Open Access Article
Open Access Articlenach0: multimodal natural and chemical languages foundation model†
Micha
Livne‡
a,
Zulfat
Miftahutdinov‡
b,
Elena
Tutubalina‡
 c,
Maksim
Kuznetsov‡
c,
Maksim
Kuznetsov‡
 b,
Daniil
Polykovskiy
b,
Daniil
Polykovskiy
 b,
Annika
Brundyn
a,
Aastha
Jhunjhunwala
a,
Anthony
Costa
a,
Alex
Aliper
d,
Alán
Aspuru-Guzik
*e and
Alex
Zhavoronkov
b,
Annika
Brundyn
a,
Aastha
Jhunjhunwala
a,
Anthony
Costa
a,
Alex
Aliper
d,
Alán
Aspuru-Guzik
*e and
Alex
Zhavoronkov
 *c
*c
aNVIDIA, 2788 San Tomas Expressway, Santa Clara, 95051, CA, USA
bInsilico Medicine Canada Inc., 3710-1250 René-Lévesque West, Montreal, Quebec, Canada
cInsilico Medicine Hong Kong Ltd., Unit 310, 3/F, Building 8W, Phase 2, Hong Kong Science Park, Pak Shek Kok, New Territories, Hong Kong. E-mail: alex@insilicomedicine.com
dInsilico Medicine AI Ltd., Level 6, Unit 08, Block A, IRENA HQ Building, Masdar City, Abu Dhabi, United Arab Emirates
eUniversity of Toronto, Lash Miller Building 80 St. George Street, Toronto, Ontario, Canada. E-mail: alan@aspuru.com
First published on 8th May 2024
Abstract
Large Language Models (LLMs) have substantially driven scientific progress in various domains, and many papers have demonstrated their ability to tackle complex problems with creative solutions. Our paper introduces a new foundation model, nach0, capable of solving various chemical and biological tasks: biomedical question answering, named entity recognition, molecular generation, molecular synthesis, attributes prediction, and others. nach0 is a multi-domain and multi-task encoder–decoder LLM pre-trained on unlabeled text from scientific literature, patents, and molecule strings to incorporate a range of chemical and linguistic knowledge. We employed instruction tuning, where specific task-related instructions are utilized to fine-tune nach0 for the final set of tasks. To train nach0 effectively, we leverage the NeMo framework, enabling efficient parallel optimization of both base and large model versions. Extensive experiments demonstrate that our model outperforms state-of-the-art baselines on single-domain and cross-domain tasks. Furthermore, it can generate high-quality outputs in molecular and textual formats, showcasing its effectiveness in multi-domain setups.
1 Introduction
Large-scale pre-training of language models (LMs), such as BERT,1 T5,2 BART3 and GPT,4 on vast amounts of text data has yielded impressive results on a variety of natural language processing (NLP) tasks. These models' success can be attributed to their ability to learn deeply contextualized representations of input tokens through self-supervision at scale.1 Recently, foundation models have built upon the concept of self-supervised learning by pre-training a single model over unlabeled data that can be easily adapted to any task.5The application of neural network architectures and LMs has significantly advanced the field of chemistry, particularly in domain-specific information retrieval, drug development, and clinical trial design.6–15 These developments include neural molecular fingerprinting, generative approaches to small molecule design,11–13 prediction of pharmacological properties, and drug repurposing.13,14 The clinical development of a drug is a time and money consuming process that typically requires several years and a billion-dollar budget to progress from phase 1 clinical trials to the patients.16 The use of state-of-the-art neural network approaches and language models has the potential to facilitate the drug development process considerably.
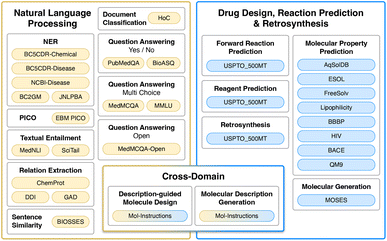
A number of LMs have been proposed for the biomedical domain, utilizing a variety of model families: for instance, researchers have developed BioBERT,17 based on BERT with 110 million parameters, and SciFive, based on T5-base and T5-large with 220 and 770 million parameters respectively, using biomedical literature from PubMed. NVIDIA has also developed BioMegatron models in the biomedical domain using a more extensive set of PubMed-derived free text, ranging from 345 million to 1.2 billion parameters. However, the datasets used in these models cover mainly biomedical natural language texts and contain biomedical named entities like drugs, genes, and cell lines names but omit important chemical structure descriptions in SMILES format. Enriching biomedical datasets with chemical structures is an important and challenging task. Recently, LMs such as Galactica,18 based on transformer architecture in a decoder-only setup19 with 120 billion parameters in its largest setup, and MolT5,20 based on T5-base and T5-large, were proposed to address this limitation. Both modes were pre-trained with natural language and chemical data, creating a shared representation space, yet were not fine-tuned on a diverse set of chemical tasks with instruction tuning in a multi-task fashion. The Venn diagram in Fig. 1 provides a summary of the existing LMs. Furthermore, simple language models trained with molecular structures can reproduce complex molecular distributions,21 and even their 3D structure of molecules, materials and proteins using a GPT framework.22
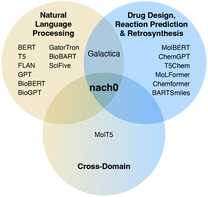 | ||
| Fig. 1 A Venn diagram that shows the relationships between fine-tuning data used in our study and related work. It is important to highlight that the majority of models typically treat the chemical space and the semantic space in the natural language domain independently. Novel cross-domain datasets such as Mol-Instructions25 and MolT5 data20 have asked whether it is possible to unify representations of natural language and molecules for NLP and molecule generation tasks within a single model. In this work, we seek to answer this question. | ||
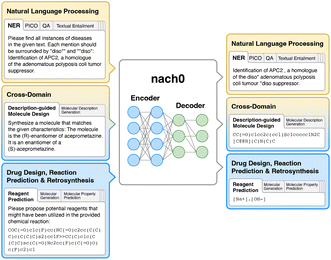
In this paper, we propose a unified encoder–decoder transformer named nach0 for natural language, chemical generalization and cross-domain tasks. We pre-train on both natural language and chemical data using self supervised learning and employ nach0 as the foundation model for a wide range of downstream tasks (Fig. 2). The tasks include well-known NLP problems such as information extraction, question answering, textual entailment, molecular structures and description generation, chemical property prediction, and reaction predictions. Inspired by Raffel et al.,2 Chung et al.,23 we follow the intuition that tasks can be described via natural language instructions, such as “What reactants could be used to synthesize O![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C(NC1CCN(Cc2ccccc2)CC1)c1c(Cl)cccc1[N+](
C(NC1CCN(Cc2ccccc2)CC1)c1c(Cl)cccc1[N+](![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)[O–]” or “describe a molecule C1
O)[O–]” or “describe a molecule C1![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CC(=CC
CC(=CC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C1C[C@H](C(
C1C[C@H](C(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)[O–])N)O”. Prompt design and instruction tuning are employed for model training using NVIDIA's Neural Modules (NeMo) framework,24 which provides scientists with a way to train and deploy LLMs using NVIDIA GPUs. Extensive evaluation in both in-domain and cross-domain setup demonstrates that nach0 is a powerful tool for the chemistry domain.
