Open Access Article
Open Access ArticleBiologically active dual functional zinc-doped biomass-derived carbon dots†
Mohammad
Tariq
a,
Mo Ahamad
Khan‡
b,
Hammad
Hasan‡
 a,
Sangeeta
Yadav‡
c,
Amaresh Kumar
Sahoo
a,
Sangeeta
Yadav‡
c,
Amaresh Kumar
Sahoo
 *c and
Md Palashuddin
Sk
*c and
Md Palashuddin
Sk
 *a
*a
aDepartment of Chemistry, Aligarh Muslim University, Aligarh-202002, Uttar Pradesh, India. E-mail: palashuddin.ch@amu.ac.in
bDepartment of Microbiology, Faculty of Medicine, Aligarh Muslim University, Aligarh-202002, Uttar Pradesh, India
cDepartment of Applied Sciences, Indian Institute of Information Technology Allahabad (IIIT Allahabad), Prayagraj-211015, Uttar Pradesh, India. E-mail: asahoo@iiita.ac.in
First published on 16th September 2024
Abstract
The resistance of bacteria to antibiotics poses a significant challenge in the current global landscape. Despite this urgency, the pace of drug development has not matched the pressing need. Addressing this gap, we have developed zinc-doped carbon dots (Zn-Cdots) using biomass as a carbon source by a simple, and eco-friendly hydrothermal method to treat bacterial infection. Plant-derived biomass serves as an excellent source of various bioactive molecules, making it a viable carbon source for synthesizing Zn-Cdots. The characterization of Zn-Cdots was performed using multiple techniques, including UV-Visible spectroscopy, photoluminescence spectroscopy, TEM analysis, XRD, FTIR and XPS. The Zn-Cdots exhibit superior antibacterial properties in combating Gram-negative and Gram-positive bacterial strains, specifically Serratia marcescens and Staphylococcus aureus compared to the precursor biomass extract. Additionally, ROS measurements revealed the antioxidant property of Zn-Cdots, while agarose gel electrophoresis studies confirmed that the interaction between pDNA and Zn-Cdots heightened the antibacterial activity of Zn-Cdots. Moreover, the ABTS assay and the TMB assay both validated the antioxidant activity of Zn-Cdots, revealing high efficacy in scavenging free radicals and further highlighting its potential in mitigating oxidative stress alongside potent antibacterial efficacy.
Sustainability spotlightWe introduce biomass-derived metal-doped carbon dots as potent antibacterial and antioxidant agents, offering a sustainable alternative with dual biomedical functions. By utilizing biomass as the carbon source, our approach promotes environmental friendliness, contrasting with conventional chemical sources. This innovative strategy not only tackles bacterial infections effectively but also reduces environmental impact. This work aligns with UN Sustainable Development Goals, particularly Good health and well-being (SDG 3) and Responsible consumption and production (SDG 12). |
Introduction
Before the advent of antibiotics, sulfa drugs were promising antibacterial drugs for combating bacterial infections.1 However, the discovery of antibiotics revolutionized the fight against infectious diseases.2,3 Nevertheless, in the decades following their discovery, bacteria have demonstrated remarkable drug resistance and evolved various strategies to survive these drugs, leading to the emergence of antimicrobial resistance as a global concern. As per the 2017 World Health Organization (WHO) report on antibiotic-resistant bacterial infections, this issue has become so severe that it poses a significant threat to modern medicine,4 implicating not only the loss of lives but also the substantial financial resources expended on treatment. The rapid rise of drug-resistant bacterial infections is outpacing the current rate of drug development, posing a significant threat. Therefore, it is paramount to design and develop a novel class of antibacterial agents that differ from traditional ones.5 Recently, nanoscience has introduced a promising approach for synthesizing innovative nano-antimicrobial agents. Nanomaterials based on metals and their oxides have been reported to demonstrate excellent antibacterial properties.6 While these metal-based nanomaterials exhibit remarkable antibacterial activities, they release metal ions that accumulate within normal cells, resulting in cytotoxic effects and cell death.7 Carbon nanomaterials (for example, carbon dots) would be promising alternatives to these metal and metal oxide-based nanomaterials owing to their good biocompatibility and low cytotoxicity.The overproduction of reactive oxygen species (ROS) generates oxidative stress in living systems, damaging regular biological processes and causing several critical diseases.8 Hence, exogenous antioxidants are promising for protecting living systems from oxidative deterioration by scavenging radical species. Various antioxidants, such as metal and metal oxide-based nanoparticles, carbon-based nanomaterials, polymer-based nanomaterials, and lipid-based nanomaterials, have been reported.9–11 However, their toxicity in biological systems remains a concern for practical applications, as these antioxidants are often developed using complex reagents. Therefore, safe nanomaterials with excellent radical species scavenging capacity are essential for protecting living systems.
