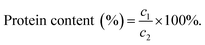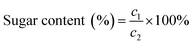Comparative extraction of antioxidant proteins from whole frogs (Rana ridibunda Pollas)
Naziermu
Dongmulati
 abc,
Ahmidin
Wali
*a,
Zi
Yang
ab,
Yusufujiang
Aili
ab,
Rexili
Kelaimu
ab,
Yanhua
Gao
a,
Abulimiti
Yili
*a and
Haji Akber
Aisa
abc,
Ahmidin
Wali
*a,
Zi
Yang
ab,
Yusufujiang
Aili
ab,
Rexili
Kelaimu
ab,
Yanhua
Gao
a,
Abulimiti
Yili
*a and
Haji Akber
Aisa
 a
a
aState Key Laboratory Basis of Xinjiang Indigenous Medicinal Plants Resource Utilization, Key Laboratory of Plants Resources and Chemistry of Arid Zone, Xinjiang Technical Institute of Physics and Chemistry, Chinese Academy of Sciences, 40-1 Beijing Road, Urumqi 830011, Xinjiang, PR China. E-mail: abu@ms.xjb.ac.cn; ahmidin@ms.xjb.ac.cn
bUniversity of Chinese Academy of Sciences, Beijing 100039, PR China
cCollege of Pharmacy, Xinjiang Medical University, Urumqi, PR China
First published on 20th November 2024
Abstract
The forest frog (Rana ridibunda Pollas) is a traditional medicinal source rich in active protein compounds. In order to extract these compounds, six extraction methods were employed, including freeze–thaw and stirring techniques. Three different solvents were utilized in this process: 0.15 M sodium chloride (NaCl), 0.05 M phosphate buffer (PB), and 0.05 M phosphate-buffered saline (PBS). The objective was to identify the most effective extraction method. The extraction efficiencies, protein content, structure, and physicochemical properties of the extracts were compared. Additionally, antioxidant activity and free amino acid composition were analyzed. The highest-scoring extract, denoted as M1, obtained through freeze–thaw extraction using 0.15 M NaCl, exhibited an extraction rate of 7.79 ± 0.71% and a protein content of 60.36 ± 2.12%. M1 also showed antioxidant activity against DPPH˙, ABTS+˙, and ˙OH free radicals, with IC50 values of 0.41, 0.41, and 0.39 mg mL−1, respectively. The freeze–thaw extraction method utilizing 0.15 M NaCl has been identified as effective for extracting proteins from dried forest frogs, confirming their potential as a source of antioxidant proteins for scientific research and application.
1 Introduction
Proteins play a crucial role in the functioning of living organisms and are the most prevalent large molecules in biological systems.1 The intricate structures and functions of proteins have made their extraction and purification a key focus of study in the fields of biochemistry and bioengineering.2,3 Currently, there are several proposed protein extraction techniques,4–6 such as ultrasonic extraction, acid–base extraction, stirring extraction, freeze–thawing extraction, and high-pressure extraction.7 These methods have their own advantages and disadvantages and yield different results depending on the experimental conditions. Recent studies on protein extraction from herbs used in animal medicine have indicated that various extraction methods differ in terms of protein yield, activity and purity.8 Some methods can effectively extract proteins, improving their purity while maintaining their biological activity. Nevertheless, further research is necessary to confirm these findings and identify the optimal extraction approach.Repeated freezing and thawing is a frequently employed technique for protein extraction,9,10 predicated on the premise that it disrupts cellular membranes and facilitates the release of proteins through successive cycles of freezing and thawing.11 This method offers the advantages of ease of operation and lower costs; however, its disadvantages include potentially lower extraction efficiency and the possibility of compromising the integrity of certain proteins. An alternative method for protein extraction is the stirring extraction technique, which involves the mechanical agitation of cells to release proteins.12 While this method is straightforward to execute and boasts high extraction efficiency, it is important to note that it carries the risk of inducing mechanical denaturation of proteins.13,14
The freeze–thaw method and the stirring method each have distinct advantages and disadvantages, and the choice of an appropriate method depends on the specific requirements and conditions of the experiment.15 For example, in industrial applications that require large-scale production, the stirring method may be more beneficial, while the freeze–thaw method may be preferred in research contexts that prioritize the preservation of protein integrity.16 In conclusion, both methods are applicable in particular scenarios and have inherent limitations in the extraction of natural protein peptides. A careful selection and optimization of these methods can enhance extraction efficiency while maintaining protein integrity.
The forest frog (Rana ridibunda Pollas) is an amphibian found in forested wetland habitats. In Chinese medicine, Rana ridibunda Pollas is a commonly used medicinal herb. According to the Chinese Materia Medica,17Rana ridibunda Pollas has the effects of clearing heat, detoxifying toxins, inducing diuresis and reducing swelling, as well as moistening the lungs and relieving coughs. Rana ridibunda Pollas with significant bioactivity is valuable for research in human health.18–20 However, there is a lack of systematic studies on the material basis of these proteins, as well as a deficiency in the application of traditional folk medicine due to the absence of a theoretical framework related to their biological activity. Therefore, various methods for extracting proteins are essential for a better understanding of the functions of Rana ridibunda Pollas proteins and for exploring their potential applications. Research on the extraction and purification methods of the proteins in the whole dried frog and its biological activity can offer a novel approach for the development and utilization of frog resources. Additionally, it can provide new insights and directions for the research and development of traditional Chinese medicine.
The primary objective of this research is to identify the most effective method for protein extraction from Rana ridibunda Pollas by combining various solvents and extraction techniques. Additionally, this study aims to conduct a thorough analysis of the physicochemical properties and antioxidant activity of the extracted proteins, a topic that has rarely been investigated in the existing literature, which may offer valuable insights for exploring the pharmacological properties and key components of Rana ridibunda Pollas in Chinese traditional medicine.
