Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Cu2+-integrated carbon dots as an efficient bioprobe for the selective sensing of guanine nucleobase†
Monalisa
Chowdhury
,
Debolina
Basu
and
Prasanta Kumar
Das
 *
*
School of Biological Sciences, Indian Association for the Cultivation of Science, Jadavpur, Kolkata – 700032, India. E-mail: bcpkd@iacs.res.in
First published on 9th July 2024
Abstract
This present work aimed to craft copper (Cu2+)-doped carbon dots (CuCDs) for the selective and sensitive detection of a guanine nucleobase. By employing a hydrothermal method, we synthesized blue-emitting CuCDs having emission maxima at 423 nm. CuCDs were used as a fluorescence turn-on ratiometric probe to detect guanine, a critical purine base in DNA involved in energy transduction, cell signalling, and metabolic processes. In the presence of guanine, the fluorescence intensity of CuCDs significantly increased due to the stable non-covalent interaction between Cu2+ and guanine. CuCDs achieved a very low limit of detection (LOD) of 0.59 nM for guanine as a highly sensitive probe. CuCDs demonstrated selectivity for guanine with no interference from other nucleobases (adenine, thymine, and cytosine) and various biomolecules and metal ions commonly found in the cellular environment. In addition, CuCDs demonstrated a higher affinity for guanine-enriched oligonucleotide cMYC G 27-mer over dsDNA 26-mer devoid of a large guanine population. Furthermore, the fluorescence intensity of CuCDs increased in guanine-treated mammalian cells and G-quadruplex-enriched cancer cells compared with that in non-cancerous cells. Hence, we developed a highly sensitive ratiometric fluorescence probe, CuCDs, for the selective detection of guanine both in vitro and within mammalian cells via a “fluorescence turn-on mechanism”.
Introduction
The human chromosome, a complex biopolymer, houses DNA that encodes the genetic blueprint crucial for life processes and protein biosynthesis.1–4 The nucleotides of DNA, i.e. guanine (G), adenine (A), thymine (T), and cytosine (C),5–7 facilitate the replication and transcription of genetic information. Guanine is vital for energy transduction, cell signalling, and metabolic cofactors.8–10 However, guanine is prone to oxidation, with its product 8-oxo-Gua serving as a biomarker for oxidative stress and DNA damage. Changes in guanine levels can lead to immune system deficiencies and diseases such as liver disease, AIDS, renal calculi, and cancer, highlighting the need for effective guanine detection methods.8,11–16 Techniques such as laser-induced fluorimetry,17 chromatography,18 capillary electrophoresis,19 high-performance liquid chromatography (HPLC),20,21 and mass spectrometry (MS)22 are used for guanine detection; however, they face challenges such as complex sample preparation and poor reproducibility.4,23,24 Fluorometric sensing is promising due to its simplicity, cost-effectiveness, and high sensitivity.25,26 Although various nanomaterials have been explored for this purpose (including metal nanoparticles, semiconductor-based quantum dots, and dye-doped materials),27–29 they have limitations such as toxicity and low photostability.30,31 Hence, there is an urgent demand for the development of simple, sensitive, and selective biosensors for guanine for both in vitro and in vivo applications.In the field of biosensing, carbon dots (CDs) have garnered considerable attention by representing a novel category of zero-dimensional carbon-based nanomaterials with diameters under 10 nm. CDs are recognized for their unique inherent fluorescence properties, cytocompatibility, facile synthesis, high photostability, water solubility, and chemical inertness.32–36 In carbon dots, surface defects play a crucial role in their optical properties. Upon photoexcitation, rapid charge separation occurs, resulting in the generation of electrons and holes. These charge carriers become trapped at various surface sites, leading to radiative recombination and the emission of light, which is a characteristic of carbon dots.37–39
In the last decade, researchers have been intrigued by nucleobase sensing, as DNA, the fundamental genetic material, comprises nucleobases (A, T, G, and C). Guanine, as discussed above, is of particular interest due to its implications under conditions such as cancer and AIDS. Researchers have used electrochemical and fluorescence methods, employing diverse tools such as quantum dots, polymers, conjugates, carbon nanotubes, and nanocapsules, to detect guanine. For example, intrinsic dual emitting ZnCdTe QDs were synthesized to selectively sense guanine having LOD 0.076 μmol L−1;23 TiO2-graphene nanocomposite was prepared by in situ hydrothermal treatment, where the electrochemical behavior of adenine and guanine at the TiO2-graphene nanocomposite modified glassy carbon electrode was investigated.40 The MgO nanoparticles (NPs) were prepared in a mechanochemical manner, and magnesium oxide and multi-walled carbon nanotubes modified carbon paste electrode (MgO-MWCNTs-MCPE) were utilized for the specific and simultaneous investigation of guanine, adenine and epinephrine using an electrochemical method.10 Surface assembly of poly(9-(2-diallylaminoethyl)adenineHCl-co-sulfurdioxide) (polyA) on silica nanoparticles form nanocapsules, which selectively sensed guanine over other nucleobases using the conjugation with curcumin.41 However, each method has limitations, such as low selectivity, the toxicity of metal-based bioprobes, and short sensor lifespans.
Considering these attributes, herein, we aim to develop copper (Cu2+)-doped carbon dots (CuCDs) for the selective and sensitive detection of guanine nucleobase. Previous reports suggest that the specific arrangement of oxygen at C6 and N7 in guanine facilitates optimal interaction with Cu2+ ions rather than other nucleobases. Considering this fact, we harnessed the blue-emission (emission maxima at 423 nm) of CuCDs prepared using the hydrothermal method. The CuCD was employed as a fluorescent turn-on bioprobe for the simple, sensitive, and selective detection of guanine. The fluorescence intensity of CuCDs significantly increased in the presence of guanine, which is attributed to the stable interaction between Cu2+ and guanine. CuCDs serve as a highly sensitive fluorescence probe for guanine with a limit of detection (LOD) of 0.59 nM. Importantly, guanine sensing by CuCDs demonstrates selectivity in solution against other nucleobases, i.e. adenine, thymine, cytosine and various biomolecules and metal ions abundant in the cellular environment with no interference. Notably, the fluorescence intensity of cytocompatible (up to 85%) CuCDs increased in guanine-enriched (B16F0, MCF-7) and guanine-treated mammalian cells (NIH3T3) compared to untreated NIH3T3 cells. Thus, we developed a highly sensitive ratiometric fluorescent probe, CuCD, for the selective detection of guanine through a “fluorescence turn-on mechanism”.
Experimental section
Materials
Ethylenediaminetetraacetic acid disodium salt (EDTA-2Na, 2H2O; purity ≥ 98.0%), copper chloride dihydrate (CuCl2·2H2O; purity ≥ 98.5%), adenine (A), guanine (G), cytosine (C), thymine (T) (all nucleobase having purity ≥ 98.0%) NaCl (purity ≥ 99.0%), KCl (purity ≥99.5%), CaCl2·2H2O (purity ≥ 98.0%), FeCl2·2H2O (purity ≥ 98.0%), ZnCl2·2H2O (purity ≥ 95.0%), L-cysteine (purity ≥ 99.0%), L-tryptophan (purity ≥ 99.0%), L-tyrosine (purity ≥ 99.0%), L-arginine (purity ≥ 99.0%), L-aspartic acid (purity ≥ 99.0%), GSH (purity ≥ 99.5%), H2O2 (purity ≥ 98.0%) and all other reagents and solvents were bought from SRL, India. cMYC G 27-mer (DTGGGGAGGGTGGGGAGGGTGGGGAAGG) and dsDNA 26-mer (DCAATCGGATCGAATTCGATCCGATTG) DNA sequences, MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide), and other deuterated solvents were purchased from Sigma-Aldrich. Dialysis tubing was procured from Thermo Scientific SnakeSkin (3.5 K MWCO). Dulbecco's modified Eagle's medium (DMEM), fetal bovine serum (FBS), and trypsin-ethylenediaminetetraacetic acid (EDTA) solution 1× (0.25% solution) were bought from Himedia. NIH3T3, B16F10 and MCF-7 cells were received from NCCS, Pune, India. All experiments were carried out using Milli-Q water. Perkin Elmer Spectrum 100 was used to record FTIR spectra. Centrifugation was performed using a Thermo Scientific Espresso centrifuge. Nano-ZS of Malvern Instruments Limited was used to measure zeta potential. A Telsonic bath sonicator was used to perform bath sonication.Synthesis of Cu2+-doped carbon dot (CuCD)
A carbon dot doped with Cu2+ (CuCD) was prepared using the hydrothermal synthesis approach. Initially, 10 mmol of ethylenediaminetetraacetic acid disodium salt (EDTA-2Na, 2H2O) and 10 mmol of dihydrated copper chloride (CuCl2·2H2O) were dissolved in 10 mL of Milli-Q water. The solution was stirred for 1 h, and the undissolved particles in the mixture were removed through filtration. The remaining solution underwent heating at 190 °C for 5 h in an autoclave Teflon chamber. The mixture was then allowed to cool to room temperature and centrifuged at 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 10 min. The obtained product was subjected to drying using a freeze dryer, affording CuCD powder with an estimated yield of around 80%. For the control experiments, we prepared another two carbon dots by taking 1
000 rpm for 10 min. The obtained product was subjected to drying using a freeze dryer, affording CuCD powder with an estimated yield of around 80%. For the control experiments, we prepared another two carbon dots by taking 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 molar ratio and 2
2 molar ratio and 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 molar ratio of EDTA-2Na, 2H2O, and CuCl2·2H2O using identical experimental conditions.
