Open Access Article
Open Access ArticleDirect measurement of PFAS levels in surface water using an engineered biosensor
Madison
Mann
a,
Victoria
Kartseva
ab,
Chelli
Stanley
c,
Maggie
Blumenthal
c,
Richard
Silliboy
d and
Bryan
Berger
 *ae
*ae
aDepartment of Chemical Engineering, University of Virginia, Charlottesville, VA, USA. E-mail: bryan.berger@virginia.edu
bDepartment of Chemistry, University of Virginia, Charlottesville, VA, USA
cUplands Grassroots, Limestone, ME, USA
dMi'kmaq Nation, Presque Isle, ME, USA
eDepartment of Biomedical Engineering, University of Virginia, Charlottesville, VA, USA
First published on 30th October 2024
Abstract
Per- and polyfluoroalkyl substances (PFAS) are a large set of emerging contaminants pervasive in the environment due to amphiphilic properties and strong carbon-fluorine bonds resistant to biodegradation. With an ever-increasing prevalence, the need for precise detection of these chemicals at low levels in drinking water is clear. However, ground and surface water as well as soil and other biosolids have become reservoirs for PFAS at extremely high levels. In fact, PFAS concentrations at part per billion and part per million levels are found in environmental samples taken near high contamination sites including industrial facilities and military bases. In this work, we demonstrate the application of a biosensor based on human liver fatty acid binding protein to detect perfluorooctanoic acid (PFOA) in surface water samples taken near Loring Airforce Base. We show this sensor can detect the high levels of PFOA found in the samples quickly and easily without the use of extensive sample pre-treatment or analytical methods. Therefore, we hope the future of this technology will better assess PFAS detection needs for a multitude of end point users.
Sustainability spotlightPer and polyfluoroalkyl substances (PFAS, otherwise known as ‘forever chemicals’) are omnipresent in our lives, where they accumulate in water, soil, crops, animals and us. Currently, PFAS are considered unsafe at any detectable level by the US EPA. We have developed fluorescent biosensors to detect these chemicals in water samples, and in our current study, demonstrate their use in a range of environmental water samples. Approaches such as ours are needed to provide rapid, on-site testing to identify samples that require LC-MS/MS for subsequent, detailed chemical analysis. It also provides key testing to help identify environmental sources of PFAS and mitigate their spread in the environment. |
Introduction
PFAS (per-/polyfluoroalkyl substances) are a group of environmental and toxicological contaminants of increasing concern. Since their development in the 1940s, over 3000 variants of PFAS have been manufactured and have entered the global market for use in industrial and consumer applications.1,2 While their fluorinated carbon chains and polar head moieties impart the oil and water repellence ideal for industrial surfactants as well as consumer goods,3–6 the unparalleled chemical stability of environmental end products like perfluoroalkyl acids (PFAAs) allow for extreme environmental accumulation.4,7 Accumulation of these recalcitrant PFAS like perfluorooctanoic acid (PFOA) and perfluorooctane sulfonic acid (PFOS) has become increasingly prevalent in humans8–10 and has been linked to a variety of health effects including endocrine dysfunction, thyroid, pancreatic, and liver diseases,9,11–13 as well as a variety of reproductive issues and cancers.9,14–17In recent years many new regulations have been developed and proposed to ensure strict limits on PFAS in drinking water.18–20 Therefore, detection of these contaminants at the extremely low (part per trillion) levels relevant for drinking water has become a growing priority for researchers. However, it is important to note that human exposure to these chemicals occurs through multiple avenues including food ingestion as well as general contact.1,4 Specifically, it is becoming increasingly relevant to be able to quickly and easily detect the presence of PFAS in the soil and groundwater around sites with large amounts of these types of contaminants.21–23 High levels of PFAS pollution are common near manufacturing sites as well as areas utilizing fluorinated AFFFs. In fact, PFOA and PFOS have been found in part per million (ppm) levels in surface and ground water.22–24
In this study we aim to show the feasibility of a protein-based biosensor to detect spiked concentrations of PFAS in complex groundwater samples collected in Aroostook County, Maine. Loring Air Force base is located in the town of Limestone within Aroostook County and is an EPA Superfunds site known to be contaminated with low levels of PFAS and other common co-contaminants including waste oils, fuels, and spent solvents.25 Adjacent to this site are the Mi'kmaq Nation Trust Lands as well as the Aroostook National Wildlife Refuge;26 prior studies have investigated use of phytoremediation to remove PFAS from soils at Loring AFB, and recent testing has detected PFAS in drinking water in Aroostook County.27,28 Our prior work developing and benchmarking an acrylodan [6-acryloyl-2-dimethylaminonaphthalene] labeled biosensor based on human liver fatty acid binding protein (hLFABP) enabled detection of PFOA and PFOS in spiked creek water samples with limits of detection in the hundreds of parts per billion.29 In this study we demonstrate the sensor can detect spiked PFOA in complex ground water samples collected adjacent to Loring AFB with minimal sample preparation in agreement with independent LC/MS-MS testing for PFAS using method EPA 1633.25 These results provide an important initial validation of our biosensor to augment LC/MS-MS testing and demonstrates the use of engineered biosensors for rapid detection of environmental contaminants.