Carbon dots (Cdots) is a class of zero-dimension nanomaterial that is composed of discrete quasispherical carbon particles sizes below 10 nm and exhibits high water dispersibility, exciting optical properties, good biocompatibility, low toxicity, and easy to synthesize from readily available resources.12–14 Due to these excellent physiochemical characteristics, Cdots have piqued the interest of researchers, and widely employed in various applications, including biological sensing, photodynamic therapy, photocatalysts, complex logic operations, antimicrobial agents, and antibacterial agents.15–18 Notably, recent studies demonstrate that strategically synthesized Cdots from various precursors are emerging as promising antibacterial agents.19–23 The common mechanism followed by the Cdots to kill bacteria is generating reactive oxygen species (ROS), infiltration into bacterial cell walls,24 and the electrostatic interaction between the surface charge of Cdots and bacterial cell walls.25 To improve the bactericidal activity of Cdots, Cdots is generally doped with different metal and non-metal atoms.26 The metal doping of heteroatoms improves the electron density and the energy gap of Cdots, which can readily transfer electrons that help in the generation of ROS.
Zinc (Zn) is one of the necessary elements in the human body after iron.27 Zn is present in trace quantities that facilitate electron transfers in different biological systems and enzymatic activities.28 ZnO nanostructures where zinc has a +2 oxidation state show excellent biocompatibility with human cells.29 Also, ZnO is documented as a safe material by the Food and Drug Administration (FDA) because of its non-toxic nature.30 However, the deficiency of Zn leads to multiple health issues. The incorporation of Zn into the Cdots would be beneficial for enhancing the bactericidal performance of Cdots.31 In this study, we have synthesized zinc-doped Cdots (Zn-Cdots) from the plant-derived biomass brand name Joshina (a mixture of bioactive compounds) via hydrothermal technique. Zn-Cdots show superior antimicrobial activities against Staphylococcus aureus and Serratia marcescens. The MIC of Staphylococcus aureus and Serratia marcescens was found to be 300 and 150 μg mL−1, respectively. The agarose gel electrophoresis study unveiled that the mechanism of bacterial cell death is due to the interaction between p-DNA and Zn-Cdots. Additionally, the antioxidant property of Zn-Cdots was confirmed through ROS measurement studies. Furthermore, the antioxidant nature of Zn-Cdots was further validated using the ABTS assay, which demonstrated significant free radical scavenging activity. Additionally, the TMB assay provided complementary evidence, reinforcing the antioxidant nature of Zn-Cdots. This comprehensive evaluation underscores the dual functionality of Zn-Cdots as both an effective antibacterial agent and a potent antioxidant.
Experimental section
Materials
Plant-derived biomass brand name Joshina (Hamdard, India) was purchased from the Unani medicine store near Tibbiya College (AMU, Aligarh). Commonly Joshina is a mixture of various bioactive-compound-containing herbs like Assyrian plum, creeping charlie, marsh mallow, jujube, sweet flag, sedge, licorice, wood violet (botanical name of these compounds Cardia myxa L., Malva sylvestris, Althaea officinalis, Ziziphus mauritiana, Acorus calamus Linn., Onosma bracteatum, Glycyrrhiza glabra, Viola odorata Linn. respectively), and zinc sulfate heptahydrate was purchased from Merck.Synthesis of Zn-Cdots
Joshina was cleansed with deionized water and dried overnight at 70 °C. Then 25 g of dry Joshina was boiled with 60 mL water at 90 °C for 5 hours. The supernatant of Joshina was filtered with Whatman filter paper and dried for four days at 70 °C to convert it into crude. The dry crude extract of Joshina (150 mg) and 0.5 mmol of ZnSO4·7H2O was taken in a beaker, and 8 mL of deionized water was added to make the concentrate solution. The solution was filled in a Teflon to heat at 160 °C for 5 hours in a furnace. The carbonized product of the reaction was extracted and purified with the 1 kDa (benzoylated dialysis bag) for 24 hours.Characterization
The UV-Visible spectra of Zn-Cdots were recorded with a PerkinElmer LAMBDA-45 spectrophotometer. The photoluminescence spectra were recorded using the HITACHI-F2500 photoluminescence spectrometer. The functional group identification of Zn-Cdots was carried out by PerkinElmer infra-red spectrometer. The morphology of Zn-Cdots was examined through a transmission electron microscope. The elemental composition of Zn-Cdots was determined by ESCA Multilab 2000 X-ray photoelectron spectroscopy. The powder XRD spectra of Zn-Cdots were recorded with the help of the Rigaku smart lab.Antimicrobial assay by agar diffusion (inhibition zones)