2 Materials and methods
2.1 Materials
The frogs were purchased from Tekes County, Yili, Xinjiang, and were identified as “Rana ridibunda Pollas” by Professor Abulimiti, Institute of Ecology and Geography, University of Chinese Academy of Sciences. Dialysis bags, all SDS-PAGE reagents and standard molecular weight proteins were purchased from Biosharp (Beijing, China).2.2 Protein extraction
To obtain degreased whole frog powder, the sun-dried whole frog should be crushed under liquid nitrogen using a crusher, sieved through a 40 mesh sieve, and degreased by stirring with petroleum ether in a ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30 (w/v). This degreasing process should be repeated three times until the supernatant becomes colorless. Subsequently, the residue left after degreasing should be air-dried to yield the desired degreased whole frog powder.21–23
30 (w/v). This degreasing process should be repeated three times until the supernatant becomes colorless. Subsequently, the residue left after degreasing should be air-dried to yield the desired degreased whole frog powder.21–23
The whole frog powder was extracted according to the method shown in Table 1. The extraction method was as follows: defatted whole frog powder was mixed with the three types of solvents (0.15 M NaCl, 0.05 M PB, and 0.05 M PBS) at a ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 (w/v). Subsequently, 1 mM PMSF was added to the mixture, which was then stirred for 30 minutes at room temperature before being placed in a refrigerator at −40 °C overnight. On the following day, the extract was removed and dissolved completely at room temperature. The supernatant was collected after centrifugation at 5000 rpm for 15 min. The supernatant was combined after three cycles of freeze–thawing. It was then desalted for 48 hours using a 3000 Da dialysis bag and freeze-dried to obtain the crude proteins extracted by freeze–thawing with three different solvents, named M1, M2, and M3, respectively. The crude proteins obtained by stirring at 4 °C for 3 hours were named M4, M5, and M6, respectively.
10 (w/v). Subsequently, 1 mM PMSF was added to the mixture, which was then stirred for 30 minutes at room temperature before being placed in a refrigerator at −40 °C overnight. On the following day, the extract was removed and dissolved completely at room temperature. The supernatant was collected after centrifugation at 5000 rpm for 15 min. The supernatant was combined after three cycles of freeze–thawing. It was then desalted for 48 hours using a 3000 Da dialysis bag and freeze-dried to obtain the crude proteins extracted by freeze–thawing with three different solvents, named M1, M2, and M3, respectively. The crude proteins obtained by stirring at 4 °C for 3 hours were named M4, M5, and M6, respectively.
| Sample | Solution | Extraction method | Material–liquid ratio (g mL−1) | Extraction temperature (°C) | Time (h) | Extraction times |
|---|---|---|---|---|---|---|
| M1 | 0.15 M NaCl | Freeze–thawing | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 10 |
−40 | 12 | 3 |
| M2 | 0.05 M PB | Freeze–thawing | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 10 |
−40 | 12 | 3 |
| M3 | 0.05 M PBS | Freeze–thawing | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 10 |
−40 | 12 | 3 |
| M4 | 0.15 M NaCl | Stirring | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 10 |
4 | 3 | 3 |
| M5 | 0.05 M PB | Stirring | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 10 |
4 | 3 | 3 |
| M6 | 0.05 M PBS | Stirring | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 10 |
4 | 3 | 3 |
2.3 Chemical composition analysis
Protein concentration was determined using the BCA method following the instructions provided in the BCA kit.24 Bovine serum protein was used as the standard to create a standard curve with protein concentration as the horizontal coordinate and absorbance as the vertical coordinate. The absorbance of the extract was measured three times in parallel and averaged. The protein content (%) was calculated using the following formula:where c1 is the concentration (μg mL−1) calculated from the standard curve and c2 is the initial concentration of the sample (μg mL−1).
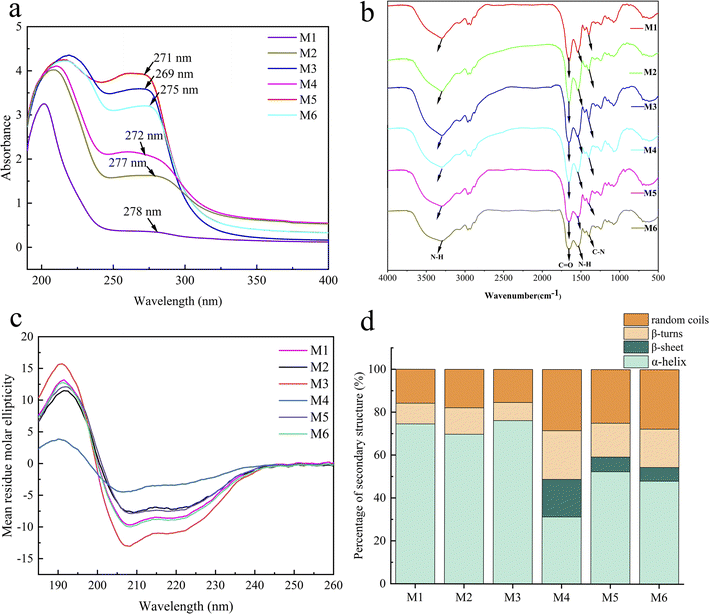
Sugar content was determined using the phenol–sulphonic acid method.25 Glucose solution was used as the standard to create the standard curve, with the concentration of glucose standard solution as the horizontal coordinate and the absorbance as the vertical coordinate. The absorbance of the extract was measured three times in parallel to obtain an average value, which was calculated using the following formula:
2.4 Spectral analysis
The samples were prepared at a concentration of 1 mg mL−1, and the UV absorption spectra were measured in the range of 200–400 nm. A small amount of each extract was mixed with an appropriate quantity of spectroscopic-grade potassium bromide powder, ground, and pressed. The infrared absorption spectra in the range of 4000–500 cm−1 were measured using a Nicolet 6700 FT-IR spectrometer (Thermo Fisher Scientific, USA).2.5 SDS-PAGE analysis
The analysis was performed under reducing, denaturing conditions using a 6% stacking gel and a 15% separating gel, and stained with Coomassie brilliant blue G-250 for 2 hours.26 2 mg of whole frog protein was dissolved in 1 mL of ultrapure water and then centrifuged. 40 μL of each extract at a concentration of 2 mg mL−1 was mixed with 10 μL of loading buffer and boiled at 95 °C for 10 min. Each lane was loaded with 30 μL of extract. The voltage was 75 V. Staining began when the dye reached the bottom of the gel, and then it was decolorized for 10 h in a decolorizing solution.2.6 Solubility study
