Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Large-scale validation of a plasmonic sensor for SARS-CoV-2 pseudo-neutralization with a cohort of food and retail workers†
Julien
Coutu
a,
Pierre
Ricard
a,
Abdelhadi
Djaïleb
b,
Étienne
Lavallée
b,
Henintsoa
Rabezanahary
cd,
Matthew
Stuible
e,
Yves
Durocher
e,
Caroline
Gilbert
cd,
Nicholas
Brousseau
cf,
Kim
Santerre
cd,
Mathieu
Thériault
cd,
Sylvie
Trottier
cd,
Denis
Boudreau
 g,
Marc-André
Langlois
hi,
Joelle N.
Pelletier
g,
Marc-André
Langlois
hi,
Joelle N.
Pelletier
 b,
Mariana
Baz
cd and
Jean-Francois
Masson
b,
Mariana
Baz
cd and
Jean-Francois
Masson
 *a
*a
aDepartment of Chemistry, Québec Centre for Advanced Materials (QCAM), Regroupement Québécois sur les Matériaux de Pointe (RQMP), and Centre interdisciplinaire de recherche sur le cerveau et l'apprentissage (CIRCA), Institut Courtois, Université de Montréal, CP 6128 Succ. Centre-Ville, Montréal, QC H3C 3J7, Canada. E-mail: jf.masson@umontreal.ca
bDepartment of Chemistry, Department of Biochemistry and PROTEO, The Québec Network for Research on Protein Function, Engineering and Applications, Université de Montréal, CP 6128 Succ. Centre-Ville, Montréal, QC H3C 3J7, Canada
cCentre de recherche du Centre hospitalier universitaire de Québec, Université Laval, 2705 boulevard Laurier, Québec, QC G1V 4G2, Canada
dAxe de Recherche Maladies Infectieuses et Immunitaires, Centre de Recherche du CHU de Québec-Université Laval, Québec, QC G1V 4G2, Canada
eMammalian Cell Expression, Human Health Therapeutics Research Centre, National Research Council Canada, Montréal, QC, Canada
fBiological Risks Department, Institut national de santé publique du Québec, Québec, QC G1V 5B3, Canada
gDepartment of Chemistry and Centre for Optics, Photonics and Lasers (COPL), Université Laval, 1045, av. de la Médecine, Québec, QC G1V 0A6, Canada
hDepartment of Biochemistry, Microbiology and Immunology, Faculty of Medicine, University of Ottawa, Ottawa, ON, Canada
iOttawa Center for Infection, Immunity and Inflammation (CI3), Ottawa, ON, Canada
First published on 9th April 2024
Abstract
Plasmonic sensors are candidates for numerous clinical applications, but few examples demonstrate their performance on large sample cohorts, a necessary step for clinical translation. The COVID-19 pandemic provided an unprecedented opportunity to validate a surface plasmon resonance (SPR) sensor for SARS-CoV-2 inhibition with a cohort of over 1000 clinical samples from the longitudinal study of a food and retail worker population. The SPR sensor provided an in vitro model to assess the level of neutralizing antibodies by measuring the inhibition of the SARS-CoV-2 spike protein interaction with ACE-2 following exposure of the spike protein to naive and immune sera (from vaccination and/or infection). In conjunction with population data on vaccination and infection, and epidemiological data from the local jurisdiction of the study cohort, it is shown that the SPR sensor performed well in assessing the level of “pseudo-neutralization” of participant sera and that the response of the SPR sensor correlates (r = 0.74) with a live virus microneutralization assay as well as with metadata of relevant events (vaccination, waves of infection, etc.) that occurred during the study period. Using these data, the article details the challenges and opportunities of using plasmonic sensors in clinical practice.
Introduction
Since the onset of the SARS-CoV-2 pandemic, a series of methods have been developed to assist in the management of the pandemic, including the assessment of active infections, and the determination of antibody levels (titer) and their ability to neutralize the interaction of viral proteins with human cells. Reference platforms rely on PCR to detect SARS-CoV-2, on ELISA to measure antibody titers and, to a much lesser extent, on the determination of antibody neutralization. While these often centralized platforms are essential for processing large volumes of samples and cohorts of individuals, they are not optimized for their rapid detection and measurement. In retrospect, the pandemic presented an unprecedented opportunity for the sensing field,1 and several platforms have been developed to enable point-of-care (POC) detection of active infection and antibodies, such as lateral flow devices, microfluidic platforms, wearable sensors, optical sensors and electrochemical sensors, among others.2–6Among them, surface plasmon resonance (SPR) sensing is a particularly tantalizing alternative to the current standard clinical assays, as SPR provides quantitative biomolecular information and is amenable to portable and POC testing formats.7 The pandemic facilitated the testing of new detection platforms in plasmonics such as SPR interferometry,8 thermoplasmonics sensing,9 the use of 2D materials such as MXenes,10 metasurfaces,11 a new background subtraction strategy,12 and the use of plasmonics for the detection of target biomarkers in saliva and diluted whole blood.13 Furthermore, the COVID-19 pandemic enabled the validation of portable sensing platforms such as fiber-optics-based SPR sensors,14 smartphone-based LSPR detection synchronized with machine learning (ML),15 and lightweight SPR sensors with simple graphical user interface.16–18 As such, SPR sensors were developed to measure antibody titer,17,19 combined antibody affinity and titer measurements,18,20,21 and viral proteins.22,23 These measurements were carried out in a variety of biofluids, including dried blood spots (DBS), serum, plasma, diluted whole blood, and saliva, showing the potential of SPR sensing for diverse clinical samples.17–23
Quantitative determination of biomolecular interactions is a hallmark of SPR sensing, which has been used to determine antibody affinity24,25 and other biomolecular binding information,26,27 and these properties were valuable during the COVID-19 pandemic. Recently, a promising SPR platform was developed to detect antibodies, viral particles and neutralized viral particles,28 providing an all-in-one sensor to address the key questions related to SARS-CoV-2 infection and the pandemic response. Similarly, SPR sensing has been applied to small molecule screening for discovery of putative therapeutic inhibitors such as peptides,29 cannabinoids,30 or other small molecule inhibitors.31–34 Plasmonic sensors are also well suited for monitoring the immune response following vaccination to facilitate vaccine development, as demonstrated in a mouse model.35 This is an area where SPR could have a huge impact, as portable SPR platforms are relatively easy to implement in a biosafety level (BSL) 3 laboratory or in animal facilities, and could be used to rapidly monitor the humoral response of test animals on site during vaccine development to detect the presence of and quantify neutralizing antibodies.
At the populational level, the detection of neutralizing antibodies provides valuable information on the humoral response of vaccinated and/or infected individuals and can assist public health in establishing vaccination policies and other measures to minimize the viral spread. Current methods to detect neutralizing antibodies include cell-free PCR,36 plaque reduction neutralization tests (PRNT, BSL-3),37 pseudovirus or microneutralization tests (BSL-2 or 3),38 ELISA (BSL-1 or 2)39 or lateral flow assays (BSL-0 or 1).40 These methods can process large numbers of samples, but are either difficult to deploy at the point of care because they require extensive infrastructure, or they are not quantitative. As such, quantitative detection of neutralizing antibodies may be an area where SPR sensors could offer an advantage in comparison to other bioanalytical platforms.
While these advances in SPR sensor development provided several proofs of concept and small-scale validations, the translation of sensors from the academic laboratories to clinical practice will require larger scale cross-validation with established techniques. Towards that goal, a recent study showed a high degree of correlation between SPR and ELISA for determination of SARS-CoV-2-specific antibodies in 115 serum samples.41 Another study also demonstrated the performance of SPR for screening antibodies on 120 samples pre-characterized using standard regulatory-approved commercial detection kits available at time of collection.42 Building on these studies, we applied a rapid BSL-2 “pseudo-neutralization” assay18 to measure the binding inhibition between the viral spike protein and ACE-2 receptor in over 1000 serum samples collected during a 18 month period in a cohort of food and retail workers. The data collected with SPR sensors were validated using a live virus microneutralization assay (BSL-3), the current gold standard assay for SARS-CoV-2-specific antibody, and captured the changes in the humoral response that were consistent with the timescale of vaccination campaigns and SARS-CoV-2 outbreaks.