The bacterial strains undergoing examination were subjected to an 18 hours sub-culturing process, followed by the preparation and adjustment of bacterial samples to attain a concentration of 0.5 McFarland Units, which is approximately equivalent to 1–2 × 108 CFU mL/1. In this experiment, 100 μL of the bacterial cultures were spread onto Muller–Hinton agar (MHA) plates using sterile cotton swabs. Then, sterile 6 mm borers were used to create wells in the agar medium, and 100 microliters of the Zn-Cdots were added to each well. The plates were incubated at 37 °C for 24 hours, and the experiment was conducted thrice to ensure consistency. The average area of inhibition and the standard deviation of the means were computed.Determination of minimum inhibitory concentrations (MICs)
Microbroth dilution was used to determine the MIC of a compound. In this method, 96 well ELISA plates were used to prepare various dilutions of the Zn-Cdots, which were then incubated at 37 ± 2 °C for 24 hours. After incubation, triphenyl tetrazolium chloride (TTC, HiMedia, India) (3 mg mL−1) was added to each well (20 μL) and left at room temperature for 20 minutes. The visual detection of a pink color change signified bacterial growth. The MIC was determined as the lowest concentration at which no color change was visible, indicating the absence of metabolically active cells.Measurement of oxidative stress by NBT assay
To probe the amount of reactive oxygen species (ROS), an NBT assay was performed where nitro blue tetrazolium salt gets reduced to blue-coloured formazan that shows absorbance at 575 nm. Overnight grown bacterial culture was treated in the presence of increasing concentration of Zn-Cdots (S1 to S3) in 0.3 mL Hank's balanced salt solution (HBSS) at 37 °C for 30 minutes along with its controls and positive control (i.e., 2 μM of H2O2). Then, 0.1 mL of 10 mg per mL NBT solution was added and kept for another 30 min. The 0.1 mL of 0.1 M HCl was added to stop the reaction. The bacterial pellet was collected (at 6000 rpm for 5 min) followed by the addition of 0.6 mL of DMSO and recorded its absorbance by a UV-Vis spectrophotometer.Agarose gel electrophoresis
The migration of plasmid DNA was performed by agarose gel electrophoresis using 0.8% gel at 5 V cm−1 following standard gel running buffer (pH 8.0). For that, two different concentrations (5 and 7 μL) of the Zn-Cdots (25 mg mL−1) were added with a fixed concentration of pDNA and incubated at 37 °C temperature for 15 min followed by agarose gel electrophoresis. The ethidium bromide staining was carried out before visualization of the same under UV trans-illuminator (excitation at 305 nm).ABTS test
The free radical scavenging activity of the Zn-Cdots was performed by considering the neutralization of ABTS+˙ radicals. In a typical experiment, 1 mL of ABTS (10 mM) was mixed with 1 mL of K2S2O8 (3.5 mM) and stored in the dark for 12 h to generate the ABTS+˙ radicals (i.e., blue color). The formed ABTS+˙ radicals were incubated with increasing Zn-Cdots concentrations (10 and 20 μL of 25 mg mL−1) for 15 min at room temperature. A time-dependent absorbance at 734 nm was recorded by UV-Vis spectroscopy.TMB assay study
To investigate the antioxidant properties of zinc-doped carbon dots (Zn-Cdots), the 3,3′,5,5′-tetramethylbenzidine (TMB) oxidation assay was conducted. In this experiment, 50 mM of TMB was prepared in 0.15 M citrate phosphate buffer (pH 3.5) and combined with 50 mM hydrogen peroxide (H2O2). The reaction was catalyzed by 10 μg mL−1 horseradish peroxidase (HRP) and produced oxidized TMB (Ox-TMB) with a deep blue-green color. To evaluate the effect of Zn-Cdots, 100 μL of 25 mg mL−1 Zn-Cdots solution was added to the Ox-TMB and the mixture was left for 15 minutes. The absorbance was recorded at 650 nm using a UV-Vis spectrophotometer for the following cases: H2O2 alone, H2O2 + TMB, H2O2 + TMB + HRP (i.e. Ox-TMB), and H2O2 + TMB + HRP + Zn-Cdots (i.e. TMB). This setup allowed us to assess the effect of Zn-Cdots on the oxidized TMB, thus demonstrating its antioxidant capability.Results and discussion
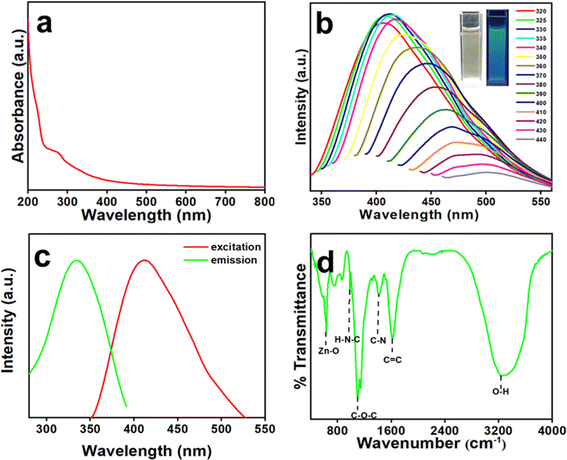