The water solubility of the extract was tested using ultra-pure water. 2 mg each of M1, M2, M3, M4, M5, and M6 were administered and dissolved in 1 mL of ultra-pure water in 6 centrifuge tubes. The mixtures were vigorously shaken and then centrifuged at a speed of 3000 rpm for 5 minutes. The clarity of the supernatant was then compared to the amount of precipitate.2.7 Amino acid composition analysis
The amino acid composition of the extracts was analyzed by pre-column derivatization with phenyl isothiocyanate (PITC). Each sample was hydrolyzed in a headspace vial with 6 mol per L hydrochloric acid containing 0.1% phenol at 110 °C for 24 hours. Subsequently, the excess hydrochloric acid was removed by distillation at 75 °C. The residue was redissolved in 0.1 mol per L hydrochloric acid and derivatized as described in the literature.27 The PITC derivatives were analyzed by RP-HPLC as described previously.82.8 Antioxidant activity assay
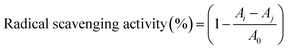 | (1) |
 | (2) |
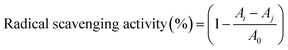 | (3) |
2.9 Statistical analysis
IC50 was calculated using GraphPad Prism 7.00.3 Results and discussion
3.1 Extraction yield and chemical composition analysis
The crude proteins extracted using various methods and solvents exhibited different extraction rates and protein contents. As shown in Table 2, the order of protein content from the largest to the smallest is M2 > M6 > M1 > M5 > M3 > M4. The extraction rates in descending order are M2 > M1 > M6 > M3 > M5 > M4. It can be observed that the extraction rate using the freeze–thawing method was higher than that of the stirring method when extracted with different solutions. The protein content of the extracts obtained through the freeze–thawing method was also higher compared to that with the stirring method when extracted with different solvents. Notably, the sugar content of the extracts obtained through the stirring method was the highest when extracted with 0.15 M NaCl. Initially, the extraction rate and protein content can be considered as key indicators. It can be concluded that the freeze–thawing method is more suitable for extracting proteins from dried frogs when considering the extraction rate and protein content as indicators.| Sample | Extraction yield% | Protein content%, m m−1 | Carbohydrate content%, m m−1 |
|---|---|---|---|
| M1 | 7.79 ± 0.7062 | 60.36 ± 2.117 | 9.94 ± 0.1054 |
| M2 | 8.01 ± 1.305 | 69.83 ± 1.066 | 6.67 ± 0.2227 |
| M3 | 6.61 ± 0.2666 | 53.56 ± 1.670 | 7.64 ± 0.1664 |
| M4 | 5.07 ± 0.9502 | 47.42 ± 1.444 | 13.00 ± 0.9590 |
| M5 | 6.51 ± 0.1300 | 59.46 ± 2.816 | 5.18 ± 0.2193 |
| M6 | 6.63 ± 0.06928 | 63.19 ± 1.725 | 7.27 ± 0.5333 |
In the research on spirulina, the repeated freeze–thaw method has shown the highest recovery rate and purity of phycocyanin, as well as the highest total protein recovery rate.32 A method utilizing freeze–thaw extraction for oat β-glucan has been proven to be simple to operate, without the requirement for enzyme agents and organic solvents, cost-effective, and capable of efficiently extracting the target substance during the freeze–thaw process.33 In essence, freeze–thaw extraction methods offer significant advantages across various fields and research areas, particularly in the extraction of bacterial DNA, microcystin toxins, cell membranes, and microalgae proteins. The primary benefits include low cost, straightforward operation, no necessity for expensive equipment and reagents, and the ability to obtain relatively pure target substances.34 Despite some limitations, the overall impact is still superior to that of traditional stirring extraction methods. This aligns with our research findings.
3.2 Spectral analysis
The maximum absorption peaks of extracts appeared at around 270 nm (Fig. 1a), and the peak shapes were narrow without any other miscellaneous peaks, indicating that valence electrons transiting in this wavelength range. The absorption peaks in this range were mainly due to the conformational changes of the aromatic amino acids in the proteins, which further confirmed the presence of the proteins in the extracts.35The infrared spectra of the six extracts are shown in Fig. 1b, illustrating the absorption characteristics of typical functional groups of proteins. The broad and strong absorption peaks between 3000 and 3670 cm−1 represent the combined absorption peaks of O–H and N–H stretching vibrations. The strong absorption peak at 1630–1670 cm−1 is the characteristic absorption peak of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O (amide I) stretching vibration. The characteristic absorption peak at 1650–1620 cm−1 corresponds to the N–H (amide II) bending vibration in primary amines. The characteristic absorption peak at 1570–1515 cm−1 is associated with free primary amine. The medium intensity absorption bands at 1420–1400 cm−1 and 1300–1260 cm−1 are the mixed frequencies of N–H bending vibration and C–N stretching vibration (amide III).36,37 In conclusion, all six types of proteins exhibit characteristic absorption peaks, but the intensities of the absorption peaks vary, indicating that different extraction methods have a certain impact on the structure of the proteins. This difference is further verified by circular dichroism analysis.
