Open Access Article
Open Access ArticleDiscovery of a lagriamide polyketide by integrated genome mining, isotopic labeling, and untargeted metabolomics†
Claire H.
Fergusson‡
a,
Julia
Saulog‡
a,
Bruno S.
Paulo
b,
Darryl M.
Wilson
a,
Dennis Y.
Liu
a,
Nicholas J.
Morehouse
c,
Samantha
Waterworth
d,
John
Barkei
d,
Christopher A.
Gray
c,
Jason C.
Kwan
 d,
Alessandra S.
Eustaquio
d,
Alessandra S.
Eustaquio
 *b and
Roger G.
Linington
*b and
Roger G.
Linington
 *a
*a
aDepartment of Chemistry, Simon Fraser University, 8888 University Drive, Burnaby, BC V5A 1S6, Canada. E-mail: rliningt@sfu.ca
bDepartment of Pharmaceutical Sciences and Center for Biomolecular Sciences, College of Pharmacy, University of Illinois at Chicago, Chicago, IL 60607, USA. E-mail: ase@uic.edu
cDepartment of Biological Sciences, University of New Brunswick, Saint John, NB, Canada
dDivision of Pharmaceutical Sciences, School of Pharmacy, University of Wisconsin, Madison, WI 53705, USA
First published on 7th May 2024
Abstract
Microorganisms from the order Burkholderiales have been the source of a number of important classes of natural products in recent years. For example, study of the beetle-associated symbiont Burkholderia gladioli led to the discovery of the antifungal polyketide lagriamide; an important molecule from the perspectives of both biotechnology and chemical ecology. As part of a wider project to sequence Burkholderiales genomes from our in-house Burkholderiales library we identified a strain containing a biosynthetic gene cluster (BGC) similar to the original lagriamide BGC. Structure prediction failed to identify any candidate masses for the products of this BGC from untargeted metabolomics mass spectrometry data. However, genome mining from publicly available databases identified fragments of this BGC from a culture collection strain of Paraburkholderia. Whole genome sequencing of this strain revealed the presence of a homologue of this BGC with very high sequence identity. Stable isotope feeding of the two strains in parallel using our newly developed IsoAnalyst platform identified the product of this lagriamide-like BGC directly from the crude fermentation extracts, affording a culturable supply of this interesting compound class. Using a combination of bioinformatic, computational and spectroscopic methods we defined the absolute configurations for all 11 chiral centers in this new metabolite, which we named lagriamide B. Biological testing of lagriamide B against a panel of 21 bacterial and fungal pathogens revealed antifungal activity against the opportunistic human pathogen Aspergillus niger, while image-based Cell Painting analysis indicated that lagriamide B also causes actin filament disruption in U2-OS osteosarcoma cells.
Introduction
To remain relevant in contemporary biomedical research, natural products programs must continue to discover and develop new classes of bioactive molecules. Yet, increasing rates of rediscovery mean that existing libraries often fail to meet this core requirement.1,2 Strains from the order Burkholderiales have been shown to contain large numbers of complex and unusual BGCs.3 By some estimates, the order Burkholderiales ranks third behind Streptomycetales and Mycobacteriales for total biosynthetic diversity.4 However, despite recent attention comparatively few compounds have been discovered from Burkholderiales strains.Currently, the Natural Products Atlas contains just 170 entries for Burkholderiales-derived compounds, compared to 5824 from Streptomycetales.5 Nevertheless, many of the compounds reported from Burkholderiales represent important discoveries. For example, recent investigation of the metagenome of the Lagria villosa beetle led to the identification of the unusual polyketide lagriamide (1) from the bacterial symbiont Burkholderia gladioli Lv-StB.6 Unfortunately it was not possible to culture the producing organism of this antifungal metabolite under laboratory conditions, preventing the determination of the relative or absolute configuration of the molecule and limiting its development potential. In this study we present the discovery of a new lagriamide variant, lagriamide B (2) using 115 whole genome sequences from an environmental Burkholderiales strain collection. A combination of spectroscopic and computational strategies defined the full absolute configuration, while biological testing against a panel of 21 microbial strains revealed antifungal activity for this new metabolite against the opportunistic pathogen Aspergillus niger.