The detail of the synthesis of Zn-Cdots using a hydrothermal process is demonstrated in the experimental section. Zn-Cdots was purified by using a 1 kDa dialysis membrane for 24 hours, and then purified Zn-Cdots was dried at 70 °C until became powdered. The dried carbonized product was dispersed into the water for optical characterization. The aqueous solubility of Zn-Cdots was investigated at room temperature. Zn-Cdots exhibited excellent water solubility and maximum solubility was noted to be 80 mg mL−1. Fig. 1a demonstrates the absorption spectra of Zn-Cdots that depict the broad absorption peak at 278 nm. This peak arises from the π–π* transition of the graphitic core of Zn-Cdots.32 The photophysical characteristics of Zn-Cdots are shown in Fig. 1b and c. The detailed photoluminescence characteristic of Zn-Cdots was examined and recorded by the fluorometer in which the sample was excited at a different wavelength to explore the emission spectra. The Zn-Cdots were excited from 320 nm to 440 nm wavelength (Fig. 1b) with a gradual increment. Notably, Fig. 1b demonstrates that the excitation tunable red shift occurred in the emission spectra at the higher excitation wavelength. This red shift of Zn-Cdots toward the higher excitation wavelength supports the doping of Zn within the carbogenic core.33,34 The excitation spectrum of Zn-Cdots corresponding to the maximum emission at 411 nm is shown in Fig. 1c at the excitation wavelength of 330 nm. The quantum yield of Zn-Cdots was found to be 2.9% with quinine sulfate used as a reference. Fourier transform infrared spectroscopy (FTIR) was used to analyze the surface functional group of Zn-Cdots (Fig. 1d). The FTIR analysis indicates different functional groups are present at the surface of Zn-Cdots. The bands of these functionals 3261 cm−1, 1624 cm−1, 1412 cm−1, 1146 cm−1, 1091 cm−1, and 982 cm−1 indicate the functional groups such as (νO–H) (νC![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), (δC–OH), (δC–N), and (δH–N–C). The 743 cm−1 and 634 cm−1 bands confirm the C–S–C asymmetric stretching and Zn–O–C bond in the Zn-Cdots.35
C), (δC–OH), (δC–N), and (δH–N–C). The 743 cm−1 and 634 cm−1 bands confirm the C–S–C asymmetric stretching and Zn–O–C bond in the Zn-Cdots.35
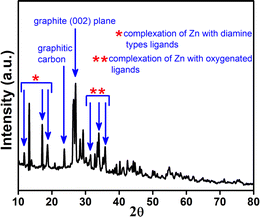
The morphological structure of Zn-Cdots was studied by using the transmission electron microscope (TEM). This study confirms the average size of Zn-Cdots 4.8 ± 0.8 nm, and the high-resolution TEM image indicates the lattice fringe present in Zn-Cdots with a d-spacing value of 0.22 nm, which indicates the (100) graphitic plane of Zn-Cdots (Fig. 2)36. The powder XRD (PXRD) of Zn-Cdots was recorded from 2θ = 10° to 80°, as shown in Fig. 3. The PXRD measurement further indicated the doping of Zn into the Cdots. The PXRD spectrum of Zn-Cdots shows a diffraction angle around 2θ = 23.7° arises because of graphitic carbon and 2θ = 26.6° indicating the graphite (002) plane of Zn-Cdots.33,37 However, different Zn phases with mixed ligands in the Zn-Cdots were noted as biomass-derived Zn-Cdots contains oxygen, nitrogen and sulfur-related functionalities. The diffraction angle observed at 2θ values of 11.7°, 17.05°, and 18.6° are related to the complexation of Zn with diamine types ligands.38 Also, diffraction angles around 31.5°, 33.9°, and 35.9° originated due to the complexation of Zn with oxygenated ligands.33,37
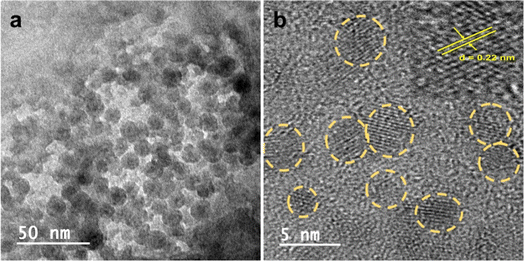 | ||
| Fig. 2 (a) TEM image of Zn-Cdots (50 nm scale bar). (b) HRTEM image showing Zn-Cdots with an average size 4.8 ± 0.8 nm (5 nm scale bar). | ||
The elemental compositions of Zn-Cdots and confirmation of Zinc doping were examined using the XPS technique, which revealed the presence of S, C, O, N, and Zn with their respective peaks at 163.25, 284.3 eV, 399.05 eV, 530.99 eV, 1022.25 eV, and 1045.25 eV (Fig. 4a). The deconvoluted high-resolution spectrum of C1s illustrates C–C/C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C (284.3 eV) bonding states of carbon in graphite core and C–N/C–S/C–O/C
C (284.3 eV) bonding states of carbon in graphite core and C–N/C–S/C–O/C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O (285.0 eV) functionalities in Fig. 4b. The deconvoluted signal of O1s reveals that Zn–O (529.4 eV), C–O (530.7 eV), and C
O (285.0 eV) functionalities in Fig. 4b. The deconvoluted signal of O1s reveals that Zn–O (529.4 eV), C–O (530.7 eV), and C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O (531.26 eV) are present in Zn-Cdots (Fig. 4c). Furthermore, high-resolution S2p spectra show two peaks of S–C (163.3 eV) and S–O (169.08 eV) (Fig. 4d). Fig. 4f shows the two deconvoluted peaks of Zn2p at 1022.7 eV and 1045.3 eV, and these are attributed to Zn2p3/22+ and Zn2p1/22+.39 Further, elemental analysis from XPS measurement demonstrated that Zn-Cdots contain (atomic%) 76.84% carbon (C), 2.26% nitrogen (N), 12.83% oxygen (O), 5.42% sulfur (S) and 2.65% zinc (Zn).