Materials and methods
Sample collection
Samples were collected in June of 2021 onsite in Aroostook County, ME and shipped to University of Virginia. Surface water samples were spiked with PFOA and filtered using a 0.2 μm polypropylene filter to remove particulates before biosensor application and shipment to Cyclopure for quantitative analysis through LC-MS-MS.LC/MS-MS quantification
Filtered samples were diluted as needed in sterile, ultrapure water. PFAS levels in filtered, diluted samples were measured using the Water Test Kit Pro (Cyclopure). The Water Test Kit utilizes DEXSORB® in passive sampling to collect PFAS. After sample application, the filter kits were shipped back to Cyclopure for analysis as outlined on their website (https://www.cyclopure.com/). Briefly, standard solid-phase extraction was used to recover PFAS, and the eluted samples were analyzed through HPLC-MS-MS (QExactive Orbitrap, ThermoFisher). The lab utilizes isotope dilution for measurement and quantification of 55 PFAS validated to the requirements of EPA methods 533, 537 and 1633. The limit of quantification (LOQ) is 1.0 ng L−1 for all PFAS except GenX which is 2.0 ng L−1.Sensor production
Acrylodan labeled hLFABP (Ac-hLFABP) was produced as previously described.29 Briefly, the single cysteine containing hLFABP F50C/C69S mutant was expressed via the pET28a(+) vector in E. coli BL21 (DE3). Cells were grown in LB media with kanamycin (50 μg mL−1) at 37 °C and induced with via addition of 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG). After 18 hours of expression at 20 °C, cells were harvested via centrifugation, resuspended in lysis buffer (50 mM Tris–Cl, 100 mM NaCl, 5% v/v glycerol, pH 8), and lysed by sonication. hLFABP F50C/C69S was purified from the clarified lysate by nickel affinity chromatography using Chelating Sepharose Fast Flow resin (Cytiva). Protein was eluted using increasing concentrations imidazole (10–500 mM), and protein containing fractions were dialyzed against PBS (pH 7.5). The N-terminal hexahistidine tag was then cleaved using a Thrombin CleanCleave Kit (Sigma-Aldrich). After cleavage, the protein was conjugated with acrylodan [6-acryloyl-2-dimethylaminonaphthalene] at a 2.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 fluorophore to protein molar ratio in denaturing conditions (8 M urea) overnight at room temperature with gentle agitation. Removal of unreacted acrylodan and refolding of the denatured protein was performed through an on-column, step-down, urea gradient. Labeled protein was loaded onto nickel charged Chelating Sepharose Fast Flow resin (Cytiva) and washed with 10 column volumes of 50 mM Tris–Cl buffer (pH 8) containing continually decreasing urea concentrations. Properly folded Ac-hLFABP was removed using 500 mM imidazole, and once again dialyzed into PBS (pH 7.6). Protein concentration was found using a BCA protein assay (Pierce), and acrylodan concentration was measured by absorption at 370 nm (extinction coefficient 16
1 fluorophore to protein molar ratio in denaturing conditions (8 M urea) overnight at room temperature with gentle agitation. Removal of unreacted acrylodan and refolding of the denatured protein was performed through an on-column, step-down, urea gradient. Labeled protein was loaded onto nickel charged Chelating Sepharose Fast Flow resin (Cytiva) and washed with 10 column volumes of 50 mM Tris–Cl buffer (pH 8) containing continually decreasing urea concentrations. Properly folded Ac-hLFABP was removed using 500 mM imidazole, and once again dialyzed into PBS (pH 7.6). Protein concentration was found using a BCA protein assay (Pierce), and acrylodan concentration was measured by absorption at 370 nm (extinction coefficient 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 400 M−1 cm−1).30 The degree of labeling for acrylodan conjugated C69S/F50C hLFABP was 1.0.