O (amide I) stretching vibration. The characteristic absorption peak at 1650–1620 cm−1 corresponds to the N–H (amide II) bending vibration in primary amines. The characteristic absorption peak at 1570–1515 cm−1 is associated with free primary amine. The medium intensity absorption bands at 1420–1400 cm−1 and 1300–1260 cm−1 are the mixed frequencies of N–H bending vibration and C–N stretching vibration (amide III).36,37 In conclusion, all six types of proteins exhibit characteristic absorption peaks, but the intensities of the absorption peaks vary, indicating that different extraction methods have a certain impact on the structure of the proteins. This difference is further verified by circular dichroism analysis.
3.3 Circular dichroism analysis
Circular dichroism (Fig. 1c) revealed variations in the intensity of the CD curves among the six extracts. Specifically, M1 exhibited higher intensity than M4, M2 was stronger than M5, and M3 surpassed M6. These results suggest that the proteins extracted using the freeze–thawing method contained higher protein content and molecular weight. All six extracts exhibited a positive Cotton effect near 195 nm, a negative Cotton effect near 208 nm and 222 nm, and a positive–negative crossover near 202 nm, which is characteristic of the α-helical structure.38 The negative Cotton effect at 218 nm and the positive Cotton effect at 195 nm, on the other hand, are characteristic of the β-helix structure. According to the Selcon3 online tool (Fig. 1d), the six whole frog proteins contained different ratios of α-helices and β-strands. Proteins M1, M2, and M3 extracted using the freeze–thawing method did not contain a β-helix. In contrast, proteins M4, M5, and M6 extracted using the stirring method contained varying amounts of the β-helix. This suggests that the different extraction methods and solutions used had an impact on the secondary structure of the proteins.3.4 Amino acid composition
In order to compare the solubility of the six extractions, the amino acid composition was analyzed (Fig. 2a). The six extracts all contained 17 common amino acids, including lysine, aspartic acid, glutamic acid, serine, glycine, histidine, arginine, threonine, alanine, proline, tyrosine, valine, methionine, cysteine, isoleucine, leucine, and phenylalanine. The amino acid compositions of the six whole frog proteins varied. M1 contained the highest hydrophobic amino acids, while M2 contained the highest hydrophilic amino acids (Fig. 2b), indicating the best solubility in water. This finding is consistent with the results of the solubility experiments. The different compositions of amino acids affect the antioxidant properties, and the relationship between them needs further analysis and verification.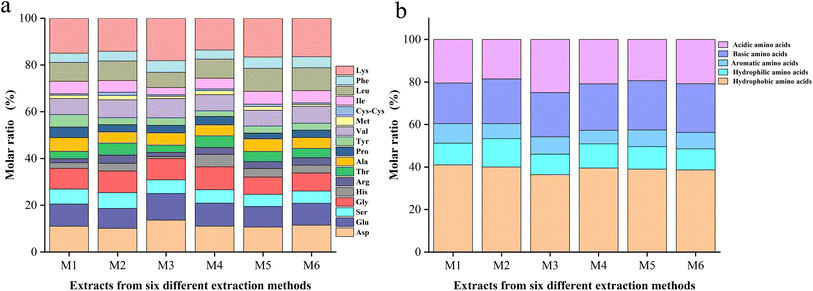 | ||
| Fig. 2 (a) Comparison of all amino acids present. (b) The five major classes of amino acids contained in M1, M2, M3, M4, M5, and M6. | ||
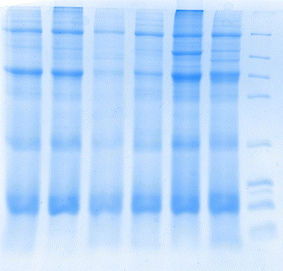
3.5 SDS-PAGE protein molecular weight
SDS-PAGE is an effective technique for separating and identifying proteins. It can separate proteins into different bands in the gel based on their molecular weight. The SDS-PAGE chromatograms of the whole frog proteins extracted by the six methods are shown in Fig. 3. The six whole frog proteins prepared showed corresponding bands in the molecular weight range of 4.1–66 kDa, and the presence of these bands indicates that the extraction process has successfully separated proteins and peptides. Proteins and peptides are crucial biomolecules in organisms, playing essential roles in the structure, function, and signal transduction of cells.3.6 Antioxidant activity
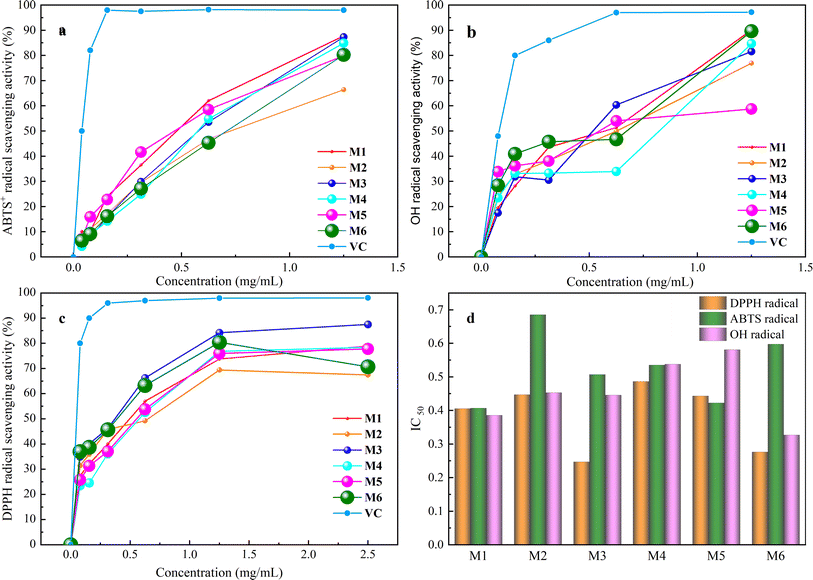
The process of antioxidation is essential, unavoidable, and vital within the body, serving a beneficial function in preserving the equilibrium of the internal milieu.39 Consequently, there is considerable interest among researchers in identifying secure and enduring natural antioxidants.40The results of antioxidant activity assay of six extractions showed (Fig. 4) that they had good scavenging ability for DPPH˙, ABTS+˙ and ˙OH radicals in the range of 0–1.5 mg mL−1 which was positively correlated with the protein concentration, and the scavenging ability was enhanced with the increase in concentration. The IC50 of DPPH˙ of the six extracts was 0.41, 0.45, 0.25, 0.49, 0.44, and 0.28 mg mL−1, and the scavenging capacity was M3 > M6 > M1 > M5 > M2 > M4. The IC50 of ABTS+˙ was 0.41, 0.69, 0.51, 0.54, 0.42, and 0.60 mg mL−1, respectively, and the magnitude of scavenging capacity was M1 > M5 > M3 > M4 > M6 > M2. The IC50 of ˙OH was 0.39, 0.45, 0.44, 0.54, 0.58 and 0.33 mg mL−1, and the magnitude of scavenging capacity was M6 > M1 > M3 > M2 > M4 > M5, which indicated that the whole frog proteins had good antioxidant properties.