O (531.26 eV) are present in Zn-Cdots (Fig. 4c). Furthermore, high-resolution S2p spectra show two peaks of S–C (163.3 eV) and S–O (169.08 eV) (Fig. 4d). Fig. 4f shows the two deconvoluted peaks of Zn2p at 1022.7 eV and 1045.3 eV, and these are attributed to Zn2p3/22+ and Zn2p1/22+.39 Further, elemental analysis from XPS measurement demonstrated that Zn-Cdots contain (atomic%) 76.84% carbon (C), 2.26% nitrogen (N), 12.83% oxygen (O), 5.42% sulfur (S) and 2.65% zinc (Zn).
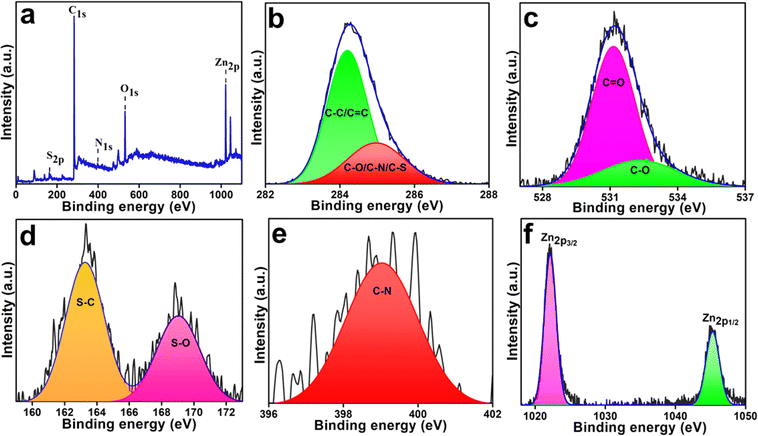 | ||
| Fig. 4 (a) XPS full scan spectrum of Zn-Cdots. Deconvoluted high-resolution (b) C1s signal (c) O1s signal (d) S2p signal (e) N1s signal (f) Zn2p signal. | ||
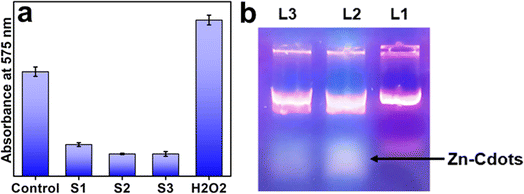
Next, we explored the bactericidal effect of Zn-Cdots against combating Gram-negative bacterial strains Serratia marcescens and Gram-positive bacterial strains Staphylococcus aureus. The detailed method of the antimicrobial study is provided in the experiment section. The results showed that the tested bacterial strains were inhibited by the Zn-Cdots effectively (Fig. 5). The mean inhibition zones were measured and found to be 17.50 ± 0.50 and 18 ± 0.5 mm for S. aureus and S. marcescens, respectively (Table 1). The control (extract of Joshina) did not exhibit any zone of inhibition, indicating that the observed effect was due to the Zn-Cdots and not from any other factors. The results suggest that the Zn-Cdots have antibacterial properties against the tested bacterial strains. Further, the MIC of the Zn-Cdots was determined using the broth micro-dilution method. The detail of MIC measurements is given in the experimental section. The MIC of the sample was measured to be 150 and 300 μg mL−1 against Gram-negative and Gram-positive bacteria, respectively, which indicates the lowest concentration of the sample at which no colour change was visible, indicating the absence of metabolically active cells. Notably, the result of the bactericidal effect of the control Cdots without zinc doping (derived from Joshina using a similar synthesis protocol as Zn-Cdots) for Gram-negative bacteria did not show prominent antibacterial activity like Zn-Cdots (Fig. S1, ESI†). The Zn-Cdots exhibited superior antibacterial properties as compared to presently reported antibacterial agents in combating Gram-negative and Gram-positive bacterial strains (Table 2). Possibly, the effectiveness of Zn-Cdots results from their small size, allowing for increased surface contact with microorganisms. Smaller Zn-Cdots might readily access bacterial cells' cytoplasmic content by interacting with the cell wall.47 One possible reason for bacterial cell death would be that Zn-Cdots entering the bacterial cell cytoplasm may cause enzymatic imbalance, which accumulates ROS. ROS may damage bacterial cytoplasmic organs and kill them.48 ROS can permeate the bacterial cell membrane and damage essential biomolecules such as DNA and proteins. The ROS study by NBT assay showed that treatment of the Zn-Cdots eventually decreased the intracellular ROS level as compared to control cells (i.e., untreated) while treatment of the H2O2 was considered as a positive control (Fig. 6a). Notably, the Zn-Cdots acted as an anti-oxidizing agent in the present case owing to its electron-donating ability that might be responsible for its antioxidant properties. Eventually, the ROS measurement experiment confirmed that ROS is not involved in bacterial cell death. Therefore, an alternative mechanism is involved in the efficacy of killing bacteria.