Experimental section
SPR pseudo-neutralization
The SPR pseudo-neutralization is based on a previously published protocol.18 In brief, the sensor chip of a portable SPR instrument (Affinité Instruments, Canada).43 The instrument has four channels that can be addressed independently. Two of these channels were used for the analytical signal, one for a positive control, and one for a negative control. A 1 mL luer-lock syringe was used for manual injection of the reagents and samples. Due to its small footprint, the instrument was operated inside a biological safety cabinet to minimize the risk of workers' exposure to aerosols of human biofluids. The SPR chip was modified with 3-mercaptopropyl-Leu-His-Asp-Leu-His-Asp-COOH (Afficoat, Affinité Instruments, Canada) was activated with N-ethyl-N′-(3-dimethylaminopropyl)carbodiimide hydrochloride (EDC, Millipore Sigma) and N-hydroxysuccinimide (NHS, Millipore Sigma) for 2 minutes, followed by the immobilization of the SARS-CoV-2 spike protein (National Research Council of Canada, cat. no. SMT1-1 for the native spike and SmT1v3 (B.1.1.529) for the Omicron BA.1 spike proteins) at a concentration of 20 μg mL−1 in acetate buffer pH 4.5 for 20 minutes. The sensor surface was finally deactivated with 1 M ethanolamine hydrochloride (Millipore Sigma) pH 8.5 for 10 minutes. The sensor was then equilibrated in running buffer (pH 7.4 PBS with 0.1% BSA and 0.005% Tween 20). Clinical serum samples were immediately assayed. A serum sample diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 in running buffer was injected for 10 minutes in three of the four channels of the SPR instrument, while a pooled pre-pandemic serum (putatively exempt of antibodies reacting with the SARS-CoV-2 spike protein) also diluted 1
5 in running buffer was injected for 10 minutes in three of the four channels of the SPR instrument, while a pooled pre-pandemic serum (putatively exempt of antibodies reacting with the SARS-CoV-2 spike protein) also diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 in running buffer was injected in the fourth channel as a positive control of the spike-ACE-2 interaction. The pre-pandemic serum was human serum from human male AB plasma and it was sterile filtered (H4522-100ML Sigma-Aldrich). A second 100 mL bottle of human serum was needed to complete the study and it was purchased in 2021. This lot was tested in ELISA to confirm the absence of anti-spike antibodies. Following a rapid wash with running buffer, a 5 μg mL−1 recombinant human ACE-2 (National Research Council of Canada, cat. no. ACE2-1, reference material) solution was injected in two of the channels exposed to the clinical samples (duplicate measurement of binding inhibition), and in the positive control channel (fourth channel) to determine the maximum ACE-2 signal on this chip. The negative control was performed in the remaining channel previously injected with the clinical sample; running buffer only was injected to measure the blank response in the absence of ACE-2. The percent inhibition (%Inhibition, eqn (1)) was calculated from the ratio of the query SPR shift to the positive control. The SPR sensor was then regenerated in 10 mM glycine pH 2.2 (Millipore Sigma) and re-equilibrated in running buffer for the analysis of the next clinical sample; each sensor was regenerated up to 10 times.
5 in running buffer was injected in the fourth channel as a positive control of the spike-ACE-2 interaction. The pre-pandemic serum was human serum from human male AB plasma and it was sterile filtered (H4522-100ML Sigma-Aldrich). A second 100 mL bottle of human serum was needed to complete the study and it was purchased in 2021. This lot was tested in ELISA to confirm the absence of anti-spike antibodies. Following a rapid wash with running buffer, a 5 μg mL−1 recombinant human ACE-2 (National Research Council of Canada, cat. no. ACE2-1, reference material) solution was injected in two of the channels exposed to the clinical samples (duplicate measurement of binding inhibition), and in the positive control channel (fourth channel) to determine the maximum ACE-2 signal on this chip. The negative control was performed in the remaining channel previously injected with the clinical sample; running buffer only was injected to measure the blank response in the absence of ACE-2. The percent inhibition (%Inhibition, eqn (1)) was calculated from the ratio of the query SPR shift to the positive control. The SPR sensor was then regenerated in 10 mM glycine pH 2.2 (Millipore Sigma) and re-equilibrated in running buffer for the analysis of the next clinical sample; each sensor was regenerated up to 10 times. | (1) |
ELISA and microneutralization assays
The in-house colorimetric ELISA to detect anti-spike (ancestral, Delta B.1.617.2 and Omicron BA.1) antibodies in clinical samples was performed as previously reported.44 The OD450 was corrected with the absorbance at 595 nm. Antibody titers (in binding antibody unit per milliliter – BAU mL−1), vaccine and natural immunity (expressed as signal to cutoff ratio – SCO ratio) were evaluated at the automated high-throughput chemiluminescent ELISA45,46 platform that has served as the reference for monitoring SARS-CoV-2 antibodies in Canada. Cell cultured-based microneutralization assays were performed against ancestral, Delta B.1.617.2, and Omicron BA.1 SARS-CoV-2 strains in a BSL3 laboratory as previously reported.47,48 A neutralizing antibody titer was determined based on the serum dilution required to completely neutralize the infectivity of 100 TCID50 of each SARS-CoV-2 strain.Cohort
Adult volunteers (n = 304) were recruited in the Capitale-Nationale and Chaudière-Appalache administrative regions (approximately delimiting the greater Quebec City region including suburbs), among workers from the food and retail business (groceries; n = 112, restaurants and bars; n = 149, and hardware stores; n = 43). All experiments were performed in accordance with the Canadian guidelines of the Tri-Council Policy Statement: Ethical Conduct for Research Involving Humans (TCPS 2), and approved by the ethics committee at Laval University in Quebec City, Canada (“Comité d'éthique de la recherche du CHU de Québec-UL”, registration number 2021-5744). Participants were recruited after informed consent at the Centre Hospitalier Universitaire de Québec – Université Laval (CHUL). Populational data were collected following the standardized Canadian Immunity Tasks Force (CITF) questionnaire.49 The participants had a mean age of 41.3 years distributed in every age group from 18 to 75 years old and were mainly Caucasian (97%), representative of the demographics of the region surveyed (circa 95% Caucasian50,51) with females making up 58% of the cohort. The participants provided blood samples at three-month intervals for a total of up to five visits. Samples were collected for an 18 month period beginning in April 2021 until early October 2022, covering the third to the seventh waves of COVID-19 infection in the province of Quebec.51,52 This period coincides with the first COVID-19 vaccination campaigns of the province of Quebec, during which the population received two primary doses and up to two booster doses of Comirnaty (RNA vaccine; Pfizer), SpikeVax (RNA vaccine; Moderna), or Vaxzevria (viral vector-based vaccine; AstraZeneca), all of which were designed to stimulate immunity against the ancestral variant of SARS-CoV-2. At the end of the surveyed period, more than 90% of the cohort had received two doses, and more than 60% and 20% had also received respectively one or two additional booster doses of the same vaccines. The study ended before the next generation of vaccines, designed against later variants of SARS-CoV-2, became locally available. A full description of the cohort is provided elsewhere.51Statistical analysis
Data analysis was performed with GraphPad Prism 9.2 using unpaired t-tests or one-way ANOVA with confidence intervals of 95% to statistically compare means.Results
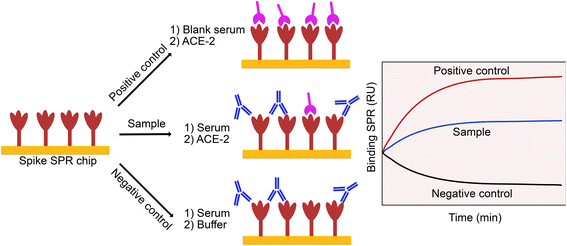
The neutralizing capacity of an antibody was defined by its ability to inhibit the interaction between the spike protein and the ACE-2 receptor, a key biomolecular interaction taking place in the SARS-CoV-2 infection process.18,29,32,53,54 To establish our model, we reproduced this interaction by binding spike proteins to a SPR chip and adding a soluble version of the ACE-2 protein on top of the chip. We then attempted to inhibit this interaction by using immune sera or competing inhibitor, providing the basis for a pseudo-neutralization assay (Fig. 1). The use of moderately diluted sera (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5) enables monitoring of pseudo-neutralization in conditions close to native, and due to the low nonspecific adsorption of the surface chemistry (3-mercaptopropyl-Leu-His-Asp-Leu-His-Asp-COOH), measurements are made with minimal interference from high concentrations of background proteins.
5) enables monitoring of pseudo-neutralization in conditions close to native, and due to the low nonspecific adsorption of the surface chemistry (3-mercaptopropyl-Leu-His-Asp-Leu-His-Asp-COOH), measurements are made with minimal interference from high concentrations of background proteins.