Various extraction techniques and solvents play a crucial role in influencing the efficiency of extracting whole frog protein, as well as its content and antioxidant properties such as DPPH˙, ABTS+˙, and ˙OH scavenging abilities. This is demonstrated by the data presented in Table 3. The findings suggest that the choice of extraction method significantly impacts both the extraction yield and the bioactivity of whole frog proteins. By assigning a weight of 20% to factors including extraction efficiency, protein content, and DPPH˙, ABTS+˙ and ˙OH scavenging capacity, composite scores were calculated as follows: M1 65.01, M2 57.74, M3 63.33, M4 60.09, M5 56.48, and M6 62.06. The ranking based on these scores is M1 > M3 > M6 > M4 > M2 > M5. Notably, the freeze–thawing method emerges as more favorable for whole frog protein extraction compared to the stirring method, likely due to its ability to disrupt cell membranes effectively and release proteins more efficiently. Moreover, PBS proves to be a more suitable solvent for extracting whole frog protein in comparison to PB, possibly owing to its optimal ionic strength for protein solubility. Upon a comprehensive evaluation, it is evident that the overall performance index of M1 whole frog protein surpasses that of the other variants. This superiority may be attributed to the 0.15 M NaCl concentration, which closely mimics physiological conditions, facilitating effective protein extraction while safeguarding them with neutral salts. Furthermore, the NaCl solvent is cost-effective, readily available, and poses no safety concerns. Consequently, a 0.15 M NaCl solution is chosen as the preferred extraction solvent, alongside the freeze–thawing method for the extraction of whole frog protein.
| Sample | Extraction yield% | Protein content%, m m−1 | Antioxidant activity | Overall score | ||
|---|---|---|---|---|---|---|
| DPPH˙ scavenging capacity% | ABTS+˙![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) scavenging capacity% scavenging capacity% |
˙OH scavenging capacity% | ||||
| M1 | 7.79 ± 0.71 | 60.36 ± 2.12 | 78.97 ± 0.82 | 87.77 ± 2.33 | 90.16 ± 0.99 | 65.01 |
| M2 | 8.01 ± 1.30 | 69.83 ± 1.07 | 67.46 ± 1.07 | 66.42 ± 1.59 | 76.98 ± 1.44 | 57.74 |
| M3 | 6.61 ± 0.27 | 53.56 ± 1.67 | 87.58 ± 0.95 | 87.34 ± 1.25 | 81.57 ± 1.01 | 63.33 |
| M4 | 5.07 ± 0.95 | 47.42 ± 1.44 | 78.44 ± 0.75 | 84.88 ± 1.01 | 84.65 ± 1.05 | 60.09 |
| M5 | 6.51 ± 0.13 | 59.46 ± 2.81 | 77.78 ± 0.49 | 79.89 ± 0.94 | 58.74 ± 0.89 | 56.48 |
| M6 | 6.63 ± 0.07 | 63.19 ± 1.72 | 70.65 ± 0.83 | 80.21 ± 0.43 | 89.61 ± 0.81 | 62.06 |
3.7 Amino acid composition and antioxidant activity correlations
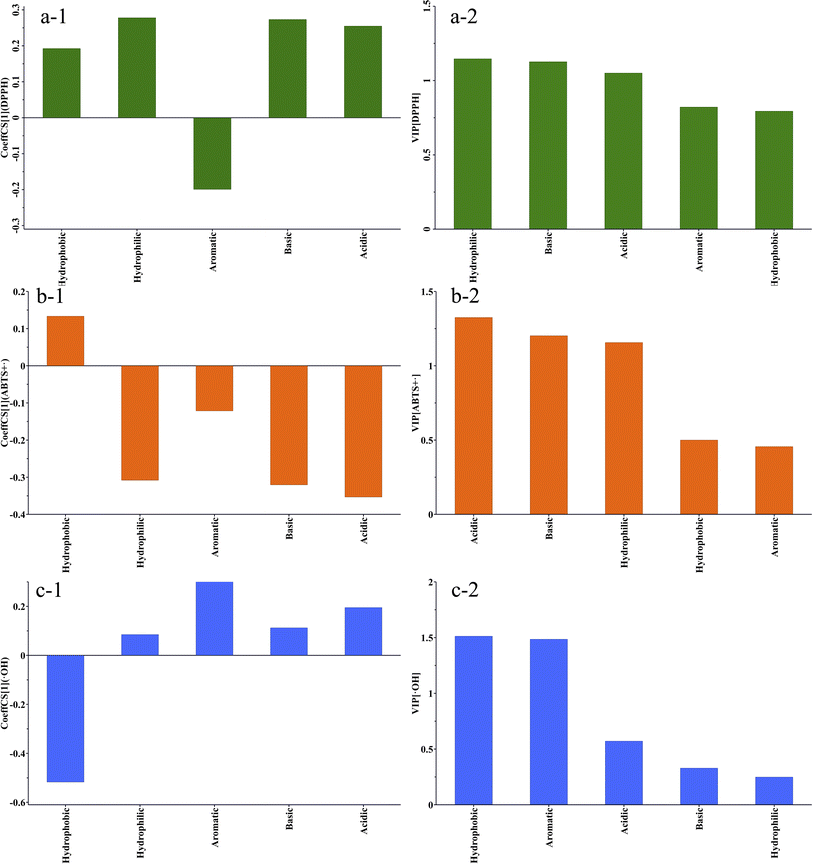
Correlation analysis is a commonly used analytical method in proteomics to examine the relationship between the amino acid composition or sequence and the activity of a compound. The correlation analysis of the amino acid composition and antioxidant activity of the extracts was conducted by utilizing the ratio of five amino acid compositions: acidic amino acids, basic amino acids, hydrophilic amino acids, hydrophobic amino acids, and aromatic amino acids, in the six extracts as the variables (X). The IC50 values of the scavenging activities of DPPH˙, ABTS+˙, and ˙OH were considered as the predicted values (Y). The results of the partial least squares regression (PLS) analysis of the amino acid composition and DPPH˙ scavenging activity are depicted in a Fig. 5a-1. The results of partial least squares regression (PLS) analysis of amino acid composition and DPPH˙ scavenging activity are shown in Fig. 5a-2. R2 = 0.999 and Q2 = 0.912. Both values are close to 1, indicating that the model has a very good fit and predictive properties. Correlation coefficient analysis revealed that hydrophobic, hydrophilic, basic, and acidic amino acids were positively correlated with the IC50 values of DPPH˙ scavenging activity. Aromatic amino acids were negatively correlated with the IC50 value. Variable Importance in Projection (VIP) is a method used to assess the relative importance of individual variables in the PLS model for predicting responses. A VIP value greater than 1 indicates strong influence, while a VIP value less than 0.5 suggests a low impact of the variable on the predicted values. The VIP score order for the impact of the five variables on the DPPH˙ scavenging activity IC50 is hydrophilic amino acids > basic amino acids > acidic amino acids > 1.0 > aromatic amino acids > hydrophobic amino acids > 0.5. The results above indicate that all variables have significant impacts on DPPH˙ scavenging activity, and low levels of hydrophilic amino acids, and high levels of aromatic amino acids and basic amino acids are associated with low IC50 values. M1 has a higher content of aromatic amino acids compared to the other extracts. Aromatic amino acids such as Tyr (tyrosine), Trp (tryptophan), and Phe (phenylalanine) can provide antioxidant protection through their aromatic groups, enhancing the ability to scavenge DPPH˙ free radicals. The aromatic groups in these amino acid residues can act as hydrogen donors, inhibiting free radical-mediated peroxidation chain reactions and thereby increasing antioxidant activity.41The PLS regression analysis results on the composition of amino acids and the ABTS+˙ scavenging activity (Fig. 5b-1) show that R2 = 0.872 > 0.6, Q2 = 0.572 > 0.5, indicating that the model has good fitting and predictive ability. The results indicate that the hydrophobic amino acids are positively correlated with the IC50 of ABTS+˙ scavenging activity, while the hydrophilic amino acids, aromatic amino acids, acidic amino acids, and basic amino acids are negatively correlated with the IC50 of ABTS+˙ scavenging activity. The VIP scores for the five variables in the order of their importance for the IC50 of ABTS+˙ scavenging activity are acidic amino acids > basic amino acids > hydrophilic amino acids > 1.0 > hydrophobic amino acids > 0.5 > aromatic amino acids.
According to reports, arginine (Arg) has a strong ability to scavenge ABTS+˙ radicals. Arginine also functions as a hydrophilic and basic amino acid. The guanidine group (–NH2) in its structure acts as a potent electron donor, capable of supplying electrons to free radicals, thereby halting free radical chain reactions.42 Compared to other extracts, M1 contains a higher amount of hydrophilic amino acids, suggesting that M1 exhibits strong antioxidant activity by efficiently interacting with free radicals and transforming them into an inactive state.
Compared to other types of amino acids, acidic amino acids also offer certain advantages in efficiently clearing ABTS+˙. According to multiple studies, acidic amino acids can chelate with metal ions because of the amino and carboxyl groups in their side chains. This process passivates oxidized metal ions and reduces the incidence of free radical chain reactions, showcasing robust antioxidant capabilities.43 For example, the acidic amino acids abundant in selenium-enriched oyster peptides exhibit high ABTS+˙ scavenging ability, with an IC50 value of 1.064 mg mL−1. Additionally, dipeptides containing tyrosine also exhibit strong free radical scavenging ability, further emphasizing the crucial role of acidic amino acids in antioxidation.44
The analysis results of the clearance activity of ˙OH show that the model's R2 = 0.978, which is close to 1, and Q2 = 0.554, which is greater than 0.5, indicating a good fit. The correlation analysis results indicate that hydrophilic amino acids, aromatic amino acids, basic amino acids, and acidic amino acids are positively correlated with the IC50 of ˙OH clearance activity, while hydrophobic amino acids are negatively correlated. The VIP scores are in the following order: hydrophobic amino acids > aromatic amino acids ≈ 1.5 > acidic amino acids > basic amino acids > hydrophilic amino acids. Hydrophobic amino acids and aromatic amino acids are closely related to the clearance ability of ˙OH. The higher the content of hydrophobic amino acids, the lower the content of aromatic amino acids, leading to higher clearance activity of ˙OH and a lower IC50 ˙OH can accept both electrons and hydrogen atoms, and can react with the heterocyclic and pentose rings of DNA, leading to damage to the DNA chain.