400 M−1 cm−1).30 The degree of labeling for acrylodan conjugated C69S/F50C hLFABP was 1.0.
Assay
Fluorescence measurements were performed using a Synergy Neo2 Hybrid Multi-Mode Microplate Reader (Biotek) at room temperature and under steady state conditions. Calibration curves and sample data were collected as previously described.29 Calibration curves were generated by titrating PFOA in deionized water into Ac-hLFABP in PBS (pH 7.6) to a final micromolar ratio of 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ligand to sensor respectively. Samples and standards were allowed to equilibrate for 5 minutes before fluorescence spectra were recorded over 400–600 nm after excitation at 360 nm. Signal was quantified as the shift in fluorescence spectra calculated as the change in the average center of mass (Δxcm) from 400 to 460 nm (eqn (1)). Calibration curve data was fit to a four-parameter log dose response model (eqn (2)), and sample PFOA concentrations were interpolated using the obtained fit parameters: Hillslope coefficient (HS), minimum and maximum signal (Δxcm,min & Δxcm,max), and half maximal effective concentration (EC50).
1 ligand to sensor respectively. Samples and standards were allowed to equilibrate for 5 minutes before fluorescence spectra were recorded over 400–600 nm after excitation at 360 nm. Signal was quantified as the shift in fluorescence spectra calculated as the change in the average center of mass (Δxcm) from 400 to 460 nm (eqn (1)). Calibration curve data was fit to a four-parameter log dose response model (eqn (2)), and sample PFOA concentrations were interpolated using the obtained fit parameters: Hillslope coefficient (HS), minimum and maximum signal (Δxcm,min & Δxcm,max), and half maximal effective concentration (EC50).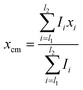 | (1) |
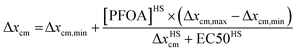 | (2) |
Results and discussion
The developed protein-based biosensor utilizes human liver fatty acid binding protein (hLFABP) as a PFAS-binding scaffold.29 hLFABP is one of a family of fatty acid binding proteins expressed cytosolically in various parts of the body with the responsibility of binding and shuttling fatty acids and other hydrophobic ligands.31 While all fatty acid binding proteins are comprised of hydrophobic β barrel binding regions, hLFABP is the only protein in the family capable of binding two cognate ligands; one in the large binding pocket and one close to an outer binding region as described in detail by others.32,33hLFABP is upregulated in response to PFOA exposure and has also been shown to bind several PFAS variants with moderate affinity due to their structural similarity to endogenous fatty acids.31,34,35 Furthermore, molecular dynamics and binding studies have concluded that these fluorinated ligands, much like fatty acids, are stabilized through hydrophobic interactions along the inner binding pocket as well as electrostatic interactions with the charged head group of the ligands in a “head-out” mode.34,36 This mode of binding is further supported as work has shown higher binding affinity of longer chain PFAS indicating increased hydrophobic stabilization.34,36,37
We previously described the binding of several PFAS to hLFABP through the introduction of tryptophan into multiple places along the inner binding pocket, and demonstrated the ability of the 50th residue to act as a probe for binding.29
By introducing a thiol conjugated solvatochromic fluorophore (acrylodan) at the 50th residue position within the ligand binding pocket, the hLFABP mutant F50C/C69S is capable of binding several PFAS including PFOA while producing a dose dependent blue-shift in acrylodan emission spectra. This shift in emission spectra to higher energy wavelengths occurs due to a polarity change in the acrylodan microenvironment upon ligand binding. The spectral shift is then quantified as the change in peak center of mass (Δxcm).