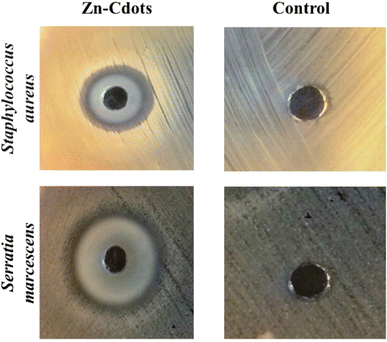 | ||
| Fig. 5 Effect of sample on pathogenic bacteria. The treated sample shows a clear zone of inhibition as compared to the control. | ||
| Bacterial strains | Zn-Cdots | Control (extract of Joshina) | ||
|---|---|---|---|---|
| Zone of inhibition (mm) | Minimum inhibitory concentration (μg mL−1) | Zone of inhibition (mm) | Minimum inhibitory concentration (μg mL−1) | |
| a (—) no activity detected. | ||||
| Staphylococcus aureus | 17.50 ± 0.50 | 300 | — | — |
| Serratia marcescens | 18 ± 0.50 | 150 | — | — |
| SN | Plant name | Method | Species | MIC | References |
|---|---|---|---|---|---|
| 1 | Figs of Ficus religiosa | Microwave | E. coli | 110 ± 5 μg mL−1 | 40 |
| B. subtilis | 120 ± 6 μg mL−1 | ||||
| 2 | Osmanthus leaves (best) tea leave milk vetch | Hydrothermal | S. aureus | 1000 μg mL−1 | 41 |
| E. coli | |||||
| 3 | Turmeric leaves (Curcuma longa) | Hydrothermal | E. coli | 0.25 mg mL−1 | 23 |
| S. aureus | 0.25 mg mL−1 | ||||
| K. pneumoniae | 1.0 mg mL−1 | ||||
| S. epidermidis | 1.0 mg mL−1 | ||||
| 4 | Artemisia argyi leaves | Ignited | E. coli | 150 μg mL−1 | 42 |
| P. aeruginosa | |||||
| P. Vulgaris | |||||
| 5 | Cannabis sativa leave | Pyrolysis | E. coli | 42 μg mL−1 | 43 |
| S. aureus | 42 μg mL−1 | ||||
| Dental microflora | 25 μg mL−1 | ||||
| 6 | Oyster mushroom | Hydrothermal | S. aureus | 30 μg mL−1 | 44 |
| K. pneumoniae | |||||
| P. aeruginosa | |||||
| 7 | Rosemary leaves | Hydrothermal | Yeast strains | 12 μg mL−1 | 45 |
| 8 | Henna leaf | Hydrothermal | S. aureus | 17 μg mL−1 | 46 |
| E. coli | 12 μg mL−1 |
Furthermore, it is imperative to elucidate the mechanisms underlying the enhanced antibacterial activity exhibited by the Zn-Cdots. To explore the interaction between Zn-Cdots and DNA, agarose gel electrophoresis experiments were conducted. The results of the gel electrophoresis revealed a significant alteration in the band pattern following treatment with Zn-Cdots, suggesting a potential interaction with the bacterial plasmid DNA (pDNA). Specifically, the treated samples exhibited band smearing and accelerated migration, indicative of DNA disintegration (as depicted in Fig. 6b). The doped Zn2+ ions within the Cdots engage in electrostatic interactions with the negatively charged pDNA.49 The presence of Zn-Cdots was visually confirmed on the gel, where they displayed blue emission and migrated alongside the pDNA. In conclusion, Zn-Cdots bind to the DNA, thereby contributing to its disintegration. This phenomenon underlies the heightened antibacterial efficacy observed with Zn-Cdots.
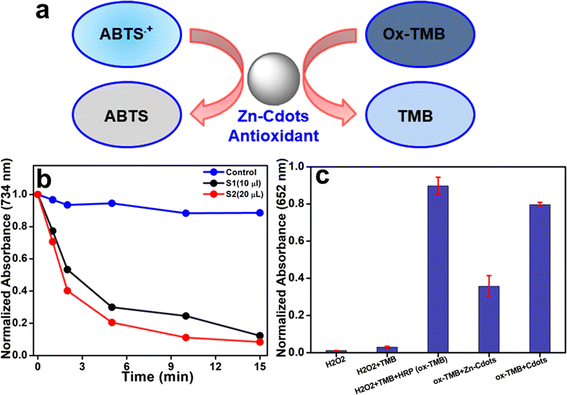
Finally, the antioxidant nature of Zn-Cdots was further confirmed using the ABTS assay (Fig. 7). The free radical scavenging activity of the Zn-Cdots was assessed by their ability to neutralize ABTS+˙ radicals. In this experiment, ABTS+˙ radicals were generated and incubated with increasing concentrations of Zn-Cdots. The time-dependent absorbance at 734 nm recorded by UV-Vis spectroscopy indicated a significant reduction in absorbance, confirming the free radical scavenging activity of Zn-Cdots.
Next, the results of the TMB assay reveal intriguing insights into the antioxidant properties of Zn-Cdots (Fig. 7c). Initially, the addition of horseradish peroxidase (HRP) to a mixture of hydrogen peroxide (H2O2) and TMB substantially increased the absorbance, indicating efficient oxidation of TMB (Ox-TMB) through the HRP-catalyzed reaction with H2O2. However, following the subsequent addition of Zn-Cdots to the Ox-TMB and allowing the mixture to incubate for 15 minutes, a notable decrease in absorbance was observed. Specifically, the absorbance decreased by approximately 60.3% compared to the mixture without Zn-Cdots. This reduction in absorbance suggests that the presence of Zn-Cdots reduced the concentration of Ox-TMB. Further, Ox-TMB with undoped Cdots showed no notable absorbance reduction, indicating a lack of antioxidant activity compared to Zn-Cdots. Such interference highlights the antioxidant activity of Zn-Cdots, as they effectively mitigate the oxidation of TMB molecules. These findings provide further evidence of the antioxidant nature of Zn-Cdots.
