A catalytic assembly triggered DNAzyme motor on spherical nucleic acids for sensitive small extracellular vesicle detection†
Xiaoying
Shi‡
a,
Tingting
Zhang‡
a,
Shisheng
Zhu
b,
Linhong
Ning
b,
Heng
Cheng
a,
Feng
Yu
 *a and
Shanshan
Tian
*c
*a and
Shanshan
Tian
*c
aDepartment of Gastroenterology, Chenjiaqiao Hospital of Shapingba District Affiliated to Chongqing Medical and Pharmaceutical College, Chongqing, 401331, China. E-mail: yufeng19790826@163.com
bCollege of Chongqing Medical and Pharmaceutical College, Chongqing, 401331, China
cPre-hospital Emergency Department, Chongqing Emergency Medical Center, Chongqing University Central Hospital, Chongqing, 400014, China. E-mail: yyg9991@sina.com
First published on 11th November 2024
Abstract
The expression levels of small extracellular vesicles (sEVs) are closely associated with several significant biological processes, which can be used as a crucial biomarker for cancer diagnosis, such as colorectal cancer. More efforts are still necessary to amplify sEV detection sensitivity, as their expression is minimal during the early stages of colorectal cancer. Through the integration of a catalytic assembly-triggered DNAzyme motor and gold nanoparticle (AuNP) aggregation, we have developed a triple signal amplified biosensor for the detection of sEVs. In this method, the catalytic assembly triggered DNAzyme motor continuously cleaved on the hairpin probe which is fixed on the surface of AuNPs, leaving a single-stranded sequence on the surface of AuNPs to induce the aggregation. This approach employs a triple signal amplification process to enhance the efficiency of the reaction and circumvent the issue of expensive and readily degradable proteases. The signal output system is based on dynamic light scattering technology, which enables ultra-sensitive detection of sEVs with a detection limit of 3.08 particles per μL. The present strategy exhibits significant potential for the analysis of a variety of additional analytes in clinical research disciplines due to its appealing analytical capabilities.
1 Introduction
Colorectal cancer is a malignant tumor that is characterized by a high incidence and mortality rate, accounting for the third most common cancer and the second leading cause of cancer-related deaths.1 The current strategies for early diagnosis of colorectal cancer primarily rely on physical and pathological methods, which are invasive, insensitive, and laborious, even though early diagnosis and nursing are beneficial for improving the five-year survival or overall survival rate.2–4 In contrast, liquid biopsy has shown significant potential for diagnosing colorectal cancer due to its non-invasiveness and high sensitivity. This is achieved by analyzing biomarkers in body fluid, including circulating tumor DNA (ctDNA), CTC (circulating tumor cell), cell-free nucleic acids (cfNAs), and small extracellular vesicles.5–7 Therefore, it is crucial to develop a liquid biopsy biomarker detection platform for colorectal cancer diagnosis and guiding nursing strategies.Small extracellular vesicles (sEVs) are nanoscale vesicles released by almost all cells and have a diameter ranging from 30 to 150 nm.8,9 According to the process by which sEVs are formed, certain biological molecules from the parent cells (such as proteins, RNA, DNA, amino acids, and metabolites) can be selectively enclosed within sEVs.10–12 These molecules then serve a crucial function by transferring them to recipient cells. Furthermore, these components have the capability to forecast the condition of parental cells. The sEVs are well recognized as a significant indicator for the liquid biopsy of tumors.13,14 Nevertheless, the abundance of sEVs during the initial phase of the disease is exceedingly limited. Therefore, the method's sensitivity in detecting sEVs is crucial for the early diagnosis of tumors. In recent years, several techniques have been developed to identify sEVs by selectively targeting various markers present on these vesicles. When compared to detecting miRNA or DNA in sEVs, detecting the surface proteins of sEVs can be a superior option for quantifying sEVs because it avoids damaging the sEVs and is more feasible as a marker.
The current approaches commonly employ western blot, enzyme-linked immunosorbent assay, and flow cytometry for the measurement of sEVs.15,16 Nevertheless, techniques that exhibit limited sensitivity and specificity tend to be both labor-intensive and costly. Although various techniques such as microfluidic platform,17 surface plasmon resonance,18,19 electrochemistry,20,21 and fluorescence22–24 have been employed to improve the analytical capabilities of sEVs, there is still a need to develop a more convenient and sensitive method for detecting exosomes in clinical applications.