Aqueous fire-fighting foams (AFFFs), such as those found at Loring AFB, can consist of over 50 different PFAS ranging from C2–C12 in length as well as many fluorotelomers that act as precursors to PFAAs such as PFOA and PFOS.7,38,39 Wide use of AFFF chemicals at military bases during training, emergency responses, and equipment maintenance make these sites high risk for PFAS contamination.38,39 In fact, AFFF fluorotelomers as well as perfluorocaboxylates and perfluorosulfonates of varying chain sizes have been found at multiple U.S. military sites, sometimes even reaching part per million levels in groundwater,21,22,40–42 and are known to have extremely high transport potential.38,43,44
Like many other military sites, Loring AFB was a site of known contamination with many hazardous toxins including PFAS, and in the years following the base's deactivation, it remains a EPA Superfunds cleanup site.25,45 Recently, concerns have been raised about persistent contamination based on PFAS found in drinking water in Aroostook County as well as in agricultural soils on land belonging to the Mi'kmaq; one potential source of this contamination is from overuse of firefighting foams that contain PFAS.27 Over 70 different types of PFAS have been detected in soil samples around the Loring AFB, with sulfonic and carboxylic acids being the primary contaminants at concentrations of up to 150 ppb in soil.45 A recent test of area schools in Aroostook County show unsafe levels of both lead and PFAS in drinking water.28,46
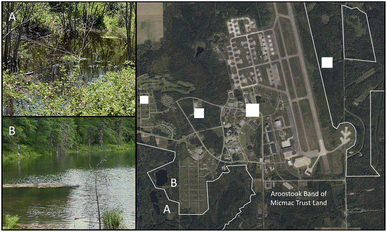
As a previous Superfunds site, the Loring base was identified as having soil and groundwater containing contaminates including volatile organic compounds (VOCs), polychlorinated biphenyls (PCBs), polyaromatic hydrocarbons (PAHs), pesticides, metals, and other petroleum related compounds. Therefore, we believe it to be important to show feasibility of PFAS detection using this biosensor in complex samples containing other co-contaminants.25 Even among AFFFs, formulations include various hydrocarbon surfactants, polymers, and organic solvents amongst the fluorinated chemicals.47–49 To demonstrate our biosensor's utility in this type of application setting, we measured total signal for PFAS binding in spiked samples collected adjacent to the Loring AFB as well as in major waterways such as the Aroostook River. Fig. 1 illustrates the sites where samples were collected. Based on LC/MS-MS analysis, all samples contained primarily PFOA as expected accounting for ∼93 and ∼60% of the total PFAS measured for sites A and B respectively. PFHxS was also found at high levels and accounted for ∼37% of total PFAS measured at site B. PFHxS was commonly used in AFFF, and PFOA is one of the most abundant PFAS found in the environment, and often is a breakdown product formed from higher molecular weight PFAS. The results of the LC/MS-MS data are summarized in Table 1. Our previous work showed the biosensor is most sensitive to PFOA at <1 ppm levels unlike PFHxS at greater than >1 ppm levels.29 Therefore, we prepared standards containing solely PFOA to calibrate the biosensor, determine apparent signal from PFAS binding in environmental samples, and compare to LC/MS-MS results.
| PFAS (ng L−1) | Chapman pit (A) | Malabeam lake (B) |
|---|---|---|
| PFBA | <LOQ | <LOQ |
| PFPeA | 112.50 | <LOQ |
| PFHxA | 183.75 | 47.73 |
| PFHpA | 1417.50 | 202.27 |
| PFOA | 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 477.50 477.50 |
78![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 840.91 840.91 |
| PFNA | <LOQ | <LOQ |
| PFDA | 86.25 | <LOQ |
| HFPO-DA (GenX) | <LOQ | <LOQ |
| PFBS | 268.75 | 831.82 |
| PFHxS | 581.25 | 47![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 920.45 920.45 |
| PFOS | 881.25 | 1379.55 |
| ∑PFAS | 54![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 008.75 008.75 |
129![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 222.73 222.73 |
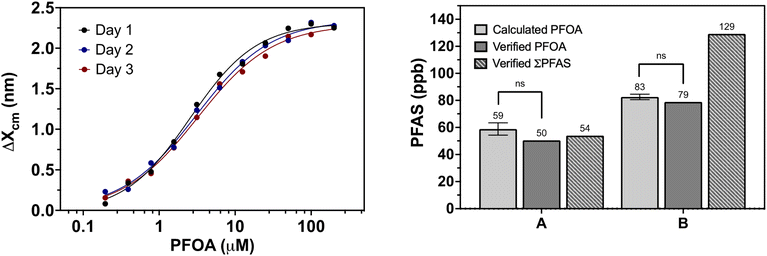
The left panel of Fig. 2 shows the calibration curves of the assay replicates utilized for sample PFOA quantification. These data were fit to four-parameter log-dose response models where Hillslope coefficient (HS), minimum and maximum signal (Δxcm,min & Δxcm,max), and half maximal effective concentration (EC50) constants were derived. Three separate sets of derived constants were used for quantifying PFOA concentration in the water samples rather than an average amongst the replicates to gauge concentrations more accurately in lower PFOA containing samples and reduce day to day error.