Results and discussion
Lagriamide B BGC discovery
As part of a program to explore the natural products chemistry from Burkholderiales we recently sequenced 115 strains of environmental Burkholderiales strains from Canada and the Western United States. Bioinformatic analysis using antiSMASH 6.0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 7 revealed the presence of one singleton BGC from Paraburkholderia acidicola RL17-388-BIF-B. This BGC was of particular interest due to its close homology to the published BGC (lga) for the antifungal polyketide lagriamide A (1) from the uncultured beetle symbiont Burkholderia gladioli Lv-StB.6
7 revealed the presence of one singleton BGC from Paraburkholderia acidicola RL17-388-BIF-B. This BGC was of particular interest due to its close homology to the published BGC (lga) for the antifungal polyketide lagriamide A (1) from the uncultured beetle symbiont Burkholderia gladioli Lv-StB.6
Annotation of the lagriamide B BGC
The two BGCs of interest are presented in Fig. 1. For clarity we named the lagriamide B BGC lgb and retained the letter level gene nomenclature from the original lagriamide A BGC lga (Table S1†). The main differences between the two gene clusters are (1) the lack of the cytochrome P450-encoding gene lgaJ in P. acidicola, (2) the presence of a phosphopantetheinyl transferase encoding gene, lgbN, in P. acidicola, (3) the lack of a KR domain in LgbD, and (4) the presence of a DH domain in LgbG.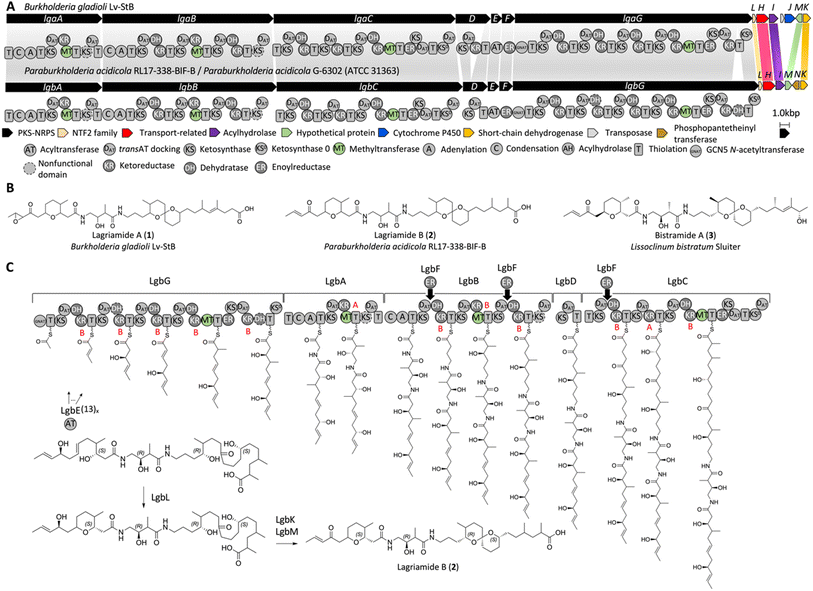 | ||
| Fig. 1 Lagriamide biosynthetic gene clusters and biosynthesis proposal. (A) The reported gene cluster for lagriamide A (top) and the identified gene cluster for lagriamide B from P. acidicola RL17-338-BIF-B and P. acidicola G-6302 (bottom). The predicted domains of the PKS-NRPS assembly line are shown. Similarities with the previously reported lagriamide A gene cluster are highlighted with shaded bars. (B) Lagriamide A, lagriamide B, and bistramide A structures from Burkholderia gladioli,6Paraburkholderia acidicola, and marine animal Lissoclinum bistratum Sluiter,16 respectively. (C) Lagriamide B biosynthesis hypothesis. The configurations of hydroxyl groups were predicted based on alignments of KR domains resulting in either D-configured alcohols (B-type KR) or L-configured alcohols (A-type KR)17 (Fig. S11†). The order of reactions for LgbKLM shown is arbitrary. | ||
As previously proposed by Flórez et al.,6 the predicted biosynthesis starts in a non-colinear manner with PKS LgbG, and proceeds with PKS-NRPSs LgbA and LgbB, and PKSs LgbD and LgbC (Fig. 1C). Both adenylation (A) domains found on LgbA and LgbB are predicted to incorporate glycine while the trans-AT LgbE is predicted to incorporate malonyl-CoA in the PKS extender modules. The enoyl reductase (ER) LgbF may act in trans on modules present on LgbB (twice) and LgbC (once). The activity of KS, DH and KR domains was predicted based on multiple sequence alignments (Fig. S11–S14 and ESI†) and the reported active site residues.8–12 LgbL belongs to the nuclear transport factor 2 (NTF2)-like superfamily, which has been shown to form tetrahydropyran and tetrahydrofuran rings in the nigericin,13 indanomycin,14 and salinomycin15 pathways, among others.
Despite the many similarities between the two BGCs several notable differences exist. The cytochrome P450 LgaJ was previously proposed to catalyze up to three transformations, i.e., epoxidation of the double bond at C2–C3, and oxidation of the hydroxyl groups at C4 and C27 to ketones.6 This gene is absent in the lgb BGC, suggesting the lack of one or more of these oxidation events in the new product. Further, it was proposed that the penultimate module in LgaC was used twice to insert two sequential acetate units. The presence of the related module in LgbC offered the opportunity to further examine this unusual biosynthetic element.