O)[O–])N)O”. Prompt design and instruction tuning are employed for model training using NVIDIA's Neural Modules (NeMo) framework,24 which provides scientists with a way to train and deploy LLMs using NVIDIA GPUs. Extensive evaluation in both in-domain and cross-domain setup demonstrates that nach0 is a powerful tool for the chemistry domain.
Contribution – our contributions are three-fold:
(1) We introduce a biochemical foundation model nach0 and pre-train base and large versions of nach0 on molecular structures and textual data from scientific articles and patents.(2) We fine-tune nach0 in a supervised and multi-task manner, using a combination of diverse tasks specified through natural language prompts.
(3) Through the experimental validation on benchmark datasets, focusing on both single-domain and cross-domain tasks, we show that our model achieves competitive results with state-of-the-art encoder–decoder models specialized for single domain.
2 Methods
2.1 Framework nach0
The aim of nach0 is to create a unified transformer capable of performing natural language, chemical generalization, and translation tasks simultaneously. Fig. 3 shows a diagram of our framework with several input/output examples. The model's representations are learned from extensive and diverse chemical SMILES data and related textual data from scientific articles and patents. Similar to Raffel et al.,2 Chung et al.,23 nach0 follows an encoder–decoder architecture that takes textual input and generates target responses. To train the model on a mixture of datasets partitioned into different tasks, we formulate all the tasks in a “text-to-text” format, where the model is given some text as a context or condition and produces the output in a text format. Each dataset is associated with multiple prompt templates used to format datasets' instances into input and target pairs. In particular, we train nach0 on three types of tasks (Fig. 2):• NLP tasks: named entity recognition (NER), PICO extraction, textual entailment, relation extraction, sentence similarity, document classification, question answering (yes/no, multi-choice, open).
• Chemistry-related (CHEM) tasks: molecular property prediction, molecular generation, forward reaction prediction, reagent prediction, retrosynthesis.
• Cross-domain (NLP ↔ CHEM) tasks: description-guided molecule design, molecular description generation.
Fig. 3 shows our model and prompt format. Details on train/test splits are presented in Table 1. Datasets' descriptions with example instances are reported in ESI, Section 2.†
| Task | Dataset | Link | Train/test split |
|---|---|---|---|
| NER | BC5CDR-chemical27 | https://huggingface.co/datasets/bigbio/blurb/viewer/bc5chem | Predefined |
| BC5CDR-disease27 | https://huggingface.co/datasets/bigbio/blurb/viewer/bc5disease | Predefined | |
| NCBI-disease28 | https://huggingface.co/datasets/bigbio/blurb/viewer/ncbi_disease/ | Predefined | |
| BC2GM29 | https://huggingface.co/datasets/bigbio/blurb/viewer/bc2gm | Predefined | |
| JNLPBA30 | https://huggingface.co/datasets/bigbio/blurb/viewer/jnlpba | Predefined | |
| PICO | EBM PICO31 | https://github.com/bigscience-workshop/biomedical | Predefined |
| Textual entailment | MedNLI32 | https://github.com/bigscience-workshop/biomedical | Predefined |
| SciTail33 | https://github.com/bigscience-workshop/biomedical | Predefined | |
| Relation extraction | ChemProt34 | https://github.com/bigscience-workshop/biomedical | Predefined |
| DDI35 | https://github.com/bigscience-workshop/biomedical | Predefined | |
| GAD36 | https://github.com/bigscience-workshop/biomedical | Predefined | |
| Sentence similarity | BIOSSES37 | https://github.com/bigscience-workshop/biomedical | Predefined |
| Document classification | HoC38 | https://github.com/bigscience-workshop/biomedical | Predefined |
| Question answering (yes/no) | PubMedQA39 | https://github.com/bigscience-workshop/biomedical | Predefined |
| BioASQ40 | https://github.com/bigscience-workshop/biomedical | Predefined | |
| Molecular property prediction | ESOL26 | https://moleculenet.org | Predefined |
| FreeSolv26 | |||
| Lipophilicity26 | |||
| BBBP26 | |||
| HIV26 | |||
| BACE26 | |||
| QM9 (ref. 25) | https://github.com/zjunlp/Mol-Instructions | Random | |
| Molecular generation | MOSES12 | https://github.com/molecularsets/moses | Predefined |
| Forward reaction prediction | Mol-Instructions25 | https://github.com/zjunlp/Mol-Instructions | Random |
| Reagent prediction | |||
| Retrosynthesis | |||
| Description-guided molecule design | Mol-Instructions25 | https://github.com/zjunlp/Mol-Instructions | Random |
| Molecular description generation |
Given the presence of textual and molecular modalities, different tokenization technique is a crucial aspect of dataset design. One way to represent molecular structures is a simplified molecular-input line-entry system (SMILES) string.41 SMILES describe a molecule as a sequence of atoms in a depth-first traversal order and uses special symbols to depict branching, cycle opening/closing, bond types, and stereochemistry. We use the following tokenization:
• Textual domain sub-word tokens adopted from FLAN-T5 (ref. 23) for natural language sequences.
• Tokenization for SMILES: we annotate each SMILES token with special symbols: <sm_{token}> and extend the vocabulary with such tokens.
2.2 Model and training configuration
In our study, we predominantly employ a model featuring the default T5 architecture, which is derived from Raffel et al.2. Our experimentation involves two model sizes: a base model consisting of 250 million parameters, characterized by 12 layers, a hidden state of 768 dimensions, a feed-forward hidden state of 3072 dimensions, and 12 attention heads; and a larger model with 780 million parameters, consisting of 24 layers, a hidden state of 1024 dimensions, a feed-forward hidden state of 4096 dimensions, and 16 attention heads.For both models, we conduct pre-training with a language modeling (LM) objective and subsequent fine-tuning. The base models were trained using NVIDIA A4000 and A5000 GPUs, while the larger models were trained on NVIDIA's DGX cloud platform. Both the pre-training and fine-tuning stages were executed using the subsequent hyperparameters: a batch size of 1024, a learning rate set to 1 × 10−4, and a weight decay of 0.01. The pre-training stage lasted for a single epoch, whereas the fine-tuning stage for 10 epochs.
To execute the pre-training phase of our model with the LM objective, we leveraged two textual data sources in addition to one chemical data source. These textual data sources encompassed abstract texts extracted from PubMed and patent descriptions derived from USPTO. All the textual data underwent a filtering process, eliminating documents that were not related to the chemistry domain. Consequently, the number of documents was curtailed to 13m for abstracts and 119k for patents. The chemical data component was sourced from the ZINC dataset, encompassing approximately 100 million documents. In aggregate, the textual data set contained 355m tokens for abstracts and 2.9b tokens for patents, whereas the chemical data encompassed 4.7b tokens.
