C pair (M14), A–T pair (M15), or C·C mismatch (M16) significantly reduced the %clv. The G11·G24 mismatch is very important, and replacing it with either the G–C pair (M17), T–A pair (M18) or A–A mismatch (M19) caused noticeable %clv reduction. The C12–G23 pair is also important as substituting it with a G–C pair (M20), a T–A pair (M22), or a A–T pair (M23) always reduced %clv. The C13–G22 pair could be changed to a G–C pair (M21) but not a T–A pair (M24). When the A14–T21 pair was mutated into a T–A pair (M25), no reduction in %clv was observed; however, if it was altered to the C–G pair (M26) or A·A mismatch (M27), then %clv dropped. For the G15–C20 pair, the substitution with a C–G pair (M28) or a T–A pair (M29) or a C·C mismatch (M30) can also result in the reduction of %clv.
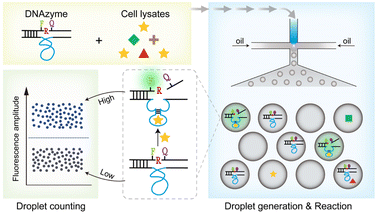
The DNAzymes-in-droplets assay for bacteria detection
We then designed a rapid and highly sensitive sensing system. Droplet microfluidic devices are well known for enabling the encapsulation of single molecules within microdroplets, making them ideal for achieving ultrasensitive biological detection in a short period.38,39 Here, we introduce the DNAzymes-in-Droplets (DID) assay, which integrates DNAzyme-based sensors, droplet encapsulation and digital counting.The fundamental operation and working principle of the DID assay are illustrated in Fig. 6. Specifically, the cell lysates from a particular viable bacterial cell are mixed with fluorogenic DNAzymes in the microfluidic chip and immediately encapsulated into up to ∼30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 droplets. Within these droplets, the following sequence of reactions takes place: (1) recognition occurs between the DNAzymes and the protein targets; (2) cleavage of an RNA-containing substrate ensues upon target binding; and (3) fluorescence dequenching occurs as a result of cleavage. Subsequently, the droplets are imaged and analysed using the on-chip counting system.
000 droplets. Within these droplets, the following sequence of reactions takes place: (1) recognition occurs between the DNAzymes and the protein targets; (2) cleavage of an RNA-containing substrate ensues upon target binding; and (3) fluorescence dequenching occurs as a result of cleavage. Subsequently, the droplets are imaged and analysed using the on-chip counting system.
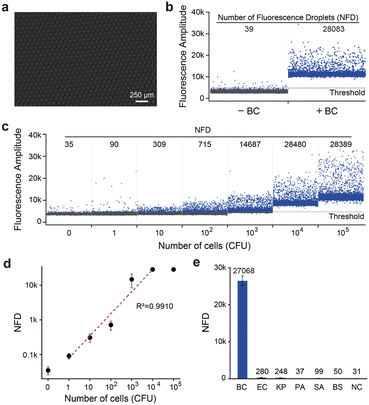
We first examined the DID assay using trans-acting RFD-BC1T1. The droplet microfluidic system generated uniform, picolitre-sized liquid droplets (typically 90 μm in diameter) (Fig. 7a). Positive droplets, containing BC cell lysates and RDS/RFD-BC1T1, displayed a high fluorescence signal (Fig. 7b), while control droplets containing only RDS/RFD-BC1T1 did not exhibit any obvious signal above the automated threshold (estimated by the Crystal Miner software that maximizes the inter-class variance and minimizes the intra-class variance). We then determined the minimal DNAzyme reaction time required in droplets. We observed the time course of the number of fluorescent droplets (NFD), which reached a plateau at approximately 60 min (Fig. S11†). The DID assay demonstrated the capability of quantifying target BC over a broad concentration range of 1 to 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 CFU mL−1 within 2 h (Fig. 7c). This time frame included the cell lysing time (<10 min), droplet generation time (∼45 min), DNAzyme reaction time (60 min), and droplet counting and analysis time (<5 min). Therefore, our strategy significantly reduced the assay time compared with the reported “Integrated Comprehensive Droplet Digital Detection” (IC 3D) method (with a total assay time of ∼4 h).38 Because a whole bacterial cell produces many thousands of potential protein targets, a large number of fluorescent droplets can be observed even at low concentrations. The DID assay exhibited single-cell sensitivity (Fig. 7d) and excellent selectivity for its cognate target (Fig. S12†). None of the CEM samples from other randomly selected live bacteria, such as EC, KP, PA, SA, and BS, produced a detectable NFD (Fig. 7e).