1 molar ratio of EDTA-2Na, 2H2O, and CuCl2·2H2O using identical experimental conditions.
Characterization
The aqueous solution of CuCDs was sonicated for a few minutes, followed by drop casting onto a grid coated with copper and drying. This was then used for examination in an EFGTEM (Transmission electron microscope, 2100F UHR microscope JEOL JEM). For the CuCD-guanine complex, first, the CuCD was sonicated for few minutes. Then, guanine was added to the CuCD for complex formation. Finally, the mixture was drop cast onto a grid and dried overnight before imaging. For atomic force microscopic (AFM) imaging, a drop of CuCD solution was air-dried on a fresh mica surface overnight and observed in an Asylum Research MFP-3D microscope in noncontact mode. In the case of X-ray photoelectron spectroscopy (XPS) (Omicron series 0571), rectangular glass plates were utilized to air-dry 8 μL of CuCD overnight. EDX (Energy-dispersive X-ray) analysis was performed using an Oxford EXTREME INCA microscope. For XRD (X-ray diffraction), the aqueous CuCD solutions (1 mg mL−1) were separately deposited over a glass slide and dried to prepare a thin film, and the spectra were acquired using a diffractometer (Bruker D8 Advance). The mean hydrodynamic diameter (Dh) of the CuCD was measured by dynamic light scattering (DLS) (Malvern Zetasizer S90 series) by dissolving 1 mg of lyophilized samples in 1 mL of deionized water. Zeta (ζ) potential measurement was conducted at room temperature using aqueous solutions of the CuCD. The UV-visible spectrum of the CuCD was measured using Agilent Cary 60, and an Agilent Cary Eclipse luminescence spectrometer was used to record the fluorescence spectra.Quantum yield (QY) measurement
The absorbance of the carbon dot solution was constrained to less than 0.1 to minimize the inner filter effect, and the integrated emission intensities of these solutions were measured using a luminescence spectrometer. The relative quantum yield (QY) was determined using the following equation: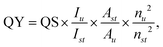 | (1) |
Fluorescence spectroscopy
Fluorescence spectra were recorded for 500 μg mL−1 of the CuCD by varying the excitation wavelength (λex) from 310 to 380 nm. Variation in the fluorescence of the CuCD (250 μg mL−1) was investigated in the presence of different concentrations of guanine nucleobase (0–20 nM). All sensing experiments were conducted in Milli-Q water. The Stern–Volmer constant (KSV) for the fluorescence enhancement of the CuCD with respect to the guanine concentration was determined using the equation F/F0 = 1 + KSV[Q], where F and F0 represent the fluorescence intensities of the CuCD in the presence and absence of guanine, respectively; and [Q] denotes the guanine concentration. The guanine detection limit was assessed from the linear curve obtained from (F − F0)/F0versus the guanine concentration. Fluorescence intensity was measured at 423 nm for the CuCD upon excitation at 350 nm.In the selectivity study, the fluorescence of the CuCD was investigated in the presence of A, T, and C (500 nM); ions, such as Na+, K+, Ca2+, Fe2+, and Zn2+; and amino acids L-cys, L-tryp, L-tyr, L-arg, L-asp, GSH and H2O2, each with a concentration of 500 nM. In addition, an interference study was performed in which the selective sensing of the guanine (20 nM) nucleobase by the CuCD (250 μg mL−1) was studied in the presence of A, T, and C (20 nM). Additionally, the fluorescence response of the CuCD (250 μg mL−1) was further examined in the presence of a guanine-enriched oligonucleotide sequence cMYC G 27mer (0.1–1.0 nM) (DTGGGGAGGGTGGGGAGGGTGGGGAAGG). The selectivity of the CuCD fluorescence probe in the presence of guanine-enriched sequence was verified through a selectivity study using the guanine-enriched oligonucleotide (cMYC G 27mer) (500 pM or 0.5 nM) against a control oligonucleotide (dsDNA 26mer) (DCAATCGGATCGAATTCGATCCGATTG) (1.0 nM) with limited presence of guanine nucleobase.
FTIR study
FTIR (Fourier-transform infrared) measurements were conducted for the CuCD and CuCD in conjunction with a guanine-enriched oligonucleotide (cMYC G 27mer) in D2O. Potassium bromide (KBr) was employed to blend with the samples, and a hydraulic press was utilized to form granules. The spectral range extended from 400 to 4000 cm−1, and the spectra underwent normalization and scaling. The experiments were carried out using a Perkin Elmer Spectrum 100 FTIR spectrometer equipped with a 1 mm CaF2 cell.Media stability of CuCD
The stability of 500 μg mL−1 aqueous solutions of the CuCD in biological media was assessed using DMEM media with FBS, with concentrations ranging from 0% to 75%. Additionally, the CuCD (500 μg mL−1) was exposed to a DMEM with FBS (10%) to examine its stability over a period of 10 days. Supernatants were collected at various time moments, and the absorbance at 365 nm was measured to calculate the suspension stability index (SSI)| SSI = At/A0 × 100, | (2) |
Cell culture
NIH3T3, B16F10 and MCF-7 cells were cultured in a medium consisting of DMEM supplemented with 10% FBS and antibiotics (streptomycin and penicillin). The cell cultures were maintained in an incubator at 37 °C with 5% CO2. After incubation, trypsinization was carried out every 3 days when the cell confluence reached 80%. The trypsinized cells were then utilized for subsequent studies.Cell viability
The cell viability of the CuCD in both cancer and non-cancer cells was assessed using the MTT assay. B16F10, MCF-7 and NIH3T3 cells were separately cultivated in a 96-well plate for 24 h in filtered media. Subsequently, the CuCD (25–200 μg mL−1) was incubated for 12 h in the cells (under 5% CO2 at 37 °C). Following the incubation period, the MTT dye was added and incubated for 4 h. In the MTT assay method, mitochondrial dehydrogenase of living cells generated hydrophobic formazan from the tetrazolium salt. The absorbance of formazan at 570 nm was then measured using a Biotek Elisa Reader, and this absorbance corresponded with the number of viable cells| The number of alive cells was expressed as percent viability = (A570(treated cells) − background)/A570(untreated cells) − background) × 100. | (3) |
Bioimaging
Non-cancerous NIH3T3 cells and cancer B16F10 and MCF-7 cell lines were cultured (1 × 104 cells per well) in a chamber slide. Initially, guanine (500 nM) was added separately to the chamber slides of NIH3T3 cells for 12 h incubation. Subsequently, the guanine-treated NIH3T3, untreated NIH3T3, and G-quadruplexes (G4s) enriched B16F10, MCF-7 cell lines were washed three times with PBS buffer (pH = 7.4). Next, the CuCD (200 μg mL−1) was separately introduced to the chamber slides of all types of cell lines, encompassing control wells and those previously incubated with guanine. For fixation, 4% paraformaldehyde solution was applied for 30 min and mounting was performed using 50% glycerol. The covered cells on the slide were left for 1 day. Imaging was conducted utilizing an Olympus IX83 inverted microscope, featuring an excitation filter within the wavelength range of BP 330–385 nm and a band absorbance filter positioned below 405 nm. The observations were recorded at a magnification of 20×.Flow cytometry
Non-cancer cells (NIH3T3) and cancer cells (B16F10, MCF-7) were cultured for 48 h in a 35 mm Petri dish. Subsequently, guanine (500 nM) was incubated with NIH3T3 cells for 12 h. The cells were then washed with PBS to eliminate excess compounds from the cell medium. Following the washing, the CuCD (200 μg mL−1) was incubated in the guanine-treated cell lines for an additional 12 h. Concurrently, the CuCD (200 μg mL−1) was also incubated in guanine non-treated NIH3T3, B16F10 and MCF-7 cell lines for 12 h. The cells incubated solely with the CuCD and those incubated with both guanine and CuCD were detached from the 35 mm Petri dish using trypsin, followed by centrifugation for 5 min. Consequently, the collected cells were resuspended in 500 μL of PBS, and a cytometric analysis was performed. A BD FACS Aria III flow cytometer was utilized to analyze the cells, employing an excitation wavelength of 405 nm.Results and discussion