In this study, we depicted a sensitive biosensor to analyze sEVs by combining a catalytic assembly-induced DNAzyme motor with the aggregation of gold nanoparticles (AuNPs). This method utilizes a sensing probe that is constructed through the hybridization of the hairpin structure sequence and the CD63 aptamer to selectively interact with sEVs and activate the catalytic assembly process. The catalytic assembly led to the formation of two fully functional DNAzyme motors within the double-stranded DNA (dsDNA), thereby initiating a triple signal amplification process. The ongoing movement of DNAzyme resulted in the continual liberation of hairpin fragments. These hairpin fragments, which were positioned on the surface of the AuNPs, were mutually complementary, leading to the aggregation of AuNPs. The DNAzyme walker experiment achieves highly sensitive signal detection by utilizing dynamic light scattering (DLS) technology as the output system for signals. Additionally, the biosensor exhibited excellent stability and performance when analyzing clinical samples, indicating significant potential for liquid biopsies.
2 Results and discussion
2.1 The principle of the catalytic assembly triggered DNAzyme for sEV detection
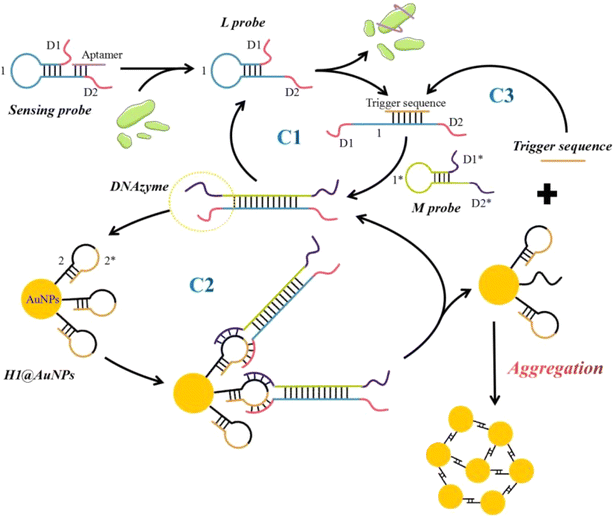
The principle of self-assembly of AuNPs, caused by the catalytic assembly triggered DNAzyme motor, for the detection of sEVs is illustrated in Scheme 1. This method involves the construction of a sensing probe by assembling a hairpin structure sequence (L probe) and a CD63 aptamer. When sEVs are present in the sensing system, the CD63 protein interacts with the CD63 aptamer on the sensing probe, exposing the CD63 aptamer binding site in the L probe. The released L probe then triggers the catalytic assembly with the help of the trigger sequence. The trigger sequence specifically causes the L probe to unfurl and create a trigger-L duplex. This duplex is then separated by the M probe, resulting in the formation of an L–M duplex. The resulting L–M hybrid consists of two catalytically active DNAzyme units positioned at each end of the duplex. Therefore, the trigger sequence initiates the efficient catalytic assembly reaction, resulting in the consecutive hybridization between the hairpin L probe and M probe, and continually producing the L–M product (C1). When magnesium ions (Mg2+) are present in the solution, the DNAzyme structure demonstrates its catalytic activity and cleaves the H1 probe. This cleavage causes the “2*” portion, which is similar to the trigger sequence, to detach from the surface of AuNPs. The released “2*” sequence binds with a subsequent L probe, resulting in the initiation of an extra signal cycle (C3). Simultaneously, the DNAzyme structure's toeholds become exposed, enabling them to connect with the subsequent H1 probe and continue the cutting process (C2). The “2” sequence remaining on the surface of AuNPs after cleavage can pair with one other, causing the AuNPs to aggregate and create a network structure with DNA. When the particle size is determined using Dynamic Light Scattering (DLS) technology, the resulting value is greater.2.2 Validation of the integrated CHA-triggered DNAzyme motor