Large hydrophobic residues, such as Phe, Ile, Leu, and Val, play a significant role in proteins primarily because of their hydrophobic nature. These residues are typically located in the nonpolar regions of proteins, contributing to the stability and functionality of proteins. Research on gelatin antioxidant peptides has shown that peptides containing higher levels of hydrophobic amino acids such as Gly, Phe, and Lys exhibit good antioxidant activity, effectively scavenging ˙OH with a clearance rate of 55.36%.45 This indicates the important role of hydrophobic amino acids in the antioxidant activity. Furthermore, studies on hydroxyproline peptides also support this point. Leu–Hyp and Ile–Hyp peptides exhibit strong abilities to scavenge hydroxyl and superoxide anion radicals. The hydrophobic amino acids, such as Ile and Pro, in these peptides may play a crucial role in their antioxidant activity.46
Therefore, multiple studies have shown that hydrophobic amino acids, by participating in the formation of peptides or proteins with antioxidant activity, can effectively scavenge hydroxyl radicals. This process exerts antioxidant effects, which aligns with our research results. From the above results, it can be inferred that the high scavenging activity of M1 against DPPH˙, ABTS˙+, and ˙OH may be attributed to its elevated levels of hydrophobic amino acids. This characteristic enables the extract to demonstrate superior activity in scavenging ˙OH compared to other extracts.
4 Conclusion
This study employed multiple extraction methods to select a suitable method for extracting native bioactive proteins from dried whole frogs and obtained six extracts. These extracts exhibited significant differences in extraction rate, protein content, sugar content, physicochemical properties, and antioxidant activity. The extract with the highest overall score was M1, obtained through freeze–thaw extraction using 0.15 M NaCl, and this was thus selected as the optimal extract. Correlation analysis results indicated that the high scavenging activity of M1 towards DPPH˙, ABTS+˙, and ˙OH may be attributed to its higher levels of hydrophobic amino acids. This characteristic allowed this extract to demonstrate superior activity in ˙OH scavenging compared to other extracts. Therefore, the method of extracting M1 is the best way to obtain proteins of native origin from dried whole frogs, which have the potential to provide antioxidant ingredients. This discovery offers new insights for the development and utilization of Rana ridibunda Pollas resources.Data availability
The data that support the findings of this study are available from the corresponding author upon reasonable request.Author contributions
Naziermu Dongmulati: conceptualization, data curation, investigation, roles and writing-original draft, visualization. Ahmidin Wali: funding acquisition, project administration, resources. Zi Yang: data curation (protein content testing). Yusufujiang Aili: data curation (purification process). Rexili Kelaimu: investigation. Yanhua Gao: resources, project administration. Abulimiti Yili: funding acquisition, writing, reviewing, editing, project administration, supervision. Haji Akber Aisa: resources.Conflicts of interest
The authors declare that they have no financial or personal relationships that could have appeared to influence the work reported in this paper.Acknowledgements
This work was supported by the Tianchi Yingcai Programme (grant number: 2024000051), Xinjiang Uygur Autonomous Region Tianshan Yingcai-Leading Talents in Scientific and Technological Innovation (grant number: 2022TSYCLJ0008), Xinjiang Uygur Autonomous Region Tianshan Cedar Plan – Science and Technology Innovation Leaders Reserve Candidate Program (grant number: 2020XS08), and the Chinese Academy of Sciences presidents international fellowship initiative (grant no. 2023VBB0005).References
- H. Frauenfelder, Proc. Natl. Acad. Sci. U. S. A., 2002, 99, 2479–2480 CrossRef PubMed.
- M. Hassan, H. Tehrani, R. Batal, M. Kamalinejad and A. Mahbubi, J. Plant Sci., 2014, 2, 70–76 Search PubMed.
- M. N. Marsh, Celiac Disease: Methods and Protocols, 2000, pp. 55–73, DOI:10.1385/1592590829.
- M. Bayram and A. Alameen, Protein Extraction Techniques from Cereals and Legumes, International gap agriculture & livestock congress, 2018 Search PubMed.
- S. Vinayashree and P. Vasu, Food Chem., 2021, 340, 128177 CrossRef CAS PubMed.
- V. M. Phan and H. H. P. Thi, SEATUC J. Sci. Eng., 2020, 1, 39–43 Search PubMed.
- B. Liu, A. Yili, H. A. Aisa and M. Aikemu, J. Food Biochem., 2017, 42, 1–9 Search PubMed.
- A. Arken, X. Zhao, Y. Gao, A. Omar, D. Tang, A. Waili, Z. Yang, Y. Wang, H. A. Aisa and A. Yili, J. Ethnopharmacol., 2023, 311, 116359 CrossRef CAS PubMed.
- W. Jun, D. Wen-bin, L. Fu-lian, G. Er-dong and G. Jing-xuan, Journal of Food and Machinery, 2015, 31, 211–215 Search PubMed.
- X. Bai, S. Shi, B. Kong, Q. Chen, Q. Liu, Z. Li, K. Wu and X. Xia, Food Chem., 2023, 399, 134020 CrossRef CAS PubMed.
- C. Ding, M. Zhang and G. Li, Int. J. Biol. Macromol., 2015, 80, 317–323 CrossRef CAS PubMed.
- R. F. Caldeira, L. d. P. Gouvêa, T. d. L. Azevedo, C. Conte, D. d. G. C. F. d. Sá, M. C. Galdeano, I. Felberg, J. R. Lima and C. G. Mellinger, Food Res. Int., 2024, 189, 114569 CrossRef CAS PubMed.