The entirety of the investigations in this paper was conducted using the multi-task model, with the exception of the ablation part. Each multi-task model underwent fine-tuning by leveraging the entire spectrum of available datasets, encompassing all domains, as elucidated in Section 1. For data mixing and balancing we followed the “Examples-proportional mixing strategy” from Raffel et al.2. The outcomes of these models are explicitly detailed in Section 3. Conversely, in the context of ablation studies, fine-tuning was specifically performed utilizing only those datasets relevant to the corresponding domain, as detailed in the discussion.
2.3 NeMo, parallel training, NVIDIA cluster
The training was performed using NVIDIA NeMo toolkit,42 which consists of pre-built modules for end-to-end workflows in Automatic Speech Recognition (ASR), NLP, and Text-to-Speech (TTS) synthesis. NeMo uses PyTorch Lightning for optimized multi-node/multi-GPU (MNMG) mixed-precision training. In this work, we leveraged the NeMo NLP collection to train and evaluate our LMs. We trained our model on a variety of tasks such as information extraction, question answering, molecular property prediction, and description-guided molecule design using the NeMo toolkit. A custom connector was added to extend the vocabulary size of the pre-trained model when continuing the training of the model with chemistry and biomedical datasets. The original vocabulary was extended to match the target vocabulary which was larger. The corresponding embedding matrix was initialized with learned embeddings of the original model. The extra tokens were initialized by re-using the first embeddings.Data was parsed using mem-map datasets from the NeMo toolkit to allow efficient data handling. The mem-map dataset relies on memory mapping directly to files, allowing the handling of very large datasets with small memory footprints and optimal reading speed. The data was loaded as raw text files and the tokenization occurred on-the-fly. Pre-fetching of the data mitigated the effects of online tokenization when compared to pre-tokenized data. The model was trained using tensor and pipeline parallelism,43 both of which are model parallel methods for distributed training and are implemented in the NeMo toolkit for efficient scaling of large language model training.
3 Results and discussion
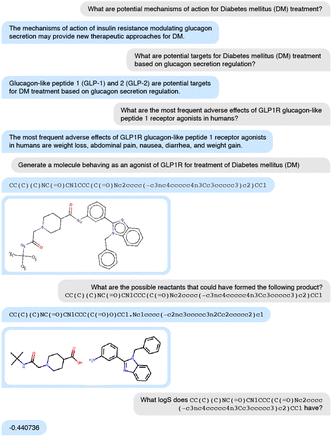
3.1 Use case: end-to-end drug discovery
In the first case study, we generate molecular structures against diabetes mellitus (DM) using just one model, nach0: discover biological targets with potential therapeutic activity, analyze the mechanism of action, generate molecular structure, propose one-step synthesis, and predict molecular properties. In a series of questions, we generate the model's responses using top-p sampling with values from 0.3 to 0.7 and step equals 0.05 and ask an expert chemist to pick the best response (Fig. 4). In total, we generate 200 SMILES on the molecule generation prompt and select one structure, CC(C)(C)NC(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)CN1CCC(C(
O)CN1CCC(C(![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O)Nc2cccc(–c3nc4ccccc4n3Cc3cc ccc3)c2)CC1, as the most promising based on a chemical expert knowledge perspective. This semi-automated approach is efficient for discovering novel molecules and assessing their properties. We predict that further iterations of this model will require less supervision, and medicinal chemists will start using it as a side-car for generating and validating ideas.
O)Nc2cccc(–c3nc4ccccc4n3Cc3cc ccc3)c2)CC1, as the most promising based on a chemical expert knowledge perspective. This semi-automated approach is efficient for discovering novel molecules and assessing their properties. We predict that further iterations of this model will require less supervision, and medicinal chemists will start using it as a side-car for generating and validating ideas.
3.2 Use case: Chemistry42 generative model
Chemistry42 is Insilico Medicine's AI drug discovery platform that efficiently generates novel active molecules using 42 generative models.44 In this experiment, we apply nach0 to one of the published case study setups available on demand at https://demo.chemistry42.com—structure-based design of Janus kinase 3 inhibitors. In Chemistry42, we use 3LXK crystal structure, pharmacophore hypothesis, and a set of physicochemical properties to set up the search space for the generative models. All generative models search the chemical space to find the best possible structures.Chemistry42 provides a set of filters ans reward modules. The 2D modules comprise of various tools including Medicinal Chemistry Filters (MCFs), Lipinski's Rule of Five (Ro5), and descriptors for drug-likeness, weighted atom-type portion, drug-likeness and novelty, the synthetic accessibility (SA) scores. Additionally, Chemistry42 use the Self-Organizing Maps (SOM) classifier module to navigate the generation of molecular structures towards a specific target class in the chemical space. The structure morphing module, another integral part of 2D modules, is utilized to tackle metabolic instability issues.
The 3D modules include the ConfGen module, which is responsible for generating conformational ensembles for each molecular structure. Subsequently, these molecules are ranked based on their intrinsic rigidity using a flexibility assessment tool. The 3D similarity between the generated structures and a reference molecule is evaluated using the 3D-Descriptors Module. The pharmacophore module is then used to find any matches with the specified pharmacophore hypothesis. The shape similarity module plays its part in evaluating the 3D shape similarity to a reference molecule. Lastly, the pocket module and the Pocket–Ligand Interaction (PLI) modules are used to assess how well the molecules fit the chosen binding site.
In this experiment, we replaced all 42 generative models with nach0 and generated a set of structures using a prompt “Generate a random druglike small inhibitor molecule for the Janus kinase 3 JAK3 that contains a classic kinase hinge binding motif”. Note that nach0 does not have access to the specific crystal structure and other required properties, so the model generated molecules using solely its knowledge about JAK3.
In Table 2, we compare generation results using a combinatorial generator,45 Chemistry42,44 and our model. In just 45 minutes (consisting of 15 minutes for generation and 30 minutes for scoring in Chemistry42), our model discovered 8 molecules satisfying all the 2D and 3D requirements; see Ivanenkov et al.44 for more details on requirements. All these structures have a hinge binder and properly bind in the active site. While our model can discover multiple molecules satisfying all constraints, the discovered structures are currently worse than those found in 72 hour generations in Chemistry42, since nach0 does not yet learn from the reinforcement learning feedback during generation and because it does not have exact knowledge of the experiment setup. In future work, we will expand our model with reinforcement learning capabilities to improve generation quality.