The results of LC/MS-MS indicated samples A and B contained 50 and 79 ppb PFOA respectively. Based on our calibration curve using PFOA, from our biosensor measurements we obtained values of 59 ± 5 ppb and 83 ± 2 ppb for these samples. One sample t-tests were conducted to determine statistical similarity between the calculated PFOA concentration and verified analytical value for both samples. Obtained p-values were 0.208 and 0.206 for samples A and B respectively showing differences in PFOA concentrations obtained through biosensor measurements and HPLC/MS-MS analysis are not statistically significant. This provides further evidence that the biosensor is robust to environmental testing and can be used for direct determination of PFOA.
PFOA was the main contaminant found in sample A which was taken from Chapman Pit. Along with PFOA, sample B, taken from Malabeam Lake, also contained 48 ppb PFHxS, a 6-carbon sulfonated perfluoroalkyl acid. The ability of the biosensor to detect PFOA in this sample with relative accuracy but not PFHxS indicates a selectivity for the 8-carbon chain carboxylic acid contaminant. This is consistent with prior work that suggests shorter chain PFAS like PFHxS, when bound to the inner binding pocket of hLFABP, provides less available contact area for specific interactions and stabilization.29,37
It is important to note challenges with sample variability; for surface water, constituents including organic matter and other co-contaminants can interfere with PFAS detection. Our prior work demonstrated that representative surfactants such as SDS which bind hLFABP do not interfere with PFAS detection.29 Furthermore, this testing on complex spiked field samples with limited sample pre-processing generated results consistent with independent LC/MS-MS detection. Future applications of this biosensor will focus on further analysis of how other pollutants or compounds might interfere with PFAS detection beyond those tested to date.
Conclusions
Overall, this study provides evidence that an engineered, protein-based biosensor can be used to detect PFAS in field samples using fluorescence readout with minimal pre-processing. With recent studies indicating widespread and significant PFAS levels in soils, water, agricultural products, food, consumer products and the human body, a multi-faceted approach to PFAS detection is needed. As illustrated in this work, biosensors have the advantage of straightforward implementation with reduced sample pre-processing; as such, they can serve as a useful initial screen for contamination to augment more sophisticated, detailed detections methods such as LC/MS-MS. As more studies show that PFAS levels in surface water and soils can reach levels greater than parts per trillion (ppt), it is also important to develop testing methods able to give reliable results in this concentration range that are cost-effective and readily available. As mentioned above, our current efforts are focused on understanding how other pollutants or environmental compounds impact detection, as well as extending detection to other compounds often found where PFAS contamination is present.Data availability
The data supporting this article has been included in the manuscript.Author contributions
Conceptualization: B. W. B., M. M. M., C. S., R. S.; investigation: B. W. B., M. M. M., V. K., C. S., M. B.; data curation and analysis: B. W. B., M. M. M., C. S.; manuscript preparation: B. W. B., M. M. M., V. K., C. S., R. S., M. B.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the Jefferson Trust (B. W. B.) and NIGMS T32GM136615 (M. M. M.).Notes and references
- Z. Wang, J. C. DeWitt, C. P. Higgins and I. T. Cousins, A Never-Ending Story of Per- and Polyfluoroalkyl Substances (PFASs)?, Environ. Sci. Technol., 2017, 51(5), 2508–2518 CrossRef CAS PubMed.
- G. M. Wickham and T. E. Shriver, Emerging contaminants, coerced ignorance and environmental health concerns: The case of per- and polyfluoroalkyl substances (PFAS), Sociol. Health Illness, 2021, 43(3), 764–778 CrossRef PubMed.
- E. Kissa, Fluorinated Surfactants and Repellents. Revised and Expanded Surfactant Science Series, Marcel Dekker, New York, 2nd edn, 2001, vol. 97, p. 615 Search PubMed.
- H. Brunn, G. Arnold, W. Körner, G. Rippen, K. G. Steinhäuser and I. Valentin, PFAS: forever chemicals—persistent, bioaccumulative and mobile. Reviewing the status and the need for their phase out and remediation of contaminated sites, Environ. Sci. Eur., 2023, 35(1), 20 CrossRef CAS.