Discovery of lagriamide B BGC homologue
Comparison of the lga and lgb BGCs suggested that P. acidicola should produce an analogue of lagriamide lacking one oxidation event. However, initial LCMS analysis of P. acidicola liquid cultures failed to identify candidate MS features for BGC products with this formula. Two possible reasons existed for this disconnect. Firstly, the product may not have been expressed under the chosen culture conditions. Alternatively, the lgb BGC product may be more distantly related to the original lagriamide structure than predicted by initial BGC analysis.One solution to these issues is to profile multiple strains containing the target BGC and to prioritize metabolites produced by both strains. Searching the NCBI database using the original lagriamide BGC revealed close relationship to genes from a Paraburkholderia acidicola strain G-6302 (ATCC 31363) isolated from a soil sample in Japan. Closer inspection revealed the presence of homologs for lga/lgb genes, split across several contigs. Attempts to improve the genome assembly using informatic or manual methods did not improve the fragmentation of the BGC region, so the strain was resequenced using MinION sequencing. Assembly using the MinION dataset in combination with the original Illumina sequence data revealed the presence of a lgb BGC on the larger of two chromosomes in the genome. This new BGC had the same gene content and synteny as lgb and 93.7% pairwise identity, making the ATCC 31363 strain an ideal candidate for parallel metabolomics investigation.
Metabolite prioritization using IsoAnalyst
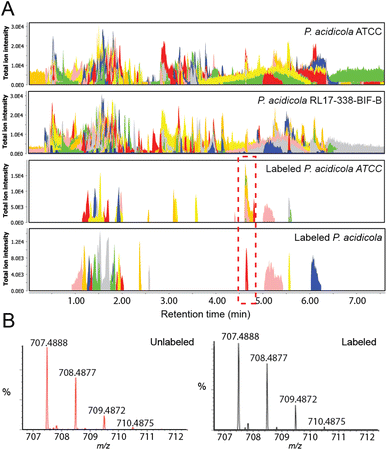
With the two strains in hand, we employed our recently developed IsoAnalyst platform to prioritize candidate BGC products present in both extracts.18 IsoAnalyst uses parallel stable isotope feeding to predict the biosynthetic building blocks incorporated into each natural product in the liquid culture, and then uses these building block distributions to connect molecules to their cognate BGCs. In this application IsoAnalyst was used to identify MS features in the untargeted metabolomics datasets that included 15N labels. This method provides a mechanism for quickly prioritizing molecules containing target elements or building blocks and is enabled by the IsoAnalyst codebase which automatically identifies isotopic labelling differences between labelled and control MS datasets. For discovery of candidate lagriamide-like molecules from the two culturable strains we performed IsoAnalyst analysis using [1-15N]-glutamate as the labelled building block. This analysis identified a suite of mass features in the 700 MW range that were common to both strains (Fig. 2A). These products possessed very similar isotope labelling patterns, consistent with the label incorporation predictions from the target BGC (Fig. 2B). Isolation and NMR analysis of the major congener yielded a molecule with a 1H NMR spectrum that was closely related to lagriamide (Fig. S16†) which we termed lagriamide B (2). Lagriamides are structurally related to bistramides (3) previously isolated from the marine tunicate animal Lissoclinum bistratum Sluiter (Fig. 1B).19–21 Although it appears likely that bistramides may have a bacterial origin, neither a bacterial producer nor the gene cluster have been identified to date.Structure elucidation and configurational analysis
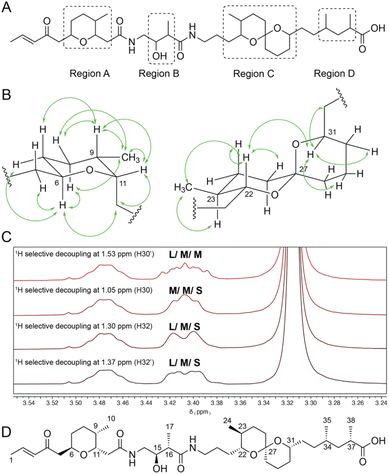
Extensive 1D and 2D NMR analyses, coupled with HRMS analysis, determined the planar structure of this new metabolite as depicted in Fig. 3A. For a full description of the structure elucidation see ESI.† Interestingly, lagriamide B contained two regions of structural variation compared with the original structure (now termed lagriamide A). At the left-hand terminus the epoxy-ketone in lagriamide A is replaced with an α-β unsaturated carbonyl in lagriamide B, consistent with the absence of the cytochrome P450-encoding gene in the lagriamide B BGC. More surprisingly, the right-hand portion of the molecule also differs between the two structures, with lagriamide B missing a two-carbon extender unit and one degree of unsaturation compared to lagriamide A. The gene responsible for this portion of lagriamide B biosynthesis (lgbC) is highly similar to the corresponding gene in the lagriamide A cluster (lgaC).Due to the limited quantities of material obtained during the original discovery of lagriamide A the configurations of the 11 chiral centers were not previously determined. With a culturable producing strain in hand, we were able to obtain multi-milligram quantities of lagriamide B, permitting a detailed configurational analysis using a suite of spectroscopic and computational methods.
Configurational analysis of lagriamide B was made challenging by the presence of four regions containing contiguous chiral centers, each separated by an achiral linker (regions A–D, Fig. 3A). To solve the full absolute configuration, we adopted a blended strategy that combined information from NMR data, molecular modelling and density functional theory-based chemical shift predictions, and genome sequence data.
The relative configurations of regions A and C were determined independently using two different approaches. Firstly, scalar and dipolar coupling NMR data were combined with selective irradiation experiments to define the relative configurations around the tetrahydropyran (A) and spiro (C) ring systems (Fig. 3B and C and ESI†). For example, to determine the relative configuration of proton 31 both protons on the adjacent position in the ring (C30) were irradiated, revealing the presence of one large and one small coupling constant (Fig. 3C). Irradiation of the protons on the exocyclic methylene (C32) revealed two medium sized coupling constants. Because the large coupling constant between H31 and H30 (10.7 Hz) indicates a 1,2-diaxial arrangement, we concluded that H31 must be in the axial position (Fig. 3B and C). This result is in line with the observed nOe signals at this position (Fig. 3B) that indicate a strong nOe correlation between protons H31 and H22.
Separately, the relative configurations of all four regions were predicted using the DP4 computational method.22 In brief, simplified models were selected for all four regions (Fig. 4A) and chemical shifts calculated for all diastereomers of each model using molecular dynamics and DFT methods (ESI†). Calculated chemical shifts for each diastereomer were then compared against the observed chemical shift values for each region, and the diastereomers ranked using the DP4 statistical package (Fig. 4B).23
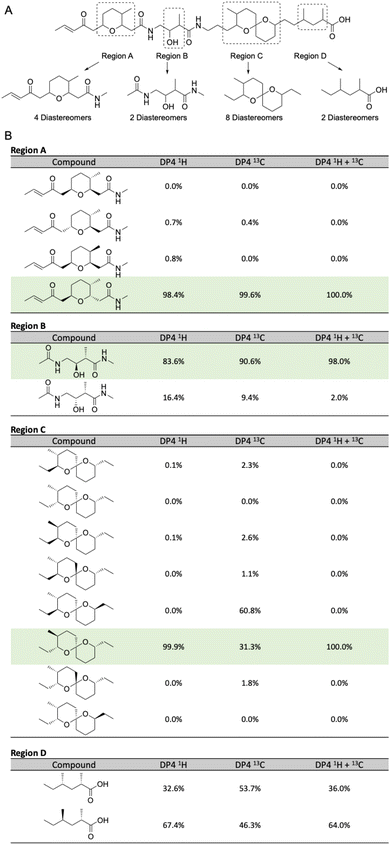 | ||
| Fig. 4 (A) Model compounds for each region containing chiral centers in lagriamide B. (B) Results of DP4 analysis for all possible diastereomers for each model compound. Predicted configurations for regions A–C highlighted in green. Note: only one set of enantiomers was calculated for each subunit. Figure therefore presents the determination of the relative configurations of each subunit, not their absolute configurations. For full depictions of both enantiomeric series see Fig. S6–S9.† | ||
For two of the four regions (A and B) this analysis afforded a single diastereomer with a very high DP4 probability when using the 1H, 13C, or a combination of 1H and 13C chemical shifts. For fragment C, the DP4 probability using only 13C chemical shifts was less conclusive and in disagreement with the DP4 analysis when using only 1H chemical shifts. However, DP4 analysis of fragment C was conclusive when using a combination of 1H and 13C chemical shifts. Similar cases of assignment of the correct diastereomer by DP4 analysis using a combination of 1H and 13C chemical shifts have been reported by Goodman and co-workers even in cases where the individual 1H and 13C DP4 probabilities disagreed.23 Unfortunately, for region D the DP4 result was inconclusive when using 1H, 13C, or a combination of 1H and 13C chemical shifts. To resolve this issue, we created a single model of the combined C + D region, setting the chiral centers in the C portion as defined in Fig. 4B, and varying the chiral centers in the D region (Table S9†). DP4 analysis for this extended model identified a single diastereomer with moderate DP4 probability. This result defined the relative configurations of the two methyl groups in region D and set the relative configurations of the full C + D region.
Finally, we compared the relative configurational assignments from the NMR and DP4 analyses. Gratifyingly, assignments for regions A and C were in full agreement between the two methods. This result provides independent evidence for these configurational assignments using two orthogonal methods, and provides support for the use of DP4 for assigning the relative configurations of regions B and D.
To determine the absolute configuration of lagriamide B we leveraged information from the genome sequence. Three of the four regions (A–C) contain chiral centers that can be predicted based on sequence information for ketoreductase (KR) domains in the BGC.10 Analysis of the LxD motif located ∼57 residues N-terminal to the catalytic tyrosine revealed that 10 KRs belong to B-type, and two to A-type (Fig. 1 and S11†). In parallel, we analysed the PKS-NRPS proteins using the webtool TransATor.24 Although the configurations imparted by three KRs were not predicted (Fig. S15†), all KR configurations that were predicted with TransATor matched our initial prediction based on sequence alignments. The KR analysis helped to define four centers as 11S, 15R, 22R, 31S. This result completed the full configurational assignment for regions A, B and C and, by extension from the combined analysis of the C + D model, region D. Together this defined the full absolute configuration of lagriamide B as 6R, 9S, 11S, 15R, 16S, 22R, 23S, 27S, 31S, 34S, 37S (Fig. 3D). This is identical to the previously reported absolute configuration for bistramide at all analogous chiral centers.21,25
Reexamination of lagriamide biosynthesis
As described above, the cytochrome P450 LgaJ was proposed to catalyze epoxidation of the double bond at C2–C3, and oxidation of the hydroxyl groups at C4 and C27.6 Because the only difference between the two lagriamide structures at these positions is the lack of the epoxide in lagriamide B, it is likely that LgaJ only catalyzes epoxidation. The possibility that the ketone at C4 would be a direct product of the PKS is not supported by in silico analysis, given that the KR at the corresponding extender module 2 of LgaG is predicted to be active based on sequence alignment (Fig. S15†). Although the possibility that the KR is inactive cannot be excluded, a plausible alternative is that one of the accessory genes may catalyze oxidation at carbon 4. A candidate oxidase is LgbK which belongs to the short-chain dehydrogenase/reductase family of proteins. Regarding C27, the lack of a KR domain on LgbD is consistent with the presence of a ketone at this position for lagriamide B. Another difference between the two clusters is that LgbG contains an additional DH domain at the C-terminus which is, however, predicted to be inactive based on modification of the HxxxGxxxxP motif (ESI Fig. S14, DH5_G†). We explored the possibility that this DH could function as a pyran synthase (PS) using in silico analyses. However, the PS characteristic replacement of a DH conserved aspartate residue in the DxxxQ motif was not observed (Fig. S14†).26 Moreover, TransATor did not predict a PS at this position either (Fig. S15†).Finally, a difference between the two lagriamide structures that is not directly apparent from the gene clusters is the presence of two additional carbons at the C-terminus of lagriamide A. Flórez et al.6 proposed that the penultimate module of LgaC is utilized twice, whereas it appears that the penultimate module of LgbC is utilized only once despite the domain organization of the two proteins being identical, which is intriguing. Since the mechanisms underlying programmed module iteration or ‘stuttering’ are not completely understood,27–30 discovery of the lagriamide B gene cluster offers an unique opportunity to investigate the mechanism of iteration or lack thereof in two highly similar synthases in the future.
Although spiroacetal formation can happen spontaneously under acidic conditions, non-enzymatic formation of spiroacetals can lead to a mixture of diastereomers.31 Different routes for stereospecific spiroacetal biosynthesis have been reported, including those catalysed by MonB (monensin),32 RevJ (reveromycin),31 and AveC (avermectin),33 SlnM (salinomycin),34 and OlmO (oligomycin).35 The only remaining, tentatively unassigned hypothetical protein in the lagriamide clusters is LgaM/LgbM. BLAST analysis of LgbM only yields seven hits, including LgaM and six other hypothetical proteins. LgaM/LgbM show no sequence similarity to known spirocyclases. However, the lack of sequence similarity between the five spirocyclases cited above is noteworthy, as they seem to have evolved independently several times, precluding their identification based on homology search. It is also noteworthy that all spirocyclases previously described are from Streptomyces. Future study of Burkholderiales and of the lagriamide clusters is expected to reveal additional sequences and mechanisms for spirocyclases.
Antimicrobial activity of lagriamide B
Lagriamide A was originally postulated to be a potent antifungal agent with protective activity against environmental pathogens that predate on beetle eggs.6 However, the original isolation yielded only 600 μg of material from 28![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 beetle eggs, precluding biological testing of the pure compound. Instead, activity was inferred from the testing of semi-pure fractions enriched in lagriamide A.
000 beetle eggs, precluding biological testing of the pure compound. Instead, activity was inferred from the testing of semi-pure fractions enriched in lagriamide A.
To examine the spectrum of activity of lagriamide B we performed antimicrobial assays against a panel of 21 strains including two species of filamentous fungi (Aspergillus niger and Purpureocillium lilacinum), two species of yeast (Saccharomyces cerevisiae and Candida albicans) and 17 species of bacteria, most of which are clinically relevant pathogens (Table S12†). Interestingly, lagriamide B possessed moderate but selective antifungal activity against A. niger (MIC = 12.5 μM), with no activity against P. lilacinum, S. cerevisiae, C. albicans or any of the 17 bacterial strains up to the highest tested concentration (128 μM).
Image-based cytological profiling of lagriamide B
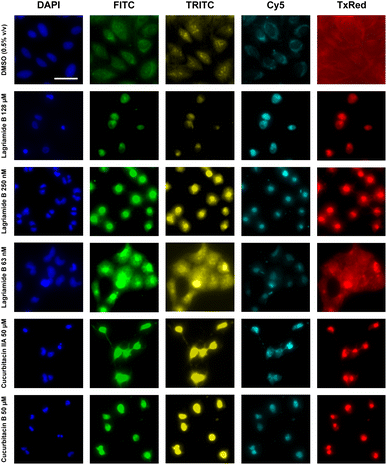
Bistramide A has been previously reported to possess potent antiproliferative activity through the disruption of actin polymerization.36 Bistramide A possesses a dual mechanism whereby the amide and spiroketal subunits enable disassembly of filamentous actin while the enone subunit causes covalent G-actin sequestration. The end result is rapid depolymerization of the actin cytoskeleton and inhibition of cell cycle progression and cytokinesis.To explore the mechanism of action of lagriamide B, which contains all three bioactive subunits found in bistramide A, we examined the morphological consequence of compound treatment in osteosarcoma epithelial cells (U2-OS) using the Cell Painting high-content imaging approach (Fig. 5 and S5†).37 Analysis of the Cell Painting profiles revealed that at low concentration (250 nM), lagriamide B induces binucleation indicative of incomplete cytokinesis as well as total disruption of actin polymerization (DAPI channel, Fig. 5). At high concentration (128 μM) lagriamide B yields lower cell counts and a small, rounded cell morphology. These observations are consistent with results from previous studies using live cell imaging to investigate natural product actin poisons38 and strongly suggests that lagriamide B's cytotoxic activity arises from the disruption of actin polymerization.
To further corroborate this hypothesis using an unbiased methodology, we combined the cell painting images from lagriamide B treatment with those previously acquired for the TargetMol library of 4400 compounds with known mechanisms of action for hierarchical clustering. Feature extraction was performed using the open-source Cell Profiler software package,39 which produced 2090 unique morphological features per compound. Clustering of a selected subset of features (Fig. S30†) resulted in a group containing all assayed lagriamide B concentrations in close proximity, as well as the two natural product standards cucurbitacin IIa and cucurbitacin B (Fig. 5). Analysis of the hierarchical clustering heatmap showed strong agreement in the features perturbed by lagriamide B and the natural product standards, with some exceptions. Notably, the cucurbitacins are secondary metabolites isolated from the Cucurbitaceae family of gourd-bearing plants with known analgesic and anticancer activities;40 the natural products are furthermore reported to disrupt actin filamentation in cells.41 The combination of evidence supports the role of actin filament disruption in the cytotoxic activity of lagriamide B.
Conclusions
Integration of complementary genome mining approaches, coupled with isotopic labelling and untargeted metabolomics, led to the discovery of lagriamide B, an antifungal polyketide with potential biomedical application. Using a combination of spectroscopic and computational methods we determined the full absolute configuration of this new metabolite. Subsequent screening in 22 separate assays (2× fungal, 2× yeast, 17× bacterial, 1× mammalian high-content assay) demonstrated antimicrobial activity selectively against a single fungal strain (A. niger), with a clear cytotoxic mechanism of action against U2-OS cells related to actin filament disruption. Importantly, this result provides the first sustainable supply of a metabolite from this class, opening the way to further investigation of this interesting new molecule.Data availability
NMR data for lagriamide B (2) have been deposited to the Natural Products Magnetic Resonance Database (https://www.np-mrd.org) under accession code NP0332828. The structure of lagriamide B has been deposited to the Natural Products Atlas (https://www.npatlas.org). The genome of P. acidicola RL17-338-BIF-B has been deposited in GenBank under accession code JAOALG010000000, and BioProject ID number PRJNA875462. The complete genome of P. acidicola G-6302 (ATCC 31363) has been deposited in GenBank under the BioProject ID number PRJNA1013544.Author contributions
Conceptualization, RGL and ASE; methodology, RGL, ASE, JCK, CHF, JS, BSP, DMW, NJM, SW, JB; investigation, CHF, JS, BSP, DMW, DYL, NJM, SW, JB; writing – original draft, RGL, CHF, JS, ASE; writing – review & editing, RGL, CHF, JS, ASE, BSP; funding acquisition, RGL, ASE, JCK, CAG; supervision, RGL, ASE, JCK, CAG.Conflicts of interest
There are no conflicts to declare.Acknowledgements
Funding was provided by the National Institutes of Health (GM129344: ASE, RGL; GM133776, JCK) and the Natural Sciences and Engineering Research Council of Canada Discovery program (RGL, CAG).References
- C. R. Pye, M. J. Bertin, R. S. Lokey, W. H. Gerwick and R. G. Linington, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, 5601–5606 CrossRef CAS PubMed.
- J. D. Hegemann, J. Birkelbach, S. Walesch and R. Müller, EMBO Rep., 2023, 24, e56184 CrossRef CAS PubMed.
- S. Kunakom and A. S. Eustáquio, J. Nat. Prod., 2019, 82, 2018–2037 CrossRef CAS PubMed.
- A. Gavriilidou, S. A. Kautsar, N. Zaburannyi, D. Krug, R. Müller, M. H. Medema and N. Ziemert, Nat. Microbiol., 2022, 7, 726–735 CrossRef CAS PubMed.
- J. A. van Santen, E. F. Poynton, D. Iskakova, E. McMann, T. A. Alsup, T. N. Clark, C. H. Fergusson, D. P. Fewer, A. H. Hughes, C. A. McCadden, J. Parra, S. Soldatou, J. D. Rudolf, E. M.-L. Janssen, K. R. Duncan and R. G. Linington, Nucleic Acids Res., 2022, 50, D1317–D1323 CrossRef CAS PubMed.
- L. V. Flórez, K. Scherlach, I. J. Miller, A. Rodrigues, J. C. Kwan, C. Hertweck and M. Kaltenpoth, Nat. Commun., 2018, 9, 2478 CrossRef PubMed.
- K. Blin, S. Shaw, Z. Charlop-Powers, G. P. van Wezel, M. H. Medema and T. Weber, Nucleic Acids Res., 2021, 49, W29–W35 CrossRef CAS PubMed.
- T. Robbins, J. Kapilivsky, D. E. Cane and C. Khosla, Biochemistry, 2016, 55, 4476–4484 CrossRef CAS PubMed.
- R. Reid, M. Piagentini, E. Rodriguez, G. Ashley, N. Viswanathan, J. Carney, D. V. Santi, C. Richard Hutchinson and R. McDaniel, Biochemistry, 2003, 42, 72–79 CrossRef CAS PubMed.
- P. Caffrey, ChemBioChem, 2003, 4, 654–657 CrossRef CAS PubMed.
- A. Keatinge-Clay, J. Mol. Biol., 2008, 384, 941–953 CrossRef CAS PubMed.
- C. R. Valenzano, Y.-O. You, A. Garg, A. Keatinge-Clay, C. Khosla and D. E. Cane, J. Am. Chem. Soc., 2010, 132, 14697–14699 CrossRef CAS PubMed.
- B. M. Harvey, T. Mironenko, Y. Sun, H. Hong, Z. Deng, P. F. Leadlay, K. J. Weissman and S. F. Haydock, Chem. Biol., 2007, 14, 703–714 CrossRef CAS PubMed.
- C. Li, K. E. Roege and W. L. Kelly, ChemBioChem, 2009, 10, 1064–1072 CrossRef CAS PubMed.
- H. Luhavaya, M. V. B. Dias, S. R. Williams, H. Hong, L. G. De Oliveira and P. F. Leadlay, Angew. Chem., Int. Ed. Engl., 2015, 54, 13622–13625 CrossRef CAS PubMed.
- D. Gouiffès, S. Moreau, N. Helbecque, J. L. Bernier, J. P. Hénichart, Y. Barbin, D. Laurent and J. F. Verbist, Tetrahedron, 1988, 44, 451–459 CrossRef.
- A. T. Keatinge-Clay, Nat. Prod. Rep., 2016, 33, 141–149 RSC.
- C. S. McCaughey, J. A. van Santen, J. J. J. van der Hooft, M. H. Medema and R. G. Linington, Nat. Chem. Biol., 2022, 18, 295–304 CrossRef CAS PubMed.
- D. Gouiffes, M. Juge, N. Grimaud, L. Welin, M. P. Sauviat, Y. Barbin, D. Laurent, C. Roussakis, J. P. Henichart and J. F. Verbist, Toxicon, 1988, 26, 1129–1136 CrossRef CAS PubMed.
- G. Zuber, M.-R. Goldsmith, T. D. Hopkins, D. N. Beratan and P. Wipf, Org. Lett., 2005, 7, 5269–5272 CrossRef CAS PubMed.
- A. V. Statsuk, D. Liu and S. A. Kozmin, J. Am. Chem. Soc., 2004, 126, 9546–9547 CrossRef CAS PubMed.