Conclusion
In this study, we present the study on the doping of zinc into biomass-derived Cdots using an eco-friendly hydrothermal method. Plant-derived biomass serves as an excellent carbon source, rich in various bioactive compounds. The resulting Zn-Cdots demonstrate remarkable antibacterial properties against both Staphylococcus aureus and Serratia marcescens, at significantly lower concentrations compared to the inherent antibacterial activities of precursor biomass extract. Notably, the MIC values of Zn-Cdots are found to be 150 μg mL−1 for Gram-negative bacteria and 300 μg mL−1 for Gram-positive bacteria. ROS measurements confirm that ROS is not involved in the mechanism of bacterial cell death, instead revealing the antioxidant property of Zn-Cdots. However, agarose gel electrophoresis studies confirm the heightened antibacterial activity resulting from the interaction between pDNA and Zn-Cdots. Furthermore, the antioxidant nature of Zn-Cdots was confirmed using the ABTS assay, which demonstrated significant free radical scavenging activity. Additionally, the TMB assay study further validated the antioxidant nature of Zn-Cdots. This study not only encourages but also paves the way for further development and investigation of metal-doped biomass-derived Cdots for potential applications.Data availability
All data is included in the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We thank the Department of Chemistry, University Sophisticated Instrument Facility (USIF), AMU, Aligarh for granting instrumental facilities, and MNIT Jaipur for their assistance with TEM and XPS facilities.References
- A. Petchiappan and D. Chatterji, ACS Omega, 2017, 2, 7400–7409 CrossRef CAS.
- A. Fleming, Clin. Infect. Dis., 1980, 2, 129–139 CrossRef CAS.
- E. D. Brown and G. D. Wright, Nature, 2016, 529, 336–343 CrossRef CAS.
- World Health Organization, Antibacterial Agents in Clinical Development, 2017 Search PubMed.
- B. N. Tse, A. A. Adalja, C. Houchens, J. Larsen, T. V Inglesby and R. Hatchett, Clin. Infect. Dis., 2017, 65, 495–500 CrossRef CAS PubMed.
- M. Morais, A. L. Teixeira, F. Dias, V. Machado, R. Medeiros and J. A. V. Prior, J. Med. Chem., 2020, 63, 14308–14335 CrossRef CAS PubMed.
- M. V. D. Z. Park, A. M. Neigh, J. P. Vermeulen, L. J. J. de la Fonteyne, H. W. Verharen, J. J. Briedé, H. van Loveren and W. H. de Jong, Biomaterials, 2011, 32, 9810–9817 CrossRef CAS PubMed.
- B. Halliwell, Nat. Rev. Mol. Cell Biol., 2024, 25, 13–33 CrossRef CAS PubMed.
- L. Valgimigli, A. Baschieri and R. Amorati, J. Mater. Chem. B, 2018, 6, 2036–2051 RSC.
- R. Priyadarshi, Z. Riahi, S. K. Tammina, A. Khan and J.-W. Rhim, Mater. Today Sustainability, 2023, 24, 100563 CrossRef.
- F. Gao, J. Liu, P. Gong, Y. Yang and Y. Jiang, Chem. Eng. J., 2023, 462, 142338 CrossRef CAS.
- C. Ding, A. Zhu and Y. Tian, Acc. Chem. Res., 2014, 47, 20–30 CrossRef CAS.
- Z. Zhong, Y. Zhang, X. Fu, S. Liu, C. Zhang, W. Guo, X. Xu and L. Liao, Nanoscale Adv., 2022, 4, 5365–5377 RSC.
- J. Dhariwal, G. K. Rao and D. Vaya, RSC Sustainability, 2024, 2, 11–36 RSC.
- M. P. Sk, S. K. Sailapu and A. Chattopadhyay, ChemPhysChem, 2015, 16, 723–727 CrossRef CAS.
- B. K. Korah and B. Mathew, Mater. Today Sustainability, 2023, 21, 100273 CrossRef.
- S. Y. Lim, W. Shen and Z. Gao, Chem. Soc. Rev., 2015, 44, 362–381 RSC.
- P. Li, L. Sun, S. Xue, D. Qu, L. An, X. Wang and Z. Sun, SmartMat, 2022, 3, 226–248 CrossRef.
- H. Zhu, N. Peng, X. Liang, S. Yang, S. Cai, Z. Chen, Y. Yang, J. Wang and Y. Wang, Biomed. Mater., 2023, 18, 062002 CrossRef.
- A. Verma, F. Arshad, K. Ahmad, U. Goswami, S. K. Samanta, A. K. Sahoo and M. P. Sk, Nanotechnology, 2020, 31, 095101 CrossRef.
- M. Tariq, S. Shivalkar, H. Hasan, A. K. Sahoo and M. P. Sk, ACS Omega, 2023, 8, 49460–49466 CrossRef PubMed.
- R. Jijie, A. Barras, J. Bouckaert, N. Dumitrascu, S. Szunerits and R. Boukherroub, Colloids Surf., B, 2018, 170, 347–354 CrossRef CAS.