3.3 Comparison of multi-task models
Table 3 compares nach0 base and large models with two existing NLP encoder–decoder models (general-domain FLAN23 and domain-specific SciFive46), and a multi-domain encoder–decoder model MolT5.20 The table contains metrics for each task and model, with the results of the top-performing base model emphasized in bold. First, FLAN base and nach0 base exhibit similar results on NLP tasks on average, demonstrating superior performance on different tasks. With single-domain models for tasks such as NER or NLI, where molecule information is not required, traditional LMs may indeed provide the best results. However, when it comes to molecular tasks that involve molecular data, nach0 has distinct advantages over similar-scale models due to its specialized architecture and ability to effectively incorporate and process molecule-related information. In particular, nach0 benefits from training on diverse datasets and the proposed tokenization approach, outperforming baselines (including FLAN) with a significant gap in molecular tasks. For regression tasks, nach0 shows the best results on both RMSE and R2 scores. Moreover, in the molecular generation task, nach0 substantially surpasses FLAN by the FCD metric, which assesses the closeness of the generated molecules distribution to the ground truth. We added this explanation to the manuscript. Second, as expected, large nach0 performed best among all the models. In terms of base models, nach0 base achieved the best results on chemical and cross-domain tasks over existing models, confirming that pre-training on two types of data with different tokens can be effective.| Dataset | Metric | MolT5 | SciFive | FLAN | nach0 | |||
|---|---|---|---|---|---|---|---|---|
| Base | Large | |||||||
| BC5-chem | F-1↑ | 77.82% | 91.02% | 88.03% | 90.96% | 92.78% | ||
| BC5-disease | 71.62% | 82.24% | 78.29% | 81.67% | 85.51% | |||
| NCBI-disease | 74.96% | 84.22% | 81.37% | 84.30% | 85.82% | |||
| BC2GM | 53.47% | 69.55% | 62.53% | 71.12% | 80.41% | |||
| JNLPBA | 63.06% | 72.99% | 70.74% | 73.70% | 79.80% | |||
| EBM PICO | F1↑ | 67.37% | 67.32% | 69.48% | 67.60% | 94.44% | ||
| MedNLI | Accuracy↑ | 58.69% | 70.29% | 79.66% | 73.40% | 89.22% | ||
| SciTail | 56.54% | 80.73% | 90.68% | 84.12% | 93.87% | |||
| ChemProt | F-1↑ | 70.52% | 75.83% | 84.38% | 83.61% | 94.46% | ||
| DDI | 56.02% | 59.53% | 85.96% | 88.69% | 93.13% | |||
| GAD | 52.10% | 64.53% | 66.93% | 75.47% | 78.24% | |||
| BIOSSES | Pearson↑ | 24.55% | 56.51% | 61.21% | 52.58% | 52.37% | ||
| HoC | F-1↑ | 70.24% | 72.49% | 72.37% | 80.40% | 85.86% | ||
| PubMedQA | F-1↑ | 49.12% | 59.44% | 62.80% | 58.76% | 74.21% | ||
| BioASQ | 61.71% | 80.29% | 87.14% | 79.43% | 89.21% | |||
| MedMCQA and MMLU | Accuracy↑ | 25.97% | 25.06% | 25.42% | 26.61% | 46.10% | ||
| MedMCQA-open | BLEU-2↑ | 4.52% | 5.83% | 5.10% | 6.30% | 2.26% | ||
| Reagent prediction | Accuracy@top1↑ | 1.10% | 3.80% | 4.00% | 6.30% | 13.08% | ||
| Retrosynthesis | Accuracy@top1↑ | 15.00% | 31.00% | 31.00% | 53.00% | 56.26% | ||
| Forward reaction prediction | Accuracy@top1↑ | 27.00% | 60.00% | 59.00% | 88.00% | 89.94% | ||
| BACE | BA↑ | 0.58 | 0.65 | 0.65 | 0.74 | 0.71 | ||
| BBBP | BA↑ | 0.55 | 0.66 | 0.6 | 0.67 | 0.68 | ||
| HIV | BA↑ | 0.5 | 0.53 | 0.53 | 0.56 | 0.60 | ||
| HFE | R 2↑ | −0.36 | 0.51 | 0.55 | 0.77 | 0.78 | ||
| RMSE↓ | 1.1 | 0.4 | 0.37 | 0.19 | 0.19 | |||
| HOMO–LUMO | R 2↑ | 0.98 | 0.99 | 0.99 | 1.00 | 1.00 | ||
| RMSE↓ | 0.0008 | 0.0003 | 0.0003 | 0.0001 | 0.0001 | |||
| LOGD | R 2↑ | −0.6 | −0.27 | −0.32 | 0.28 | 0.28 | ||
| RMSE↓ | 2.4 | 1.9 | 1.9 | 1.1 | 1.1 | |||
| LOGS | R 2↑ | −0.49 | 0.31 | 0.001 | 0.48 | 0.48 | ||
| RMSE↓ | 1.4 | 0.63 | 0.91 | 0.48 | 0.48 | |||
| MOSES | Valid↑ | 98.30% | 95.79% | 97.63% | 99.86% | 99.93% | ||
| Unique@10000↑ | 99.93% | 99.94% | 99.95% | 99.92% | 99.97% | |||
| FCD/test↓ | 0.5212 | 0.5778 | 0.5289 | 0.3106 | 0.3038 | |||
| SNN/test↑ | 0.5745 | 0.5688 | 0.5742 | 0.6118 | 0.6222 | |||
| Frag/test↑ | 0.9974 | 0.9967 | 0.9965 | 0.9985 | 1.00 | |||
| Scaf/test↑ | 0.8748 | 0.8737 | 0.8823 | 0.9205 | 0.9292 | |||
| IntDiv↑ | 0.8460 | 0.8464 | 0.8462 | 0.8478 | 0.8585 | |||
| Filters↑ | 98.89% | 98.67% | 98.68% | 99.54% | 99.67% | |||
| Novelty↑ | 93.92% | 93.98% | 93.67% | 87.60% | 93.87% | |||
| Description-guided molecule design | BLEU-2↑ | 30.32% | 44.17% | 43.64% | 48.97% | 48.76% | ||
| Molecular description generation | BLEU-2↑ | 35.61% | 39.56% | 38.58% | 43.91% | 41.73% | ||
Furthermore, we conducted zero-shot experiments involving nach0, FLAN, and SciFive (all base versions) in an information retrieval task. The objective was to detect whether an abstract is relevant to a given disease or gene query. The dataset used for these experiments, along with its specific details, can be found in Tutubalina et al.47. In these experiments, we employed the following prompt: “Given the following passage, answer the question: is the following text related to the synonym? Passage: text”. To evaluate the models' performance, we utilized precision (P), recall (R), and F-measure (F1). Our findings indicate that nach0 achieved an F1 score of 82.24% (with a recall of 96.32% and precision of 71.76%), while FLAN and SciFive achieved F1 scores of 82.24% and 77.20%, respectively. However, it is worth noting that the supervised BERT-based pipeline from Tutubalina et al.47 achieved a higher F1 score of 88.81%. Based on these results, we can conclude that these models exhibit the ability to perform slightly different NLP tasks in a zero-shot setup. However, they still fall significantly behind supervised models in terms of performance.
3.4 Ablations
To examine the impact of cross-domain data on multi-task fine-tuning, we conducted training on mono-domain data. The results of four pre-trained checkpoints (SciFive, FLAN, MolT5, nach0) fine-tuned exclusively on NLP data are presented in ESI, Section 1.† When considering average performance on the NLP group, nach0, SciFive, and FLAN exhibit similar results, MolT5 achieves lower scores compared to the other models.Next, we investigate how chemical tasks groups combination effects on joint model performance in comparison with individual models trained on each separate chemical tasks group—on predictive tasks group, on reaction tasks group and molecular generation/cross-domain tasks group. We perform the same experiments with MolT5 model to elaborate on how pretraining data and special chemical tokens affect the quality of the model on chemical tasks.
The results of this ablation study can be found in Table 4 and show that nach0 benefits from combining chemical tasks group—model trained on the whole set of chemical data without NLP outperforms in total set of metrics models trained on distinct task groups. It is important to mention that despite the joint model showing worse metrics than the model trained only on molecular generation and cross-domain tasks, it works better since it does not overfit on training data—the novelty metric is more prevail here over all other molecule generation metrics.
| Dataset | Metric | nach0 | MolT5 | ||||||
|---|---|---|---|---|---|---|---|---|---|
| All | Pred. | React. | Mol. gen. | All | Pred. | React. | Mol. gen. | ||
| Prediction tasks | |||||||||
| BACE | BA↑ | 0.74 | 0.67 | — | — | 0.58 | 0.52 | — | — |
| BBBP | BA↑ | 0.67 | 0.62 | — | — | 0.55 | 0.57 | — | — |
| HIV | BA↑ | 0.56 | 0.65 | — | — | 0.5 | 0.51 | — | — |
| HFE | R 2↑ | 0.77 | 0.015 | — | — | −0.36 | −0.74 | — | — |
| RMSE↓ | 0.19 | 0.81 | — | — | 1.1 | 1.4 | — | — | |
| HOMO–LUMO | R 2↑ | 1.0 | 1.0 | — | — | 0.98 | 0.94 | — | — |
| RMSE↓ | 1 × 10−4 | 1 × 10− 5 | — | — | 7 × 10−4 | 2 × 10−4 | — | — | |
| LOGD | R 2↑ | 0.28 | 0.27 | — | — | −0.6 | −2.9 | — | — |
| RMSE↓ | 1.1 | 1.1 | — | — | 2.4 | 5.7 | — | — | |
| LOGS | R 2↑ | 0.48 | 0.32 | — | — | −0.49 | −1.2 | — | — |
| RMSE↓ | 0.48 | 0.62 | — | — | 1.4 | 2.0 | — | — | |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||||||||
| Reaction tasks | |||||||||
| Reagent prediction | Accuracy↑ | 0.063 | — | 0.14 | — | 0.011 | — | 0.13 | — |
| Retrosynthesis | Accuracy↑ | 0.53 | — | 0.39 | — | 0.15 | — | 0.39 | — |
| Forward reaction prediction | Accuracy↑ | 0.88 | — | 0.89 | — | 0.27 | — | 0.89 | — |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) |
|||||||||
| Molecular generation and cross-domain tasks | |||||||||
| Molecule generation | Validity↑ | 99.86% | — | — | 99.99% | 98.3% | — | — | 0.0% |
Unique@10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000↑ 000↑ |
99.92% | — | — | 99.81% | 99.93% | — | — | N/A | |
| FCD/test↓ | 0.3106 | — | — | 0.2411 | 0.5212 | — | — | N/A | |
| SNN/test↑ | 0.6118 | — | — | 0.6551 | 0.5745 | — | — | N/A | |
| Frag/test↑ | 0.9985 | — | — | 0.9988 | 0.9974 | — | — | N/A | |
| Scaf/test↑ | 0.9205 | — | — | 0.9403 | 0.8748 | — | — | N/A | |
| IntDiv↑ | 0.8478 | — | — | 0.8493 | 0.846 | — | — | N/A | |
| Filters↑ | 99.54% | — | — | 99.95% | 98.89% | — | — | N/A | |
| Novelty↑ | 87.6% | — | — | 64.34% | 93.92% | — | — | N/A | |
| Description-guided molecule gen. | BLEU-2↑ | 48.97% | — | — | 52.90% | 30.32% | — | — | 30.78% |
| Molecular description generation | BLEU-2↑ | 43.91% | — | — | 46.22% | 35.61% | — | — | 31.32% |
Also, experiments show that the special chemical tokens and pre-training on both natural language and chemical data improve the model quality—nach0 outperforms MolT5 baseline or show equal metrics on each chemical task group. We miss some MolT5 metrics on molecule generation task since it produces non-valid SMILES sequences.
3.5 Comparison with ChatGPT
Recently, a comprehensive benchmark for biomedical text generation and mining problems with ChatGPT was conducted, revealing its poor performance on several biomedical NLP benchmark datasets.48,49 Chen et al.49 specifically evaluated ChatGPT on a BLURB benchmark,50 which encompasses BC5-chem, BC5-disease, NCBI-disease, BC2GM, JNLPBA, EMB-PICO, ChemProt, DDI, GAD, BIOSSES, HoC, PubMedQA, BioASQ. In particular, ChatGPT got an average BLURB score of 48.27 on NER, while fine-tuned BERT achieved 86.27. For more details on evaluation scores, please refer to Chen et al.49.In our evaluation setup, we focus on three specific datasets: EMB-PICO, MedMCQA-open, and molecular description generation (Mol-Instructions). The inclusion of EMB-PICO dataset was driven by its practical importance. This dataset involves the task of identifying and extracting specific fragments of text related to the population/patient/problem (P), intervention (I), comparator (C), and outcome (O) elements from unstructured biomedical texts, such as research articles and clinical trial reports. It is worth noting that the clinical trial domain holds particular significance for inClinico, a transformer-based artificial intelligence software platform designed to predict the outcome of phase II clinical trials.10 The molecular generation task is relevant to the Chemistry42 platform.44
To evaluate the zero-shot performance, we had to limit the evaluation to a subset of 2000 samples from the test set for each of the three datasets, considering the computational constraints of ChatGPT. As well we utilized the GPT-3.5-turbo model through the OpenAI API and multi-task nach0 base for evaluation purposes. In the case of the PICO dataset, ChatGPT achieved a word-level F1 score of 64.43%, comparable to the results obtained by fine-tuned nach0 base on this subset (F1 score of 67.60%). For MedMCQA-open, ChatGPT achieved a BLEU2 score of 1.68%, while the fine-tuned nach0 base attained a BLEU2 score of 6.30%. In the molecular description generation task, ChatGPT achieved a BLEU2 score of 2.23%, whereas the fine-tuned nach0 base excelled with a BLEU2 score of 42.80%. Based on our preliminary findings, it is evident that utilizing ChatGPT directly leads to subpar performance compared to models trained specifically on the domain-specific dataset, how it was done in nach0.
3.6 Discussion
In this study, we pretrained and fine-tuned T5 models, which have an encoder–decoder architecture. Nevertheless, a broad range of model families, including T5, BERT-based BioMegatron,51 decoder-only PaLM52 and GPT,4 exist. To determine the most suitable architecture for pre-training and fine-tuning on chemical-related data, it may be necessary to evaluate these alternatives. We suggest it as a potential topic for future research.There have been several efforts to train large language models (LLMs) on biomedical corpora, particularly on PubMed. Notable examples include BioGPT (347m and 1.5b),53 PubMedGPT (2.7b),54 and Galactica (120b).18 Through our experiments with scaling from a base model (250m) to a large model (780m), we demonstrated the benefits of scale on several datasets. Based on our findings, we can conclude that scaling can further enhance the chemical capabilities of models, particularly in terms of generation and reasoning skills.
Key LLM capabilities for chemistry. Although our LM was able to reach state-of-the-art performance on several chemistry-related benchmarks, our human evaluations clearly suggested that these models are not at the chemist expert level. In order to bridge this gap, several new LLM capabilities need to be researched and developed including (i) knowledge alignment between textual and chemical sources as well as domain-specific knowledge graphs; (ii) ability to perform chemical reasoning and provide explanations for their predictions; (iii) ability to learn from and adapt to feedback from human experts, (iv) ability to generate novel chemical reactions and materials.
Molecular representations. One limitation of our LM is its focus on string representations of molecules, specifically the SMILES notation. Although SMILES is a widely used notation for representing molecules, it provides only 2D information of the molecule, missing the 3D geometry and spatial arrangement of atoms and bonds in a molecule. This can result in inaccuracies in predicting molecular properties and interactions. To address these limitations, it would be beneficial to incorporate additional modalities of molecules, such as the molecular graphs in terms of 2D or 3D representations, in the training of the language model.
Another significant drawback of the SMILES format is the absence of a one-to-one translation between molecules and SMILES strings. Typically, a molecule can have multiple SMILES representations that differ from each other due to factors such as the starting atom, molecular graph traversal, and kekulization. In practice, SMILES strings are often converted to a canonical form using an unambiguous algorithm. A molecular representation called SELFIES55,56 was defined from scratch to be attractive as a sequential representation for molecules. All random SELFIES are valid molecular representations. SELFIES was extended to treat molecular groups as well.57 As SELFIES have been repeatedly shown to have advantages over other representations in the context of generative models, exploring their use as the main representation for a language model is a future potential direction.
Prompt design. Our language model has a limitation in that it heavily relies on the quality and specificity of the prompts, as well as the potential for biases in both the training data and the prompts themselves. To enhance the performance of the model, incorporating domain-specific and information-rich prompts is essential. One potential approach to achieving this is by leveraging the knowledge of domain experts to design effective biomedical prompts. Yet, over-reliance on domain-specific prompts may lead to a lack of diversity in the model's responses, which can limit its usefulness.
Chemical diversity. Mol-Instructions includes cross-domain datasets that consist of compounds and their corresponding descriptions collected from PubChem. PubChem is a publicly available database administered by the National Center for Biotechnology Information (NCBI). It is important to note that the datasets primarily encompass current drugs and known chemical probes, representing only a fraction of the vast predicted chemical space. Furthermore, these datasets do not encompass testing on novel chemical diversity distinct from molecules documented in the literature.
4 Conclusion
Our study integrates a diverse range of one-domain and multi-domain task types and biomolecular text instructions to address the landscape of chemical research on drug design, reaction prediction, and retrosynthesis and leverage the advancements in NLP and LLMs. The multi-domain training approach allows our model, nach0, to leverage a broader understanding of both chemical and linguistic knowledge. Extensive experiments and two case studies demonstrate that nach0's capabilities in translating between natural language and chemical language enable it to tackle tasks effectively. Considering the unique training methodology and the broader scope of tasks that our model can effectively handle, we believe our work presents a significant contribution to the field.Based on our findings, we foresee several promising directions for future research. One direction could involve such as protein sequences, which would require adding special tokens into the model similar to SMILES. This task could be easily achieved with group SELFIES. New modalities require collecting diverse tasks with natural language prompts for fine-tuning. A second direction involves extending NLP datasets and conducting zero-shot evaluations to assess the reasoning and generalization capabilities of nach0. Finally, exploring the fusion of information from textual sequences and relevant knowledge graphs as input in a self-supervised approach remains an area to be explored.
Code availability
The nach0 framework is available for research purposes: nach0 base is available viahttps://huggingface.co/insilicomedicine/nach0_base; nach0 large is available viahttps://huggingface.co/insilicomedicine/nach0_large; for pre-processing scripts, see https://github.com/insilicomedicine/nach0.Data availability
All datasets used in the study for pre-training and fine-tuning are publicly available.Author contributions
These authors contributed equally: Micha Livne, Zulfat Miftahutdinov, Elena Tutubalina, Maksim Kuznetsov. ET, DP, AA, and AZ contributed to the conception and design of the work. ET, ZM, and MK contributed to the data acquisition and curation. ZM, MK, ML, AC, AB, and AJ contributed to the technical implementation with the NeMo framework, provided technical and infrastructure guidance. ET, ZM, and MK contributed to the evaluation framework used in the study. All authors contributed to the drafting and revising of the manuscript.Conflicts of interest
The authors declare no competing interests. This study is a collaboration of NVIDIA and Insilico Medicine employees.Notes and references
- J. Devlin, M.-W. Chang, K. Lee and K. Toutanova, Proceedings of the 2019 Conference of the North American Chapter of the Association for Computational Linguistics: Human Language Technologies, Volume 1 (Long and Short Papers), Minneapolis, Minnesota, 2019, pp. 4171–4186 Search PubMed.
- C. Raffel, N. Shazeer, A. Roberts, K. Lee, S. Narang, M. Matena, Y. Zhou, W. Li and P. J. Liu, J. Mach. Learn. Res., 2020, 21, 1–67 Search PubMed.
- M. Lewis, Y. Liu, N. Goyal, M. Ghazvininejad, A. Mohamed, O. Levy, V. Stoyanov and L. Zettlemoyer, Proceedings of the 58th Annual Meeting of the Association for Computational Linguistics, Online, 2020, pp. 7871–7880 Search PubMed.
- T. Brown, B. Mann, N. Ryder, M. Subbiah, J. D. Kaplan, P. Dhariwal, A. Neelakantan, P. Shyam, G. Sastry, A. Askell, S. Agarwal, A. Herbert-Voss, G. Krueger, T. Henighan, R. Child, A. Ramesh, D. Ziegler, J. Wu, C. Winter, C. Hesse, M. Chen, E. Sigler, M. Litwin, S. Gray, B. Chess, J. Clark, C. Berner, S. McCandlish, A. Radford, I. Sutskever and D. Amodei, Advances in Neural Information Processing Systems, 2020, pp. 1877–1901 Search PubMed.
- R. Bommasani, D. A. Hudson, E. Adeli, R. Altman, S. Arora, S. von Arx, M. S. Bernstein, J. Bohg, A. Bosselut and E. Brunskill, et al., arXiv, 2021, preprint, arXiv:2108.07258 Search PubMed.
- E. Tutubalina, Z. Miftahutdinov, V. Muravlev and A. Shneyderman, Proceedings of the 2022 Conference on Empirical Methods in Natural Language Processing (EMNLP): Industry Track, Abu Dhabi, UAE, 2022, pp. 596–605 Search PubMed.
- Z. Miftahutdinov, A. Kadurin, R. Kudrin and E. Tutubalina, Bioinformatics, 2021, 37, 3856–3864 CrossRef CAS PubMed.
- Z. Miftahutdinov, A. Kadurin, R. Kudrin and E. Tutubalina, Lecture Notes in Computer Science (Including Subseries Lecture Notes in Artificial Intelligence and Lecture Notes in Bioinformatics), 12656 LNCS, 2021, pp. 451–466 Search PubMed.
- E. Tutubalina, A. Kadurin and Z. Miftahutdinov, COLING 2020 – 28th International Conference on Computational Linguistics, Proceedings of the Conference, 2020, pp. 6710–6716 Search PubMed.
- A. Aliper, R. Kudrin, D. Polykovskiy, P. Kamya, E. Tutubalina, S. Chen, F. Ren and A. Zhavoronkov, Clin. Pharmacol. Ther., 2023, 114(5), 972–980 CrossRef PubMed.
- E. Putin, A. Asadulaev, Q. Vanhaelen, Y. Ivanenkov, A. V. Aladinskaya, A. Aliper and A. Zhavoronkov, Mol. Pharmaceutics, 2018, 15, 4386–4397 CrossRef CAS PubMed.
- D. Polykovskiy, A. Zhebrak, D. Vetrov, Y. Ivanenkov, V. Aladinskiy, P. Mamoshina, M. Bozdaganyan, A. Aliper, A. Zhavoronkov and A. Kadurin, Mol. Pharmaceutics, 2018, 15, 4398–4405 CrossRef CAS PubMed.
- R. Shayakhmetov, M. Kuznetsov, A. Zhebrak, A. Kadurin, S. Nikolenko, A. Aliper and D. Polykovskiy, Front. Pharmacol, 2020, 11, 269 CrossRef CAS PubMed.
- A. Aliper, S. Plis, A. Artemov, A. Ulloa, P. Mamoshina and A. Zhavoronkov, Mol. Pharmaceutics, 2016, 13, 2524–2530 CrossRef CAS PubMed.
- M. Kuznetsov and D. Polykovskiy, Proceedings of the AAAI Conference on Artificial Intelligence, 2021, vol. 35, pp. 8226–8234 Search PubMed.
- H. Dowden and J. Munro, Nat. Rev. Drug Discovery, 2019, 18, 495–496 CrossRef CAS PubMed.
- J. Lee, W. Yoon, S. Kim, D. Kim, S. Kim, C. H. So and J. Kang, Bioinformatics, 2020, 36, 1234–1240 CrossRef CAS PubMed.
- R. Taylor, M. Kardas, G. Cucurull, T. Scialom, A. Hartshorn, E. Saravia, A. Poulton, V. Kerkez and R. Stojnic, Galactica: A Large Language Model for Science, arXiv, 2022, preprint, arXiv:2211.09085, DOI:10.48550/arXiv.2211.09085.
- A. Vaswani, N. Shazeer, N. Parmar, J. Uszkoreit, L. Jones, A. N. Gomez, L. u. Kaiser and I. Polosukhin, Advances in Neural Information Processing Systems, 2017 Search PubMed.
- C. Edwards, T. Lai, K. Ros, G. Honke, K. Cho and H. Ji, Proceedings of the 2022 Conference on Empirical Methods in Natural Language Processing, 2022, pp. 375–413 Search PubMed.
- D. Flam-Shepherd, K. Zhu and A. Aspuru-Guzik, Nat. Commun., 2022, 13, 3293 CrossRef CAS PubMed.
- D. Flam-Shepherd and A. Aspuru-Guzik, arXiv, 2023, preprint, arXiv:2305.05708, DOI:10.48550/arXiv.2305.05708.
- H. W. Chung, L. Hou, S. Longpre, B. Zoph, Y. Tay, W. Fedus, E. Li, X. Wang, M. Dehghani and S. Brahmaet al., arXiv, 2022, preprint, arXiv:2210.11416 Search PubMed.
- O. Kuchaiev, J. Li, H. Nguyen, O. Hrinchuk, R. Leary, B. Ginsburg, S. Kriman, S. Beliaev, V. Lavrukhin and J. Cooket al., arXiv, 2019, preprint, arXiv:1909.09577 Search PubMed.
- Y. Fang, X. Liang, N. Zhang, K. Liu, R. Huang, Z. Chen, X. Fan and H. Chen, Mol-Instructions: A Large-Scale Biomolecular Instruction Dataset for Large Language Models, The Twelfth International Conference on Learning Representations, 2024 Search PubMed.
- Z. Wu, B. Ramsundar, E. N. Feinberg, J. Gomes, C. Geniesse, A. S. Pappu, K. Leswing and V. Pande, Chem. Sci., 2018, 9, 513–530 RSC.
- J. Li, Y. Sun, R. J. Johnson, D. Sciaky, C.-H. Wei, R. Leaman, A. P. Davis, C. J. Mattingly, T. C. Wiegers and Z. Lu, Database, 2016, 2016, baw068 CrossRef PubMed.
- R. I. Doğan, R. Leaman and Z. Lu, J. Biomed. Inf., 2014, 47, 1–10 CrossRef PubMed.
- L. Smith, L. Tanabe and R. Ando, Genome Biol., 2008, 9, 1–9 CrossRef PubMed.
- N. Collier, T. Ohta, Y. Tsuruoka, Y. Tateisi and J.-D. Kim, Proceedings of the International Joint Workshop on Natural Language Processing in Biomedicine and its Applications (NLPBA/BioNLP), Geneva, Switzerland, 2004, pp. 73–78 Search PubMed.
- B. Nye, J. J. Li, R. Patel, Y. Yang, I. J. Marshall, A. Nenkova and B. C. Wallace, Proceedings of the conference, Meeting, Association for Computational Linguistics, 2018, p. 197 Search PubMed.
- C. Shivade, et al., Proceedings of the 2018 Conference on Empirical Methods in Natural Language Processing, Association for Computational Linguistics, Brussels, Belgium. 2019, pp. 1586–1596 Search PubMed.
- T. Khot, A. Sabharwal and P. Clark, Proceedings of the AAAI Conference on Artificial Intelligence, 2018 Search PubMed.
- M. Krallinger, O. Rabal, S. A. Akhondi, M. P. Pérez, J. Santamaría, G. P. Rodríguez, G. Tsatsaronis, A. Intxaurrondo, J. A. López and U. Nandalet al., Proceedings of the sixth BioCreative challenge evaluation workshop, 2017, pp. 141–146 Search PubMed.