In the current version of the assay, binding inhibition was monitored from exposing the surface-bound spike protein (from the native or Omicron BA.1 strains) to immune sera, where immunity was acquired in the participants by: (1) vaccination, (2) infection or (3) a combination of infection and vaccination. Neutralizing antibodies present in the serum sample bind to the epitope on the spike protein and prevent the interaction with the soluble ACE-2 proteins injected immediately after, thereby inhibiting (or neutralizing) spike binding and providing the basis for calculating a %Inhibition (eqn (1)). Values closer to 0% Inhibition are expected for unvaccinated or virus naive individuals, while values closer to 100% indicate presence of neutralizing antibodies elicited by vaccination and/or infection (they are undistinguishable). Initial validation with a cohort of 32 infected individuals provided a proof-of-concept that the assay was reliable.18 However, such a small cohort fell short of providing unequivocal proof that the SPR assay can serve the intended clinical purpose. Furthermore, the cross-validation had been done only with ELISA, rarely used for the measurement of binding inhibition especially in the case of COVID-19.
Scaling up the validation of the pseudo-neutralization was thus the focus of this study, in addition to demonstrating that SPR sensors can provide robust data on the longitudinal survey of a population of individuals and cross-validating the assay with a reference live virus microneutralization platform. The cohort of participants provided a total of 1285 serum samples (visit 1: n = 304, visit 2: n = 297, visit 3: n = 292, visit 4: n = 198, and visit 5: n = 194), where the inhibition was measured for the native (visit 1: n = 304, visit 2: n = 143, visit 3: n = 105, visit 4: n = 194, and visit 5: n = 193), Delta B.1.617.2 (visit 1: n = 152, visit 2: n = 142, and visit 3: n = 82) and Omicron BA.1 (visit 2: n = 143, visit 3: n = 96, visit 4: n = 194, and visit 5: n = 193) spike proteins. Taken together, a total of nearly 2000 unique measurements (n = 1941) were taken. All samples were also analyzed by ELISA (antibody titer) and in a microneutralization assay with live virus. The collective data set was correlated to populational data collected from the survey questionnaire which included, among many other aspects, the vaccination and infection events for each participant.
Cross-validation with established techniques
Validation of SPR sensors requires comparison with established techniques. We compared the SPR method to ELISA because it is the most widely used method of reporting antibody levels. We used two ELISA platforms, a chemiluminescence ELISA widely used in Canada to study the pandemic45 and an in-house colorimetric ELISA44 that was recently shown to correlate with the former.55 Experimental details, including the optimal dilution factors, were the same as those reported in previous studies. Antibody levels as measured with ELISA varied temporally as the pandemic progressed showing concordance with vaccination and emergence of new variants. However, while these ELISAs measured antibody titers from which could be calculated antibody counts, they were not designed to detect the neutralization capacity of those antibodies against the interaction between spike proteins and ACE-2 receptors or between viruses and cells.We therefore also compared the SPR method to a live virus microneutralization assay where infection of cells with SARS-CoV-2 can be inhibited by immune sera.48 In this assay, the viral neutralization is measured as a dilution factor of the serum required to completely inhibit infection of cells by SARS-CoV-2. As this assay uses live virus, it must be performed in a BSL3 laboratory, strictly limiting its use to laboratories with the proper expertise and infrastructure. As such, both techniques report on the spike protein inhibition resulting from an exposition to immune sera. But while microneutralization use cells and live virus, the SPR pseudo-neutralization only use the viral protein and the human ACE-2 receptor, rendering the test much safer and thus less constrained in its use.
The comparison was carried out on the full set of nearly 1000 serum samples, using the native spike protein or viral strain in all assays. The microneutralization assay correlated well with SPR (r = 0.74), while a moderate correlation was observed for SPR with in-house colorimetric ELISA (r = 0.41) and the chemiluminescence ELISA on the automated platform (r = 0.32, Fig. S1†). Despite the nonlinearities in the plots of Fig. S1,† the ‘r’ coefficient provides an indication (although imperfect) of collinearity between methods. A similar trend was observed in the correlation plots of microneutralization with both ELISA methods (Fig. S2†), where nonlinearities were observed for the in-house colorimetric ELISA (r = 0.37) and chemiluminescence ELISA (r = 0.35). While both ELISA platforms were found to correlate well with each other,55 lower correlation with the SPR assay is expected, because ELISA measures antibody titers irrespective of their ability to neutralize binding of the spike protein to the ACE-2 receptor. The results indicate that antibody level is a contributing factor in the response of the SPR assay, but not necessarily a major driver. This is in agreement with a previous report that did not find significant correlation between antibody titers measured by ELISA and neutralizing antibodies obtained from a live-virus neutralization assay.56 Conversely, since both the SPR and microneutralization assays measure inhibition, a high correlation between these assays was expected. Stratifying the SPR data according to the results of the microneutralization assays or ELISA revealed a higher correlation for lower microneutralization titers and ELISA OD450 or BAU mL−1 (Fig. 2). The SPR signal saturated for the samples that gave the highest response in the microneutralization assay or in ELISA. This difference in sensitivity is likely due to the dilution factors used for each assay. SPR was optimized for a serum dilution factor of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5, the microneutralization assays were performed at a dilution of 1
5, the microneutralization assays were performed at a dilution of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 or greater, the in-house colorimetric ELISA used a dilution factor of 1
10 or greater, the in-house colorimetric ELISA used a dilution factor of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 15
15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000, and the chemiluminescence ELISA had dilutions of 1
000, and the chemiluminescence ELISA had dilutions of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 and 1
100 and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10
10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000. As such, the range in sensitivity overlapped, but with some difference between techniques. This partly explains the correlation factors, but the high variability of clinical samples accounts for most of the signal variability. Only with large sample cohorts can these trends be seen, highlighting the need for large sample sizes that include a wide span of natural immune responses to robustly cross-validate SPR sensing to other techniques.
000. As such, the range in sensitivity overlapped, but with some difference between techniques. This partly explains the correlation factors, but the high variability of clinical samples accounts for most of the signal variability. Only with large sample cohorts can these trends be seen, highlighting the need for large sample sizes that include a wide span of natural immune responses to robustly cross-validate SPR sensing to other techniques.
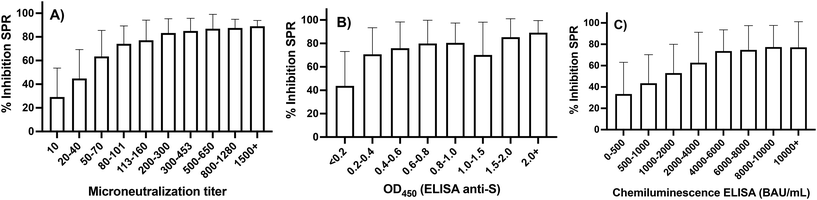 | ||
| Fig. 2 SPR correlation with the microneutralization (A), the in-house colorimetric ELISA (B) and the automated chemiluminescence ELISA (C). Error bars correspond to one standard deviation. | ||
Extracting populational data from the data set
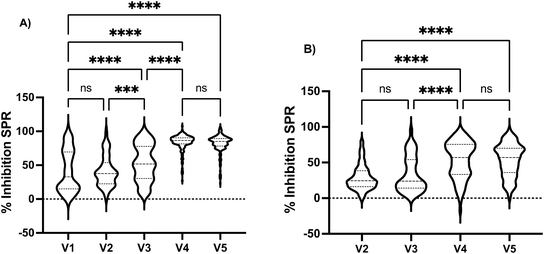
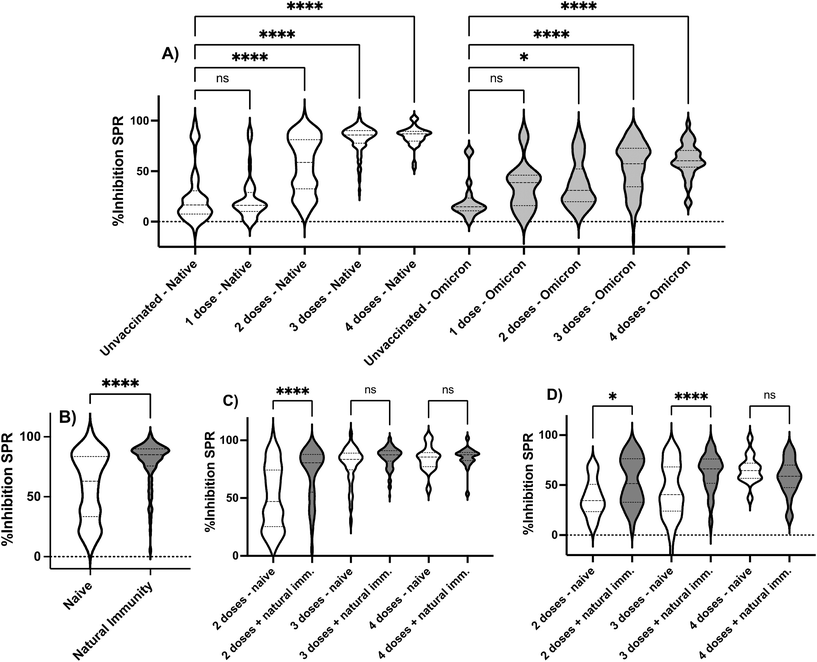
Following SPR sensor validation, the vast amount of data collected allowed us to extract valuable information in the surveyed period to demonstrate the ability of SPR sensors to capture descriptive information in a cohort. Stratification according to visits showed a progressive increase in %Inhibition SPR when assayed with the native spike protein from visits 2 to 4, where the first two and two last visits were not statistically different (Fig. 3). Similarly, a significant increase in %Inhibition SPR was observed between the 3rd and 4th visits when assayed with the Omicron BA.1 spike protein. Stratifying the cohort according to sex revealed no statistical difference (Fig. S3†).10,44 Similarly, no statistical difference was observed when comparing the workers' groups, where individuals working in grocery stores, restaurants/bars or hardware stores had indistinguishable responses for the %Inhibition SPR with the native spike protein (Fig. S4†). These results are in agreement with the ELISA55 and the microneutralization48 data for the same cohort. This is interesting because grocery stores remained open throughout the entire study period, whereas restaurants/bars and hardware stores were required to either temporarily close or exclusively offer drive-through/delivery services during the phases of peak contagion in that district. The differences in potential exposure to contagious clients between the groups of workers had no impact on our results; it is possible that the application of barrier measures such as mask wearing, hand washing and use of Plexiglas separators at the workplace, as recommended by the local government at that time, were successful in reducing viral transmission.The impact of vaccination on the results obtained by SPR was then assessed. Vaccination progressed over the course of the longitudinal study, as cohort recruitment coincided with mass vaccination in Quebec City. The cohort became highly vaccinated (>60% 3 doses or more; >90% 2 doses or more) and thus provided an opportunity to demonstrate the ability of the SPR sensor to monitor the progress of the immune response in the individuals. Stratifying the cohort according to the number of vaccine doses revealed that unvaccinated and individuals having received one dose had the lowest %Inhibition SPR and the response of these subgroups was statistically identical when assayed with the native and Omicron BA.1 spike proteins (Fig. 4A). The second dose increased the %Inhibition SPR, but the response remained heterogeneous in the cohort as illustrated by the large distribution of the %Inhibition SPR. A third dose was required to elicit a sustained humoral response and high %Inhibition SPR for both the native and Omicron BA.1 spike protein. Once again, the SPR data agrees with ELISA55 and microneutralization,48 where a statistically different and higher ELISA responses or geometric mean titer (GMT) for microneutralization were observed for individuals having received more doses of vaccines.