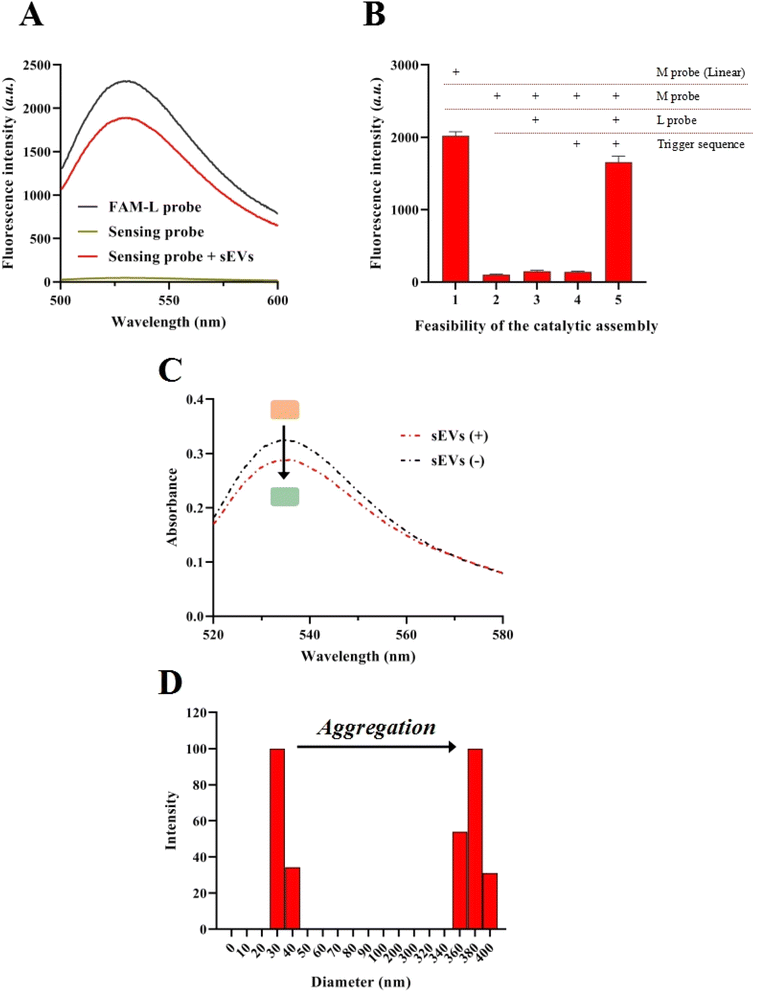
A fluorescence assay was conducted to investigate the assembly of the sensing probe and the sEV identification. In detail, the 3′ terminal of the L probe and the 5′ terminal of the aptamer sequence were labeled with FAM (FAM-L probe) and DABCYL. When the L probe was assembled with the aptamer sequence, the FAM signal was significantly quenched by the DABCYL (sensing probe). When the sensing probe is mixed with sEVs, the FAM signal reappeared (sensing probe + sEVs), indicating that the aptamer sequence was released from the sensing probe (Fig. 1A). The feasibility of the catalytic assembly was tested by using the synthesized L probe. Moreover, the FAM and DABCYL were labeled on the two terminals of the M probe. In the linear state of the M probe, the FAM signal was high. When the M probe was assembled into a hairpin structure, the FAM signals significantly decreased because the FAM signal was quenched by the DABCYL. When the M probe was incubated with the trigger sequence, the FAM signal remained at a low level, implying no non-specific hybridization between the M probe and trigger sequence. Meanwhile, no significant increase in the FAM signal could be observed when the M probe was incubated with the L probe. Only when the M probe, trigger sequence, and L probe existed in the sensing system, a greatly increased FAM signal could be recorded, indicating the feasibility of the catalytic assembly process (Fig. 1B).To further verify the cleavage effect of DNAzyme motor-mediated aggregation of the AuNPs, ultraviolet absorption spectroscopy and DLS studies were conducted. Fig. 1C demonstrates that when the target is present, the color of the sample solution changes from red to blue-black, indicating the aggregation of AuNPs. The maximum intensity of the UV absorption spectra of the solution reduced following the reaction, indicating the aggregation of AuNPs. While the UV spectra had a small drop, the DLS detection exhibited a highly substantial alteration in the signal. In the absence of the target, the hydrodynamic diameter of AuNPs in the post-reaction solution was 40.1 nm and the size distribution of the AuNPs alone was narrow (Fig. 1D). However, when the target was added, the average hydrodynamic diameter of AuNPs in the post-reaction solution increased to 384.3 nm, showing that the AuNPs aggregated but with a wider size distribution because the hybridization of the AuNPs is random. TEM results also demonstrated the appearance and aggregation of the AuNPs (Fig. S1†).