Carbon dots (CDs) exhibit notable advantages in biosensing due to their distinctive physicochemical properties, including stable photoluminescence (PL), non-toxicity, and chemical inertness. Their remarkable feature lies in converting abstract information related to target analytes into discernible optical signals, such as changes in fluorescence intensity, wavelength, or color, thereby establishing significance in biosensing.42 Interestingly, the PL property of CDs can be tuned by selecting appropriate carbon precursors,43 adjusting synthesis conditions,44 and introducing heteroatom doping.45 Doping introduces novel chemical reactivities and potential applications for CDs. Metal atom doping is particularly noteworthy due to its different band structures that make CDs potent nanoprobes for the desired molecule sensing.46,47 Copper (Cu2+) complexes, along with other transition metals, such as Fe2+, Co2+, and Ni2+, have garnered significant interest owing to their biocompatible properties and oxidative characteristics.48 The unique features of Cu2+ make it particularly intriguing in both biological applications and industrial processes. There are extensive reports on various Cu2+ complexes, showcasing various mixed ligand donors or Schiff Base donors.49 Notably, Cu2+ forms strong binding with the guanine nucleobase of DNA.50,51 Various models describing the binding of Cu2+ to DNA have been proposed in the literature.50,51 One such model suggests a charge transfer type complex, where two adjacent guanine molecules in the same strand act as donors and Cu2+ acts as the acceptor. In this configuration, the ion is intercalated between the two G-C pairs. The primary binding sites of Cu2+ on the DNA's guanine base are the N7 and O6 of guanine.50,51 Another model proposes a chelation scenario, where there is a potential interaction between the accessible N7 of guanine and the nearest phosphate group of the same DNA strand, and so forth.50,51 These proposed binding models depict the affinity of Cu2+ with guanine. Guanine, which is a crucial component of DNA, plays a pivotal role in various biological processes, such as mutation, tumorigenesis, and cancer. Therefore, any alteration in its concentration can induce mutations, tumorigenesis and cell death. Hence, the accurate detection of guanine in biological samples is crucial for the early identification of disease indicators and for monitoring the cellular environment.52 Many researchers have attempted to sense guanine mostly using the electrochemical method4 and fluorescence method,53 quantum dot,23 conjugate systems,53 ionic liquids,54 nanosheets,55 and polymers using transition metals,56,57 carbon nanotubes,58etc. (Table 1). Each method has its advantages and limitations, such as low selectivity, toxicity of components, and short or limited life for electrochemical sensors.| Sl no. | Probes | LOD | Response type | References |
|---|---|---|---|---|
| 1 | MoS2-pencil graphite electrode4 | 0.76 mM | Electrochemical sensor | N. Vishnu et al.4 |
| 2 | Self-assembled monolayer (SAM) of 1,8,15,22-tetraaminophthalocyanatonickel(II)8 | 3 × 10−8 M | Electrochemical sensor | A. J. Jeevagan et al.8 |
| 3 | Magnesium oxide and multi-walled carbon nanotubes (MgO-MWCNTs-MCPE)10 | 0.92 mM | Electrochemical sensor | K. Chetankumar et al.10 |
| 4 | Dual-emitting ZnCdTe quantum dots23 | 0.076 μmol L−1 | Fluorescent sensing | X. Xua et al.23 |
| 5 | TiO2-graphene nanocomposite40 | 0.15 mM | Electrochemical sensor | Y. Fan et al.40 |
| 6 | Curcumin conjugated nanocapsules using silica nanoparticles41 | 90 μM | Fluorescent sensing | M. Mouslmani et al.41 |
| 7 | Carbon nanodot and Cu2+ conjugate53 | 0.67 × 10−8 mol L−1 | Fluorescence turn off- turn on | S. Pang et al.53 |
| 8 | An imidazolium-based ionic liquid54 | 45 nM | Electrochemical sensing by cyclic voltammetry | A. Singh et al.54 |
| 9 | Boron-doped graphene (B–G) nanosheets55 | 3.9 × 10−7 mol L−1 | Electrochemical sensor | J. Borowiec et al.55 |
| 10 | AgNCs-NFR + Cu2+ complex56 | 1.85 μM | Fluorescent sensing | Y. Li et al.56 |
| 11 | Polymer nanoparticles (LCPNPs) composed of terbiumions (Tb3+) and citrate(Cit)57 | 100 nM | Fluorescent sensing | D. R. Kumar et al.57 |
| 12 | Oleylamine-capped CuO nanopar-ticles on MWCNTs58 | 0.084 μM | Fluorescent sensing | L. Fu et al.58 |
| 13 | Cu 2+ doped carbon dot (CuCD) | 0.59 nM | Fluorescence turn on | Present study |
Motivated by these considerations, we endeavored to develop a selective, sensitive, and easily synthesizable nanoprobe for guanine sensing. Our approach involved synthesizing a Cu2+-doped carbon dot (CuCD) exhibiting blue emission under UV light irradiation.59–61 Cu2+ was included as an integral component of the synthesized carbon dot to make it a selective sensor for guanine (Scheme 1).
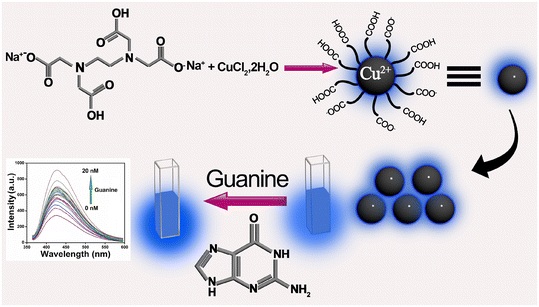 | ||
| Scheme 1 Schematic representation of guanine sensing by utilizing the intrinsic fluorescence property of Cu2+-doped carbon dot (CuCD). | ||
Synthesis and characterization of Cu2+-doped carbon dot (CuCD)
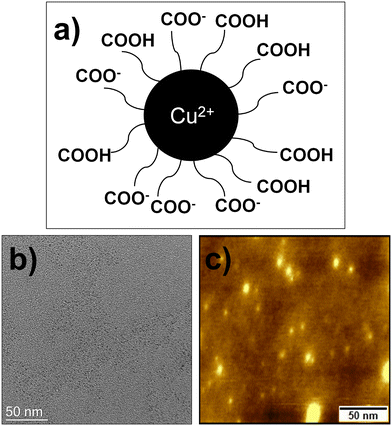
Cu2+-integrated carbon dot (CuCD) preparation involved using the hydrothermal method. The hydrothermal method is a simple bottom-up approach for synthesizing carbon dots. In general, the water solution of the mixtures was enclosed in a Teflon container in an oven and hydrothermally reacted at high pressure and high temperature. The decomposition and polymerization strongly influenced carbon dots derived by the hydrothermal treatment of precursors. Precursors are decomposed into smaller organic compounds, and these organic compounds are then further polymerized into larger molecules, which finally lead to the formation of the carbon dots in the hydrothermal reactor. Herein, an equivalent amount of EDTA-2Na, 2H2O, and CuCl2·2H2O was used to prepare the Cu2+-integrated carbon dot (CuCD). EDTA-2Na, 2H2O was used as a source of carbon core and carboxylic acid surface functionalization (Scheme S1, (ESI†)) while Cu2+ as the doping element (Fig. 1a). We utilized an aqueous saturated solution of the precursors and enclosed it with a Teflon container in an oven with a temperature of 190 °C for 5 h. For control experiments, we prepared another two carbon dots by taking a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 molar ratio and a 2
2 molar ratio and a 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 molar ratio of EDTA-2Na, 2H2O, and CuCl2·2H2O (Schemes S2 and S3, ESI†) using identical experimental conditions. The fluorescence intensity for these two carbon dots was significantly low (Quantum yield (QY) ∼1–2%). However, the QY of the carbon dot prepared from a 1
1 molar ratio of EDTA-2Na, 2H2O, and CuCl2·2H2O (Schemes S2 and S3, ESI†) using identical experimental conditions. The fluorescence intensity for these two carbon dots was significantly low (Quantum yield (QY) ∼1–2%). However, the QY of the carbon dot prepared from a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 molar ratio of EDTA-2Na, 2H2O, and CuCl2·2H2O was observed at ∼5%. Thus, we chose the CuCD, which was synthesized by an equivalent amount of both precursors because it was more suitable for sensing purposes due to its higher QY. The synthesized CuCD was found to be soluble in water, and its corresponding zeta potential (ζ) value was −23.3 mV, suggesting considerable stability in an aqueous medium. Microscopic characterization of the CuCD was conducted using TEM and AFM. The TEM images affirmed the size of CuCD to be ∼5 nm (Fig. 1b). AFM images (Fig. 1c) also indicated that the size of CuCD was between 2 and 5 nm, which was similar to the result obtained from the TEM image. In addition, another two FEGTEM high-resolution images of the CuCD were captured, where uniformly spherical nanoparticles with diameters of around 5 nm were observed (Fig. S1, ESI†).
1 molar ratio of EDTA-2Na, 2H2O, and CuCl2·2H2O was observed at ∼5%. Thus, we chose the CuCD, which was synthesized by an equivalent amount of both precursors because it was more suitable for sensing purposes due to its higher QY. The synthesized CuCD was found to be soluble in water, and its corresponding zeta potential (ζ) value was −23.3 mV, suggesting considerable stability in an aqueous medium. Microscopic characterization of the CuCD was conducted using TEM and AFM. The TEM images affirmed the size of CuCD to be ∼5 nm (Fig. 1b). AFM images (Fig. 1c) also indicated that the size of CuCD was between 2 and 5 nm, which was similar to the result obtained from the TEM image. In addition, another two FEGTEM high-resolution images of the CuCD were captured, where uniformly spherical nanoparticles with diameters of around 5 nm were observed (Fig. S1, ESI†).
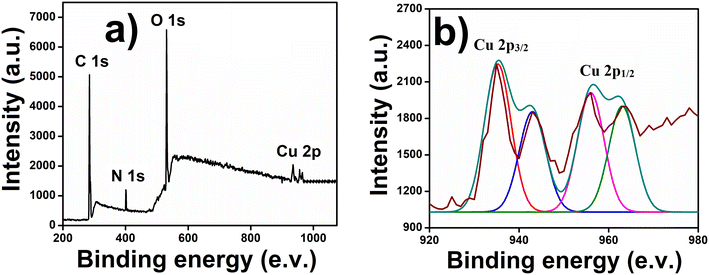
In accordance with the microscopic observations, the hydrodynamic diameter of the synthesized CuCDs was determined by the DLS study in the solution phase and was found to be around 7–9 nm (Fig. S2, ESI†). The size of CuCD was observed to be slightly higher in the DLS study because the nanoparticles were present in the hydrated state, resulting in a hydrodynamic diameter typically larger than the particle diameter determined by TEM and AFM in the dried state. In accordance with the XPS analysis of CuCD, peaks at 284, 406, and 529 eV confirmed the presence of C (1s), N (1s), and O (1s) orbitals, respectively (Fig. 2a).