The cohort was further stratified based on the type of vaccine received by individuals who had received two doses of either adenovirus vector vaccine (COVISHIELD from AstraZeneca) or mRNA vaccines (Moderna Spikevax or Pfizer-BioNTech Comirnaty), or a combination of these vaccines. Due to limited supplies and government recommendations, some individuals in the cohort received a mixed vaccination schedule, especially those who received the AstraZeneca vaccine early in the campaign. Only three statistically significant differences were observed in the study. The dual AstraZeneca dosed individuals (n = 11, mean age = 58 ± 1 years old) had a lower %Inhibition SPR (p = 0.04) compared to those who received two doses of Moderna (n = 140, mean age = 38 ± 13 years old). The study also compared the %Inhibition SPR of individuals who received mixed schedules of AstraZeneca/Moderna (n = 70, mean age = 58 ± 6 years old) to those who received dual Moderna (n = 140, mean age = 38 ± 13 years old) and Pfizer-BioNTech (n = 256, mean age = 39 ± 17 years old) vaccines (Fig. S5†). The results showed that the %Inhibition SPR was lower in the group that received the mixed schedule. However, it is important to note that age and the delay between vaccination and the blood draw are confounding factors that may have influenced the results. Individuals who have received at least one dose of AstraZeneca were, on average, over 55 years old, while those who completed the vaccination regime with either Moderna or Pfizer-BioNTech were around 40 years old on average. There was no difference in the average number of days post-vaccination that could explain differences in %Inhibition SPR. All vaccine subgroups were vaccinated on average between 94 and 143 days before the blood draw. This emphasizes that the immune response is influenced by various factors, such as the interval between doses, the time elapsed since the last dose, the age and health status of individuals, and the type of vaccine. SPR sensing can capture these confounding factors.
The %Inhibition SPR was then compared between individuals who were fully vaccinated (two or more vaccine doses) but naive to the virus with individuals having a hybrid protection (infection + full vaccination). A stronger response was observed for individuals with a hybrid immunity (Fig. 4B), where two vaccine doses were sufficient to boost hybrid immunity in most individuals, when assessed with the native spike protein, yet three doses were required to achieve a similar boost to hybrid immunity when assessed with the Omicron BA.1 spike protein (Fig. 4C and D). Once again, the stronger response observed in SPR for hybrid immunity was also in agreement with microneutralization.48 No statistical difference was seen with SPR between virus-naive and previously infected individuals having received three or more doses when assayed with the native spike protein (Fig. 4C) and four doses with the Omicron BA.1 spike protein (Fig. 4D).
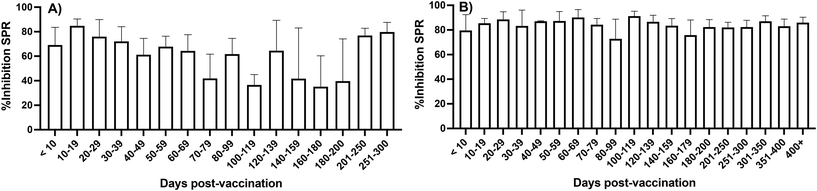
The data set allowed observing temporal changes in the %Inhibition SPR following vaccination since the visits were scheduled irrespective of the vaccination schedule. The large cohort size and number of visits (up to five) provided sufficient time points post-vaccination to observe trends (Fig. 5). Virus-naïve individuals had lower %Inhibition SPR for the period ranging between 70 and 200 days post-vaccination, whereas individuals with hybrid immunity showed a sustained level of %Inhibition throughout the study period (up to 400 days after the last dose). Furthermore, stratifying the cohort based on the period of sample collection allowed correlating changes in cohort immunity with populational events (Fig. 6). The %Inhibition SPR increased until the summer of 2021, by which time the majority of study participants had received their second vaccine dose. It then slowly waned until the beginning of 2022. This period, coinciding with the 3rd dose vaccination campaign, also correlated the Omicron BA.1 strain becoming dominant and infecting an increasing number of the participants (Fig. 6). These results clearly demonstrate the ability of SPR to track biomolecular changes related to SARS-CoV-2 infection in a population over the course of a longitudinal study, which has never been demonstrated before.
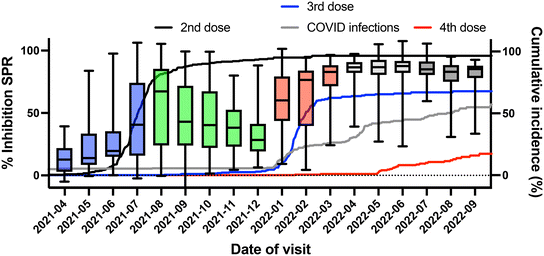 | ||
| Fig. 6 Temporal trend of the SPR inhibition for the cohort. The first rise in inhibition coincides with the 2nd vaccination campaign in the district under study and the sharp increase in inhibition in 2022-01 coincides with a wave of infections (onset of Omicron BA.1 in that district) and administration of the first booster shots (3rd dose). The different waves of infection (blue boxes: 3rd wave, green boxes: 4th wave; red boxes: 5th wave, light gray boxes: 6th wave and dark gray boxes: 7th wave) and event timeline are detailed by the Institut National de Santé Publique du Québec.52 The 5th to the 7th waves were dominated by the Omicron BA.1 strain. Box plots represent the 25th to the 75th percentile, while error bars delineate the minimum and maximum range of observed responses. Underlaid are traces of the cumulative vaccination incidence (2nd dose: black; 3rd dose: blue; and 4th dose: red) and COVID infections reported (gray) by individuals in the cohort. | ||
Discussion
This study was performed on a portable SPR platform that allowed the duplicate measurement of a single clinical sample and its controls per run, consistent with the intended purpose of point-of-care SPR sensing. Hence, this study provides a robust account of the limitations and opportunities of portable or POC SPR sensing. Given that one SPR sensor allowed a maximum of 10 samples to be analyzed before performance dropped, study completion required more than 200 SPR chips. Immobilization of the spike protein was performed over a fixed time and monitoring the resulting binding shift provided a data set of a size sufficient to evaluate the reproducibility of the surface chemistry. The binding shifts for 160 sensorgrams (40 chips × 4 channels) gave an average peak surface area of 1640 RU with a 32% coefficient of variation. The coefficient of variation is higher than what is typically observed in SPR. A value closer to 10% is more common. This higher coefficient of variation could be due to several factors, such as the large number of chips prepared in this study, which may indicate long-term drifts in some of the processes, instability of reagents, or inherent variations in surface chemistry. However, the use of a normalization process with positive and negative controls for each sample analyzed minimized the impact of chip preparation. Variations around the typical 10% range were observed in samples analyzed on different chips. The measurements were conducted by two full-time technicians, for a total period of a little over two years of labor time within the 18 calendar months of the project. Despite this important effort, we were unable to analyze all serum samples collected in the study with all the variants of interest due to cost, time limitations and rapidly shifting priorities from the epidemiological standpoint. For example, following its emergence about halfway through the longitudinal study, we integrated the Omicron BA.1 variant to our workflow from the analysis of the second visit onward (baseline point corresponding to the last visit before the emergence of the variant) at the expense of the Delta B.1.617.2 variant, which had rapidly lost its importance from an epidemiological standpoint. Thus, results presented here were mostly acquired with the native and Omicron BA.1 strains, with a partial data set acquired for the Delta B.1.617.2 variant.The throughput of different methods for the analysis of large cohorts of samples was compared in the project. Microneutralization assays and ELISA measurements were performed in multi-well plates and, in the case of the chemiluminescence ELISA, with a fully automated platform providing larger analytical capabilities than the SPR pseudo-neutralization. Thus, all samples were analyzed with microneutralization and ELISA. In addition, more antigenic proteins were analyzed (variant spike or nucleocapsid proteins) by ELISA during the same period. This is an important conclusion of the study: although SPR inhibition results correlated moderately well with established techniques including ELISA and correlated well with live virus microneutralization, its throughput in the POC or portable format does not rival automated platforms such as ELISA due to greater labor cost and lower throughput. Nonetheless, it compares favorably with live virus microneutralization because the read-out occurs in minutes and requires only a BSL2 lab which lowers the barrier for users without access to a BSL3 lab. As a consequence, POC SPR-type of platforms see their greatest advantage in studies where the number of samples is not large enough to run ELISA, or in the case of neutralization assays, where BSL-3 infrastructure is not available.