2.3 Optimization of assay conditions
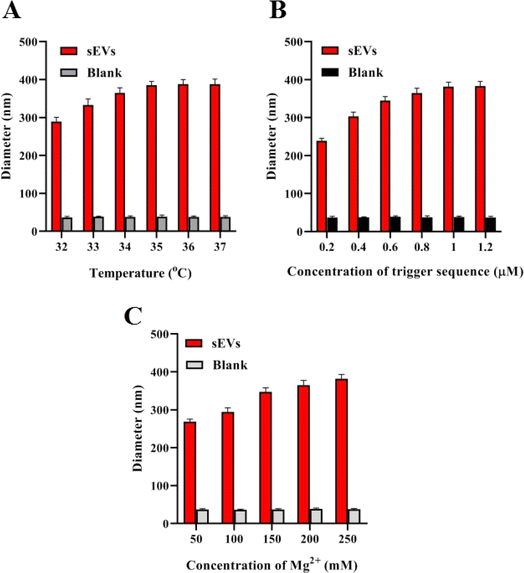
Once the feasibility of the catalytic assembly triggered DNAzyme was confirmed, the important parameters that could potentially impact the experimental sensitivity were optimized. The sensitivity of the method is potentially influenced by the hybridization temperature, the concentration of the trigger sequence, and the concentration of Mg2+ ions. The optimal parameters are determined by applying the proposed method for detecting 105 particles per μL of sEVs and the difference in diameter (Δdiameter) between the two solutions, one without the target and one with the target, was compared. The hybridization temperature of 32–37 °C was initially investigated, as depicted in Fig. 2A. The results indicated that the average diameter difference of AuNPs in the solution progressively rose within the temperature range of 32–35 °C, reaching its peak at 35 °C. Therefore, 35 °C was chosen as the best temperature for hybridization. Next, the impact of the concentration of the trigger sequence on the detection performance was investigated (Fig. 2B). Observations indicate that when the concentration of the trigger sequence ranged from 0 to 1.0 μM, the disparity in particle size progressively grew. However, when the concentration exceeded 1.0 μM, the particle size difference appeared to stabilize. Thus, a concentration of 1.0 μM trigger sequence was determined to be the optimal condition. The study focused on analyzing the concentration of Mg2+ ions. Lastly, the concentration of the Mg2+ ion was investigated (Fig. 2C). The findings indicated that the particle size difference of AuNPs increases progressively between 50 and 200 mM for Mg2+ ions and tends to remain constant when the concentration exceeds 200 mM. The particle size difference attained its maximum at a concentration of 200 mM of Mg2+ ions. Consequently, 200 mM was selected as the optimal condition. This is in accordance with the optimal reaction conditions that were previously reported for DNAzyme.2.4 Sensitivity of the assay
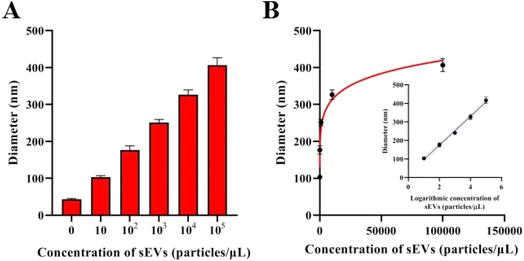
To assess the sensitivity of this method, various concentrations of sEVs were detected by the proposed method. Given the best experimental conditions mentioned above, the average diameter of particles consistently grows as the concentration of sEVs increases. Fig. 3A demonstrates that there is a linear relationship between the hydrodynamic size of particles and the logarithm of sEV concentration when the concentration falls between the range of 10 particles per μL–105 particles per μL. The equation describing the relationship between the diameter of the AuNPs (represented by D) and the concentration of sEVs (represented by C) is D = 77.65 × log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) C + 19.77. The correlation coefficient for this relationship is 0.9973, as shown in Fig. 3B. The LOD achieved according to the 3δ rule is 3.08 particles per μL.
C + 19.77. The correlation coefficient for this relationship is 0.9973, as shown in Fig. 3B. The LOD achieved according to the 3δ rule is 3.08 particles per μL.