- A. Saravanan, M. Maruthapandi, P. Das, J. H. T. Luong and A. Gedanken, Nanomaterials, 2021, 11, 369 CrossRef CAS.
- W. Bing, H. Sun, Z. Yan, J. Ren and X. Qu, Small, 2016, 12, 4713–4718 Search PubMed.
- W. Wu, L. Zhan, W. Fan, J. Song, X. Li, Z. Li, R. Wang, J. Zhang, J. Zheng, M. Wu and H. Zeng, Angew. Chem., Int. Ed., 2015, 54, 6540–6544 Search PubMed.
- S. Lubna and R. Ahmad, J. Trace Elem. Med. Biol., 2023, 77, 127130 CrossRef CAS PubMed.
- M. Arifuzzaman and Y. Zhao, ACS Catal., 2018, 8, 8154–8161 CrossRef CAS.
- J. Pulit-Prociak, J. Chwastowski, A. Kucharski and M. Banach, Appl. Surf. Sci., 2016, 385, 543–553 CrossRef CAS.
- Z. Li, R. Yang, M. Yu, F. Bai, C. Li and Z. L. Wang, J. Phys. Chem. C, 2008, 112, 20114–20117 CrossRef CAS.
- M. Mishra, J. S. Paliwal, S. K. Singh, E. Selvarajan, C. Subathradevi and V. Mohanasrinivasan, J. Pure Appl. Microbiol., 2013, 7, 1263–1268 Search PubMed.
- M. Ahamad Khan, S. A. Lone, M. Shahid, M. T. Zeyad, A. Syed, A. Ehtram, A. M. Elgorban, M. Verma and M. Danish, Toxics, 2023, 11, 452 Search PubMed.
- F. Arshad, A. Pal and M. P. Sk, ECS J. Solid State Sci. Technol., 2021, 10, 021001 CrossRef CAS.
- P. Khare, A. Bhati, S. R. Anand, Gunture and S. K. Sonkar, ACS Omega, 2018, 3, 5187–5194 CrossRef CAS.
- Q. Xu, W. Cai, M. Zhang, R. Su, Y. Ye, Y. Li, L. Zhang, Y. Guo, Z. Yu, S. Li, X. Lin, Y. Chen, Y. Luo, J. Street and M. Xu, RSC Adv., 2018, 8, 17254–17262 RSC.
- D. Gao, P. Zhao, B. Lyu, Y. Li, Y. Hou and J. Ma, Appl. Organomet. Chem., 2020, 34(8), e5665 CrossRef CAS.
- K. Holá, M. Sudolská, S. Kalytchuk, D. Nachtigallová, A. L. Rogach, M. Otyepka and R. Zbořil, ACS Nano, 2017, 11, 12402–12410 CrossRef.
- E. K. Adotey, M. Amouei Torkmahalleh, P. K. Hopke and M. P. Balanay, Sensors, 2023, 23, 1632 CrossRef CAS PubMed.
- M. J. Brabha and M. A. Malbi, Chem. Phys. Impact, 2023, 7, 100248 CrossRef.
- J. Jana, J. S. Chung and S. H. Hur, ACS Omega, 2019, 4, 17031–17038 CrossRef CAS PubMed.
- M. Tariq, A. Singh, N. Varshney, S. K. Samanta and M. P. Sk, Mater. Today Commun., 2022, 33, 104347 Search PubMed.
- Y. Ma, M. Zhang, H. Wang, B. Wang, H. Huang, Y. Liu and Z. Kang, Mater. Today Commun., 2020, 24, 101222 Search PubMed.
- H. Wang, M. Zhang, Y. Ma, B. Wang, M. Shao, H. Huang, Y. Liu and Z. Kang, J. Mater. Chem. B, 2020, 8, 2666–2672 Search PubMed.
- S. Raina, A. Thakur, A. Sharma, D. Pooja and A. P. Minhas, Mater. Lett., 2020, 262, 127122 CrossRef CAS.
- T. Boobalan, M. Sethupathi, N. Sengottuvelan, P. Kumar, P. Balaji, B. Gulyás, P. Padmanabhan, S. T. Selvan and A. Arun, ACS Appl. Nano Mater., 2020, 3, 5910–5919 Search PubMed.
- H. Eskalen, M. Çeşme, S. Kerli and Ş. Özğan, J. Chem. Res., 2021, 45, 428–435 Search PubMed.
- M. Shahshahanipour, B. Rezaei, A. A. Ensafi and Z. Etemadifar, Mater. Sci. Eng., C, 2019, 98, 826–833 Search PubMed.
- Y. Meng, M. Yang, X. Liu, W. Yu and B. Yang, Nano, 2019, 14, 1950029 Search PubMed.
- A. K. Sahoo, M. P. Sk, S. S. Ghosh and A. Chattopadhyay, Nanoscale, 2011, 3, 4226 Search PubMed.
- M. R. Rodríguez, M. J. Lavecchia, B. S. Parajón-Costa, A. C. González-Baró, M. R. González-Baró and E. R. Cattáneo, Biochimie, 2021, 186, 43–50 CrossRef.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4su00439f |
| ‡ Authors equally contributed to this work. |
| This journal is © The Royal Society of Chemistry 2024 |