000 CFU mL−1 within 2 h (Fig. 7c). This time frame included the cell lysing time (<10 min), droplet generation time (∼45 min), DNAzyme reaction time (60 min), and droplet counting and analysis time (<5 min). Therefore, our strategy significantly reduced the assay time compared with the reported “Integrated Comprehensive Droplet Digital Detection” (IC 3D) method (with a total assay time of ∼4 h).38 Because a whole bacterial cell produces many thousands of potential protein targets, a large number of fluorescent droplets can be observed even at low concentrations. The DID assay exhibited single-cell sensitivity (Fig. 7d) and excellent selectivity for its cognate target (Fig. S12†). None of the CEM samples from other randomly selected live bacteria, such as EC, KP, PA, SA, and BS, produced a detectable NFD (Fig. 7e).
To challenge the developed assay, B. cocovenenans was analysed in a matrix of the Tremella fuciformis mushroom, a traditional Chinese food associated with B. cocovenenans food poisoning outbreaks. Samples spiked with 50, 500, and 5000 living BC cells were tested in this experiment (Fig. S13†). The results exhibited a good correlation between the detected number of bacterial cells and the actual concentration of target bacteria spiked into the sample, with calculated recovery values ranging from 76% to 107%. These results demonstrated the capability of our assay to accurately detect target bacteria in food samples.
Conclusions
In this study, we present the first successful derivation of RNA-cleaving DNAzymes that are activated by B. cocovenenans. We identified a set of DNAzymes with specific recognition for B. cocovenenans, among which one notable DNAzyme is RFD-BC1. Under the reaction conditions of 50 mM citrate (pH 5.0 at 25 °C), 15 mM MgCl2, and 0.01% Tween 20, RFD-BC1 exhibited a kobs of 0.0052 min−1. Moreover, RFD-BC1 can be used as a sensitive DNAzyme sensor capable of producing a detectable signal at concentrations as low as 103 CFU mL−1. In addition, we found that truncating RFD-BC1 produced a trans-acting DNAzyme system, termed RFD-BC1T1, which displays a kobs of 0.01 min−1. Finally, we developed an on-chip droplet counting system that integrates DNAzyme-based molecular recognition and droplet microfluidics, enabling the rapid and selective detection of live B. cocovenenans with single-cell sensitivity. The high-resolution structural studies of RFD-BC1 will be the subject of a future investigation, which will enable us to understand the detailed mechanisms of catalysis.40,41 We believe that the strategies demonstrated here have the potential to expand the practical utility of DNAzymes in combating various food-borne and medically important pathogens. These findings mark a significant advancement in developing sensitive and efficient detection methods for bacterial pathogens, thus contributing to efforts to prevent outbreaks and minimize the spread of epidemics caused by such pathogens.Data availability
Experimental data is available in the ESI online.†Author contributions
M. L. applied for the funding. M. L. conceived the idea and supervised the project. X. Q. L. performed the experiments and data collection. X. Q. L., Y. Y. C., Y. P. W., and M. L. analyzed the data. X. Q. L., Y. Y. C., Y. P. W., and M. L. wrote the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the the National Key R&D Program of China (No. 2023YFC3205804) and Dalian Science and Technology Innovation Fund (2023YGZD06).Notes and references
- World Health Organization, WHO Estimates of the Global Burden of Foodborne Diseases: Foodborne Disease Burden Epidemiology Reference Group 2007-2015, WHO, 2015, p. 255 Search PubMed.
- P. A. Vandamme, A. G. Johannes, H. C. Cox and W. Berends, Recl. Trav. Chim. Pays-Bas, 1960, 79, 255–267 CrossRef CAS.
- B. Dose, C. Ross, S. P. Niehs, K. Scherlach, J. P. Bauer and C. Hertweck, Angew. Chem., Int. Ed., 2020, 59, 21535–21540 CrossRef CAS PubMed.