2.5 Stability and repeatability of the assay in constructed clinical samples
To assess the stability of the suggested method, a commercial serum solution containing 104 particles per μL sEV sample was subjected to a reaction and stored for 0, 12, 24, 36, 48, and 60 hours. Subsequently, the diameter of the nanoparticles in the solution was determined. The findings are depicted in Fig. 4A, and the detected diameters were relatively stable. The relative standard deviation (RSD) of the samples was used to assess the strategy's repeatability. In this study, the RSD was determined by repeating a constructed clinical sample with 100 particles per μL sEVs six times (n = 6). The experimental results are depicted in Fig. 4B. The calculated RSD value of the method for the detection of six sEV samples was 3.12%, suggesting that the method has good repeatability.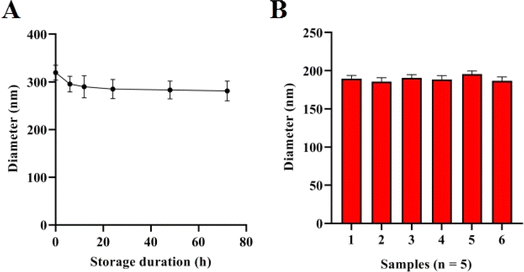 | ||
| Fig. 4 Stability analysis of the method. (A) Diameters of the method with different storage durations for sEV detection. (B) Diameter of the method when detecting sEVs from six sample duplicates. | ||
3 Conclusion
In conclusion, we propose a sensitive and reliable approach for sEV detection based on catalytic assembly-triggered DNAzyme motor-induced AuNP self-assembly. This technique has several advantages (Table S2†).22–24 First, it utilizes the enzyme cleavage features of DNAzyme to achieve signal amplification without the need for expensive and quickly destroyed protein enzymes typically utilized in conventional signal amplification processes. Second, the approach is highly sensitive due to the triple signal amplification process. Lastly, by employing DLS technology as the signal output system, the method achieves a highly sensitive and reliable signal output. Based on this, the proposed method exhibited a low limit of detection of 3.08 particles per μL and a wide detection range covering 5 orders of magnitudes. Overall, this approach shows significant promise in accurately detecting and identifying sEVs with a high level of sensitivity and specificity.4 Materials and methods
4.1 Chemicals and reagents
Table S1† lists the DNA sequences that were acquired from TaKaRa Biotech Company (Dalian, China). The hairpin probe was modified with a 3-mercapto group for HPLC purification to modify AuNPs. The DNAzyme and protecting probe were purified using polyacrylamide gel electrophoresis (PAGE). Dynamic light scattering (DLS) experiments were conducted on a Malvern Nano ZS/Mastersizer 2000 E. The DLS instrument operated under the following conditions: a scattering angle of 90° and a temperature of 25 °C.4.2 Preparation of AuNPs modified with a hairpin probe
AuNPs were synthesized using sodium citrate reduction. First, a volume of 25 μL of a 20 μM H1 probe was mixed with 100 μL of AuNPs. After extensive agitation, the mixture was stored at a temperature of −20 °C for 2 hours and subsequently thawed at the ambient room temperature. Subsequently, the mixture was centrifuged at a speed of 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 20 minutes at a temperature of 4 °C. Following centrifugation, the liquid portion above the sediment was discarded, and the underlying layer of oil was dissolved in 100 μL of sterilized water. The above centrifugation procedure was done twice for washing. Ultimately, the oil was evenly distributed in a volume of 20 μL of water.
000 rpm for 20 minutes at a temperature of 4 °C. Following centrifugation, the liquid portion above the sediment was discarded, and the underlying layer of oil was dissolved in 100 μL of sterilized water. The above centrifugation procedure was done twice for washing. Ultimately, the oil was evenly distributed in a volume of 20 μL of water.
4.3 Detection procedures
Ethical statement
Ethical Approval This article does not contain any studies with human participants or animals performed by any of the authors.Data availability
All data generated and analyzed during this study are included in this article.Author contributions
F. Y. and S. T. were the supervisors of the team in all research steps including designing, data analysis and manuscript writing. X. S. and T. Z., as the first authors, played the main role in experimental data collection, data gathering, preparation of results, and data analysis. S. Z., L. N., and H. C. assisted in the data analysis.Conflicts of interest
The authors declare no competing interests.Acknowledgements
The authors acknowledge financial and technical support from the Chongqing science and health joint medical research project fund (no. 2023MSXM074).References
- L. Klimeck, T. Heisser, M. Hoffmeister and H. Brenner, Colorectal cancer: A health and economic problem, Best Pract. Res. Clin. Gastroenterol., 2023, 66, 101839 CrossRef.
- E. Dekker, P. J. Tanis, J. L. A. Vleugels, P. M. Kasi and M. B. Wallace, Colorectal cancer, Lancet, 2019, 394(10207), 1467–1480 CrossRef.
- N. N. Mahmoud, Colorectal Cancer: Preoperative Evaluation and Staging, Surg. