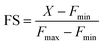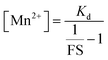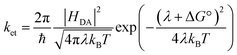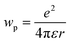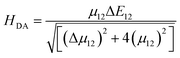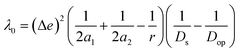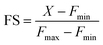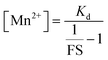Open Access Article
Open Access ArticleA water-soluble, cell-permeable Mn(II) sensor enables visualization of manganese dynamics in live mammalian cells†
Smitaroopa
Kahali‡
 a,
Sujit Kumar
Das‡
a,
Sujit Kumar
Das‡
 a,
Ravinder
Kumar
a,
Kunika
Gupta
a,
Ravinder
Kumar
a,
Kunika
Gupta
 a,
Rajasree
Kundu
a,
Rajasree
Kundu
 a,
Baivabi
Bhattacharya
b,
Arnab
Nath
b,
Ravindra
Venkatramani
a,
Baivabi
Bhattacharya
b,
Arnab
Nath
b,
Ravindra
Venkatramani
 a and
Ankona
Datta
a and
Ankona
Datta
 *a
*a
aDepartment of Chemical Sciences, Tata Institute of Fundamental Research, 1 Homi Bhabha Road, Mumbai 400005, India. E-mail: ankona@tifr.res.in
bDepartment of Developmental Biology and Genetics, Indian Institute of Science, Bangalore 560012, India
First published on 13th June 2024
Abstract
Central roles of Mn2+ ions in immunity, brain function, and photosynthesis necessitate probes for tracking this essential metal ion in living systems. However, developing a cell-permeable, fluorescent sensor for selective imaging of Mn2+ ions in the aqueous cellular milieu has remained a challenge. This is because Mn2+ is a weak binder to ligand-scaffolds and Mn2+ ions quench fluorescent dyes leading to turn-off sensors that are not applicable for in vivo imaging. Sensors with a unique combination of Mn2+ selectivity, μM sensitivity, and response in aqueous media are necessary for not only visualizing labile cellular Mn2+ ions live, but also for measuring Mn2+ concentrations in living cells. No sensor has achieved this combination thus far. Here we report a novel, completely water-soluble, reversible, fluorescent turn-on, Mn2+ selective sensor, M4, with a Kd of 1.4 μM for Mn2+ ions. M4 entered cells within 15 min of direct incubation and was applied to image Mn2+ ions in living mammalian cells in both confocal fluorescence intensity and lifetime-based set-ups. The probe was able to visualize Mn2+ dynamics in live cells revealing differential Mn2+ localization and uptake dynamics under pathophysiological versus physiological conditions. In a key experiment, we generated an in-cell Mn2+ response curve for the sensor which allowed the measurement of the endogenous labile Mn2+ concentration in HeLa cells as 1.14 ± 0.15 μM. Thus, our computationally designed, selective, sensitive, and cell-permeable sensor with a 620 nM limit of detection for Mn2+ in water provides the first estimate of endogenous labile Mn2+ levels in mammalian cells.
Introduction
The biological importance of manganese (Mn) is undisputed, evident from its distinct roles in brain function, immunity against pathogens, all the way to one of the most crucial processes on the planet, photosynthesis.1–5 Since Mn is an essential nutrient, the uptake, distribution, and removal of Mn ions are tightly regulated in all forms of life, ranging from bacteria to animals and plants.5 The playground of Mn cell biology encompasses the Mn ion labile pool, Mn dependent proteins and enzymes, and Mn transporters.1,2,4–13 The concerted interplay of these players maintains the total mammalian cellular levels of Mn ions between low μM to sub mM levels.5,14 Due to the reducing cellular environment, the major oxidation state of the labile Mn ion pool is +2.15 The labile Mn2+ pool can act as an antioxidant as per in vitro studies.2,15 Recent reports have shown that the antioxidant properties of the labile pool are linked to the mobilization of Mn2+ ions during cancer and the early stages of microbial infection.3,4,16–18 Further, mutations in Mn2+ transporters can lead to early-onset Parkinsonism, due to accumulation of Mn ions in the blood, brain, and liver.19–23 Mn-induced Parkinsonism is also observed in miners and welders who are occupationally exposed to Mn.5,24–26 In this intricate backdrop of Mn essentiality and pathophysiology, Mn2+ sensitive fluorescent sensors are key chemical tools that can help chalk out Mn regulatory pathways in both physiological and disease contexts. Importantly, in theory fluorescent Mn2+ sensors should be able to afford estimates of labile Mn2+ ions in living mammalian cells; however, no sensor has achieved this goal to date.The requirements for an ideal Mn2+ sensor for tracking, imaging, and estimating endogenous Mn2+ ions in living systems are: (1) selectivity over other biologically essential metal ions especially Ca2+ and Mg2+ ions which have very similar coordination properties to Mn2+ ions but are present in overlapping to higher concentration ranges in living systems;27 (2) sensitivity to detect μM levels of the labile Mn2+ ion pool; (3) cell permeability; and finally (4) water-solubility to generate in vitro/in-cell fluorescence calibration curves and obtain dissociation constants for the sensor in aqueous buffer replicating the aqueous cellular milieu. The last feature would allow the key estimation of labile Mn2+ levels in living cells. To date no Mn2+ sensor has shown the combination of all of the aforementioned properties (Fig. 1). The very few small molecule-based Mn2+ sensors either suffer from selectivity issues or have low water-solubility.28–31 The only two macromolecular sensors, one protein-based and another DNA-based, either rely on transfection for cell incorporation or are cell impermeable.32,33 In addition, the protein-based sensor did not have the desired selectivity toward Mn2+ over Ca2+ and Mg2+ and especially given that the intra-cellular concentration of Mg2+ ions is in the mM range, competition would be expected. Thus far the protein-based sensor has only been applied to detect Mn2+ in bacterial cells (Fig. 1a).33 The DNA-based cell-impermeable sensor is selective toward Mn2+ but has low sensitivity allowing imaging of only high, mM levels of Mn2+ ions (Fig. 1a).32 A reason for the lack of fluorescent sensors for detecting Mn2+ ions is the fact that Mn2+ ions lie at the bottom of the Irving–Williams series which ranks the stability of metal complexes of divalent transition metal ions in ascending order from Mn2+ to Cu2+ in water.34 Apart from the Irving–Williams series, a more significant challenge is to design selective binders for Mn2+ over Ca2+ and Mg2+ ions which are present in sub μM (Ca2+) to mM (Mg2+) concentrations in cells within mammalian systems. Finally, Mn2+ ions can quench the fluorescence of fluorophores leading to turn-off sensors which are not suitable for tracking metals in living systems.35
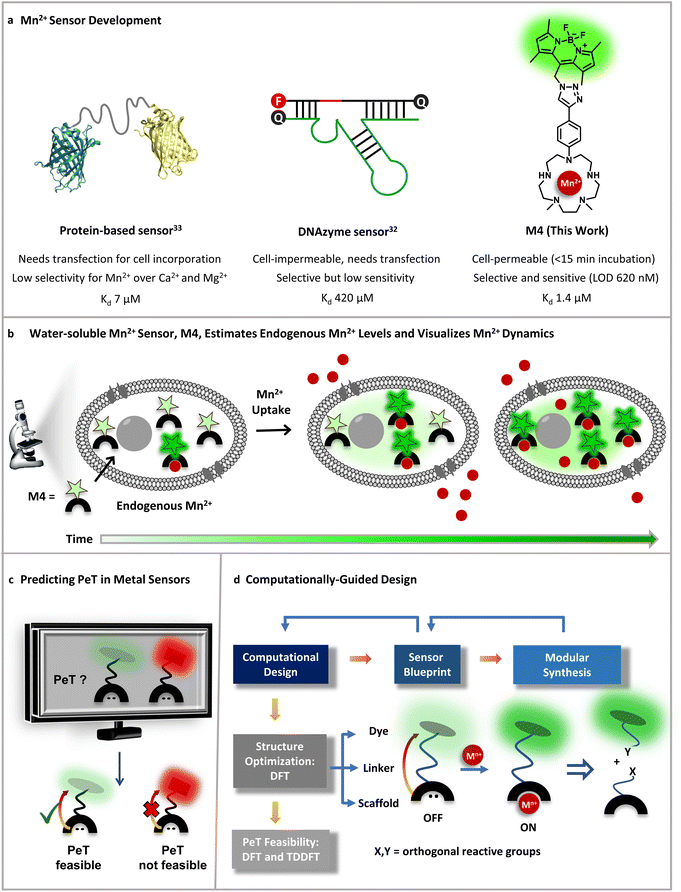 | ||
| Fig. 1 Designing an Mn2+ sensor. (a) Comparison of previous Mn2+ probes with this work. A fluorescent protein-based sensor requires transfection and has limited selectivity for Mn2+ over Ca2+ making it only applicable to Mn2+ detection in bacterial cells thus far.33 A DNAzyme sensor exhibits Mn2+ selectivity, but is cell-impermeable and has low sensitivity only allowing imaging of high, mM levels of Mn2+ ions.32 A novel small-molecule based turn-on fluorescent water-soluble sensor, in this work, directly permeates cell-membranes and exhibits Mn2+ selectivity and sensitivity to enable imaging of Mn2+ dynamics in living mammalian cells. (b) Schematic depicting the cell-permeable small-molecule based sensor described in (a), which is sensitive to and can estimate physiological levels of Mn2+ and can also visualize Mn2+ uptake dynamics in live cells. (c) A computational workflow devised to predict PeT, central to the design of turn-on fluorescent metal ion sensors. (d) Schematic representing the utilization of the computational workflow to predict PeT in any metal ion sensor blueprint, highlighting the feedback between design guided by chemical intuition, synthetic feasibility, and computations. The general workflow was leveraged to develop a water-soluble, cell-permeable Mn2+ sensor. | ||
To address all of the aforementioned challenges and develop a cell-permeable, water-soluble, fluorescent sensor for Mn2+ ions we devised a general computational workflow to design a turn-on fluorescent sensor for any metal ion. Specifically, we noted that none of the previously reported sensors had a combination of water-solubility, selectivity, and sensitivity toward μM levels of Mn2+ ions. In order to visualize endogenous levels of Mn2+ ions in living cells and estimate the levels of labile Mn2+ ions in mammalian cells we needed a ‘turn-on’ fluorescent sensor that would respond to μM levels of Mn2+ ions in aqueous buffer with high selectivity. We thus went to the drawing board and designed putative water-soluble Mn2+ sensors that could be synthesized via modular synthetic strategies amenable to easy modifications. The computational workflow was applied to these sensor blueprints (Fig. 1c and d). In this proof-of-concept study, we successfully combined our predictive computational workflow in conjunction with a modular synthetic approach, to obtain a novel, selective, sensitive, completely water-soluble, and cell-permeable turn-on fluorescent sensor for Mn2+ ions (Fig. 1a and b). The sensor had a dissociation constant of 1.4 ± 0.2 μM toward Mn2+ ions in aqueous buffer and a limit of detection (LOD) of 620 ± 70 nM for Mn2+ ions. The sensor, henceforth referred to as M4, entered mammalian cells within 15 min of direct incubation in aqueous media. M4 showed a distinct selective increase in fluorescence intensity (Fig. 1b) and fluorescence lifetime in the presence of Mn2+ ions. The cell-permeable sensor was applied to image Mn2+ ions in confocal fluorescence microscopy setups in both fluorescence intensity and lifetime modes, a first for Mn2+ imaging. Importantly, we were able to apply the sensor to not only visualize endogenous Mn2+ ions but also performed an in-cell calibration of the fluorescence response of the sensor. By utilizing the information on the dissociation constant of the sensor in aqueous buffer and performing a key live cell Mn2+ uptake imaging experiment we provide the first estimate of labile Mn2+ ions in mammalian cells as 1.14 ± 0.15 μM.
Finally, the probe was used to visualize live Mn2+ uptake dynamics in mammalian cells lacking a critical Mn2+ ion transporter implicated in early onset Parkinsonism via excess Mn accumulation. Comparison of cells with and without the transporter revealed significant differences in Mn2+ uptake rates and visual differences in localization in physiological versus pathophysiological contexts. The study indicates that the sensor would be clearly applicable to the elucidation of Mn2+ regulation not only under physiological conditions but also in the context of neurodegeneration, immunity, and cancers in the future.
Results and discussion
A computational workflow for designing metal ion sensors
Small-molecule based cell-permeable metal ion sensors are attractive chemical tools for deciphering the cell-biology of metal ions. However, the lack of predictability in design principles leads to emphasis on intuitive design which might often lead to unpredictable outcomes. A popular approach to develop turn-on fluorescent metal ion sensors relies upon photo-induced electron transfer (PeT).35,36 The basic blueprint of any PeT based metal ion sensor is a metal-binding ligand conjugated to a fluorescent dye via a linker (Fig. 1c). The sensor should remain in the quenched or off state in the absence of metal ions due to PeT from the metal binding ligand to the dye (Fig. 1c). Upon metal binding, PeT should be abolished leading to an increase in emission. Key to the design of PeT based sensors is the initial off state of the probe. Any random dye–scaffold combination would not lead to a fluorescence quenched apo probe which is a necessary condition for turn-on fluorescence sensors. Realizing that a major challenge in the field of metal ion probe design, including Mn2+ ion sensor design, is predictability in the sensing strategy, we devised a computational workflow to predict PeT in the metal unbound state of a sensor (Fig. 1c, d and 2).The requirements for PeT to occur in a metal unbound initial state or apo sensor are: (1) no electron delocalization between the metal-binding ligand and the dye, as PeT is a through-space electron transfer process;37 (2) favorable driving force between PeT donor and acceptor states that have electron densities localized on the metal-binding scaffold and dye, respectively. To predict if PeT would be feasible in a candidate metal ion sensor, we employed density functional theory (DFT) to optimize the proposed sensor geometries in appropriate solvents and obtain electronic energies and wavefunctions (Fig. 1d). Following DFT calculations, time-dependent DFT (TD-DFT) calculations were performed to identify the most probable electronic transitions within the sensor. The workflow that we devised to determine the feasibility of PeT in any designed metal ion sensor (Fig. 1c and d) therefore included: (1) identifying dye-centered orbitals within the sensor conducive to photo-absorption which can act as PeT acceptors; (2) identifying ligand-centered orbitals with favorable driving forces to act as PeT donors. To test the efficacy of the computational workflow, we decided to apply it toward solving a long-standing challenge in metal ion imaging: the development of a water-soluble, cell-permeable Mn2+ ion probe.
Design of a water-soluble, turn-on fluorescent Mn2+ selective probe
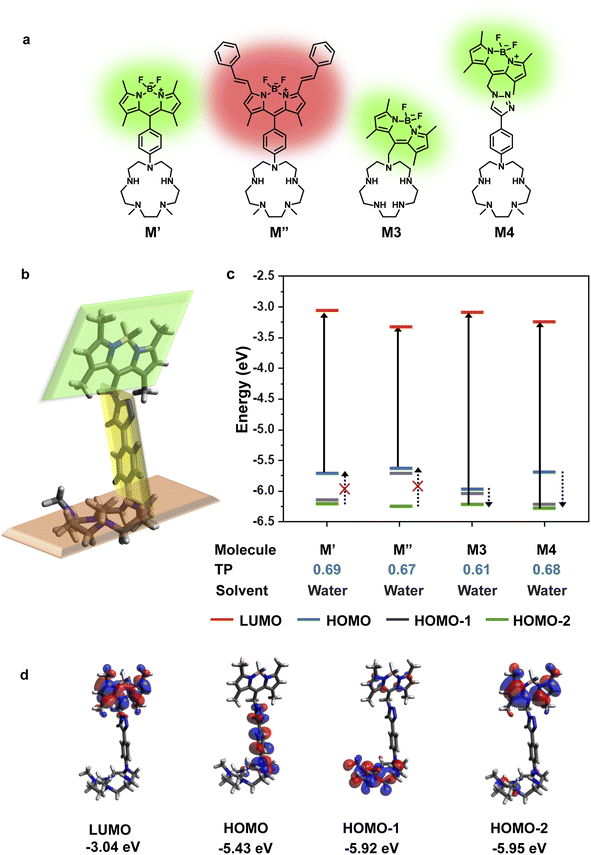
To design a water-soluble, cell-permeable, turn-on fluorescent Mn2+ ion sensor, we applied the computational workflow to blueprints of potential Mn2+ ion sensors (Fig. 2). Blueprints of Mn2+ selective, water-soluble fluorescent sensors were chalked out based on the following considerations: (1) an appropriate Mn2+ binding scaffold with high aqueous solubility; (2) a visible emitting fluorescent dye; (3) feasibility of PeT in the final sensor design as predicted by our computational workflow. For the Mn2+ binding scaffold design, we selected a penta-aza macrocyclic ligand (Fig. 2a). The design was based on the possibility of attaining a hepta-coordinate Mn2+ binding geometry via equatorial N-atom coordination and axial coordination to O-atom donor containing molecules like water or anions like phosphate or citrate2,13,15 that would be present in the cellular milieu. An aza-macrocycle with amine moieties would have inherently high water-solubility. The preference of Mn2+ ions toward a hepta-coordinate geometry has been observed in Mn2+ binding proteins and small-molecule based complexes.38,39 Further, Mn2+ is a borderline ion, that is, it is neither as hard as Ca2+ ions nor as soft as Zn2+ ions.5 Hence, we reckoned that a planar pentagonal N-atom coordination sphere providing a suitable metal-binding core size to fit Mn2+ ions along with axial O-donor coordination would provide the appropriate N,O donor mix for selective Mn2+ ion coordination. We designed four potential water-soluble Mn2+ sensors, M′, M′′, M3, and M4 (Fig. 2a), by combining synthetically feasible pentaaza macrocycles with BODIPY based visible emitting dyes via either aliphatic or aromatic linkers. Amongst these designs, M3 would be attainable via a modular SN2 based scheme to join the metal-binding scaffold to the dye. M4 would be accessible via a “click” reaction based modular synthetic strategy involving conjugation of an alkyne-tagged metal binding scaffold to an azide-tagged dye. Modular synthetic schemes (Fig. 1d) were more preferable since modifications in either the dye or scaffold would be easily accessible in the future.Before employing the computational workflow on the newly designed water-soluble sensor blueprints, the workflow was validated on a previously reported PeT-based Mn2+ ion sensor, M1, that had afforded a selective turn-on response toward Mn2+ ions in acetonitrile (Fig. S1 and Table S1†).30 We employed Gaussian D.01 for all electronic structure calculations, Gaussview 6.0.16 for initial modelling, Avogadro 1.1.1 for visualization, and Gaussview and Avogadro for analysis, for all computations described henceforth. The results from the DFT calculations of M1 in acetonitrile indicated that HOMO−2 and LUMO had electron densities centered on the dye (Fig. S1†). The DFT optimized structure of M1 showed that the dye and the ligand were not in the same plane, thereby indicating a possibility of through-space PeT from the ligand to the dye (Fig. S1†). The most probable transition was from HOMO−2 to LUMO (Table S1†), resulting in two energetically favorable donor orbitals, HOMO and HOMO−1 which were dimethylaniline (DMA) and macrocycle-centered respectively, to fill the hole in HOMO−2 (Fig. S1†). We note here that the DMA amine is a part of the metal binding scaffold. Hence, the computations indicated that PeT would occur in apo M1. Indeed, PeT had been found to be operational in M1, based on the low quantum yield, 0.0038, of the apo sensor in acetonitrile.30 Having validated our workflow, we carried out calculations for all the designed water-soluble Mn2+ sensor molecules with water as solvent (Fig. 2c and S2–S4, and Table S1†). For M′ and M′′ the most probable transitions were from the HOMO to LUMO (Fig. 2c, S2, and S3, and Table S1†). Hence, no favorable PeT donor levels were available. For M3, the most probable transition was from HOMO−2 to LUMO, allowing HOMO and HOMO−1 to act as potential PeT donor levels (Fig. 2c and S4, and Table S1†). However, MOs were delocalized over the dye and the scaffold, making PeT less likely in this molecule.
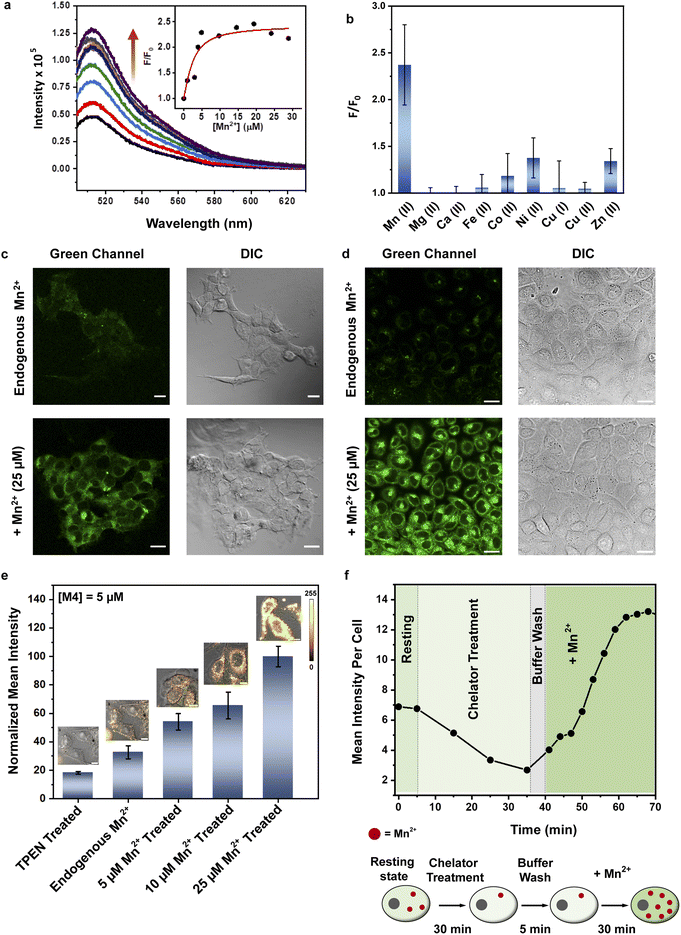
Finally, for M4, we observed that the dye and the ligand were distinctly out-of-plane with respect to each other which was a favorable configuration for PeT (Fig. 2b). The most probable transition was from dye-centered HOMO−2 to LUMO (Fig. 2c and d, and Table S1†). Hence, the DMA-centered HOMO or macrocycle-centered HOMO−1 would act as PeT donor orbitals (Fig. 2c and d). Therefore, based on computations, M4 should exhibit PeT based quenching of the BODIPY unit in its metal unbound state, fulfilling the necessary initial criterion for turn-on sensing. The thermodynamic driving force associated with PeT (ΔG°) and electron transfer rates (ket) were calculated using the Rehm–Weller equation40 and Marcus theory,41 respectively (details in Computational methods, Fig. S5 and Tables S2–S4†). For M4, during excitation, the most probable transition took place from the dye-centered HOMO−2 to LUMO with a hole created on the former, enabling it to act as an electron acceptor state. The potential donor states for PeT were the scaffold centered HOMO and HOMO−1. ΔG° for electron transfer from HOMO to HOMO−2 was calculated to be −2.27 eV and ket was 4.09 × 1012 s−1. ΔG° for electron transfer from HOMO−1 to HOMO−2 was −2.77 eV and ket was 9.13 × 109 s−1. The negative ΔG° values indicated that PeT would be functional in apo M4. The electron transfer rates calculated for M4 were also similar to experimental values obtained for a previously reported BODIPY based Ca2+ ion sensor that functioned via PeT in water.42 Since the computations convincingly predicted that PeT would be feasible in apo M4, we decided to proceed with the evaluation of M4 as a potential water-soluble Mn2+ sensor.
M4 is a water-soluble, Mn2+ ion selective and sensitive turn-on fluorescent sensor
The probe M4, shortlisted from the computational predictions, was next synthesized (Fig. S6†). The synthesis involved a Cu ion assisted “click” reaction to conjugate the BODIPY azide 10 to the Boc-protected alkyne-tagged penta-aza macrocycle 7 (Fig. S7†) to afford the Boc-protected sensor 11 (Fig. S8†) which was then deprotected to afford M4 (Fig. S9†). M4 was characterized by LC-ESI-MS for purity (Fig. S9†), followed by HR-ESI-MS, 1H-NMR, and 13C NMR (Fig. S32–S41†). The molecule was completely water-soluble as designed, with an experimentally determined log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P value of −0.2 (Fig. S10†). Hence, all further experiments were performed in aqueous buffer without the addition of any organic solvent.
P value of −0.2 (Fig. S10†). Hence, all further experiments were performed in aqueous buffer without the addition of any organic solvent.
The presence of the secondary amines in the macrocycle along with the triazole moiety in the linker in M4 (Fig. 2a), we believe, are responsible for the high water-solubility of the molecule. We observed that an aqueous solution of the M4 sensor showed very low fluorescence intensity indicating that PeT was occurring in the apo sensor (Fig. 3a). Importantly, the quantum yield of the apo sensor was 0.027 ± 0.005 (Fig. S11†). Hence, our experimental observations were in line with the computational predictions for M4 indicating that the workflow (Fig. 1d) could be employed successfully to predict PeT in metal ion sensors.
The absorption spectrum of M4 showed a maxima at 508 nm in HEPES buffer (Fig. S9b†) with an extinction coefficient of 2673 ± 127 M−1 cm−1 (Fig. S12†). When excited at 480 nm, the sensor afforded an emission spectrum with a maxima at 514 nm (Fig. 3a). Upon addition of Mn2+ ions in the form of MnCl2, we observed a 2.4-fold increase in emission intensity of M4 at pH 7.1, in aqueous buffer (Fig. 3a and b). The quantum yield of the sensor in the presence of Mn2+ was 0.055 ± 0.005 (Fig. S11†) which was also twice that of the apo sensor matching with the observed fluorescence enhancement in the presence of Mn2+ ions. The extent of enhancement remained unaltered even when other Mn2+ salts, perchlorate and acetate, were used as Mn2+ ion sources, indicating that the response of M4 was not altered when the counterion was changed (Fig. S13†). Further, the response of the sensor toward Mn2+ ions remained unchanged in acetonitrile (Fig. S14†) indicating that the response would not be affected by the polarity of the medium. Studies in other organic solvents could not be performed since the sensor was highly water-soluble and could not be dissolved in other organic solvents. The pH response of M4 was a critical parameter that would govern its applicability in physiological contexts. pH dependent studies of M4 indicated that the emission of the apo sensor remained constant within the physiological pH range of 5–8 (Fig. S15†).
We next tested the response of M4 in the presence of other biologically essential metal ions (Fig. 3b and S16†). The sensor did not respond at all to Mg2+ (up to a tested concentration of 1 mM, Fig. S16a†) and Ca2+ ions (up to a tested concentration of 15 μM) (Fig. 3b) which was a necessary criterion for any designed probe to be applied for imaging Mn2+ ions in living cells especially mammalian cells. M4 also barely responded to all other tested metal ions (Fig. 3b). The only other metal ions that showed minimal response were Ni2+ (maximum 1.4-times at saturation concentrations) and Zn2+ (maximum 1.3-times at saturation concentrations) ions. Notably, for both these ions the emission increase was significantly lower compared to that afforded by Mn2+ ions (Fig. 3b). The dissociation constant of M4 toward Mn2+ ions was obtained as 1.4 ± 0.2 μM (Fig. 3a, inset, and Table S5†). Ni2+ ions afforded a 2-times higher Kd value while Zn2+ ions showed a 6-times higher Kd value when compared to the dissociation constant of M4 toward Mn2+ ions (Table S5 and Fig. S17†). The dissociation constant values for Ni2+ and Zn2+ were further validated by performing fluorescence competition titrations with the Mn2+ complex of M4 (Fig. S18†). The putative levels of labile Zn2+ ions within cellular systems are pM and intracellular labile Ni2+ levels are in the sub nM range.35,43–45 Hence, we expect no interference from these ions when imaging Mn2+ ions which are present in μM levels in cellular systems. Na+ and K+ ions were already present in high mM amounts in the buffer. No change in the fluorescence response of M4 was observed after addition of excess buffer to an aqueous solution of M4 (Fig. S16b†). Any possibility of interference from Na+ and K+ ions was thus eliminated. Finally, the response of M4 toward Mn2+ ions remained unaffected in the presence of a representative protein, bovine serum albumin (Fig. S19†). Therefore, no interference from the proteins in the biological context is expected. Hence, M4 was an Mn2+ selective sensor that afforded a distinct turn-on response in the presence of Mn2+ ions in water. Importantly, based on the binding affinity (0.7 × 106 M−1) of M4 toward Mn2+ ions and a LOD for Mn2+ ions of 620 ± 70 nM in water (Fig. S20†), M4 had the apt sensitivity required to detect Mn2+ ions in biological systems.
The cell-permeable, Mn2+ ion selective probe images physiologically relevant levels of Mn2+ ions in living cells
We next tested whether M4 could be applied to image Mn2+ ions in living mammalian cells (Fig. 3c–f). An MTT assay for cellular toxicity in HEK293T cells indicated that the sensor was not toxic up to a tested concentration of 15 μM, till the maximum tested incubation time of 1 h (Fig. S21†). Hence, concentrations lower than 15 μM and incubation times less than 1 h were used for all cell studies. When living HEK293T and HeLa cells were treated with aqueous solutions of M4 (5 μM), we observed that the sensor efficiently entered cells within 15 min of direct incubation (Fig. 3c and d, S22, and S23†). Fluorescence emission from cells that were not exogenously treated with Mn2+ indicated that M4 could be used to detect Mn2+ ions inherently present in living cells (Fig. 3c and d, top panels, and Fig. 3e). A distinct increase in fluorescence emission was observed in both HEK293T and HeLa cells exogenously treated with low μM (25 μM) levels of Mn2+ ions (Fig. 3c and d, bottom panels). Intensity analysis of cell images indicated significant 2.2-fold overall emission enhancement in Mn-treated HEK293T cells and 2.7-fold emission enhancement in Mn-treated HeLa cells versus untreated cells. Furthermore, when N,N,N′,N′-tetrakis(2-pyridylmethyl)ethylenediamine (TPEN), a cell-permeable, divalent metal ion chelator that can bind to Mn2+, was added to cells that were not exogenously treated with Mn2+, a clear decrease in fluorescence intensity was observed (Fig. 3e and f, and S24†). A similar decrease in fluorescence intensity was also observed when TPEN was added to cells that were exogenously treated with Mn2+ (Fig. S25†), thus establishing M4 as a reversible sensor for detecting Mn2+ ions within living cells. Hence, M4 was a water-soluble, reversible, cell-permeable sensor that could clearly image μM, physiologically relevant levels of Mn2+ ions in living mammalian cells.In both cell types, increased emission was observed in the cytoplasm and in the perinuclear regions, in Mn-treated cells. The presence of Mn in these regions of Mn-treated mammalian cells has been reported previously via Mn elemental maps generated using nano-synchrotron X-ray fluorescence imaging.23,46,47 This fact further validated the ability of M4 toward Mn2+ ion imaging in living cells.
Although the labile Zn2+ levels in mammalian cells are in pM,35,43,45 to further confirm that there was no interference from Zn2+ ions in the in-cell response of M4, a control experiment was performed on Zn2+ treated cells. No increase in fluorescence intensity was observed in cells exogenously treated with Zn2+ (Fig. S26†), again confirming the in-cell Mn2+ specificity of M4. Finally, we wanted to confirm that M4 was not localized in lysosomes as an acidic environment might lead to fluorescence enhancement in PeT based sensors. No colocalization was observed between M4 and LysoTracker Red (Fig. S27†). These experiments distinctly confirmed the in-cell Mn2+ specificity of M4.
In order to test whether M4 could detect changes in Mn2+ concentration inside living mammalian cells in the physiologically relevant Mn2+ concentration range and if the sensor could afford an Mn2+ concentration dependent response in living mammalian cells, different sets of HeLa cells were incubated with varying concentrations of Mn2+ ions. The lowest intensity was observed when cells were treated with the chelator TPEN (Fig. 3e). A step-wise increase in cellular Mn2+ levels from untreated cells containing endogenous Mn2+ to cells treated with up to 25 μM Mn2+ could be distinctly observed via a concomitant increase in the fluorescence intensity of the sensor. Therefore, M4 could report on endogenous levels of Mn2+ and importantly track changes in Mn2+ ion concentrations inside living cells (Fig. 3e).
Encouraged by this result we next set forth to apply the water-soluble, cell-permeable sensor, M4, to estimate labile Mn2+ levels by a method first introduced by Tsien et al.48 and applied by Palmer et al.49 to metal-based sensors to determine the levels of intracellular Zn2+ ions. The method involved determining the fractional saturation of the sensor in resting cells that had not been externally treated with any Mn2+ ions. The fractional saturation could then be plugged in to an equation relating the dissociation constant of the sensor and fractional saturation to the concentration of the labile metal ion. In brief, the experiment involved temporal imaging of fluorescence intensity within living mammalian cells starting from the resting state, chelator treatment, and wash, followed by Mn2+ addition (Fig. 3f). The experiment was repeated and analyzed for multiple individual cells to afford the average minimum fluorescence intensity (Fmin), fluorescence intensity in resting cells (X), and maximum fluorescence intensity (Fmax) in living cells. To obtain the minimum in-cell fluorescence intensity of apo-M4, cells were treated with TPEN, a chelator that can bind to Mn2+ ions. Subsequently, the maximum fluorescence intensity corresponding to the bound form of M4 inside the cells was obtained by adding Mn2+ ions to the same set of cells. The titration curves (Fig. 3f and S28†) indicate that M4 responds to both the decrease in Mn2+ levels upon chelator treatment and increase in Mn2+ levels due to exogenously added Mn2+. Fractional saturation FS was calculated from:
Since we could measure the dissociation constant of the sensor for Mn2+ ions in aqueous media owing to the water-solubility of the sensor, the concentration of labile Mn2+ ions in resting cells could be estimated from the equation
Thus, by generating in-cell Mn2+ responses we obtained the average labile Mn2+ concentration in living mammalian HeLa cells as 1.14 ± 0.15 μM which to the best of our knowledge is the first estimate of labile Mn2+ ion concentration in a living mammalian cell.
Fluorescence lifetime imaging of Mn2+ ions in living cells
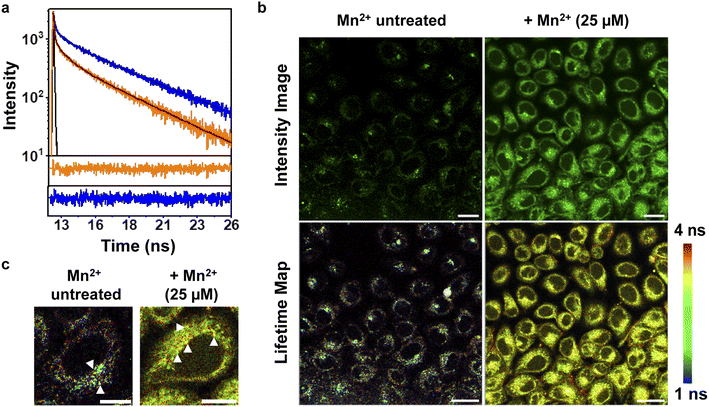
To further increase the repertoire of applications of M4 toward imaging biological Mn2+ in living cells, we next explored fluorescence lifetime imaging microscopy (FLIM) (Fig. 4). Intensity based fluorescence imaging with a turn-on fluorescence sensor cannot distinguish between intracellular regions of high sensor accumulation versus regions with high analyte concentration. Since FLIM is inherently concentration and intensity independent, this technique in conjunction with intensity images can provide distinct information on intracellular analyte-localization.50,51 Hence, we imaged Mn2+ untreated and exogenously Mn2+ treated mammalian cells on a FLIM setup in the presence of M4 (Fig. 4b, and S29†). We first performed in vitro lifetime measurements of M4 in the absence and presence of Mn2+ ions and observed a 2.3-fold increase in the lifetime value of the sensor in the presence of Mn2+ ions (Fig. 4a, and Table S6†). This result was clearly in line with the in vitro fluorescence titration results in Fig. 3a. In live cell FLIM experiments, we observed significantly lower lifetime values in certain regions in Mn2+ untreated cells (Fig. 4b, bottom left panel, Fig. 4c, left panel) compared to Mn2+ treated cells (Fig. 4b, bottom right panel, Fig. 4c, right panel) indicating the presence of the apo sensor in cells that were not exogenously treated with Mn2+. We note that the IRFs of the TCSPC measurement setup and FLIM setup were different, with the FLIM IRF (224 ps) being significantly longer than the TCSPC IRF (68 ps). Hence, we have not compared the absolute average lifetime values between in vitro TCSPC and FLIM and reported the relative increase in average lifetime values of the probe in the absence and presence of Mn2+ ions. The lifetime values were uniformly higher throughout the cytoplasm and peri-nuclear regions in the Mn2+ treated cells showing the localization of Mn2+ ions in these regions (Fig. 4c, white arrows, right panel). In contrast, in Mn2+ untreated cells, few punctate regions showed high lifetime values attributable to the bound sensor, hinting toward perhaps a more compartmentalized Mn2+ ion localization (Fig. 4c, white arrows, left panel). Hence, M4 provides an unprecedented handle into imaging Mn2+ localization in living cells via both intensity and lifetime image maps (Fig. 4b).Visualization of Mn2+ uptake dynamics in live mammalian cells reveals pathophysiologically relevant differences
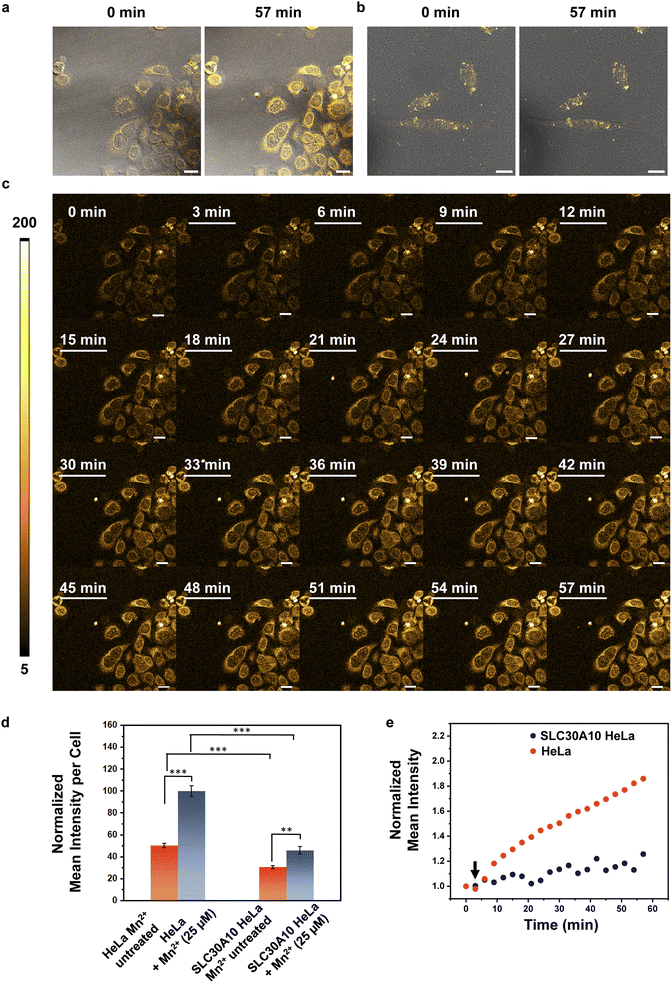
Finally, since M4 is a turn-on, cell-permeable, fluorescent sensor for Mn2+ ions that can detect Mn2+ ions in aqueous media, we wanted to challenge the sensor by testing if the probe could be used to image Mn2+ dynamics in living mammalian cells (Fig. 5, ESI Videos S1 and S2†). Mutations in the cell-surface localized Mn2+ ion efflux transporter SLC30A10 causes hypermanganesemia, leading to liver damage and Parkinson's like dystonia.19–21,23 We reasoned that tracking differences in Mn2+ ion uptake rates in cells with low levels of SLC30A10 mRNA versus cells overexpressing SLC30A10 would afford information on the spatio-temporal dynamics of Mn2+ ions and its link to Mn-transport disorders reported in children and adults. HeLa cells have inherently very low levels of SLC30A10 and can be used to study disorders caused by deactivating mutations in the Mn2+ efflux transporter SLC30A10.20 For comparison, SLC30A10 wild-type (WT) was expressed in HeLa cells, based on a previously reported procedure.20 To apply M4 for spatiotemporal imaging of Mn2+ uptake, we first tested the photostability of the sensor under a fluorescence confocal microscope and we found that the probe was photostable up to the tested time of 1 h (Fig. S30†). For imaging Mn2+ uptake dynamics, cells were first treated with M4 for 15 min in aqueous media. Excess M4 was carefully washed out. The M4 treated cells were placed under a confocal microscope and imaged. Following the acquisition of the first image, Mn2+ ions were added to the media and images were recorded every 3 min for 1 h (Fig. 5c). We could distinctly image the live uptake dynamics of Mn2+ ions in living HeLa cells which mimic pathophysiological conditions in which the SLC30A10 efflux transporter does not function (Fig. 5a and c, and ESI Video S1†). A sequential increase in the fluorescence intensity was observed within these cells along with a significant 1.9-fold increase in emission intensity over 1 h (Fig. 5a and c–e, and ESI Video S1†). Importantly, the basal fluorescence intensity in HeLa cells (Fig. 5a and d) was 1.6 times higher than that of HeLa cells expressing SLC30A10-WT (Fig. 5b and d) indicating that the presence of the transporter was necessary to regulate endogenous Mn2+ ion levels. When the identical Mn2+ uptake experiment was performed on HeLa cells expressing SLC30A10-WT, we observed fluctuating fluorescence intensities, with a significantly slower, gradual increase in emission intensity (Fig. 5b and e, and ESI Video S2†). The final intensity in SLC30A10-WT expressing HeLa cells was significantly lower (2.2 times) than control HeLa cells (Fig. 5d and e). These results proved that M4 could be applied to visualize Mn2+ dynamics in living mammalian cells. Further, since SLC30A10 is a proven Mn2+ efflux transporter, this study validated the exquisite in-cell selectivity of the M4 probe toward sensing, tracking, and imaging Mn2+ ions.We also observed visual differences in the fluorescence intensity distribution under pathophysiologically relevant conditions with low expression of an Mn2+ efflux transporter versus cells expressing the transporter (Fig. S31†). In the case of HeLa cells, the emission intensity of M4 increased in the cytoplasm, peri-nuclear, and nuclear regions revealing regions of Mn2+ ion accumulation (Fig. S31a†). In HeLa cells expressing SLC30A10-WT, a more punctate intensity distribution was observed, with a significantly lower increase in overall emission intensity and barely any Mn2+ ion accumulation in the regions where Mn2+ ions had accumulated in the HeLa cells (Fig. S31b†). These results indicated that M4 was applicable for spatiotemporal imaging of Mn2+ ions in living mammalian cells, in both physiological and pathophysiological contexts. This study places M4 in a unique vantage position as a chemical tool for deciphering Mn2+ ion regulation in living systems, finally filling a critical gap by affording a water-soluble, cell-permeable Mn2+ ion selective probe.
Conclusions
In conclusion, we have successfully devised and employed a general computational workflow to design a novel, highly water-soluble, selective, and sensitive, turn-on fluorescent sensor, M4, for detecting Mn2+ ions in aqueous media. The sensor entered living mammalian cells within 15 min of direct incubation, hence was distinctly cell-permeable, and could image Mn2+ ion localization in living cells. M4 was used to image Mn2+ ions in both intensity and significantly in lifetime fluorescence imaging setups, making it a valuable chemical tool for probing Mn2+ ion regulation in both turn-on intensity-based and ratiometric lifetime-based approaches. Importantly, M4 could not only be applied to image Mn2+ ion localization but also afforded live spatiotemporal visualization of Mn2+ ion uptake dynamics in mammalian cells. The experiments revealed visual differences in Mn2+ ion localization and significant differences in uptake rates in cells with differential expression levels of an Mn2+ transporter implicated in Mn2+-induced early-onset Parkinsonism. Hence, these results open the gateway to tracking and imaging Mn2+ ion dynamics in living systems with our easy to apply, cell-permeable probe. In a key experiment, we generated an in-cell Mn2+ fluorescence response curve for the M4 sensor which afforded imaging of endogenous Mn2+ ions within living mammalian cells thus providing the first estimate of labile Mn2+ ions to be 1.14 ± 0.15 μM in a mammalian cell.Looking into the future, the probe design principle is modular and provides a route for easy modifications to incorporate features of sub-cellular targetability and further tuning of probe sensitivity, based on the biological setting. Any design modification can be wetted through our predictive computational workflow. Therefore, our study starting from the computational design of a sensor, to synthesis, all the way to biological applicability in visualizing live intracellular dynamics forms a general rational sensor design workflow and provides a feedback loop between computational design and experiments. We believe that this workflow can guide the design and development of fluorescent sensors for any metal ion and can be further developed to specifically aid fine-tuning of sensors based on biological contexts and compartments which is the next challenge in deciphering metallobiology.
Experimental section
Computational workflow
The following workflow (Fig. 1d), based on DFT and TD-DFT calculations, for predicting PeT in the designed water-soluble Mn2+ sensors in the metal unbound or apo state, was drafted: (1) the ground state geometries of the molecules were optimized using DFT calculations in a suitable solvent model (acetonitrile or water depending on the predicted solubility of the candidate sensor molecule); (2) the ground state energy levels and orbital maps of the molecules were obtained from DFT calculations using a suitable solvent model; (3) the localization of molecular orbitals in the ground state sensor geometry, on the dye or on the scaffold unit of the molecule, was investigated. Delocalization of orbitals over the dye and scaffold may lead to a decreased possibility of PeT since PeT is a through-space process; (4) the most probable transitions during excitation of the molecule were identified using TD-DFT calculations (transitions corresponding to wavelengths close to the reported absorption of dye were chosen; 400–550 nm for M1, M′, M3 and M4 and 500–650 nm for M′′). Transitions with the maximum oscillator strength were considered as the most probable transition in each case; (5) the orbitals (localized on the dye) involved in the most probable transition were identified, thereby also locating the orbital vacated during excitation, i.e., PeT acceptor orbital; (6) potential PeT donor orbitals, i.e., orbitals localized on either the linker or the scaffold with favorable energies from where electron transfer to the dye-centered orbital vacated during excitation was possible, were identified; (7) ΔG° and ket for the molecules in which potential PeT donor orbitals were available were calculated using the Rehm–Weller equation40 and Marcus theory,41 respectively (see details below).Geometry optimizations and electronic structure calculations
The starting point of the geometry optimizations was a planar geometry of the sensor generated by GaussView 6.0.16 software. DFT optimized ground state geometries and electronic structure of the molecules were generated in a vacuum and other suitable dielectric media modelled via the polarizable continuum model (PCM) using the B3LYP exchange–correlation functional and the 6-311++G** basis set in Gaussian D.01 software. Solvents, water and acetonitrile, were modelled using PCM. Frontier molecular orbitals (MOs) of the molecules were visualized in Avogadro 1.1.1. software for the optimized ground state geometries. TD-DFT calculations were performed on each DFT optimized sensor geometry to extract the most probable (highest oscillator strength) transition and its MO compositions showing the initial and final electronic states following photoexcitation. A total of 150 singlet transitions were considered for each sensor.PeT rate calculations
The electron transfer rates were estimated using the semiclassical Marcus theory in the nonadiabatic limit as41:which depends on the thermodynamic driving force ΔG° of the reaction, the electronic coupling HDA between donor and acceptor states, and the reorganization energy λ. In the above equation, kB, T, and ℏ are Boltzmann's constant, temperature (298 K), and the reduced Planck's constant respectively.
The free energy changes (driving force) associated with PeT were calculated according to the Rehm–Weller equation:40
| ΔG° = ED − EA − ΔE0,0 − wp |
where e is the electronic charge, r is the distance between the centroids of the donor and acceptor orbitals and ε is the permittivity of the suitable solvent (ACN for M1 and water for M4). The value of r was calculated using Multiwfn 3.8.52
H
DA is the electronic coupling matrix element between the donor and the acceptor states that can be expressed as:
| λ = λi + λ0 |
where Δe is the charge of one electron. Here, we have assumed the potential donor and acceptor orbitals as spherical moieties described by radii a1 and a2 respectively. To calculate a1 and a2, we determined the spatial distribution of electron density of each orbital and manually calculated the distance between the atoms at the extremes of this spatial distribution in both horizontal and vertical directions. We then took an average of the horizontal and vertical values to get 2a1 and 2a2, respectively for each orbital (Fig. S5†). r is the distance between the centroids of the donor and acceptor orbitals. r has been obtained from Multiwfn 3.8.52Ds and Dop are the static and optical dielectric constants of the suitable solvent (ACN for M1 and water for M4).
Synthesis and characterization of molecules
Details of synthetic procedures are available in the ESI.† Novel molecules were characterized via liquid chromatography (LC), low resolution mass spectrometry (LRMS), high resolution mass spectrometry (HRMS), 1H NMR and 13C NMR. Data are provided in the ESI.†In vitro fluorescence measurements
All spectroscopic measurements were performed at room temperature. Since M4 was completely water-soluble, all stock solutions were prepared in aqueous media. A solution of 2 μM M4 was prepared from a primary stock (3 mM) in water, by further dissolving in aqueous buffer consisting of HEPES (20 mM) and KNO3 (100 mM); pH was adjusted to 7.1. Fluorescence spectra were recorded on a FluoroLog®-3 (Horiba Jobin Yvon Inc.) spectrofluorometer using quartz cuvettes with 10 mm × 2 mm inner dimensions (Hellma® Analytics). Fluorescence spectra were obtained by excitation at 480 nm with a slit width of 5 nm for both excitation and emission, for M4. Mn2+ was used in the form of MnCl2·4H2O from a stock solution of 1 mM MnCl2·4H2O in water. Other biologically relevant metal ions were used to test the selectivity of M4 towards Mn2+ ions. Mg2+, Ca2+, Fe2+, Co2+, Ni2+, and Cu2+ ions were used in the form of their chlorides as MgCl2·6H2O, CaCl2·2H2O, FeCl2·4H2O, CoCl2, NiCl2, and CuCl2 from 1 mM stock solutions prepared in water. Zn2+ ions were used in the form of nitrate as Zn(NO3)2·6H2O from 1 mM stock solution prepared in water. Cu+ ions were used in the form of tetrakis(acetonitrile)copper(I) hexafluorophosphate from 1 mM stock in degassed acetonitrile. In order to check the effect of anions on the fluorescence response of M4 toward Mn2+ ions, Mn2+ was also used in the form of Mn(ClO4)2·H2O and Mn(OAc)2. All stock solutions were diluted in aqueous buffer consisting of HEPES (20 mM) and KNO3 (100 mM) (pH was adjusted to 7.1) when required, to prepare 0.2 mM secondary stocks. The stock solution of Cu+ was diluted in degassed buffer. The apparent dissociation constants (Kd) corresponding to each metal ion were determined from a plot of observed fluorescence intensity (F) over fluorescence intensity of the sensor in the absence of any metal ion (F0) at 514 nm versus [M2+]. The data were fitted to eqn (1) (ESI†). For fluorescence studies of the probe with varying pH, experiments were carried out with probe M4 (2 μM) prepared from the primary stock (3 mM) in water, further dissolved in aqueous buffer consisting of HEPES (20 mM) and KNO3 (100 mM) (pH 4, 4.5, 5, 6, 7, 8). Buffers of different pH were made by adding concentrated HCl or NaOH solutions.Cell viability experiments
For testing whether M4 affected the viability of cells, HEK293T cells were seeded and grown for 36 h in 96 well plates (cell density 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per well) in Dulbecco's Modified Eagle's Medium (DMEM, Sigma-Aldrich®) with phenol red supplemented with fetal bovine serum (10%, Gibco®), and antibiotic (100×, 10 mL L−1) at 37 °C under humidified air containing 5% CO2. Solutions of different concentrations (2.5 μM, 5 μM, 10 μM, 15 μM) of the sensor were prepared from a stock in water. Cells were incubated with the abovementioned concentrations of M4 in aqueous buffer (pH 7.4) (200 μL per well). The incubation with the sensor was continued for 15 min, 30 min and 1 h at 37 °C in an incubator. As control, cells were incubated with media containing no sensor. After incubation, the medium containing the sensor was removed and the wells were filled with 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) solution (0.5 mg mL−1, 180 μL per well) in DMEM, pH 7.4. MTT treated cells were kept at 37 °C in an incubator for 3–4 h for the formation of formazan crystals. Then the medium from each well was removed and the wells were filled with DMSO (200 μL per well) and the plates were shaken for 5 min to dissolve the crystal. Absorption values at 570 nm of the resultant purple solutions were recorded for each well using Cytation 5 Cell Imaging Multimode Reader (Biotek, Agilent). As background control, absorbance of only DMSO was also recorded.
000 cells per well) in Dulbecco's Modified Eagle's Medium (DMEM, Sigma-Aldrich®) with phenol red supplemented with fetal bovine serum (10%, Gibco®), and antibiotic (100×, 10 mL L−1) at 37 °C under humidified air containing 5% CO2. Solutions of different concentrations (2.5 μM, 5 μM, 10 μM, 15 μM) of the sensor were prepared from a stock in water. Cells were incubated with the abovementioned concentrations of M4 in aqueous buffer (pH 7.4) (200 μL per well). The incubation with the sensor was continued for 15 min, 30 min and 1 h at 37 °C in an incubator. As control, cells were incubated with media containing no sensor. After incubation, the medium containing the sensor was removed and the wells were filled with 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) solution (0.5 mg mL−1, 180 μL per well) in DMEM, pH 7.4. MTT treated cells were kept at 37 °C in an incubator for 3–4 h for the formation of formazan crystals. Then the medium from each well was removed and the wells were filled with DMSO (200 μL per well) and the plates were shaken for 5 min to dissolve the crystal. Absorption values at 570 nm of the resultant purple solutions were recorded for each well using Cytation 5 Cell Imaging Multimode Reader (Biotek, Agilent). As background control, absorbance of only DMSO was also recorded.
Cell viability was calculated as follows:
| % cell viability = [(experimental value − background control)/(control − background control)] × 100 |
Confocal fluorescence imaging
HEK293T and HeLa cells were cultured in DMEM (Sigma-Aldrich®), supplemented with fetal bovine serum (10%, Gibco®), and antibiotic (100×, 10 mL L−1) in T25 culture plates at 37 °C under humidified air containing 5% CO2. For HeLa cells, additional glucose (3.5 mg L−1) was added to the medium. A day before the imaging, the cells were plated on home-made glass coverslip bottomed Petri plates (35 mm diameter, Tarsons) coated with polylysine (200 μg mL−1). Fluorescence images of the cells were recorded on either a confocal microscope (LSM 510, Carl Zeiss, Germany) using 40× oil immersion objectives and a 488 nm laser (argon source) as the excitation source for M4 and emission was collected from 500 to 600 nm or confocal microscope (LEICA TCS SP8 X) using 60× oil immersion objectives and a white light laser set at 514 nm as the excitation source for M4 and emission was collected from 540 to 650 nm. Modified Thomson's buffer (TB) consisting of sodium HEPES (20 mM), NaCl (146 mM), KCl (5.4 mM), MgSO4 (0.8 mM), KH2PO4 (0.4 mM), Na2HPO4 (0.3 mM) and glucose (5.5 mM) (pH adjusted to 7.4) was used during the confocal studies. A stock solution of M4 (5 mM) was prepared in water. The cells were washed with TB and incubated with M4 (5 μM in TB) for 15 min at 37 °C under humidified air containing 5% CO2. After staining, the cells were washed with TB and imaged. For Mn treatment, cells were incubated with MnCl2 (25 μM in TB) for 1 h at 37 °C under humidified air containing 5% CO2. Then the cells were washed with TB to remove excess MnCl2, incubated with M4 as mentioned above and imaged. For concentration dependent experiments, five sets of cells were used. The first set of cells were incubated with M4 (5 μM) in TB for 15 min and washed, and TPEN (50 μM) was added and the cells were imaged 40 min after addition of TPEN. The second set of cells were incubated with M4 (5 μM) in TB for 15 min, washed with TB and imaged. The third, fourth and fifth sets were incubated with Mn2+ (5 μM (third), 10 μM (fourth) and 25 μM (fifth) respectively) for 1 h, washed and then incubated with M4 (5 μM) in TB for 15 min, washed with TB and imaged.Estimation of intracellular labile Mn2+ concentration
For estimation of intracellular labile Mn2+ concentration, analysis was done using ImageJ software. Background-subtracted mean fluorescence intensity was calculated taking individual cells as regions of interest (ROIs) during the resting state, 30 min after TPEN addition, and at the saturation level after Mn2+ addition respectively.Fractional saturation (FS) was calculated according to the following equation:49
From the FS, intracellular labile Mn2+ concentration was estimated according to the following equation:49
FS and [Mn2+] for 12 individual unique cells from three different independent experiments (4 from each experiment) were calculated and the average value has been reported as an estimate of the intracellular labile Mn2+ concentration.
Time resolved fluorescence measurements
For fluorescence lifetime measurements, the probe solution was excited at 480 nm by using a DeltaFlex Time Correlated Single Photon Counting (TCSPC) spectrometer. The vertically polarized second harmonic of the output of a Mai-Tai laser, with a repetition rate of 10 MHz, was used to excite the samples. Emission was collected (at 90° to the excitation beam) at 514 nm using a 32 nm slit width. No filter was used. The instrument response function (IRF) was taken at 480 nm and was calculated to be 68 ps (FWHM). For all the fluorescence lifetime measurements, peak counts of 3000 were collected with the emission polarizer oriented at the magic angle (54.7°) with respect to the excitation polarizer. Fluorescence lifetimes of samples containing only M4 (2 μM) and samples containing both M4 (2 μM) and Mn2+ (20 μM) were recorded. The decay was de-convoluted with respect to the IRF and fitted using a three-exponential model.I(t) = Σαi![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) exp(−t/τi) exp(−t/τi) |
Average lifetime (τm) values were calculated from the equation:
| τm = Σαiτi |
All lifetime data for M4 and M4 with Mn2+ were recorded in triplicate. Error bars represent standard deviations.
Fluorescence lifetime imaging microscopy
Fluorescence lifetime images were recorded in a Leica TCS SP8 X equipped with a FLIM module. Live cells were excited using a white light laser set at 514 nm. The confocal image was captured with a 60× oil objective and then the FLIM images were recorded by switching the excitation laser to a pulsed laser. For FLIM imaging, a 514 nm picosecond pulse laser at 80 MHz pulse frequency was used for excitation. Emission was collected from 540 to 650 nm until the photon count reached 500 per pixel. The instrument response function (IRF) was taken at 514 nm and was calculated to be 224 ps (FWHM). The cells were washed with TB and incubated with M4 (5 μM in TB) for 15 min at 37 °C. After incubation, the cells were washed with TB and imaged. For Mn treatment, cells were incubated with MnCl2 (25 μM in TB) for 1 h at 37 °C. Then the cells were washed with TB to remove excess MnCl2, stained with M4 as mentioned above and imaged. Fluorescence lifetime distributions in cells were analyzed on Leica Lax X software. Data from each image pixel was fitted to a three-component exponential decay function to extract the average fluorescence lifetime at each pixel. Color-coded representation of the average lifetime distribution within living cells under both Mn2+ untreated and exogenously treated conditions is presented in Fig. 4b.Imaging Mn2+ dynamics in living cells
HeLa cells were used in a pathophysiological context mimicking conditions of hypermanganesemia and Mn induced Parkinsonism caused by deactivating mutations in SLC30A10. To generate cells expressing the transporter, SLC30A10 plasmid with an ampicillin resistant insert was amplified in DH5α cells in agar medium containing ampicillin. The extracted, amplified SLC30A10 plasmid was then transiently transfected into a suspension of HeLa cells, using Lipofectamine 3000 in OptiMEM. The suspension was then plated on home-made glass coverslip bottomed Petri plates (35 mm diameter, Tarsons) coated with polylysine (200 μg mL−1). To check whether M4 had the requisite photo-stability for prolonged time-dependent dynamic uptake studies, cells were incubated with M4 (5 μM in TB) for 15 min. After 15 min, cells were washed with TB and imaged for 1 h at intervals of 3 min. For time-dependent Mn2+ uptake imaging studies, cells were incubated with M4 (5 μM in TB) for 15 min. After 15 min, cells were washed with TB three times and imaged. MnCl2 (25 μM in TB) was then added and time-dependent imaging was started immediately after addition. Images were recorded at an interval of 3 min for 1 h. The image recorded before addition of MnCl2 was taken as the image at 0 min.Data availability
All datasets supporting this article have either been uploaded as part of the ESI† or are available upon request to the corresponding author.Author contributions
A. D., S.·K., and S. K. D. designed and conceptualized the project, performed the experiments, analyzed the data, and wrote the paper. K. G. participated in optimizations of control experiments. Raj. K. participated in cell experiments. R. V., S. K., S. K. D., Rav. K., and K. G. participated in computational studies. A. N. and B.·B. participated in FLIM experiments. All authors checked the results and the manuscript and approved the final version of the manuscript.Conflicts of interest
The authors declare no competing interest.Acknowledgements
A. D. acknowledges the support of the Department of Atomic Energy, Government of India, under Project Identification No. RTI4003 and Indo-French Centre for the Promotion of Advanced Research – CEFIPRA under Project No. 6505-1. The authors acknowledge Prof. Anindya Datta, Arkaprava Chowdhury and the TCSPC facility, S.A.I.F, I.I.T. Bombay for help with TCSPC experiments; Prof. Deepak Saini, Suchetha K. S. and the Bio-Imaging facility, Department of Biological Sciences, IISc, Bangalore for help with FLIM experiments; Prof. Somshuvra Mukhopadhyay for sharing SLC30A10 WT plasmids and Ms Deepika Furtado for help with plasmid amplification and isolation; Ms Mamata Joshi and Mr Devidas Jadhav, National NMR facility, TIFR, and NMR facility, I.I.T. Bombay; DCS Cell Culture facility; and Mass Laboratory, Chemistry Dept., I.I.T. Bombay and I.I.S.E.R., Pune.References
- Y. Umena, K. Kawakami, J.-R. Shen and N. Kamiya, Crystal structure of oxygen-evolving photosystem II at a resolution of 1.9 Å, Nature, 2011, 473(7345), 55–60, DOI:10.1038/nature09913.
- K. Barnese, E. B. Gralla, J. S. Valentine and D. E. Cabelli, Biologically relevant mechanism for catalytic superoxide removal by simple manganese compounds, Proc. Natl. Acad. Sci. U.S.A., 2012, 109(18), 6892–6897, DOI:10.1073/pnas.1203051109.
- S. M. Damo, T. E. Kehl-Fie, N. Sugitani, M. E. Holt, S. Rathi, W. J. Murphy, Y. Zhang, C. Betz, L. Hench and G. Fritz, et al., Molecular basis for manganese sequestration by calprotectin and roles in the innate immune response to invading bacterial pathogens, Proc. Natl. Acad. Sci. U.S.A., 2013, 110(10), 3841–3846, DOI:10.1073/pnas.1220341110.
- M. B. Brophy and E. M. Nolan, Manganese and Microbial Pathogenesis: Sequestration by the Mammalian Immune System and Utilization by Microorganisms, ACS Chem. Biol., 2015, 10(3), 641–651, DOI:10.1021/cb500792b.
- S. Das, K. Khatua, A. Rakshit, A. Carmona, A. Sarkar, S. Bakthavatsalam, R. Ortega and A. Datta, Emerging chemical tools and techniques for tracking biological manganese, Dalton Trans., 2019, 48(21), 7047–7061, 10.1039/C9DT00508K.
- Y. Chiba, T. Miyakawa, Y. Shimane, K. Takai, M. Tanokura and T. Nozaki, Structural comparisons of phosphoenolpyruvate carboxykinases reveal the evolutionary trajectories of these phosphodiester energy conversion enzymes, J. Biol. Chem., 2019, 294(50), 19269–19278, DOI:10.1074/jbc.RA119.010920.
- V. C. Culotta, M. Yang and T. V. O'Halloran, Activation of superoxide dismutases: putting the metal to the pedal, Biochim. Biophys. Acta Mol. Cell Res., 2006, 1763(7), 747–758, DOI:10.1016/j.bbamcr.2006.05.003.
- M. J. Daly, E. K. Gaidamakova, V. Y. Matrosova, A. Vasilenko, M. Zhai, A. Venkateswaran, M. Hess, M. V. Omelchenko, H. M. Kostandarithes and K. S. Makarova, et al., Accumulation of Mn(II) in Deinococcus radiodurans Facilitates Gamma-Radiation Resistance, Science, 2004, 306(5698), 1025–1028, DOI:10.1126/science.1103185.
- L. Di Costanzo, M. E. Pique and D. W. Christianson, Crystal Structure of Human Arginase I Complexed with Thiosemicarbazide Reveals an Unusual Thiocarbonyl μ-Sulfide Ligand in the Binuclear Manganese Cluster, J. Am. Chem. Soc., 2007, 129(20), 6388–6389, DOI:10.1021/ja071567j.
- A. R. Reddi, L. T. Jensen and V. C. Culotta, Manganese Homeostasis in Saccharomyces cerevisiae, Chem. Rev., 2009, 109(10), 4722–4732, DOI:10.1021/cr900031u.
- A. Sharma, E. K. Gaidamakova, V. Y. Matrosova, B. Bennett, M. J. Daly and B. M. Hoffman, Responses of Mn2+ speciation in Deinococcus radiodurans and Escherichia coli to γ-radiation by advanced paramagnetic resonance methods, Proc. Natl. Acad. Sci. U.S.A., 2013, 110(15), 5945–5950, DOI:10.1073/pnas.1303376110.
- Y. Sheng, I. A. Abreu, D. E. Cabelli, M. J. Maroney, A.-F. Miller, M. Teixeira and J. S. Valentine, Superoxide Dismutases and Superoxide Reductases, Chem. Rev., 2014, 114(7), 3854–3918, DOI:10.1021/cr4005296.
- L. C. Tabares and S. Un, In situ Determination of Manganese(II) Speciation in Deinococcus radiodurans by High Magnetic Field EPR: Detection of High Levels of Mn(II) Bound to Proteins, J. Biol. Chem., 2013, 288(7), 5050–5055, DOI:10.1074/jbc.C112.444992.
- A. B. Bowman and M. Aschner, Considerations on manganese (Mn) treatments for in vitro studies, Neurotoxicology, 2014, 41, 141–142, DOI:10.1016/j.neuro.2014.01.010.
- R. L. McNaughton, A. R. Reddi, M. H. S. Clement, A. Sharma, K. Barnese, L. Rosenfeld, E. B. Gralla, J. S. Valentine, V. C. Culotta and B. M. Hoffman, Probing in vivo Mn2+ speciation and oxidative stress resistance in yeast cells with electron-nuclear double resonance spectroscopy, Proc. Natl. Acad. Sci. U.S.A., 2010, 107(35), 15335–15339, DOI:10.1073/pnas.1009648107.
- P. A. Doble and G. L. G. Miklos, Distributions of manganese in diverse human cancers provide insights into tumour radioresistance, Metallomics, 2018, 10(9), 1191–1210, 10.1039/c8mt00110c.
- M. Lv, M. Chen, R. Zhang, W. Zhang, C. Wang, Y. Zhang, X. Wei, Y. Guan, J. Liu and K. Feng, et al., Manganese is critical for antitumor immune responses via cGAS-STING and improves the efficacy of clinical immunotherapy, Cell Res., 2020, 30(11), 966–979, DOI:10.1038/s41422-020-00395-4.
- J. M. Rozenberg, M. Kamynina, M. Sorokin, M. Zolotovskaia, E. Koroleva, K. Kremenchutckaya, A. Gudkov, A. Buzdin and N. Borisov, The Role of the Metabolism of Zinc and Manganese Ions in Human Cancerogenesis, Biomedicines, 2022, 10(5), 1072, DOI:10.3390/biomedicines10051072.
- K. Tuschl, P. T. Clayton, S. M. Gospe, S. Gulab, S. Ibrahim, P. Singhi, R. Aulakh, R. T. Ribeiro, O. G. Barsottini and M. S. Zaki, et al., Syndrome of Hepatic Cirrhosis, Dystonia, Polycythemia, and Hypermanganesemia Caused by Mutations in SLC30A10, a Manganese Transporter in Man, Am. J. Hum. Genet., 2012, 90(3), 457–466, DOI:10.1016/j.ajhg.2012.01.018.
- L.-I. Dinorah, C. Pan, E. Z. Charles, H. Steven, M. M. Jonathan, D. S. Caleb, A. M. Richard, B. B. Aaron, A. Michael and M. Somshuvra, SLC30A10 Is a Cell Surface-Localized Manganese Efflux Transporter, and Parkinsonism-Causing Mutations Block Its Intracellular Trafficking and Efflux Activity, J. Neurosci., 2014, 34(42), 14079, DOI:10.1523/JNEUROSCI.2329-14.2014.
- K. Tuschl, E. Meyer, L. E. Valdivia, N. Zhao, C. Dadswell, A. Abdul-Sada, C. Y. Hung, M. A. Simpson, W. K. Chong and T. S. Jacques, et al., Mutations in SLC39A14 disrupt manganese homeostasis and cause childhood-onset parkinsonism–dystonia, Nat. Commun., 2016, 7(1), 11601, DOI:10.1038/ncomms11601.
- C. E. Zogzas, M. Aschner and S. Mukhopadhyay, Structural Elements in the Transmembrane and Cytoplasmic Domains of the Metal Transporter SLC30A10 Are Required for Its Manganese Efflux Activity, J. Biol. Chem., 2016, 291(31), 15940–15957, DOI:10.1074/jbc.M116.726935.
- A. Carmona, C. E. Zogzas, S. Roudeau, F. Porcaro, J. Garrevoet, K. M. Spiers, M. Salomé, P. Cloetens, S. Mukhopadhyay and R. Ortega, SLC30A10 Mutation Involved in Parkinsonism Results in Manganese Accumulation within Nanovesicles of the Golgi Apparatus, ACS Chem. Neurosci., 2019, 10(1), 599–609, DOI:10.1021/acschemneuro.8b00451.
- C. J. da Silva, A. José da Rocha, M. F. Mendes, A. P. S. d. M. Braga and S. Jeronymo, Brain Manganese Deposition Depicted by Magnetic Resonance Imaging in a Welder, Arch. Neurol., 2008, 65(7), 983, DOI:10.1001/archneur.65.7.983.
- S. Bakthavatsalam, S. Das Sharma, M. Sonawane, V. Thirumalai and A. Datta, A zebrafish model of manganism reveals reversible and treatable symptoms that are independent of neurotoxicity, Dis. Models Mech., 2014, 7(11), 1239–1251, DOI:10.1242/dmm.016683.
- S. L. O'Neal and W. Zheng, Manganese Toxicity Upon Overexposure: a Decade in Review, Curr. Environ. Health Rep., 2015, 2(3), 315–328, DOI:10.1007/s40572-015-0056-x.
- R. Bagur and G. Hajnóczky, Intracellular Ca2+ Sensing: Its Role in Calcium Homeostasis and Signaling, Mol. Cell, 2017, 66(6), 780–788, DOI:10.1016/j.molcel.2017.05.028.
- J. Liang and J. W. Canary, Discrimination between Hard Metals with Soft Ligand Donor Atoms: An On-Fluorescence Probe for Manganese(II), Angew. Chem., Int. Ed., 2010, 49(42), 7710–7713, DOI:10.1002/anie.201002853.
- F. Gruppi, J. Liang, B. B. Bartelle, M. Royzen, D. H. Turnbull and J. W. Canary, Supramolecular metal displacement allows on-fluorescence analysis of manganese(II) in living cells, Chem. Commun., 2012, 48(87), 10778–10780, 10.1039/C2CC34742C.
- S. Bakthavatsalam, A. Sarkar, A. Rakshit, S. Jain, A. Kumar and A. Datta, Tuning macrocycles to design ‘turn-on’ fluorescence probes for manganese(II) sensing in live cells, Chem. Commun., 2015, 51(13), 2605–2608, 10.1039/C4CC09542A.
- S. Adhikari, A. Ghosh, A. Sahana, S. Guria and D. Das, Tailoring Ligand Environment toward Development of Colorimetric and Fluorescence Indicator for Biological Mn(II) Imaging, Anal. Chem., 2016, 88(2), 1106–1110, DOI:10.1021/acs.analchem.5b03551.
- H. Fan, C. E. McGhee, R. J. Lake, Z. Yang, Z. Guo, X.-B. Zhang and Y. Lu, A Highly Selective Mn(II)-Specific DNAzyme and Its Application in Intracellular Sensing, JACS Au, 2023, 3(6), 1615–1622, DOI:10.1021/jacsau.3c00062.
- J. Park, M. B. Cleary, D. Li, J. A. Mattocks, J. Xu, H. Wang, S. Mukhopadhyay, E. M. Gale and J. A. Cotruvo, A genetically encoded fluorescent sensor for manganese(II), engineered from lanmodulin, Proc. Natl. Acad. Sci. U.S.A., 2022, 119(51), e2212723119, DOI:10.1073/pnas.2212723119.
- H. Irving and R. J. P. Williams, 637. The stability of transition-metal complexes, J. Chem. Soc., 1953, 3192–3210, 10.1039/JR9530003192.
- K. P. Carter, A. M. Young and A. E. Palmer, Fluorescent sensors for measuring metal ions in living systems, Chem. Rev., 2014, 114(8), 4564–4601, DOI:10.1021/cr400546e.
- J. Cody, S. Mandal, L. Yang and C. J. Fahrni, Differential Tuning of the Electron Transfer Parameters in 1,3,5-Triarylpyrazolines: A Rational Design Approach for Optimizing the Contrast Ratio of Fluorescent Probes, J. Am. Chem. Soc., 2008, 130(39), 13023–13032, DOI:10.1021/ja803074y.
- S. Das, A. Sarkar, A. Rakshit and A. Datta, A Sensitive Water-Soluble Reversible Optical Probe for Hg2+ Detection, Inorg. Chem., 2018, 57(9), 5273–5281, DOI:10.1021/acs.inorgchem.8b00310.
- A. M. McGuire, B. J. Cuthbert, Z. Ma, K. D. Grauer-Gray, M. Brunjes Brophy, K. A. Spear, S. Soonsanga, J. I. Kliegman, S. L. Griner and J. D. Helmann, et al., Roles of the A and C Sites in the Manganese-Specific Activation of MntR, Biochemistry, 2013, 52(4), 701–713, DOI:10.1021/bi301550t.
- D. Salvemini, D. P. Riley and S. Cuzzocrea, Sod mimetics are coming of age, Nat. Rev. Drug Discovery, 2002, 1(5), 367–374, DOI:10.1038/nrd796.
- D. Rehm and A. Weller, Kinetics of Fluorescence Quenching by Electron and H-Atom Transfer, Isr. J. Chem., 1970, 8, 259–271, DOI:10.1002/ijch.197000029.
- R. A. Marcus and N. Sutin, Electron transfers in chemistry and biology, Biochim. Biophys. Acta Bioenerg., 1985, 811(3), 265–322, DOI:10.1016/0304-4173(85)90014-X.
- P. Batat, G. Vives, R. Bofinger, R.-W. Chang, B. Kauffmann, R. Oda, G. Jonusauskas and N. D. McClenaghan, Dynamics of ion-regulated photoinduced electron transfer in BODIPY-BAPTA conjugates, Photochem. Photobiol. Sci., 2012, 11(11), 1666–1674, 10.1039/c2pp25130b.
- C. E. Outten and T. V. O’Halloran, Femtomolar Sensitivity of Metalloregulatory Proteins Controlling Zinc Homeostasis, Science, 2001, 292(5526), 2488–2492, DOI:10.1126/science.1060331.
- A. W. Foster, R. Pernil, C. J. Patterson, A. J. P. Scott, L.-O. Pålsson, R. Pal, I. Cummins, P. T. Chivers, E. Pohl and N. J. Robinson, A tight tunable range for Ni(II) sensing and buffering in cells, Nat. Chem. Biol., 2017, 13(4), 409–414, DOI:10.1038/nchembio.2310.
- E. L. Que, D. W. Domaille and C. J. Chang, Metals in Neurobiology: Probing Their Chemistry and Biology with Molecular Imaging, Chem. Rev., 2008, 108(5), 1517–1549, DOI:10.1021/cr078203u.
- A. Carmona, S. Roudeau, L. Perrin, G. Veronesi and R. Ortega, Environmental manganese compounds accumulate as Mn(II) within the Golgi apparatus of dopamine cells: relationship between speciation, subcellular distribution, and cytotoxicity, Metallomics, 2014, 6(4), 822–832, 10.1039/c4mt00012a.
- S. Das, A. Carmona, K. Khatua, F. Porcaro, A. Somogyi, R. Ortega and A. Datta, Manganese Mapping Using a Fluorescent Mn2+ Sensor and Nanosynchrotron X-ray Fluorescence Reveals the Role of the Golgi Apparatus as a Manganese Storage Site, Inorg. Chem., 2019, 58(20), 13724–13732, DOI:10.1021/acs.inorgchem.9b01389.
- R. Tsien and T. Pozzan, Measurement of cytosolic free Ca2+ with quin2, Methods Enzymol., 1989, 172, 230–262, DOI:10.1016/s0076-6879(89)72017-6.
- L. Sanford and A. E. Palmer, Dissociated Hippocampal Neurons Exhibit Distinct Zn2+ Dynamics in a Stimulation-Method-Dependent Manner, ACS Chem. Neurosci., 2020, 11(4), 508–514, DOI:10.1021/acschemneuro.0c00006.
- A. Kaur, M. A. Haghighatbin, C. F. Hogan and E. J. New, A FRET-based ratiometric redox probe for detecting oxidative stress by confocal microscopy, FLIM and flow cytometry, Chem. Commun., 2015, 51(52), 10510–10513, 10.1039/C5CC03394B.
- D. Rupsa, G. Amani, S. Matthew and C. S. Melissa, Recent innovations in fluorescence lifetime imaging microscopy for biology and medicine, J. Biomed. Opt., 2021, 26(7), 070603, DOI:10.1117/1.JBO.26.7.070603.
- T. Lu and F. Chen, Multiwfn: A multifunctional wavefunction analyzer, J. Comput. Chem., 2012, 33(5), 580–592, DOI:10.1002/jcc.22885.
- R. J. Cave and M. D. Newton, Generalization of the Mulliken-Hush treatment for the calculation of electron transfer matrix elements, Chem. Phys. Lett., 1996, 249(1), 15–19, DOI:10.1016/0009-2614(95)01310-5.
Footnotes |
| † Electronic supplementary information (ESI) available: Details of DFT and TDDFT calculations, synthesis, and characterization of novel molecules via LC-ESI-MS, 1H-NMR and 13C-NMR, fluorescence experiments, cytotoxicity assay, cell studies, photostability assay, Mn2+ uptake studies, TCSPC and FLIM experiments (PDF). Supporting videos of live imaging of Mn2+ dynamics: Videos S1 and S2 (MP4). See DOI: https://doi.org/10.1039/d4sc00907j |
| ‡ Equal contribution. |
| This journal is © The Royal Society of Chemistry 2024 |