The project provided us the opportunity to evaluate the actual labor time and cost of running assays on a larger scale with POC-type of SPR technologies. As technical staff performed all SPR measurements, it provided a rare opportunity in the academic realm to accurately evaluate the cost of assays that could be expected at scale and in an industrial environment. Given the labor contract in place in our institution (35 h per week and an average of 47 work weeks per year), sample and SPR chip preparation time, SPR analysis and reporting required approximately 3300 hours, or about 1.6 hours per sample. The total cost for the SPR aspects of this project was $213![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 222 + overhead (total of $251
222 + overhead (total of $251![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 602 Canadian dollars, approximately $186
602 Canadian dollars, approximately $186![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 USD or €174
000 USD or €174![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500), solely dedicated to the salaries of the technical staff (75% of total) and consumables for the SPR assays (25% of total). We calculate that the total cost of analysis per sample at $126 Canadian dollars ($93 USD or €87) including cost of goods of about $31 Canadian dollars ($23 USD or €22) per test, of this about 80% being cost of proteins and reagents. This analysis excludes the cost of acquisition of two SPR instruments (circa $25
500), solely dedicated to the salaries of the technical staff (75% of total) and consumables for the SPR assays (25% of total). We calculate that the total cost of analysis per sample at $126 Canadian dollars ($93 USD or €87) including cost of goods of about $31 Canadian dollars ($23 USD or €22) per test, of this about 80% being cost of proteins and reagents. This analysis excludes the cost of acquisition of two SPR instruments (circa $25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 per instrument), costs related to participant recruitment and serum sample acquisition, and all costs related to ELISA ($36 Canadian dollars per sample, where three measurements at $12 each are necessary to evaluate the infection and vaccinal status of individuals) and microneutralization assays ($109 per sample). One can envision cost reduction by automating SPR chip preparation and sample analysis. This was highlighted in a recent paper, which reported the analysis of 115 serum samples for SARS-CoV-2 antibodies using an automated SPR platform. They showed that the cost could be significantly decreased to sub-dollars of cost of goods,41 further demonstrating the potential of SPR for clinical analysis. The cost of the instrument and chips must be reduced, pre-functionalized chip storage must be demonstrated, and regulatory approval must be obtained for point-of-care (POC) applications. However, this study demonstrates that SPR sensors provide reliable clinical data, and POC SPR instruments have the potential for clinical analysis.
000 per instrument), costs related to participant recruitment and serum sample acquisition, and all costs related to ELISA ($36 Canadian dollars per sample, where three measurements at $12 each are necessary to evaluate the infection and vaccinal status of individuals) and microneutralization assays ($109 per sample). One can envision cost reduction by automating SPR chip preparation and sample analysis. This was highlighted in a recent paper, which reported the analysis of 115 serum samples for SARS-CoV-2 antibodies using an automated SPR platform. They showed that the cost could be significantly decreased to sub-dollars of cost of goods,41 further demonstrating the potential of SPR for clinical analysis. The cost of the instrument and chips must be reduced, pre-functionalized chip storage must be demonstrated, and regulatory approval must be obtained for point-of-care (POC) applications. However, this study demonstrates that SPR sensors provide reliable clinical data, and POC SPR instruments have the potential for clinical analysis.
The main findings of this study are in agreement with published literature and comparisons made within this study are internally coherent. One of these main findings is that the %Inhibition peaks at around 2–3 weeks and wanes within 3–4 months post-vaccination (Fig. 5). This agrees with literature reports indicating that the levels of antibodies, including neutralizing antibodies, are maximal within 30 days and decrease thereafter.57,58 Hybrid immunity, a combination of immunity from vaccination and infection, has been reported to be more potent than vaccination or infection alone.56,59 We also found that hybrid immunity with 2 vaccine doses resulted in statistically higher SPR % immunity than twice vaccinated individuals naive to the virus (Fig. 4), also in agreement with literature.20,56,60,61
Cohort-specific data was validated against data from live-virus microneutralization and ELISA. The longitudinal trend observed in the %Inhibition SPR (Fig. 6) was similar to the ones observed for microneutralization and ELISA (Fig. S6†), where a rise in %Inhibition, microneutralization and the colorimetric ELISA OD450 was observed around July 2021. This period corresponded to the initial vaccination campaign when participants received their 1st and 2nd vaccine doses (Fig. 6) along with the general population,52 as clearly observed in the increased response for each technique. Waning antibody titers, %Inhibition SPR and microneutralization titers were all evident between July and December 2021, in agreement with other reports of post-vaccination waning and consistent with low infection rates in the region under study at that time.57,58 In January 2022, the Quebec government authorized an additional booster dose to all residents of the province,52 and a significant fraction of the cohort received it (Fig. 6). Coincidentally, the Omicron BA.1 wave had hit the province one month prior (December 2021) and infections increased rapidly (Fig. 6), observed from a spike in the antibody titer, %Inhibition SPR and microneutralization titer (Fig. 6 and S6†). As an important fraction of the cohort was vaccinated either three (>60%) or four times (>20%), or had reported an infection (117 of the 304 participants were declared positive to COVID from an antigen or PCR test, including 99 from December 2021 onward), or showed evidence of natural immunity (>50% of the cohort had eventually been declared positive for natural immunity by ELISA during the longitudinal study55), this surge in antigenic response was sustained until the end of the surveyed period. The data clearly demonstrate the ability of SPR sensing to capture changes in the immune response from individuals in the cohort. Taken together, the results presented here are a solid demonstration of the ability of portable SPR sensing to address clinical questions and to longitudinally study a cohort of individuals.