- A. G. Paul, K. C. Jones and A. J. Sweetman, A First Global Production, Emission, And Environmental Inventory For Perfluorooctane Sulfonate, Environ. Sci. Technol., 2009, 43(2), 386–392 CrossRef CAS PubMed.
- K. Prevedouros, I. T. Cousins, R. C. Buck and S. H. Korzeniowski, Sources, Fate and Transport of Perfluorocarboxylates, Environ. Sci. Technol., 2006, 40(1), 32–44 CrossRef CAS PubMed.
- E. F. Houtz, C. P. Higgins, J. A. Field and D. L. Sedlak, Persistence of Perfluoroalkyl Acid Precursors in AFFF-Impacted Groundwater and Soil, Environ. Sci. Technol., 2013, 47(15), 8187–8195 CrossRef CAS PubMed.
- A. M. Calafat, Z. Kuklenyik, J. A. Reidy, S. P. Caudill, J. S. Tully and L. L. Needham, Serum Concentrations of 11 Polyfluoroalkyl Compounds in the U.S. Population: Data from the National Health and Nutrition Examination Survey (NHANES) 1999–2000, Environ. Sci. Technol., 2007, 41(7), 2237–2242 CrossRef CAS PubMed.
- S. E. Fenton, A. Ducatman, A. Boobis, J. C. DeWitt, C. Lau and C. Ng, et al., Per- and Polyfluoroalkyl Substance Toxicity and Human Health Review: Current State of Knowledge and Strategies for Informing Future Research, Environ. Toxicol. Chem., 2021, 40(3), 606–630 CrossRef CAS PubMed.
- J. M. Jian, D. Chen, F. J. Han, Y. Guo, L. Zeng and X. Lu, et al., A short review on human exposure to and tissue distribution of per- and polyfluoroalkyl substances (PFASs), Sci. Total Environ., 2018, 636, 1058–1069 CrossRef CAS PubMed.
- J. E. Lee and K. Choi, Perfluoroalkyl substances exposure and thyroid hormones in humans: epidemiological observations and implications, Ann. Pediatr. Endocrinol. Metab., 2017, 22(1), 6 CrossRef PubMed.
- T. C. Guillette, J. McCord, M. Guillette, M. E. Polera, K. T. Rachels and C. Morgeson, et al., Elevated levels of per- and polyfluoroalkyl substances in Cape Fear River Striped Bass (Morone saxatilis) are associated with biomarkers of altered immune and liver function, Environ. Int., 2020, 136, 105358 CrossRef CAS PubMed.
- K. P. Das, B. E. Grey, M. B. Rosen, C. R. Wood, K. R. Tatum-Gibbs and R. D. Zehr, et al., Developmental toxicity of perfluorononanoic acid in mice, Reprod. Toxicol., 2015, 51, 133–144 CrossRef CAS PubMed.
- B. E. Blake and S. E. Fenton, Early life exposure to per- and polyfluoroalkyl substances (PFAS) and latent health outcomes: A review including the placenta as a target tissue and possible driver of peri- and postnatal effects, Toxicology, 2020, 443, 152565 CrossRef CAS PubMed.
- C. Fei, J. K. McLaughlin, L. Lipworth and J. Olsen, Maternal levels of perfluorinated chemicals and subfecundity, Hum. Reprod., 2009, 24(5), 1200–1205 CrossRef CAS PubMed.
- B. N. VanNoy, J. Lam and A. R. Zota, Breastfeeding as a Predictor of Serum Concentrations of Per- and Polyfluorinated Alkyl Substances in Reproductive-Aged Women and Young Children: A Rapid Systematic Review, Curr. Environ. Health Rep., 2018, 5(2), 213–224 CrossRef CAS PubMed.
- I. Šabović, I. Cosci, L. De Toni, A. Ferramosca, M. Stornaiuolo and A. Di Nisio, et al., Perfluoro-octanoic acid impairs sperm motility through the alteration of plasma membrane, J. Endocrinol. Invest., 2020, 43(5), 641–652 CrossRef PubMed.
- United States Environmental Protection Agency, Drinking Water Health Advisory for Perfluorooctane Sulfonate (PFOS) EPA Document Number 822-R-16-004, US EPA Federal Registry, 2016.
- United States Environmental Protection Agency, Drinking Water Health Advisory for Perfluorooctanoic Acid (PFOA) EPA Document Number: 822-R-16-005, US EPA Federal Registry, 2016.