Along with these peaks, a peak with a doublet character was observed in the region of 935–963 eV, corresponding to the presence of Cu in the moiety. The deconvoluted spectra of Cu2+ showed two typical peaks at 935.24 eV and 956.02 eV with satellite peaks (943.13 eV and 962.98 eV, respectively) that were associated with Cu 2p3/2 and Cu 2p1/2 electronic configurations, respectively, indicating the existence of copper as Cu2+ within the CuCD (Fig. 2b).62 We also deconvoluted spectra of C 1s, N 1s and O 1s. A high-resolution XPS spectrum of C 1s can be deconvoluted into four peaks at 283.7, 285.1, 287, and 288.6 eV, which provides proof of the presence of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C, C–C, C–N/C–O, and HO–C
C, C–C, C–N/C–O, and HO–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O bonds, respectively (Fig. S3a, ESI†). The deconvoluted high-resolution O 1s spectrum of CuCD shows three peaks at 530.2, 532.2, and 534.8 eV, which can be attributed to the C
O bonds, respectively (Fig. S3a, ESI†). The deconvoluted high-resolution O 1s spectrum of CuCD shows three peaks at 530.2, 532.2, and 534.8 eV, which can be attributed to the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, C–OH/C–O–C, and O
O, C–OH/C–O–C, and O![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–OH groups, respectively (Fig. S3b, ESI†). Two deconvoluted peaks at 399.1 and 401.3 eV in the N 1s level of CuCD can be assigned to the C–N–C and N–(C3) bonds, respectively (Fig. S3c, ESI†). The atomic percentages of C1s, N1s, O1s and Cu were 24.02%, 8.92%, 60.8%, and 6.1%, respectively. The XRD analysis was carried out for the characterization of CuCD. The presence of peaks in the range of 20–25° confirmed the amorphous nature of CuCD, and we also obtained a peak at 49.7°, which confirmed the presence of Cu in the CuCD (ref. 63) (Fig. S4a, ESI†). Further, the EDX analysis of CuCD corroborated the analysis of XPS that Cu was present in the CuCD along with other elements (C, O, and N) (Fig. S4b, ESI†).
C–OH groups, respectively (Fig. S3b, ESI†). Two deconvoluted peaks at 399.1 and 401.3 eV in the N 1s level of CuCD can be assigned to the C–N–C and N–(C3) bonds, respectively (Fig. S3c, ESI†). The atomic percentages of C1s, N1s, O1s and Cu were 24.02%, 8.92%, 60.8%, and 6.1%, respectively. The XRD analysis was carried out for the characterization of CuCD. The presence of peaks in the range of 20–25° confirmed the amorphous nature of CuCD, and we also obtained a peak at 49.7°, which confirmed the presence of Cu in the CuCD (ref. 63) (Fig. S4a, ESI†). Further, the EDX analysis of CuCD corroborated the analysis of XPS that Cu was present in the CuCD along with other elements (C, O, and N) (Fig. S4b, ESI†).
UV-vis and fluorescence spectroscopy of CuCD
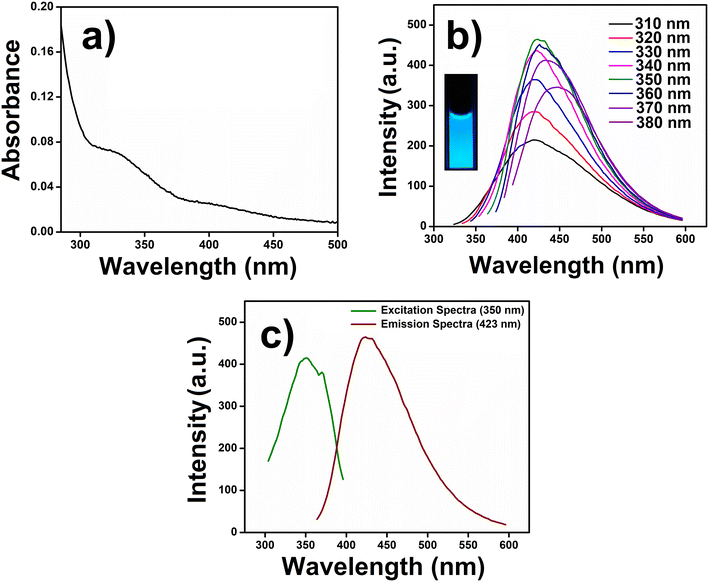
The UV-visible spectrum of CuCD aqueous solution showed one prominent feature centered at 334 nm (Fig. 3a) due to n → π* transitions attributed to the oxygen containing functional groups. The optical band gap of the CuCDs was determined using a Tauc plot derived from the UV-vis spectrum.64 The optical band gap, Eg, and absorption coefficient, α, are related through the Tauc equation (αhυ)1/n = B(hυ − Eg), where hυ is the photon energy, B is a proportionality constant, n is an exponent that is equal to 1/2 for direct transitions, and Plank's constant (h) = 6.626 × 10−34 Joule s and velocity of light (c) = 2.99 × 108 meter per s. The Tauc plot in Fig. S5† shows the relation between (αhυ)1/2versus hυ from which the optical band gap of the CuCDs was found to be ∼4.22 eV (Fig. S5, ESI†). Such band gap energy illustrates the quantum confinement effect in CuCDs.In the fluorescence study, the CuCD showed an excitation-dependent emission. We varied the excitation wavelength from 310 nm to 380 nm and observed a red shift (∼29 nm) from 419.7 nm to 448.7 nm in the emission spectra. The CuCD showed emission maxima at 423 nm (λmax) upon excitation at 350 nm (Fig. 3b). The aqueous solution of CuCD showed bright blue fluorescence (inset, Fig. 3b), which remained consistent with the emission behavior of CuCD under UV light irradiation at 365 nm. The excitation spectrum (350 nm) of CuCD for λmax (423 nm) was plotted along with the emission spectrum (423 nm), as shown in Fig. 3c. The QY of CuCD was observed to be around 5% with respect to quinine sulfate. Thus, the intrinsic fluorescence properties of CuCD may be explored for the guanine.
Guanine sensing by CuCD
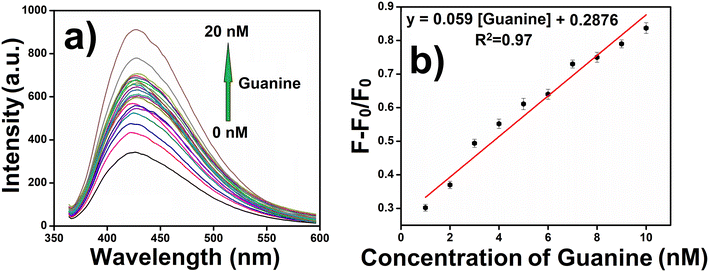
After the successful preparation and photophysical characterization of CuCD, we intend to explore its emissive properties in sensing one of the nucleobases of DNA, guanine. Guanine is one of the two essential purine bases of DNA, which plays a major role in coronary blood flow regulation, energy transduction, and cell proliferation; the modulation of its concentration leads to mutation and multiple diseases.10 For the past decade, nucleobase sensing has been an interesting topic among researchers as DNA, and the key to the genetic materials of all beings was created by these nucleobases (A, T, G, and C). Sensing and investigation of these nucleobases will open a gateway for new domains regarding the diagnosis and treatment of various dreadful diseases. Among all nucleobases, guanine is particularly susceptible to oxidation because it is responsible for many diseases, such as cancer and AIDS. Many researchers have attempted to sense guanine primarily using electrochemical and fluorescence methods. Various materials and techniques have been employed, including quantum dots, conjugate systems, ionic liquids, nanosheets, polymers using transition metals, and carbon nanotubes (Table 1). Each method has unique advantages and limitations. For example, electrochemical sensors often suffer from low selectivity, component toxicity, and limited lifespan. However, the CuCD has cost-effective synthesis, low toxicity, high stability and high sensitivity towards guanine. Hence, our objective is to use the intrinsic fluorescence property of CuCD for the selective sensing of guanine.In this regard, the intensity of the fluorescence spectra of CuCD (250 μg mL−1) at 423 nm was recorded in the presence of varying concentrations of guanine (0–20 nM). Interestingly, as the concentration of guanine gradually increased, there was a corresponding increase in the emission intensity of CuCD (Fig. 4a).