Conclusions
This study provides one of the largest data sets to validate the use of SPR sensors in clinical studies, and clearly demonstrate the utility of SPR sensors on a portable platform to provide valuable clinical information. The SPR serum inhibition data of the spike protein correlated well with a microneutralization assay and fully coincided with vaccination events and outbreaks of SARS-CoV-2 in the surveyed population. The results also agreed with established literature on COVID-19 regarding the benefit of hybrid immunity (higher %Inhibition SPR), the improvement of the %Inhibition in fully vaccinated individuals, waning response following vaccination and increased response for fully vaccinated individuals who received one or more booster shots of an RNA vaccine. Most importantly, this study provided the opportunity to reflect on the best uses of a portable SPR platform with a large cohort of clinical samples. The quality of the cohort-specific data and cross-correlation with microneutralization provided compelling evidence of the utility of portable SPR sensors to analyze clinical samples. Although the cost of goods was reasonable at $23 USD per test, we calculated that the labor cost far exceeded the cost of goods due to labor intensity of the workflow (total of 1.6 hours per sample). This cost analysis highlights the future opportunities to implement automated sample and reagents handling to reduce the workflow labor needs. Taken together, the results presented in this article constitute a significant step towards clinical translation of SPR sensors.Author contributions
Investigation and experiments were carried by JC, PR, AD, EL, HR, and MS. Methodology was established by YD, CG, KS, ST, DB, MAL, JNP, MB, and JFM. Formal analysis was done by NB, MT, MAL, MB, and JFM. Data curation was done by KS and MT. Funding was acquired by DB, ST, CG, JNP, MB and JFM. Writing of the original draft was done by JFM and review and editing was done by NB, JNP, DB, ST, MB, and JFM.Conflicts of interest
JFM and JNP have financial interest in Affinité Instruments.Acknowledgements
The authors thank Christian Gervais from the National Research Council Canada for assistance in some experiments. The authors acknowledge the financial support of the Public Health Agency of Canada through the Canadian Immunity Task Force, and of the Natural Science and Engineering Research Council of Canada.References
- N. Bhalla, Y. Pan, Z. Yang and A. F. Payam, Opportunities and Challenges for Biosensors and Nanoscale Analytical Tools for Pandemics: COVID-19, ACS Nano, 2020, 14(7), 7783–7807 CrossRef CAS PubMed.
- A. K. Kaushik, J. S. Dhau, H. Gohel, Y. K. Mishra, B. Kateb, N.-Y. Kim and D. Y. Goswami, Electrochemical SARS-CoV-2 Sensing at Point-of-Care and Artificial Intelligence for Intelligent COVID-19 Management, ACS Appl. Bio Mater., 2020, 3(11), 7306–7325 CrossRef CAS PubMed.
- Q. Song, X. Sun, Z. Dai, Y. Gao, X. Gong, B. Zhou, J. Wu and W. Wen, Point-of-care testing detection methods for COVID-19, Lab Chip, 2021, 21(9), 1634–1660 RSC.
- A. Sena-Torralba, R. Álvarez-Diduk, C. Parolo, A. Piper and A. Merkoçi, Toward Next Generation Lateral Flow Assays: Integration of Nanomaterials, Chem. Rev., 2022, 122(18), 14881–14910 CrossRef CAS PubMed.
- D. Li, C. Sun, X. Mei and L. Yang, Achieving broad availability of SARS-CoV-2 detections via smartphone-based analysis, TrAC, Trends Anal. Chem., 2023, 158, 116878 CrossRef CAS PubMed.
- G. Chen, S. Shen, T. Tat, X. Zhao, Y. Zhou, Y. Fang and J. Chen, Wearable respiratory sensors for COVID-19 monitoring, View, 2022, 3(5), 20220024 CrossRef PubMed.
- J. F. Masson, Surface Plasmon Resonance Clinical Biosensors for Medical Diagnostics, ACS Sens., 2017, 2(1), 16–30 CrossRef CAS PubMed.
- Z. Dai, X. Xu, Y. Wang, M. Li, K. Zhou, L. Zhang and Y. Tan, Surface plasmon resonance biosensor with laser heterodyne feedback for highly-sensitive and rapid detection of COVID-19 spike antigen, Biosens. Bioelectron., 2022, 206, 114163 CrossRef CAS PubMed.
- G. Qiu, Z. Gai, Y. Tao, J. Schmitt, G. A. Kullak-Ublick and J. Wang, Dual-Functional Plasmonic Photothermal Biosensors for Highly Accurate Severe Acute Respiratory Syndrome Coronavirus 2 Detection, ACS Nano, 2020, 14(5), 5268–5277 CrossRef CAS PubMed.
- Q. Wu, W. Wu, F. Chen and P. Ren, Highly sensitive and selective surface plasmon resonance biosensor for the detection of SARS-CoV-2 spike S1 protein, Analyst, 2022, 147(12), 2809–2818 RSC.
- R. Ahmed, C. F. Guimarães, J. Wang, F. Soto, A. H. Karim, Z. Zhang, R. L. Reis, D. Akin, R. Paulmurugan and U. Demirci, Large-Scale Functionalized Metasurface-Based SARS-CoV-2 Detection and Quantification, ACS Nano, 2022, 16(10), 15946–15958 CrossRef CAS PubMed.
- C. Han, T. Dong, P. Wang and F. Zhou, Microfluidically Partitioned Dual Channels for Accurate Background Subtraction in Cellular Binding Studies by Surface Plasmon Resonance Microscopy, Anal. Chem., 2022, 94(49), 17303–17311 CrossRef CAS PubMed.
- J.-H. Qu, K. Leirs, W. Maes, M. Imbrechts, N. Callewaert, K. Lagrou, N. Geukens, J. Lammertyn and D. Spasic, Innovative FO-SPR Label-free Strategy for Detecting Anti-RBD Antibodies in COVID-19 Patient Serum and Whole Blood, ACS Sens., 2022, 7(2), 477–487 CrossRef CAS PubMed.
- Y. Saad, M. H. Gazzah, K. Mougin, M. Selmi and H. Belmabrouk, Sensitive Detection of SARS-CoV-2 Using a Novel Plasmonic Fiber Optic Biosensor Design, Plasmonics, 2022, 17(4), 1489–1500 CrossRef CAS PubMed.
- E. M. Materón, F. R. Gómez, M. B. Almeida, F. M. Shimizu, A. Wong, K. B. R. Teodoro, F. S. R. Silva, M. J. A. Lima, M. K. S. C. Angelim, M. E. Melendez, N. Porras, P. M. Vieira, D. S. Correa, E. Carrilho, O. N. Oliveira Jr, R. B. Azevedo and D. Goncalves, Colorimetric Detection of SARS-CoV-2 Using Plasmonic Biosensors and Smartphones, ACS Appl. Mater. Interfaces, 2022, 14(49), 54527–54538 CrossRef PubMed.
- A. Djaileb, B. Charron, M. H. Jodaylami, V. Thibault, J. Coutu, K. Stevenson, S. Forest, L. S. Live, D. Boudreau, J. Pelletier and J. Masson, A Rapid and Quantitative Serum Test for SARS-CoV-2 Antibodies with Portable Surface Plasmon Resonance Sensing, ChemRxiv, 2020, preprint, DOI:10.26434/chemrxiv.12118914.v1.
- A. Djaileb, M. Hojjat Jodaylami, J. Coutu, P. Ricard, M. Lamarre, L. Rochet, S. Cellier-Goetghebeur, D. Macaulay, B. Charron, É. Lavallée, V. Thibault, K. Stevenson, S. Forest, L. S. Live, N. Abonnenc, A. Guedon, P. Quessy, J.-F. Lemay, O. Farnós, A. Kamen, M. Stuible, C. Gervais, Y. Durocher, F. Cholette, C. Mesa, J. Kim, M.-P. Cayer, M.-J. de Grandmont, D. Brouard, S. Trottier, D. Boudreau, J. N. Pelletier and J.-F. Masson, Cross-validation of ELISA and a portable surface plasmon resonance instrument for IgG antibody serology with SARS-CoV-2 positive individuals, Analyst, 2021, 146(15), 4905–4917 RSC.
- M. Hojjat Jodaylami, A. Djaïleb, P. Ricard, É. Lavallée, S. Cellier-Goetghebeur, M.-F. Parker, J. Coutu, M. Stuible, C. Gervais, Y. Durocher, F. Desautels, M.-P. Cayer, M. J. de Grandmont, S. Rochette, D. Brouard, S. Trottier, D. Boudreau, J. N. Pelletier and J.-F. Masson, Cross-reactivity of antibodies from non-hospitalized COVID-19 positive individuals against the native, B.1.351, B.1.617.2, and P.1 SARS-CoV-2 spike proteins, Sci. Rep., 2021, 11(1), 21601 CrossRef CAS PubMed.
- R. Funari, K.-Y. Chu and A. Q. Shen, Detection of antibodies against SARS-CoV-2 spike protein by gold nanospikes in an opto-microfluidic chip, Biosens. Bioelectron., 2020, 169, 112578 CrossRef CAS PubMed.
- L. Bellusci, G. Grubbs, F. T. Zahra, D. Forgacs, H. Golding, T. M. Ross and S. Khurana, Antibody affinity and cross-variant neutralization of SARS-CoV-2 Omicron BA.1, BA.2 and BA.3 following third mRNA vaccination, Nat. Commun., 2022, 13(1), 4617 CrossRef CAS PubMed.
- C. R. Basso, C. D. Malossi, A. Haisi, V. de Albuquerque Pedrosa, A. N. Barbosa, R. T. Grotto and J. P. Araujo Junior, Fast and reliable detection of SARS-CoV-2 antibodies based on surface plasmon resonance, Anal. Methods, 2021, 13(29), 3297–3306 RSC.