- United States Environmental Protection Agency, PFAS National Primary Drinking Water Regulation Rulemaking, 2023, pp. 18638–18754, Document Citation: 88 FR 18638, Report No.: 2023-05471, https://www.federalregister.gov/documents/2023/03/29/2023-05471/pfas-national-primary-drinking-water-regulation-rulemaking#addresses.
- M. L. Brusseau, R. H. Anderson and B. Guo, PFAS concentrations in soils: Background levels versus contaminated sites, Sci. Total Environ., 2020, 740, 140017 CrossRef CAS PubMed.
- X. Dauchy, V. Boiteux, C. Bach, C. Rosin and J. F. Munoz, Per- and polyfluoroalkyl substances in firefighting foam concentrates and water samples collected near sites impacted by the use of these foams, Chemosphere, 2017, 183, 53–61 CrossRef CAS PubMed.
- X. Dauchy, V. Boiteux, A. Colin, J. Hémard, C. Bach and C. Rosin, et al., Deep seepage of per- and polyfluoroalkyl substances through the soil of a firefighter training site and subsequent groundwater contamination, Chemosphere, 2019, 214, 729–737 CrossRef CAS PubMed.
- D. Q. Andrews, J. Hayes, T. Stoiber, B. Brewer, C. Campbell and O. V. Naidenko, Identification of point source dischargers of per- and polyfluoroalkyl substances in the United States, AWWA Water Sci., 2021, 3(5), 1–16 Search PubMed , [cited 2023 Jun 15], https://www.onlinelibrary.wiley.com/doi/10.1002/aws2.1252.
- United States Environmental Protection Agency, Loring Air Force Base Limestone, ME; Announcements and Key Topics, [cited 2023 Jun 6], https://www.epa.gov/superfund/loring.
- United States Airforce, Former Loring Airforce Base, Maine. Official United States Air Force Website, 2012, https://www.afcec.af.mil/About-Us/Fact-Sheets/Display/Article/466134/former-loring-air-force-base-maine/.
- S. L. Nason, C. J. Stanley, C. E. PeterPaul, M. F. Blumenthal, N. Zuverza-Mena and R. J. Silliboy, A community based PFAS phytoremediation project at the former Loring Airforce Base, iScience, 2021, 24(7), 102777 CrossRef PubMed.
- PFAS in Public Water Systems. Maine CDC Drinking Water Program, 2022, https://www.maine.gov/dhhs/mecdc/environmental-health/dwp/pws/pfas.shtml.
- M. M. Mann, J. D. Tang and B. W. Berger, Engineering human liver fatty acid binding protein for detection of poly- and perfluoroalkyl substances, Biotechnol. Bioeng., 2022, 119(2), 513–522 CrossRef CAS PubMed.
- F. G. Prendergast, M. Meyer, G. L. Carlson, S. Iida and J. D. Potter, Synthesis, spectral properties, and use of 6-acryloyl-2-dimethylaminonaphthalene (Acrylodan). A thiol-selective, polarity-sensitive fluorescent probe, J. Biol. Chem., 1983, 258(12), 7541–7544 CrossRef CAS PubMed.
- R. L. Smathers and D. R. Petersen, The human fatty acid-binding protein family: Evolutionary divergences and functions, Hum. Genomics, 2011, 5(3), 170 CrossRef CAS PubMed.
- A. Sharma and A. Sharma, Fatty acid induced remodeling within the human liver fatty acid-binding protein, J. Biol. Chem., 2011, 286(36), 31924–31928 CrossRef CAS PubMed.
- Y. He, R. Estephan, X. Yang, A. Vela, H. Wang and C. Bernard, et al., An NMR-Based Structural Rationale for Contrasting Stoichiometry and Ligand Binding Site(s) in Fatty Acid-binding Proteins, Biochemistry, 2011, 50(8), 1283–1295 CrossRef CAS PubMed.
- N. Sheng, J. Li, H. Liu, A. Zhang and J. Dai, Interaction of perfluoroalkyl acids with human liver fatty acid-binding protein, Arch. Toxicol., 2016, 90(1), 217–227 CrossRef CAS PubMed.
- C. Y. Ku, Y. H. Liu, H. Y. Lin, S. C. Lu and J. Y. Lin, Liver fatty acid-binding protein (L-FABP) promotes cellular angiogenesis and migration in hepatocellular carcinoma, Oncotarget, 2016, 7(14), 18229–18246 CrossRef PubMed.