- S. G. Smith and J. M. Goodman, J. Am. Chem. Soc., 2010, 132, 12946–12959 CrossRef CAS PubMed.
- K. Ermanis, K. E. B. Parkes, T. Agback and J. M. Goodman, Org. Biomol. Chem., 2019, 17, 5886–5890 RSC.
- E. J. N. Helfrich, R. Ueoka, A. Dolev, M. Rust, R. A. Meoded, A. Bhushan, G. Califano, R. Costa, M. Gugger, C. Steinbeck, P. Moreno and J. Piel, Nat. Chem. Biol., 2019, 15, 813–821 CrossRef CAS PubMed.
- M. T. Crimmins and A. C. DeBaillie, J. Am. Chem. Soc., 2006, 128, 4936–4937 CrossRef CAS PubMed.
- P. Pöplau, S. Frank, B. I. Morinaka and J. Piel, Angew. Chem., Int. Ed. Engl., 2013, 52, 13215–13218 CrossRef PubMed.
- K. M. Guzman, K. P. Yuet, S. R. Lynch, C. W. Liu and C. Khosla, J. Org. Chem., 2021, 86, 11100–11106 CrossRef CAS PubMed.
- S. J. Moss, C. J. Martin and B. Wilkinson, Nat. Prod. Rep., 2004, 21, 575–593 RSC.
- Y. Sugimoto, K. Ishida, N. Traitcheva, B. Busch, H.-M. Dahse and C. Hertweck, Chem. Biol., 2015, 22, 229–240 CrossRef CAS PubMed.
- N. Traitcheva, H. Jenke-Kodama, J. He, E. Dittmann and C. Hertweck, ChemBioChem, 2007, 8, 1841–1849 CrossRef CAS PubMed.
- S. Takahashi, A. Toyoda, Y. Sekiyama, H. Takagi, T. Nogawa, M. Uramoto, R. Suzuki, H. Koshino, T. Kumano, S. Panthee, T. Dairi, J. Ishikawa, H. Ikeda, Y. Sakaki and H. Osada, Nat. Chem. Biol., 2011, 7, 461–468 CrossRef CAS PubMed.
- A. R. Gallimore, C. B. W. Stark, A. Bhatt, B. M. Harvey, Y. Demydchuk, V. Bolanos-Garcia, D. J. Fowler, J. Staunton, P. F. Leadlay and J. B. Spencer, Chem. Biol., 2006, 13, 453–460 CrossRef CAS PubMed.
- P. Sun, Q. Zhao, F. Yu, H. Zhang, Z. Wu, Y. Wang, Y. Wang, Q. Zhang and W. Liu, J. Am. Chem. Soc., 2013, 135, 1540–1548 CrossRef CAS PubMed.
- C. Jiang, Z. Qi, Q. Kang, J. Liu, M. Jiang and L. Bai, Angew. Chem., Int. Ed. Engl., 2015, 54, 9097–9100 CrossRef CAS PubMed.
- O. Bilyk, G. S. Oliveira, R. M. de Angelo, M. O. Almeida, K. M. Honório, F. J. Leeper, M. V. B. Dias and P. F. Leadlay, J. Am. Chem. Soc., 2022, 144, 14555–14563 CrossRef CAS PubMed.
- S. A. Rizvi, D. S. Courson, V. A. Keller, R. S. Rock and S. A. Kozmin, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 4088–4092 CrossRef CAS PubMed.
- M.-A. Bray, S. Singh, H. Han, C. T. Davis, B. Borgeson, C. Hartland, M. Kost-Alimova, S. M. Gustafsdottir, C. C. Gibson and A. E. Carpenter, Nat. Protoc., 2016, 11, 1757–1774 CrossRef CAS PubMed.
- Y. Hayashi-Takanaka, Y. Kina, F. Nakamura, S. Yamazaki, M. Harata, R. W. M. van Soest, H. Kimura and Y. Nakao, Sci. Rep., 2019, 9, 7540 CrossRef PubMed.
- D. R. Stirling, M. J. Swain-Bowden, A. M. Lucas, A. E. Carpenter, B. A. Cimini and A. Goodman, BMC Bioinf., 2021, 22, 433 CrossRef PubMed.
- J. C. Chen, M. H. Chiu, R. L. Nie, G. A. Cordell and S. X. Qiu, Nat. Prod. Rep., 2005, 22, 386–399 RSC.
- E. E. Delgado-Tiburcio, J. Cadena-Iñiguez, E. Santiago-Osorio, L. D. M. Ruiz-Posadas, I. Castillo-Juárez, I. Aguiñiga-Sánchez and M. Soto-Hernández, Pharmaceuticals, 2022, 15, 1325 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: NMR spectra, screening data and sequence data for lagriamide B. See DOI: https://doi.org/10.1039/d4sc00825a |
| ‡ Authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2024 |