- M. A. Mohammad, I. M. Grimsey, R. T. Forbes, I. S. Blagbrough and B. R. Conway, Colloids Surf., B, 2016, 146, 700–706 CrossRef CAS PubMed.
- A. Singh, V. Upadhyay, A. K. Upadhyay, S. M. Singh and A. K. Panda, Microb. Cell Factories, 2015, 14, 41 CrossRef PubMed.
- S. A. Bernal-Chávez, A. Romero-Montero, H. Hernández-Parra, S. I. Peña-Corona, M. L. Del Prado-Audelo, S. Alcalá-Alcalá, H. Cortés, L. Kiyekbayeva, J. Sharifi-Rad and G. Leyva-Gómez, J. Biol. Eng., 2023, 17, 35 CrossRef.
- K. Jain, N. Salamat-Miller and K. Taylor, Sci. Rep., 2021, 11, 11332 CrossRef CAS PubMed.
- H. M. Chang, P. H. But, S. C. Yao, L. L. Wang and C. S. Yeung, Pharmacology and Applications of Chinese Materia Medica, 1986 Search PubMed.
- T. H. Joseph, B. Kim, Y. J. Basir, J. Dulka, F. C. Knoop, P. W. Abel and J. Michael Conlon, Regul. Pept., 2000, 90, 53–60 CrossRef PubMed.
- M. Al-Halbosiy, Iraqi J. Agric. Sci., 2018, 49, 623–631 Search PubMed.
- M. F. Ali, F. C. Knoop, H. Vaudry and J. M. Conlon, Peptides, 2003, 24, 955–961 CrossRef CAS.
- A. Wali, Y. Mijiti, G. Yanhua, A. Yili, H. A. Aisa and A. Kawuli, Int. J. Pept. Res. Ther., 2020, 27, 219–227 CrossRef.
- A. Wali, A. Wubulikasimu, S. Mirzaakhmedov, Y. Gao, A. Omar, A. Arken, A. Yili and H. A. Aisa, Molecules, 2019, 24, 4103–4118 CrossRef PubMed.
- H. Wang, Y. Lu, X. Zhang, Y. Hu, H. Yu, J. Liu and J. Sun, Peptides, 2009, 30, 273–282 Search PubMed.
- I. Arcan and A. Yemenicioglu, Food Res. Int., 2010, 43, 140–147 Search PubMed.
- T. Masuko, A. Minamii, N. Iwasaki, T. Majima, S. Nishimura and Y. Lee, Anal. Biochem., 2005, 339, 69–72 CrossRef PubMed.
- U. K. Laemmli, Nature, 1970, 227, 680–685 Search PubMed.
- V. Barkholt and A. L. Jensen, Anal. Biochem., 1989, 177, 318–322 CrossRef PubMed.
- C. Yi, X. M. Yong and G. X. Feng, Nat. Prod. Res. Dev., 2006, 917–921 Search PubMed.
- J. W. Dong, L. Cai, Y. Xing, J. Yu and Z. T. Ding, Nat. Prod. Commun., 2015, 10, 2169 Search PubMed.
- I. Valent, D. Topoľská, K. Valachová, J. Bujdák and L. Šoltés, Biophys. Chem., 2016, 212, 9–16 CrossRef PubMed.
- U. Venkatachalam and S. Muthukrishnan, J. Acute Med., 2012, 2, 36–42 CrossRef.
- Z. Shaobin, Y. Xiaonan, L. Ying and L. Hui, J. Jilin Agric. Univ., 2007, 29, 381–383 Search PubMed.
- J. Wu, X. Lin, D. Huang and B. Xie, J. Chin. Inst. Food Sci. Technol., 2011, 11, 48–54 Search PubMed.
- G. Feng, M. Chen, S. Yang and H. Zhu, J. South China Agric. Univ., 2013, 34, 439–442 Search PubMed.
- C. Li, G. Bu, F. Chen and T. Li, CyTA--J. Food, 2020, 18, 409–416 CrossRef CAS.
- A. Arken, X. M. Zhao, Y. H. Gao, A. Omar, D. Tang, A. Waili, Z. Yang, Y. H. Wang, H. A. Aisa and A. Yili, J. Ethnopharmacol., 2023, 311, 116359 CrossRef CAS PubMed.
- K. Sitthiya, L. Devkota, M. B. Sadiq and A. K. Anal, J. Food Sci. Technol., 2018, 55, 658–666 CrossRef CAS PubMed.
- Y. Zhou, Y. Ma, L. Li and X. Yang, Food Chem., 2020, 309, 125671 CrossRef CAS.
- İ. Gulcin, Arch. Toxicol., 2020, 94, 651–715 CrossRef.
- S. Egerton, S. Culloty, J. Whooley, C. Stanton and R. P. Ross, Food Chem., 2018, 245, 698–706 CrossRef CAS.
- X. HU, M. Zeng and j. Chen, Food Sci., 2012, 33, 6–10 CAS.
- C. L. Hawkins and M. J. Davies, Biochim. Biophys. Acta, Bioenerg., 2001, 1504, 196–219 CrossRef CAS PubMed.
- Y. Zhang, W. Chen, P. Zou and Y. Xu, Food and Fermentation Industries Editorial Staff, 2024, 50, 197–205 Search PubMed.
- Z. Xia, B. Chen, W. Huang, A. Kang, X. Liang, Y. Yi, D. Tang, L. Meng, G. Liu and Y. Cao, Food and Fermentation Industries Editorial Staff, 2023, 49, 120–128 Search PubMed.
- X. Zhang and X. Chen, Food Res. Dev., 2014, 35, 5 Search PubMed.
- Z. Ruiya, Z. Xiaohua and D. U. Shouying, Chin. J. Biotechnol., 2007, 23, 704–709 Search PubMed.
| This journal is © The Royal Society of Chemistry 2025 |