- T. Lewis, E. Giroux, M. Jovic and S. Martic-Milne, Localized surface plasmon resonance aptasensor for selective detection of SARS-CoV-2 S1 protein, Analyst, 2021, 146(23), 7207–7217 RSC.
- Y. Yang, J. Murray, J. Haverstick, R. A. Tripp and Y. Zhao, Silver nanotriangle array based LSPR sensor for rapid coronavirus detection, Sens. Actuators, B, 2022, 359, 131604 CrossRef CAS PubMed.
- R. B. M. Schasfoort, J. van Weperen, M. van Amsterdam, J. Parisot, J. Hendriks, M. Koerselman, M. Karperien, A. Mentink, M. Bennink, H. Krabbe, L. W. M. M. Terstappen and A. H. L. Mulder, Presence and strength of binding of IgM, IgG and IgA antibodies against SARS-CoV-2 during CoViD-19 infection, Biosens. Bioelectron., 2021, 183, 113165 CrossRef CAS PubMed.
- J. Hendriks, R. Schasfoort, M. Koerselman, M. Dannenberg, A. D. Cornet, A. Beishuizen, J. van der Palen, J. Krabbe, A. H. L. Mulder and M. Karperien, High Titers of Low Affinity Antibodies in COVID-19 Patients Are Associated With Disease Severity, Front. Immunol., 2022, 13, 867716 CrossRef CAS PubMed.
- A. G. Savitt, S. Manimala, T. White, M. Fandaros, W. Yin, H. Duan, X. Xu, B. V. Geisbrecht, D. A. Rubenstein, A. P. Kaplan, E. I. Peerschke and B. Ghebrehiwet, SARS-CoV-2 Exacerbates COVID-19 Pathology Through Activation of the Complement and Kinin Systems, Front. Immunol., 2021, 12, 767347 CrossRef CAS PubMed.
- F. Zhang, P. He, A. L. Rodrigues, W. Jeske, R. Tandon, J. T. Bates, M. A. Bierdeman, J. Fareed, J. Dordick and R. J. Linhardt, Potential Anti-SARS-CoV-2 Activity of Pentosan Polysulfate and Mucopolysaccharide Polysulfate, Pharmaceuticals, 2022, 15(2), 258 CrossRef CAS PubMed.
- T. Dong, C. Han, M. Jiang, T. Zhang, Q. Kang, P. Wang and F. Zhou, A Four-Channel Surface Plasmon Resonance Sensor Functionalized Online for Simultaneous Detections of Anti-SARS-CoV-2 Antibody, Free Viral Particles, and Neutralized Viral Particles, ACS Sens., 2022, 7(11), 3560–3570 CrossRef CAS PubMed.
- F. Abouhajar, R. Chaudhuri, S. N. Valiulis, D. D. Stuart, A. S. Malinick, M. Xue and Q. Cheng, Label-Free Analysis of Binding and Inhibition of SARS-Cov-19 Spike Proteins to ACE2 Receptor with ACE2-Derived Peptides by Surface Plasmon Resonance, ACS Appl. Bio Mater., 2023, 6(1), 182–190 CrossRef CAS PubMed.
- C. Liu, T. Puopolo, H. Li, A. Cai, N. P. Seeram and H. Ma, Identification of SARS-CoV-2 Main Protease Inhibitors from a Library of Minor Cannabinoids by Biochemical Inhibition Assay and Surface Plasmon Resonance Characterized Binding Affinity, Molecules, 2022, 27(18), 6127 CrossRef CAS PubMed.
- Z.-L. Zhu, X.-D. Qiu, S. Wu, Y.-T. Liu, T. Zhao, Z.-H. Sun, Z.-R. Li and G.-Z. Shan, Blocking Effect of Demethylzeylasteral on the Interaction between Human ACE2 Protein and SARS-CoV-2 RBD Protein Discovered Using SPR Technology, Molecules, 2021, 26(1), 57 CrossRef CAS PubMed.
- J. Mei, Y. Zhou, X. Yang, F. Zhang, X. Liu and B. Yu, Active components in Ephedra sinica stapf disrupt the interaction between ACE2 and SARS-CoV-2 RBD: Potent COVID-19 therapeutic agents, J. Ethnopharmacol., 2021, 278, 114303 CrossRef CAS PubMed.
- Y. Hou, S. Ge, X. Li, C. Wang, H. He and L. He, Testing of the inhibitory effects of loratadine and desloratadine on SARS-CoV-2 spike pseudotyped virus viropexis, Chem.-Biol. Interact., 2021, 338, 109420 CrossRef CAS PubMed.
- J. Lu, Y. Hou, S. Ge, X. Wang, J. Wang, T. Hu, Y. Lv, H. He and C. Wang, Screened antipsychotic drugs inhibit SARS-CoV-2 binding with ACE2 in vitro, Life Sci., 2021, 266, 118889 CrossRef CAS PubMed.
- R. Funari, H. Fukuyama and A. Q. Shen, Nanoplasmonic multiplex biosensing for COVID-19 vaccines, Biosens. Bioelectron., 2022, 208, 114193 CrossRef CAS PubMed.
- K. Danh, D. G. Karp, M. Singhal, A. Tankasala, D. Gebhart, F. de Jesus Cortez, D. Tandel, P. V. Robinson, D. Seftel, M. Stone, G. Simmons, A. Bagri, M. A. Schreiber, A. Buser, A. Holbro, M. Battegay, M. K. Morris, C. Hanson, J. R. Mills, D. Granger, E. S. Theel, J. R. Stubbs, L. M. Corash and C.-t. Tsai, Detection of neutralizing antibodies against multiple SARS-CoV-2 strains in dried blood spots using cell-free PCR, Nat. Commun., 2022, 13(1), 4212 CrossRef CAS PubMed.
- E. H. Y. Lau, O. T. Y. Tsang, D. S. C. Hui, M. Y. W. Kwan, W.-H. Chan, S. S. Chiu, R. L. W. Ko, K. H. Chan, S. M. S. Cheng, R. A. P. M. Perera, B. J. Cowling, L. L. M. Poon and M. Peiris, Neutralizing antibody titres in SARS-CoV-2 infections, Nat. Commun., 2021, 12(1), 63 CrossRef CAS PubMed.
- J. Nie, Q. Li, J. Wu, C. Zhao, H. Hao, H. Liu, L. Zhang, L. Nie, H. Qin, M. Wang, Q. Lu, X. Li, Q. Sun, J. Liu, C. Fan, W. Huang, M. Xu and Y. Wang, Establishment and validation of a pseudovirus neutralization assay for SARS-CoV-2, Emerging Microbes Infect., 2020, 9(1), 680–686 CrossRef CAS PubMed.
- C. W. Tan, W. N. Chia, X. Qin, P. Liu, M. I. C. Chen, C. Tiu, Z. Hu, V. C.-W. Chen, B. E. Young, W. R. Sia, Y.-J. Tan, R. Foo, Y. Yi, D. C. Lye, D. E. Anderson and L.-F. Wang, A SARS-CoV-2 surrogate virus neutralization test based on antibody-mediated blockage of ACE2–spike protein–protein interaction, Nat. Biotechnol., 2020, 38(9), 1073–1078 CrossRef CAS PubMed.
- J. J. Wang, N. Zhang, S. A. Richardson and J. V. Wu, Rapid lateral flow tests for the detection of SARS-CoV-2 neutralizing antibodies, Expert Rev. Mol. Diagn., 2021, 21(4), 363–370 CrossRef CAS PubMed.
- M. Jiang, T. Dong, C. Han, L. Liu, T. Zhang, Q. Kang, P. Wang and F. Zhou, Regenerable and high-throughput surface plasmon resonance assay for rapid screening of anti-SARS-CoV-2 antibody in serum samples, Anal. Chim. Acta, 2022, 1208, 339830 CrossRef CAS PubMed.
- O. Calvo-Lozano, M. Sierra, M. Soler, M. C. Estévez, L. Chiscano-Camón, A. Ruiz-Sanmartin, J. C. Ruiz-Rodriguez, R. Ferrer, J. J. González-López, J. Esperalba, C. Fernández-Naval, L. Bueno, R. López-Aladid, A. Torres, L. Fernández-Barat, S. Attoumani, R. Charrel, B. Coutard and L. M. Lechuga, Label-Free Plasmonic Biosensor for Rapid, Quantitative, and Highly Sensitive COVID-19 Serology: Implementation and Clinical Validation, Anal. Chem., 2022, 94(2), 975–984 CrossRef CAS PubMed.
- S. S. Zhao, N. Bukar, J. L. Toulouse, D. Pelechacz, R. Robitaille, J. N. Pelletier and J. F. Masson, Miniature multi-channel SPR instrument for methotrexate monitoring in clinical samples, Biosens. Bioelectron., 2015, 64, 664–670 CrossRef CAS PubMed.
- A. Djaïleb, É. Lavallée, M.-F. Parker, M.-P. Cayer, F. Desautels, M. J. de Grandmont, M. Stuible, C. Gervais, Y. Durocher, S. Trottier, D. Boudreau, J.-F. Masson, D. Brouard and J. N. Pelletier, Assessment of the longitudinal humoral response in non-hospitalized SARS-CoV-2-positive individuals at decentralized sites: Outcomes and concordance, Front. Immunol., 2023, 13, 1052424 CrossRef PubMed.
- K. Colwill, Y. Galipeau, M. Stuible, C. Gervais, C. Arnold, B. Rathod, K. T. Abe, J. H. Wang, A. Pasculescu, M. Maltseva, L. Rocheleau, M. Pelchat, M. Fazel-Zarandi, M. Iskilova, M. Barrios-Rodiles, L. Bennett, K. Yau, F. Cholette, C. Mesa, A. X. Li, A. Paterson, M. A. Hladunewich, P. J. Goodwin, J. L. Wrana, S. J. Drews, S. Mubareka, A. J. McGeer, J. Kim, M.-A. Langlois, A.-C. Gingras and Y. Durocher, A scalable serology solution for profiling humoral immune responses to SARS-CoV-2 infection and vaccination, Clin. Transl. Immunol., 2022, 11(3), e1380 CrossRef CAS PubMed.
- F. Cholette, R. Fabia, A. Harris, H. Ellis, K. Cachero, L. Schroeder, C. Mesa, P. Lacap, C. Arnold, Y. Galipeau, M.-A. Langlois, K. Colwill, A.-C. Gingras, A. McGeer, E. Giles, J. Day, C. Osiowy, Y. Durocher, C. Hankins, B. Mazer, M. Drebot and J. Kim, Comparative performance data for multiplex SARS-CoV-2 serological assays from a large panel of dried blood spot specimens, Heliyon, 2022, 8(9), e10270 CrossRef CAS PubMed.
- J. Pedersen, I. H. Koumakpayi, G. Babuadze, M. Baz, O. Ndiaye, O. Faye, C. T. Diagne, N. Dia, M. Naghibosadat, A. McGeer, S. Muberaka, I. P. Moukandja, S. Ndidi, C. B. Tauil, J.-B. Lekana-Douki, C. Loucoubar, O. Faye, A. Sall, K. G. Magalhães, N. Weis, R. Kozak, G. P. Kobinger and H. Fausther-Bovendo, Cross-reactive immunity against SARS-CoV-2 N protein in Central and West Africa precedes the COVID-19 pandemic, Sci. Rep., 2022, 12(1), 12962 CrossRef CAS PubMed.
- H. Rabezanahary, C. Gilbert, K. Santerre, M. Scarrone, M. Gilbert, M. Thériault, N. Brousseau, J.-F. Masson, J. N. Pelletier, D. Boudreau, S. Trottier and M. Baz, Live virus neutralizing antibodies against pre and post Omicron strains in food and retail workers in Québec, Canada, medRxiv, 2023, preprint, DOI:10.1101/2023.09.03.23294976.
- COVID-19 Immunity Task Force standardized core survey data elements, https://www.covid19immunitytaskforce.ca/covid-19-immunity-task-force-releases-standardized-core-survey-data-elements/ (accessed February 15th, 2023).
- Census profile, 2016 census, Québec City, https://www12.statcan.gc.ca/census-recensement/2016/dp-pd/prof/details/page.cfm?Lang=E&Geo1=CSD&Code1=2423027&Geo2=CD&Code2=2423&Data=Count&SearchText=quebec&SearchType=Begins&SearchPR=01&B1=All&TABID=1 (accessed February 16th, 2023).
- K. Santerre, M. Thériault, N. Brousseau, S. Rochette, J. N. Pelletier, C. Gilbert, J.-F. Masson, M. Baz, D. Boudreau and S. Trottier, Cohort Profile: Prospective Cohort to Study the COVID-19 Immune Response in Retail Workers in Québec, Canada (CISACOV), medRxiv, 2023, preprint, DOI:10.1101/2023.08.18.23294172.
- COVID-19 timeline in Quebec. https://www.inspq.qc.ca/covid-19/donnees/ligne-du-temps (accessed February 23rd, 2023).
- S. A. Andrade, J. V. Batalha-Carvalho, R. Curi, F. H. Wen, D. T. Covas, A. M. Chudzinski-Tavassi and A. M. Moro, Equine Anti-SARS-CoV-2 Serum (ECIG) Binds to Mutated RBDs and N Proteins of Variants of Concern and Inhibits the Binding of RBDs to ACE-2 Receptor, Front. Immunol., 2022, 13, 871874 CrossRef CAS PubMed.
- J. C. Xiong, Y. S. Xiang, Z. M. Huang, X. H. Liu, M. G. Wang, G. B. Ge, H. Z. Chen, J. R. Xu, M. Y. Zheng and L. L. Chen, Structure-Based Virtual Screening and Identification of Potential Inhibitors of SARS-CoV-2 S-RBD and ACE2 Interaction, Front. Chem., 2021, 9, 740702 CrossRef CAS PubMed.
- A. Djaïleb, M.-F. Parker, É. Lavallée, M. Stuible, C. Gervais, Y. Durocher, M. Thériault, D. Boudreau, M. Baz, J.-F. Masson, S. Trottier, D. Quaglia and J. N. Pelletier, SARS-CoV-2 Longitudinal Study on Incidence, Seroprevalence, and Immune Response in a Highly Vaccinated Population of Food and Retail Workers in the Québec City Region Through Ad-Hoc Calibration and Transformation of ELISA Datasets, MedRXiv, 2024, preprint, DOI:10.1101/2024.01.27.24301877.
- P. R. Wratil, M. Stern, A. Priller, A. Willmann, G. Almanzar, E. Vogel, M. Feuerherd, C. C. Cheng, S. Yazici, C. Christa, S. Jeske, G. Lupoli, T. Vogt, M. Albanese, E. Mejias-Perez, S. Bauernfried, N. Graf, H. Mijocevic, M. Vu, K. Tinnefeld, J. Wettengel, D. Hoffmann, M. Muenchhoff, C. Daechert, H. Mairhofer, S. Krebs, V. Fingerle, A. Graf, P. Steininger, H. Blum, V. Hornung, B. Liebl, K. Uberla, M. Prelog, P. Knolle, O. T. Keppler and U. Protzer, Three exposures to the spike protein of SARS-CoV-2 by either infection or vaccination elicit superior neutralizing immunity to all variants of concern, Nat. Med., 2022, 28(3), 496–503 CrossRef CAS PubMed.
- E. G. Levin, Y. Lustig, C. Cohen, R. Fluss, V. Indenbaum, S. Amit, R. Doolman, K. Asraf, E. Mendelson, A. Ziv, C. Rubin, L. Freedman, Y. Kreiss and G. Regev-Yochay, Waning Immune Humoral Response to BNT162b2 Covid-19 Vaccine over 6 Months, N. Engl. J. Med., 2021, 385(24), e84 CrossRef CAS PubMed.
- M. Gilboa, G. Regev-Yochay, M. Mandelboim, V. Indenbaum, K. Asraf, R. Fluss, S. Amit, E. Mendelson, R. Doolman, A. Afek, L. S. Freedman, Y. Kreiss and Y. Lustig, Durability of Immune Response After COVID-19 Booster Vaccination and Association With COVID-19 Omicron Infection, JAMA Netw. Open, 2022, 5(9), e2231778–e2231778 CrossRef PubMed.
- S. Crotty, Hybrid immunity, Science, 2021, 372(6549), 1392–1393 CrossRef CAS.
- T. A. Bates, S. K. McBride, H. C. Leier, G. Guzman, Z. L. Lyski, D. Schoen, B. Winders, J.-Y. Lee, D. X. Lee, W. B. Messer, M. E. Curlin and F. G. Tafesse, Vaccination before or after SARS-CoV-2 infection leads to robust humoral response and antibodies that effectively neutralize variants, Sci. Immunol., 2022, 7(68), eabn8014 CrossRef CAS PubMed.
- Y. Goldberg, M. Mandel, Y. M. Bar-On, O. Bodenheimer, L. S. Freedman, N. Ash, S. Alroy-Preis, A. Huppert and R. Milo, Protection and Waning of Natural and Hybrid Immunity to SARS-CoV-2, N. Engl. J. Med., 2022, 386(23), 2201–2212 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Figures reporting raw data point correlation plots, cohort stratified data and temporal evolution of ELISA and microneutralization response. See DOI: https://doi.org/10.1039/d3sd00333g |
| This journal is © The Royal Society of Chemistry 2024 |