Because the CuCD exhibited a sensitive response to guanine, we determined the detection limit of guanine. The emission intensity of CuCD (250 μg mL−1) at 423 nm showed a steady increase with the concentration of guanine in the range of 1–20 nM (Fig. 4a). Sensitivity was assessed by examining the relationships between the ratios of the emission intensities of CuCD at different guanine concentrations. The fluorescence response curve demonstrated a well-defined linear behavior, as depicted in the plot. (F − F0)/F0, where F0 is the emission intensity of the native CuCD and F is the emission intensity of CuCD in the presence of guanine against the concentration of guanine demonstrated a linear relationship in the concentration range of 1–10 nM, indicating efficient guanine sensing at low concentrations (Fig. 4b). The detection limit of guanine was found to be 0.59 nM for the CuCD as determined based on 3σ/S, where σ is the standard deviation and S is the slope of the calibration curve. The linear regression equation was (F − F0)/F0 = 0.059x + 0.2876, where ‘x’ is the concentration of guanine (nM), with a correlation coefficient (R2) of 0.97. The increase in the fluorescence intensity of CuCD by guanine was analyzed quantitatively using the Stern–Volmer equation:
| F/F0 = 1 + KSV[Q] | (4) |
This highly sensitive and simple sensing mechanism employed by this CuCD for guanine nucleobase is deemed superior in comparison to some previously reported methods (Table 1).
Selective guanine sensing by CuCD
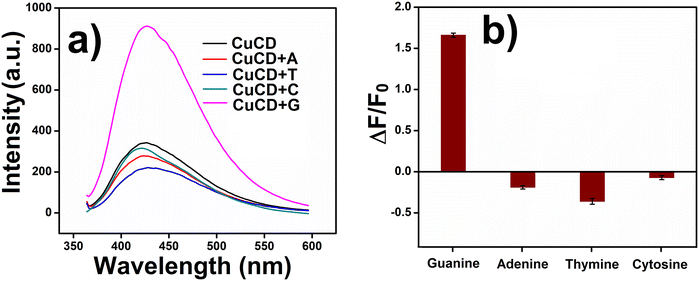
The next obvious intriguing issue is the influence of other nucleobases on the emission property of CuCD. Other than guanine (G), DNA has another three nucleobases, i.e., adenine (A), cytosine (C) and thymine (T). We investigated the sensitivity and selectivity of CuCD towards other nucleobase bases (A, T, and C). Selectivity is essential and crucial for the credibility of any new sensor. To this end, in different sets of experiments, A, T, and C (500 nM) were separately added to 1 mL of CuCD (250 μg mL−1) solution. Interestingly, in comparison to the native emission intensity of CuCD, very meager changes (quenching) in the fluorescence intensity of CuCD were noted in the presence of very high concentrations (500 nM) of adenine, cytosine and thymine (Fig. 5a). However, the increase in fluorescence intensity in the presence of only 20 nM of guanine was overwhelmingly significant compared to that in the presence of A, T and C (Fig. 5a). In contrast, 500 nM of A, T and C could not influence any significant change in the emission intensity of CuCD. A lesser amount of guanine is capable of increasing the emission of CuCD far more significantly than that of the 25-times higher concentration of other nucleobases. This observation undoubtedly increases the reliability of the newly developed CuCD as a selective and highly sensitive guanine detector, among other nucleobases (Fig. 5a and b).Molecular sensing always depends on its interaction with a particular sensor. Simultaneously, the specific interaction influences the selectivity of a sensor towards the particular molecule. It is well-established that Cu2+ can make strong interactions with Schiff bases, oxygen, nitrogen, and sulphur containing units specially mixed ligand donor type moieties.47 All the nucleobases of DNA have one of these structural features within them (Fig. S7, ESI†). A, G, and C have Schiff bases, while T has oxygen as a donor ligand in its structure. Within the nucleobases, only adenine (A) and guanine (G) feature Schiff bases as donor ligands. Additionally, guanine employs oxygen, and adenine utilizes nitrogen as additional donor ligands. This configuration enables them to establish a more robust interaction with CuCD compared to cytosine (C) and thymine (T). It is interesting that despite having similar structural properties of adenine and guanine, the CuCD was more selective towards guanine. According to previous reports, guanine oxygen at C6 and N7 has the perfect arrangement for making suitable interaction with Cu2+ of CuCD.48,49,65,66 Primary amine at C2 of guanine can engage in the resonance; consequently, the lone pair of oxygen at C6 could be readily available for creating impactful interactions with Cu2+ of CuCD. The abundant surface defects of carbon dots play a major role in their optical properties. Upon photoexcitation, rapid charge separation occurs in the carbon dots, leading to the formation of electrons and holes. These charge carriers are ‘trapped’ at various surface sites, and radiative recombination of the electrons and holes contributes to their intrinsic emission.66,67 The interaction of carbon dots with the analyte can influence the recombination between the surfaces of electron–hole pairs, thereby affecting the fluorescence properties of carbon dots. A more effective interaction with analytes in carbon dots may stabilize the surface sites for electrons and holes, facilitating efficient radiative recombination, which results in higher fluorescence. Considering these facts, it can be concluded that a possible stronger interaction of guanine with Cu2+ doped in carbon dots might have significantly enhanced the emission of CuCD (Fig. S8, ESI†).66,67 Thus, it may be assumed that adenine with primary amine at C6 and N7 has the optimal arrangement for making suitable interaction with Cu2+ of CuCD. However, the lone pair of primary amine at C6 was not available for greater interactions with Cu2+ because it was possibly engaged in extended resonance within the ring of adenine (Fig. S7, ESI†). Consequently, Cu2+ of CuCD could not have a strong interaction with adenine in comparison to that of guanine. Therefore, adenine did not affect the inherent emission intensity of CuCD.
Furthermore, an interference study was performed to investigate the selective sensing of guanine even in the presence of other nucleobases. The fluorescence intensities of the CuCD (250 μg mL−1) solution in the presence of 20 nM of all the other nucleobases (A, T, and C) were separately recorded in the absence of guanine. As expected, there was no change in the emission intensity of CuCD in the presence of A, T and C (Fig. S9, ESI†). Subsequently, guanine (20 nM) was added to each sample of the nucleobase and CuCD mixture, and the emission intensities were further recorded. In each case, the fluorescence intensity significantly increased upon the addition of guanine in that mixture (Fig. S9, ESI†). These observations further established that CuCD could selectively sense guanine even in the presence of other nucleobases (A, C, and T).
Apart from being a major part of DNA, guanine is also an essential biomolecule for cells. Thus, selective guanine sensing with respect to different monovalent ions, divalent ions, amino acids and biomolecules, which are abundant in the cell environment, is very important. To this end, the fluorescence intensity of CuCD (250 μg mL−1) was recorded in the presence of 500 nM (each component) of monovalent ion (Na+ and K+), divalent ion (Ca2+, Fe2+, and Zn2+), amino acids (L-cys, L-trp, L-tyr, L-arg, and L-asp acids) and biomolecules (GSH and H2O2). For monovalent and divalent ions, there were no changes in the fluorescence intensity of CuCD with respect to native emission (Fig. S10a and b, ESI†). In the case of amino acids and biomolecules, there were no significant changes in the fluorescence intensity of CuCD compared to that in the presence of guanine (20 nM) (Fig. S10a and b, ESI†). L-Arginine among other interferons showed little impact because Cu2+ can strongly interact with Schiff bases (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N–). These results once again affirmed the strong interaction of CuCD with guanine compared to all other investigated molecules. Furthermore, we represented the selectivity bar graph of CuCD, where the selectivity of guanine with other nucleobases and different metal ions, amino acids, and biomolecules were depicted altogether (Fig. S10c†). Therefore, we can certainly assume that the newly synthesized CuCD detects guanine with high selectivity and sensitivity against all other DNA nucleobases, different ions, amino acids and biomolecules.
N–). These results once again affirmed the strong interaction of CuCD with guanine compared to all other investigated molecules. Furthermore, we represented the selectivity bar graph of CuCD, where the selectivity of guanine with other nucleobases and different metal ions, amino acids, and biomolecules were depicted altogether (Fig. S10c†). Therefore, we can certainly assume that the newly synthesized CuCD detects guanine with high selectivity and sensitivity against all other DNA nucleobases, different ions, amino acids and biomolecules.
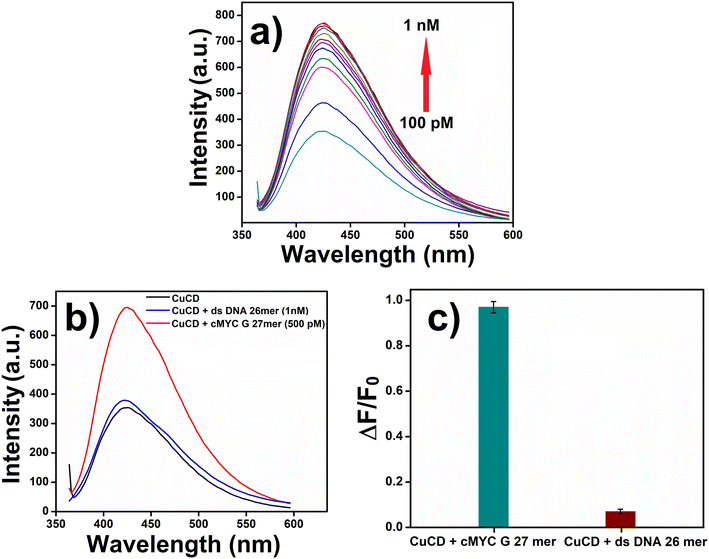
Oligonucleotides are short DNA or RNA molecules, that have a wide range of applications in bio-medical research. Desired oligonucleotides can be prepared by modifying the precursor nucleotides. These modifications give new properties to the oligonucleotides and make them key elements in diverse research domains. Herein, we used two oligonucleotides: cMYC G 27mer and ds DNA 26mer. cMYC G 27mer (DTGGGGAGGGTGGGGAGGGTGGGGAAGG) was the guanine-enriched oligonucleotide, and ds DNA 26mer (DCAATCGGATCGAATTCGATCCGATTG) was the control oligonucleotide. A stock solution (100 μM) of both oligonucleotides was prepared. Subsequently, we investigated the fluorescence intensity of CuCD (250 μg mL−1) in the presence of cMYC G 27mer (0.1–1.0 nM). Interestingly, with increasing concentrations of cMYC G 27mer, the fluorescence intensity of CuCD gradually increased (Fig. 6a). Additionally, we carried out a similar experiment on the fluorescence property of CuCD in the presence of both cMYC G 27mer (500 pM/0.5 nM) and ds DNA 26mer (1.0 nM). The fluorescence intensity of CuCD increased in the presence of cMYC G 27mer but remained unchanged in the presence of ds DNA 26mer (Fig. 6b and c). Guanine enrichment in the cMYC G 27mer oligonucleotide made it susceptible to the CuCD to influence its emission property more significantly in contrast to the control oligonucleotide, ds DNA 26mer. These observations further validated the utilization of CuCD as a potential biomarker that is highly specific to guanine.
IR spectra study
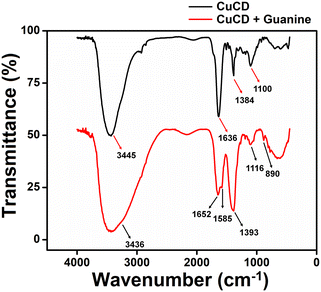
After highly sensitive and selective sensing of guanine by CuCD, we investigated the change in the vibrational frequency of CuCD in the presence of guanine using IR spectra. The IR spectrum of CuCD showed O–H stretching of the –COOH group at around 3445 cm−1, and a sharp –C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O spectrum for the –COOH group merged with the C
O spectrum for the –COOH group merged with the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C bending of alkyne at 1636 cm−1 and the O–H bending of the –COOH group at 1384 cm−1 (Fig. 7(i)). The above frequency peaks in the IR spectrum of CuCD indicate the presence of carboxylic acid on the surface of the newly synthesized carbon dot. Next, we investigated the IR spectra of CuCD in the presence of guanine. The FTIR spectrum of the CuCD-guanine complex showed O–H stretching of the –COOH group at around 3436 cm−1, and the sharp peak due to the presence of –C
C bending of alkyne at 1636 cm−1 and the O–H bending of the –COOH group at 1384 cm−1 (Fig. 7(i)). The above frequency peaks in the IR spectrum of CuCD indicate the presence of carboxylic acid on the surface of the newly synthesized carbon dot. Next, we investigated the IR spectra of CuCD in the presence of guanine. The FTIR spectrum of the CuCD-guanine complex showed O–H stretching of the –COOH group at around 3436 cm−1, and the sharp peak due to the presence of –C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O frequency for the –COOH group was split into two peaks. One was at 1652 cm−1 in the presence of a –C
O frequency for the –COOH group was split into two peaks. One was at 1652 cm−1 in the presence of a –C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O bond and another at 1580 cm−1 possibly due to the rise of a complex formation between –C
O bond and another at 1580 cm−1 possibly due to the rise of a complex formation between –C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N– of guanine and Cu2+.49 The peak at 1393 cm−1 was sharper than the peak of CuCD possibly due to the merging of the different vibrational frequencies of the heterocyclic ring of guanine along with the frequency of the O–H bond of the –COOH group. The peak at 890 cm−1 in the spectrum of CuCD in the presence of guanine may arise from the bond formation between Cu2+ and guanine, which was not properly visible only in the CuCD spectrum (Fig. 7).68 These characteristic peaks in the FTIR spectra conclude that the CuCD successfully formed a complex with guanine, resulting in a dramatic increase in the fluorescence intensity of CuCD. Moreover, we carried out a FEGTEM microscopic experiment of the CuCD-guanine complex to find out if there was any change in the microscopic property of carbon dots (Fig. S11, ESI†). The FEGTEM image of the CuCD-guanine complex showed assimilating structures of carbon dots (Fig. S11b and d, ESI†) in comparison to the dispersed FEGTEM image of the CuCD (Fig. S11a and c, ESI†). A high-resolution FEGTEM image showed the presence of big size particles along with small size carbon dots in the assimilated structure, which may be proof of the formation of the CuCD-guanine complex (Fig. S11d†). This type of assimilating structure along with the different size particles was absent in the FEGTEM image of only CuCD (Fig. S11c†). Hence, in the presence of guanine, the CuCD forms a complex that might have facilitated the increase in the fluorescence intensity of CuCD, thereby sensing the guanine.
N– of guanine and Cu2+.49 The peak at 1393 cm−1 was sharper than the peak of CuCD possibly due to the merging of the different vibrational frequencies of the heterocyclic ring of guanine along with the frequency of the O–H bond of the –COOH group. The peak at 890 cm−1 in the spectrum of CuCD in the presence of guanine may arise from the bond formation between Cu2+ and guanine, which was not properly visible only in the CuCD spectrum (Fig. 7).68 These characteristic peaks in the FTIR spectra conclude that the CuCD successfully formed a complex with guanine, resulting in a dramatic increase in the fluorescence intensity of CuCD. Moreover, we carried out a FEGTEM microscopic experiment of the CuCD-guanine complex to find out if there was any change in the microscopic property of carbon dots (Fig. S11, ESI†). The FEGTEM image of the CuCD-guanine complex showed assimilating structures of carbon dots (Fig. S11b and d, ESI†) in comparison to the dispersed FEGTEM image of the CuCD (Fig. S11a and c, ESI†). A high-resolution FEGTEM image showed the presence of big size particles along with small size carbon dots in the assimilated structure, which may be proof of the formation of the CuCD-guanine complex (Fig. S11d†). This type of assimilating structure along with the different size particles was absent in the FEGTEM image of only CuCD (Fig. S11c†). Hence, in the presence of guanine, the CuCD forms a complex that might have facilitated the increase in the fluorescence intensity of CuCD, thereby sensing the guanine.
Media stability of CuCD
Before deploying the newly synthesized CuCD, as a potential bioprobe for guanine sensing in a cellular environment, we conducted an assessment of its (i) stability in biological media over time and (ii) cytocompatibility with mammalian cells. In FBS-DMEM media, 500 μg mL−1 of CuCD was introduced with varying concentrations of FBS (up to 75%) and incubated for 48 h (Fig. S12a, ESI†). The prolonged stability of CuCD was also evaluated by maintaining mixtures of CuCD (500 μg mL−1)-FBS (10%)-DMEM media for 10 days (Fig. S12b, ESI†). In both cases, the suspension stability index (SSI) of CuCD in the media was approximately 90 ± 2% for different concentrations of FBS (0–75%), (Fig. S12a and b, ESI†). The stability of CuCD in biological media was corroborated by visual images (Fig. S12a, ESI†). Furthermore, we examined the stability of CuCD under UV light exposure (365 nm and 12 W), where no photobleaching property was observed, and the emission characteristics of the fluorescent bioprobes remained unchanged even after 200 min of UV irradiation (Fig. S12c, ESI†).Cytocompatibility of CuCD
After confirming the stability of CuCD in the biological milieu, we investigated the cytocompatibility of CuCD against mammalian cells with the aim of utilizing this bioprobe for cellular imaging and guanine detection inside cells. We assessed the cytocompatibility of CuCD within B16F10, MCF-7 and NIH3T3 cells using MTT assay. Various concentrations of CuCD (25–200 μg mL−1) were incubated with B16F10 melanoma cells, MCF-7 breast cancer cells and noncancerous NIH3T3 cells. Remarkably, the results indicated that the cell viability of CuCD was approximately 85% for all the investigated mammalian cells after 12 h of incubation. (Fig. S13, ESI†).Bioimaging
Detecting guanine in the cellular environment is crucial for the early diagnosis of various diseases and for monitoring human health.10 Disruptions in the normal levels of guanine within nucleic acids can lead to deficiencies and mutations in the immune system, potentially contributing to a range of diseases such as carcinoma, Parkinson's disease, epilepsy, and liver diseases.50 Therefore, we aimed to exploit the fluorescence property of CuCD in bioimaging. The observed increase in the fluorescence intensity of CuCD in the presence of guanine also prompted us to delve deeper into utilizing this probe to distinguish between guanine-enriched cells and the respective control cells. In this search, we identified G-quadruplexes (G4s), which are non-canonical secondary structures found in DNA and RNA sequences rich in guanine, as potential targets for investigation.69 They play crucial roles in cancer development and progression. G4s consist of two or more planar G-tetrads held together by Hoogsteen hydrogen bonds and pi-stacking.69 They are associated with telomere shortening, DNA replication, and the expression of oncogenes and proto-oncogenes.69 The presence of G4s is linked to genomic instability and carcinogenesis, with a significant association with oncogenes and tumor suppressors. G4s have become attractive targets for gene regulation, and various small molecules are being explored even in clinical trials because of their potential to target G4s in oncogene promoters.69 Considering this guanine-enriched G4 structure, we planned to diagnose two cancer cell lines, B16F10 and MCF-7, that have overexpressed G469,70 against the noncancer cell line NIH3T3 with and without guanine incubation by investigating the change in the fluorescence of CuCD.Native fluorescence, also known as autofluorescence (AF), occurs when biological substrates emit light in the UV-visible or near-IR spectral range upon excitation with suitable wavelengths of light. The CuCD exhibits fluorescence when excited at 350 nm. Following the incubation of CuCD in various cell lines, we initially examined the cells under fluorescence microscopy. Subsequently, upon capturing images of the cell lines incubated with CuCD, we performed background subtraction to generate the final CuCD-incubated cell line images. This process effectively eliminated the autofluorescence present in the image background, resulting in clear cell images exhibiting blue fluorescence.
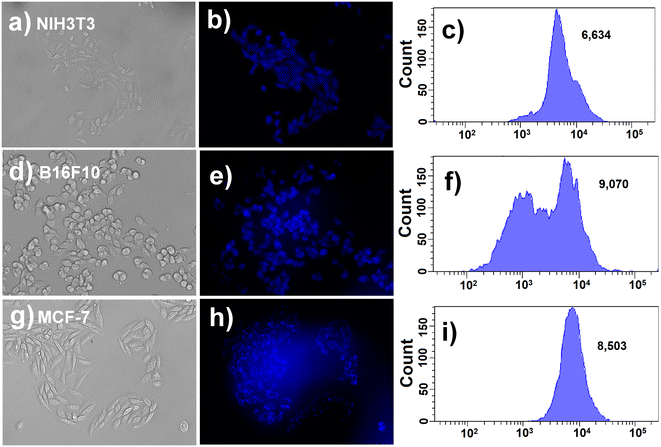
In this context, we incubated guanine (500 nM) with NIH3T3 cell lines for 12 h. After 12 h, the guanine-incubated NIH3T3 cell line was washed three times with PBS. Then, guanine-treated NIH3T3 and guanine-non-treated NIH3T3 cells were incubated with CuCD (200 μg mL−1) for 12 h. In addition, the G4-enriched B16F10 and MCF-7 cell lines were incubated with CuCD (200 μg mL−1) for 12 h. Following the incubation period, both guanine-treated and -non-treated cells NIH3T3 and B16F10, and MCF-7 were examined under a fluorescence microscope. Notably, guanine-treated NIH3T3 cells displayed vivid blue fluorescence, indicating internalized CuCD (Fig. S14a and b, ESI†) with a mean fluorescence intensity of 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 235 (Fig. S14c, ESI†). In contrast, comparatively poor fluorescence was observed in guanine-non-treated NIH3T3 cells (Fig. 8a and b) with a mean fluorescence intensity of 6634 (Fig. 8c) under identical experimental conditions. Interestingly, in the case of G4-enriched B16F10 (Fig. 8d and e) and MCF-7 cells (Fig. 8g and h), bright blue fluorescence of internalized CuCD was observed with mean fluorescence intensities of 9070 (Fig. 8f) and 8500, respectively, (Fig. 8i) without the addition of any external guanine. The difference between the mean fluorescence intensity of the observed cellular internalized CuCD in guanine-treated NIH3T3 cells and G4-enriched B16F10 and MCF-7 cells compared to that of guanine-non-treated NIH3TE cell lines demonstrated its utility as a potential biomarker for guanine nucleobase both in vitro and within mammalian cells.
235 (Fig. S14c, ESI†). In contrast, comparatively poor fluorescence was observed in guanine-non-treated NIH3T3 cells (Fig. 8a and b) with a mean fluorescence intensity of 6634 (Fig. 8c) under identical experimental conditions. Interestingly, in the case of G4-enriched B16F10 (Fig. 8d and e) and MCF-7 cells (Fig. 8g and h), bright blue fluorescence of internalized CuCD was observed with mean fluorescence intensities of 9070 (Fig. 8f) and 8500, respectively, (Fig. 8i) without the addition of any external guanine. The difference between the mean fluorescence intensity of the observed cellular internalized CuCD in guanine-treated NIH3T3 cells and G4-enriched B16F10 and MCF-7 cells compared to that of guanine-non-treated NIH3TE cell lines demonstrated its utility as a potential biomarker for guanine nucleobase both in vitro and within mammalian cells.
Conclusion
In this work, we synthesized a Cu+2-doped carbon dot (CuCD) using the hydrothermal method, which exhibited excitation-dependent blue fluorescence at 423 nm when excited at 350 nm. All microscopic and spectroscopic characterizations were carried out for this newly synthesized CuCD. The size of the synthesized CuCD was found to be ∼5 nm, as observed from TEM, AFM and DLS. XPS confirmed the presence of Cu2+ in the CuCD. The CuCD can efficiently detect the guanine nucleobase through the enhancement of its intrinsic fluorescence with a limit of detection of 0.59 nM. The formation of a complex with a guanine nucleobase by the CuCD was substantiated by FTIR spectroscopy. This Cu+2-doped carbon dot can also selectively detect guanine with high sensitivity against other nucleobases of DNA (A, T and C) and with respect to monovalent and divalent ions, amino acids and biomolecules. The selective affinity of CuCD for guanine was also observed particularly in a guanine-enriched oligonucleotide, cMYC G 27-mer (DTGGGGAGGGTGGGGAGGGTGGGGAAGG) in contrast to oligonucleotide, dsDNA 26-mer (DCAATCGGATCGAATTCGATCCGATTG) devoid of high guanine content. Concurrently, the CuCD was successfully employed for bioimaging guanine-enriched B16F10, MCF-7 cells and guanine-treated NIH3T3 cell lines via an increase in fluorescence intensity within living cells with respect to guanine non-treated NIH3T3 cells. Hence, the CuCD can be used as a selective and sensitive diagnostic probe for guanine sensing both in vitro and within mammalian cells.Data availability
The data supporting this article have been included as part of the ESI.†Author contributions
Monalisa Chowdhury: conceptualization, methodology, investigation, formal analysis, analysis and interpretation of data, software, writing – original draft preparation. Debolina Basu: methodology, investigation, data curation, validation, formal analysis, software, editing. Prasanta Kumar Das: supervision, design of the study, analysis and interpretation of data, project administration, funding acquisition, writing – review & editing.Conflicts of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.Acknowledgements
P. K. D. is thankful to Science and Engineering Research Board (SERB), Department of Science and Technology (DST), India for financial assistance (No. CRG/2021/000235). M. C. acknowledges IACS, India and for her research fellowships and D. B. acknowledges DST-INSPIRE for her research fellowship. The authors thank Mr. Gopal Manna for assisting in preparing the TOC graphic.References
- A. H. Kamel, F. T. C. Moreira, C. Delerue-Matos and M. G. F. Sales, Biosens. Bioelectron., 2008, 24, 591–599 CrossRef CAS.
- D. Wang, B. Huang, J. Liu, X. Guo, G. Abudukeyoumu, Y. Zhang, B. Ye and Y. Li, Biosens. Bioelectron., 2018, 102, 389–395 CrossRef CAS PubMed.
- E. Palecek, Bioelectrochem. Bioenerg., 1986, 15, 275–295 CrossRef CAS.
- N. Vishnu and S. Badhulika, Biosens. Bioelectron., 2019, 124-125, 122–128 CrossRef CAS.
- K. L. Ng and S. M. Khor, Anal. Chem., 2017, 89, 10004–10012 CrossRef CAS PubMed.
- I. Hirao, M. Kimoto and R. Yamashige, Acc. Chem. Res., 2012, 45, 2055–2065 CrossRef CAS.
- J. G. Wetmur, Crit. Rev. Biochem. Mol. Biol., 1991, 26, 227–259 CrossRef CAS PubMed.
- A. J. Jeevagan and S. A. John, Anal. Biochem., 2012, 424, 21–26 CrossRef PubMed.
- W. Saenger and C. R. Cantor, Principles of nucleic acid structure, Springer, New York, 1984 Search PubMed.
- K. Chetankumar, B. E. Kumara Swamy and H. S. Bhojya Naik, Mater. Chem. Phys., 2021, 267, 124610 CrossRef CAS.
- S. S. Wallace, Free Radical Biol. Med., 2002, 33, 1–14 CrossRef CAS PubMed.
- X. Wang, J. Zhang, Y. Wei, T. Xing, T. Cao, S. Wu and F. Zhu, Analyst, 2020, 145, 1933–1942 RSC.
- A. Ferancova, S. Rengaraj, Y. Kim, J. Labuda and M. Sillanpaa, Biosens. Bioelectron., 2010, 26, 314–320 CrossRef CAS.
- M. Arvand, N. Ghodsi and M. A. Zanjanchi, Biosens. Bioelectron., 2016, 77, 837–844 CrossRef CAS.
- S. Jesny, S. Menon and K. G. Kumar, RSC Adv., 2016, 6, 75741–75748 RSC.
- C. C. Marvel, J. Delrowe, E. G. Bremer and J. R. Moskal, Mol. Chem. Neuropathol., 1994, 21, 353–368 CrossRef CAS PubMed.
- C. Li and H. Wang, J. Chromatogr. A, 2015, 1406, 324–330 CrossRef CAS.
- P. Graven, M. Tambalo, L. Scapozza and R. Perozzo, Exp. Parasitol., 2014, 141, 28–38 CrossRef CAS.
- G. Chen, Q. C. Chu, L. Y. Zhang and J. N. Ye, Anal. Chim. Acta, 2002, 457, 225–233 CrossRef CAS.
- B. D. Gill and H. E. Indyk, Int. Dairy J., 2007, 17, 596–605 CrossRef CAS.