- M. Anwar, A. Kasper, A. R. Steck and J. G. Schier, J. Med. Toxicol., 2017, 13, 173–179 CrossRef CAS PubMed.
- N. Moebius, C. Ross, K. Scherlach, B. Rohm, M. Roth and C. Hertweck, Chem. Biol., 2012, 19, 1164–1174 CrossRef CAS PubMed.
- S. Fujita, M. Suyama, K. Matsumoto, A. Yamamoto, T. Yamamoto, Y. Hiroshima, T. Iwata, A. Kano, Y. Shinohara and M. Shindo, Tetrahedron, 2018, 74, 962–969 CrossRef CAS.
- E. S. Gudo, K. Cook, A. M. Kasper, A. Vergara, C. Salomao, F. Oliveira, H. Ismael, C. Saeze, C. Mosse, Q. Fernandes, S. O. Viegas, C. S. Baltazar, T. J. Doyle, E. Yard, A. Steck, M. Serret, T. M. Falconer, S. E. Kern, J. L. Brzezinski, J. A. Turner, B. L. Boyd, I. V. Jani and Chitima Investigation Group, Clin. Infect. Dis., 2018, 66, 1400–1406 CrossRef PubMed.
- J. Li, L. Zhou, C. Long, L. Fang, Q. Chen, Q. Chen, J. Liang, Y. Yang, H. Zhu, Z. Chen, S. Gao, Z. Li, Q. Li, Q. Huang and Y. Zhang, J. Food Prot., 2019, 82, 1650–1654 CrossRef CAS PubMed.
- Z. Peng, T. Dottorini, Y. Hu, M. Li, S. Yan, S. Fanning, M. Baker, J. Xu and F. Li, Front. Microbiol., 2021, 12, 628538 CrossRef PubMed.
- Y. Yuan, R. Gao, Q. Liang, L. Song, J. Huang, N. Lang and J. Zhou, China CDC Weekly (CCDC Weekly), 2020, 2, 975–978 Search PubMed.
- M. Zourob, S. Elwary and A. Turner, Principles of Bacterial Detection: Biosensors, Recognition Receptors and Microsystems.Springer, NY, USA, 2008 Search PubMed.
- V. Velusamy, K. Arshak, O. Korostynska, K. Oliwa and C. Adley, Biotechnol. Adv., 2010, 28, 232–254 CrossRef CAS PubMed.
- R. R. Breaker and G. F. Joyce, Chem. Biol., 1994, 1, 223–229 CrossRef CAS PubMed.
- M. V. Sednev, V. Mykhailiuk, P. Choudhury, J. Halang, K. E. Sloan, M. T. Bohnsack and C. Höbartner, Angew. Chem., Int. Ed., 2018, 57, 15117–15121 CrossRef CAS PubMed.
- V. Dokukina and S. K. Silverman, Chem. Sci., 2012, 3, 1707–1714 RSC.
- Y. Wang, Y. Wang, D. Song, X. Sun, Z. Li, J.-Y. Chen and H. Yu, Nat. Chem., 2022, 14, 350–359 CrossRef PubMed.
- R. Pandey, D. Chang, M. Smieja, T. Hoare, Y. Li and L. Soleymani, Nat. Chem., 2021, 13, 895–901 CrossRef CAS PubMed.
- S. K. Silverman, Acc. Chem. Res., 2009, 42, 1521–1531 CrossRef CAS PubMed.
- E. M. McConnell, I. Cozma, Q. Mou, J. D. Brennan, Y. Lu and Y. Li, Chem. Soc. Rev., 2021, 50, 8954–8994 RSC.
- H. Wang, H. Wang, Q. Wu, M. Liang, X. Liu and F. Wang, Chem. Sci., 2019, 10, 9597–9604 RSC.
- Y. Xiang, Z. Wang, H. Xing and Y. Lu, Chem. Sci., 2013, 4, 398–404 RSC.
- K. Chiba, T. Yamaguchi and S. Obika, Chem. Sci., 2023, 14, 7620–7629 RSC.
- D. Chang, S. Zakaria, S. E. Samani, Y. Chang, C. D. M. Filipe, L. Soleymani, J. D. Brennan, M. Liu and Y. Li, Acc. Chem. Res., 2021, 54, 3540–3549 CrossRef CAS PubMed.
- M. M. Ali, S. D. Aguirre, H. Lazim and Y. Li, Angew. Chem., Int. Ed., 2011, 50, 3751–3754 CrossRef CAS PubMed.
- Z. Shen, Z. Wu, D. Chang, W. Zhang, T. Kha, C. Lee, P. Kim, B. J. Salena and Y. Li, Angew. Chem., Int. Ed., 2016, 55, 2431–2434 CrossRef CAS PubMed.
- M. M. Ali, R. Silva, D. White, S. Mohammadi, Y. Li, A. Capretta and J. D. Brennan, Angew. Chem., Int. Ed., 2022, 61, e202112346 CrossRef CAS PubMed.
- M. M. Ali, M. Wolfe, K. Tram, J. Gu, C. D. M. Filipe, Y. Li and J. D. Brennan, Angew. Chem., Int. Ed., 2019, 58, 9907–9911 CrossRef CAS PubMed.
- M. Rothenbroker, E. M. McConnell, J. Gu, M. L. Urbanus, S. E. Samani, A. W. Ensminger, C. D. M. Filipe and Y. Li, Angew. Chem., Int. Ed., 2021, 60, 4782–4788 CrossRef CAS PubMed.
- M. M. Ali, A. Slepenkin, E. Peterson and W. Zhao, ChemBioChem, 2019, 20, 906–910 CrossRef CAS PubMed.
- J. Li, S. Khan, J. Gu, C. D. M. Filipe, T. F. Didar and Y. Li, Angew. Chem., Int. Ed., 2023, 62, e202300828 CrossRef CAS PubMed.
- Q. Feng, S. Zakaria, D. Morrison, K. Tram, J. Gu, B. Salena and Y. Li, Angew. Chem., Int. Ed., 2023, 62, e202306272 CrossRef CAS PubMed.
- Z. J. Liu, S. H. J. Mei, J. D. Brennan and Y. Li, J. Am. Chem. Soc., 2003, 125, 7539–7545 CrossRef CAS PubMed.
- S. A. Kandadai and Y. Li, Nucleic Acids Res., 2005, 33, 7164–7175 CrossRef CAS PubMed.
- S. A. Kandadai, W. W. Mok, M. M. Ali and Y. Li, Biochemistry, 2009, 48, 7383–7391 CrossRef CAS PubMed.
- V. K. Jayasena and L. Gold, Proc. Natl. Acad. Sci. U. S. A., 1997, 94, 10612–10617 CrossRef CAS PubMed.
- A. Ponce-Salvatierra, K. Wawrzyniak-Turek, U. Steuerwald, C. Hoebartner and V. Pena, Nature, 2016, 529, 231–234 CrossRef CAS PubMed.
- J. Borggraefe, J. Victor, H. Rosenbach, A. Viegas, C. G. W. Gertzen, C. Wuebben, H. Kovacs, M. Gopalswamy, D. Riesner, G. Steger, O. Schiemann, H. Gohlke, I. Span and M. Etzkorn, Nature, 2022, 601, 144–149 CrossRef CAS PubMed.
- D. K. Kang, M. M. Ali, K. X. Zhang, S. S. Huang, E. Peterson, M. A. Digman, E. Gratton and W. A. Zhao, Nat. Commun., 2014, 5, 5427 CrossRef CAS PubMed.
- L. Shang, Y. Cheng and Y. Zhao, Chem. Rev., 2017, 117, 7964–8040 CrossRef CAS PubMed; Y. Ding, P. D. Howes and A. J. deMello, Anal. Chem., 2020, 92, 132–149 CrossRef PubMed.
- A. Ponce-Salvatierra, K. Wawrzyniak-Turek, U. Steuerwald, C. Hoebartner and V. Pena, Nature, 2016, 529, 231–234 CrossRef CAS PubMed.
- J. Borggraefe, J. Victor, H. Rosenbach, A. Viegas, C. G. W. Gertzen, C. Wuebben, H. Kovacs, M. Gopalswamy, D. Riesner, G. Steger, O. Schiemann, H. Gohlke, I. Span and M. Etzkorn, Nature, 2022, 601, 144–149 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3sc05874c |
| This journal is © The Royal Society of Chemistry 2024 |