- M. Herrero-Zazo, I. Segura-Bedmar, P. Martínez and T. Declerck, J. Biomed. Inf., 2013, 46, 914–920 CrossRef PubMed.
- À. Bravo, J. Piñero, N. Queralt-Rosinach, M. Rautschka and L. I. Furlong, BMC Bioinf., 2015, 16, 1–17 CrossRef PubMed.
- G. Soğancıoğlu, H. Öztürk and A. Özgür, Bioinformatics, 2017, 33, i49–i58 CrossRef PubMed.
- D. Hanahan and R. A. Weinberg, Cell, 2000, 100, 57–70 CrossRef CAS PubMed.
- Q. Jin, B. Dhingra, Z. Liu, W. Cohen and X. Lu, Proceedings of the 2019 Conference on Empirical Methods in Natural Language Processing and the 9th International Joint Conference on Natural Language Processing (EMNLP-IJCNLP), 2019, pp. 2567–2577 Search PubMed.
- A. Nentidis, K. Bougiatiotis, A. Krithara and G. Paliouras, Machine Learning and Knowledge Discovery in Databases: International Workshops of ECML PKDD 2019, Würzburg, Germany, September 16–20, 2019, Proceedings, Part II, 2020, pp. 553–568 Search PubMed.
- D. Weininger, J. Chem. Inf. Comput. Sci., 1988, 28, 31–36 CrossRef CAS.
- E. Harper, S. Majumdar, O. Kuchaiev, L. Jason, Y. Zhang, E. Bakhturina, V. Noroozi, S. Subramanian, K. Nithin, H. Jocelyn, F. Jia, J. Balam, X. Yang, M. Livne, Y. Dong, S. Naren and B. Ginsburg, NeMo: A Toolkit for Conversational AI and Large Language Models, 2019, https://github.com/NVIDIA/NeMo Search PubMed.
- D. Narayanan, M. Shoeybi, J. Casper, P. LeGresley, M. Patwary, V. Korthikanti, D. Vainbrand, P. Kashinkunti, J. Bernauer, B. Catanzaro, A. Phanishayee and M. Zaharia, Proceedings of the International Conference for High Performance Computing, Networking, Storage and Analysis, New York, NY, USA, 2021 Search PubMed.
- Y. A. Ivanenkov, D. Polykovskiy, D. Bezrukov, B. Zagribelnyy, V. Aladinskiy, P. Kamya, A. Aliper, F. Ren and A. Zhavoronkov, J. Chem. Inf. Model., 2023, 63, 695–701 CrossRef CAS PubMed.
- D. Polykovskiy, A. Zhebrak, B. Sanchez-Lengeling, S. Golovanov, O. Tatanov, S. Belyaev, R. Kurbanov, A. Artamonov, V. Aladinskiy and M. Veselov, et al. , Front. Pharmacol., 2020, 11, 565644 CrossRef CAS PubMed.
- L. N. Phan, J. T. Anibal, H. Tran, S. Chanana, E. Bahadroglu, A. Peltekian and G. Altan-Bonnet, arXiv, 2021, preprint, arXiv:2106.03598, DOI:10.48550/arXiv.2106.03598.
- E. Tutubalina, Z. Miftahutdinov, V. Muravlev and A. Shneyderman, Proceedings of the 2022 Conference on Empirical Methods in Natural Language Processing: Industry Track, 2022, pp. 596–605 Search PubMed.
- R. Tang, X. Han, X. Jiang and X. Hu, arXiv, 2023, preprint, arXiv:2303.04360, DOI:10.48550/arXiv.2303.04360.
- Q. Chen, H. Sun, H. Liu, Y. Jiang, T. Ran, X. Jin, X. Xiao, Z. Lin, Z. Niu and H. Chen, Bioinformatics, 2023, 39(9), btad557 CrossRef CAS PubMed.
- Y. Gu, R. Tinn, H. Cheng, M. Lucas, N. Usuyama, X. Liu, T. Naumann, J. Gao and H. Poon, ACM Trans. Comput. Healthc., 2021, 3, 1–23 CrossRef.
- H.-C. Shin, Y. Zhang, E. Bakhturina, R. Puri, M. Patwary, M. Shoeybi and R. Mani, Proceedings of the 2020 Conference on Empirical Methods in Natural Language Processing (EMNLP), 2020, pp. 4700–4706 Search PubMed.
- A. Chowdhery, S. Narang, J. Devlin, M. Bosma, G. Mishra, A. Roberts, P. Barham, H. W. Chung, C. Sutton, S. Gehrmann, P. Schuh, K. Shi, S. Tsvyashchenko, J. Maynez, A. Rao, P. Barnes, Y. Tay, N. Shazeer, V. Prabhakaran, E. Reif, N. Du, B. Hutchinson, R. Pope, J. Bradbury, J. Austin, M. Isard, G. Gur-Ari, P. Yin, T. Duke, A. Levskaya, S. Ghemawat, S. Dev, H. Michalewski, X. Garcia, V. Misra, K. Robinson, L. Fedus, D. Zhou, D. Ippolito, D. Luan, H. Lim, B. Zoph, A. Spiridonov, R. Sepassi, D. Dohan, S. Agrawal, M. Omernick, A. M. Dai, T. S. Pillai, M. Pellat, A. Lewkowycz, E. Moreira, R. Child, O. Polozov, K. Lee, Z. Zhou, X. Wang, B. Saeta, M. Diaz, O. Firat, M. Catasta, J. Wei, K. Meier-Hellstern, D. Eck, J. Dean, S. Petrov and N. Fiedel, J. Mach. Learn. Res., 2023, 24, 113 Search PubMed.
- R. Luo, L. Sun, Y. Xia, T. Qin, S. Zhang, H. Poon and T.-Y. Liu, Briefings Bioinf., 2022, 23, bbac409 CrossRef PubMed.
- E. Bolton, StanfordCRFM introduces PubMedGPT 2.7B, Stanford University, 2022 Search PubMed.
- M. Krenn, F. Häse, A. Nigam, P. Friederich and A. Aspuru-Guzik, Mach. Learn.: Sci. Technol., 2020, 1, 045024 Search PubMed.
- M. Krenn, Q. Ai, S. Barthel, N. Carson, A. Frei, N. C. Frey, P. Friederich, T. Gaudin, A. A. Gayle, K. M. Jablonka, R. F. Lameiro, D. Lemm, A. Lo, S. M. Moosavi, J. M. Nápoles-Duarte, A. Nigam, R. Pollice, K. Rajan, U. Schatzschneider, P. Schwaller, M. Skreta, B. Smit, F. Strieth-Kalthoff, C. Sun, G. Tom, G. Falk von Rudorff, A. Wang, A. D. White, A. Young, R. Yu and A. Aspuru-Guzik, Patterns, 2022, 3, 100588 CrossRef CAS PubMed.
- A. H. Cheng, A. Cai, S. Miret, G. Malkomes, M. Phielipp and A. Aspuru-Guzik, Digital Discovery, 2023, 2(3), 748–758 RSC.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4sc00966e |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2024 |