- W. Cheng and C. A. Ng, Predicting Relative Protein Affinity of Novel Per- and Polyfluoroalkyl Substances (PFASs) by An Efficient Molecular Dynamics Approach, Environ. Sci. Technol., 2018, 52(14), 7972–7980 CrossRef CAS PubMed.
- L. Zhang, X. M. Ren and L. H. Guo, Structure-Based Investigation on the Interaction of Perfluorinated Compounds with Human Liver Fatty Acid Binding Protein, Environ. Sci. Technol., 2013, 47(19), 11293–11301 CrossRef CAS PubMed.
- R. H. Anderson, G. C. Long, R. C. Porter and J. K. Anderson, Occurrence of select perfluoroalkyl substances at U.S. Air Force aqueous film-forming foam release sites other than fire-training areas: Field-validation of critical fate and transport properties, Chemosphere, 2016, 150, 678–685 CrossRef CAS PubMed.
- B. J. Place and J. A. Field, Identification of Novel Fluorochemicals in Aqueous Film-Forming Foams Used by the US Military, Environ. Sci. Technol., 2012, 46(13), 7120–7127 CrossRef CAS PubMed.
- A. Nickerson, A. E. Rodowa, D. T. Adamson, J. A. Field, P. R. Kulkarni and J. J. Kornuc, et al., Spatial Trends of Anionic, Zwitterionic, and Cationic PFASs at an AFFF-Impacted Site, Environ. Sci. Technol., 2021, 55(1), 313–323 CrossRef CAS PubMed.
- M. E. McGuire, C. Schaefer, T. Richards, W. J. Backe, J. A. Field and E. Houtz, et al., Evidence of Remediation-Induced Alteration of Subsurface Poly- and Perfluoroalkyl Substance Distribution at a Former Firefighter Training Area, Environ. Sci. Technol., 2014, 48(12), 6644–6652 CrossRef CAS PubMed.
- C. A. Moody and J. A. Field, Determination of Perfluorocarboxylates in Groundwater Impacted by Fire-Fighting Activity, Environ. Sci. Technol., 1999, 33(16), 2800–2806 CrossRef CAS.
- J. L. Guelfo and C. P. Higgins, Subsurface Transport Potential of Perfluoroalkyl Acids at Aqueous Film-Forming Foam (AFFF)-Impacted Sites, Environ. Sci. Technol., 2013, 47(9), 4164–4171 CrossRef CAS PubMed.
- J. Reinikainen, N. Perkola, L. Äystö and J. Sorvari, The occurrence, distribution, and risks of PFAS at AFFF-impacted sites in Finland, Sci. Total Environ., 2022, 829, 154237 CrossRef CAS PubMed.
- S. L. Nason, J. Koelmel, N. Zuverza-Mena, C. Stanley, C. Tamez and J. A. Bowden, et al., Software Comparison for Nontargeted Analysis of PFAS in AFFF-Contaminated Soil, J. Am. Soc. Mass Spectrom., 2021, 32(4), 840–846 CrossRef CAS PubMed.
- Testing for Lead in K-12 School Drinking Water. Maine CDC Drinking Water Program, 2023, https://www.maine.gov/dhhs/mecdc/environmental-health/dwp/pws/testingLeadSchoolWater.shtml.
- J. A. Field, M. Schultz and D. Barofsky, Identifying Hydrocarbon and Fluorocarbon Surfactants in Specialty Chemical Formulations of Environmental Interest by Fast Atom Bombardment/Mass Spectrometry, Chimia, 2003, 57(9), 556 CrossRef CAS.
- M. K. Moe, S. Huber, J. Svenson, A. Hagenaars, M. Pabon and M. Trümper, et al., The structure of the fire fighting foam surfactant Forafac®1157 and its biological and photolytic transformation products, Chemosphere, 2012, 89(7), 869–875 CrossRef CAS PubMed.
- R. A. García, A. C. Chiaia-Hernández, P. A. Lara-Martin, M. Loos, J. Hollender and K. Oetjen, et al., Suspect Screening of Hydrocarbon Surfactants in AFFFs and AFFF-Contaminated Groundwater by High-Resolution Mass Spectrometry, Environ. Sci. Technol., 2019, 53(14), 8068–8077 CrossRef PubMed.
| This journal is © The Royal Society of Chemistry 2024 |