- F. Yang, J. Guan and S. P. Li, Talanta, 2007, 73, 269–273 CrossRef CAS.
- Y. Huang and H. Chang, Anal. Chem., 2007, 79, 4852–4859 CrossRef CAS PubMed.
- X. Xua, L. Hea, Y. Longa, S. Pana, H. Liu, J. Yangc and X. Hu, Sens. Actuators, B, 2019, 279, 44–52 CrossRef.
- L. Zou, Y. Li and B. Ye, Microchim. Acta, 2011, 173, 285–291 CrossRef CAS.
- X. Li, X. Gao, W. Shi and H. Ma, Chem. Rev., 2014, 114, 590–659 CrossRef CAS PubMed.
- Y. Xu, W. Shi, X. He, X. Wu, X. Li and H. Ma, Anal. Chem., 2017, 89, 10980–10984 CrossRef CAS.
- G. Li, Nano-inspired biosensors for protein assay with clinical applications, Elsevier, United Kingdom, 2019 Search PubMed.
- Z. Wang, X. Xiao, T. Zou, Y. Yang, X. Xing, R. Zhao, Z. Wang and Y. Wang, Nanomaterials, 2019, 9, 32 CrossRef.
- V. Gubala, G. Giovannini, F. Kunc, M. P. Monopoli and C. J. Moore, Cancer Nanotechnol., 2020, 11, 1–43 CrossRef CAS.
- B. H. Lee, S. Suresh and A. Ekpenyong, J. Biophotonics, 2018, 12, e201800172 CrossRef.
- G. Yang, S. Z. F. Phua, A. K. Bindra and Y. Zhao, Adv. Mater., 2019, 31, 1805730 CrossRef PubMed.
- S. N. Baker and G. A. Baker, Angew. Chem., Int. Ed., 2010, 49, 6726–6744 CrossRef CAS.
- S. Sarkar, K. Das and P. K. Das, ACS Sustainable Chem. Eng., 2017, 5, 8356–8369 CrossRef CAS.
- K. K. R. Datta, G. Qi, R. Zboril and E. P. Giannelis, J. Mater. Chem. C, 2016, 4, 9798–9803 RSC.
- B. D. Mansuriya and Z. Altintas, Nanomaterials, 2021, 11, 2525 CrossRef CAS PubMed.
- J. C. G. Esteves da Silva and H. M. R. Gonçalves, TrAC, Trends Anal. Chem., 2011, 30, 1327–1336 CrossRef CAS.
- F. Ahmed, S. Iqbal, L. Zhao and H. Xiong, Anal. Chim. Acta, 2021, 1183, 338977 CrossRef CAS.
- F. Ahmed, S. Iqbal and H. Xiong, Environ. Sci.: Nano, 2022, 9, 2624–2637 RSC.
- F. Ahmed, W. Xu, M. M. Hussain, W. U. Khan and H. Xiong, Chem. Eng. J., 2023, 477, 147300 CrossRef CAS.
- Y. Fan, K. J. Huang, D. J. Niu, C. P. Yang and Q. S. Jing, Electrochim. Acta, 2011, 56, 4685–4690 CrossRef CAS.
- M. Mouslmani, K. H. Bouhadir and D. Patra, Biosens. Bioelectron., 2015, 68, 181–188 CrossRef CAS.
- C. Ji, Y. Zhou, R. M. Leblanc and Z. Peng, ACS Sens., 2020, 5, 2724–2741 CrossRef CAS.
- J. Ge, Q. Jia, W. Liu, L. Guo, Q. Liu, M. Lan, H. Zhang, X. Meng and P. Wang, Adv. Mater., 2015, 27, 4169–4177 CrossRef CAS PubMed.
- E. Loi, R. W. C. Ng, M. M. F. Chang, J. F. Y. Fong, Y. H. Ng and S. M. Ng, Luminescence, 2017, 32, 114–118 CrossRef CAS PubMed.
- L. Jiang, H. Z. Ding, S. Y. Lu, T. Geng, G. J. Xiao, B. Zou and H. Bi, Angew. Chem., Int. Ed., 2020, 59, 9986–9991 CrossRef CAS PubMed.
- M. Vázquez-González, W. C. Liao, R. Cazelles, S. Wang, X. Yu, V. Gutkin and I. Willner, ACS Nano, 2017, 11, 3247–3253 CrossRef PubMed.
- J. Du, Y. Zhao, J. Chen, P. Zhang, L. Gao, M. Wang, C. Cao, W. Wen and C. Zhu, RSC Adv., 2017, 7, 33929–33936 RSC.
- T. Y. Han, T. S. Guan, M. A. Iqbal, R. A. Haque, K. S. Rajeswari, M. B. K. Ahamed and A. M. S. A. Majid, Med. Chem. Res., 2014, 23, 2347–2359 CrossRef CAS.
- S. Jaina, T. A. Khana, Y. P. Patil, D. Pagariya, N. Kishore, S. Tapryal, A. D. Naike and S. G. Naik, J. Photochem. Photobiol., B, 2017, 174, 35–43 CrossRef.
- I. Sissoeff, J. Grisvard and E. Guillé, Prog. Biophys. Mol. Biol., 1976, 31, 165–199 CrossRef CAS.
- V. Andrushchenko, J. H. Van De Sande and H. Wieser, Biopolymers, 2003, 72, 374–390 CrossRef CAS.
- S. Pradhan, S. Biswas, D. K. Das, R. Bhar, R. Bandyopadhyay and P. Pramanik, New J. Chem., 2018, 42, 564–573 RSC.
- S. Pang, Y. Zhang, C. Wu and S. Feng, Sens. Actuators, B, 2016, 222, 857–863 CrossRef CAS.
- A. Singh, A. Singha and N. A. Singh, Dalton Trans., 2014, 43, 16283 RSC.
- J. Borowiec and J. Zhang, J. Electrochem. Soc., 2015, 162, B332–B336 CrossRef CAS.
- Y. Li and J. Liu, Analyst, 2020, 145, 6753–6768 RSC.
- D. R. Kumar, D. Manoj and J. Santhanalakshmi, Sens. Actuators, B, 2013, 188, 603–612 CrossRef.
- L. Fu, L. Yan, G. Wang, H. Ren and L. Jin, Talanta, 2019, 193, 161–167 CrossRef CAS PubMed.
- Q. Xu, J. Wei, J. Wang, Y. Liu, N. Li, Y. Chen, C. Gao, W. Zhangd and T. Sreeprasede, RSC Adv., 2016, 6, 28745–28750 RSC.
- J. Guo, W. Lu, H. Zhang, Y. Meng, F. Du, S. Shuang and C. Dong, Sens. Actuators, B, 2021, 330, 129360 CrossRef CAS.
- Y. Wang, J. Yao, Z. Cao, P. Fu, C. Deng, S. Yan, S. Shi and J. Zheng, Chem. – Eur. J., 2022, 28, e202104174 CrossRef CAS PubMed.
- J. Wang, M. Xu, D. Wang, Z. Li, F. L. Primo, A. C. Tedesco and H. Bi, Inorg. Chem., 2019, 58, 13394–13402 CrossRef CAS PubMed.
- M. Najaflu, M. Shahgolzari, F. Bani and A. Y. Khosroushahi, ACS Omega, 2022, 7, 34573–34582 CrossRef CAS.
- M. Masteri-Farahani and F. Askari, Spectrochim. Acta, Part A, 2019, 206, 448–453 CrossRef CAS PubMed.
- D. X. Xing, X. J. Tan, X. B. Jiang and B. Wang, Comput. Theor. Chem., 2011, 963, 490–496 CrossRef CAS.
- Y. Liu, P. Wang, K. A. S. Fernando, G. E. LeCroy, H. Maimaiti, B. A. Harruff-Miller, W. K. Lewis, C. E. Bunker, Z. L. Houa and Y. P. Sun, J. Mater. Chem. C, 2016, 4, 6967–6974 RSC.
- J. Zuo, T. Jiang, X. Zhao, X. Xiong, S. Xiao and Z. Zhu, J. Nanomater., 2015, 787862 Search PubMed.
- Y. Liu, P. Wu, X. Wu, C. Ma, S. Luo, M. Xu, W. Li and S. Liu, Talanta, 2020, 210, 120649 CrossRef CAS.
- U. Bhatt, A. L. Kretzmann, A. Guedin, A. Ou, S. Kobelke, C. S. Bond, C. W. Evans, L. H. Hurley, J. L. Mergny, K. S. Iyer, A. H. Fox and N. M. Smith, Biochimie, 2021, 190, 124–131 CrossRef CAS PubMed.
- U. Chilakamarthi, D. Koteshwar, S. Jinka, N. V. Krishna, K. Sridharan, N. Nagesh and L. Giribabu, Biochemistry, 2018, 57, 6514–6527 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Synthetic schemes, high-resolution FEGTEM images of CuCD, DLS study, deconvoluted spectra of C 1s, O 1s, N 1s, XRD and EDX of CuCD, Tauc's plot, Stern–Volmer plot, structure of nucleobases, proposed binding model of CuCD-guanine complex, Stern–Volmer plot, interference study, FEGTEM image of CuCD-guanine complex, stability index of CuCD, cytocompatibility of CuCD, bioimaging of guanine-treated CuCD-incubated NIH3T3 cells. See DOI: https://doi.org/10.1039/d4sd00137k |
| This journal is © The Royal Society of Chemistry 2024 |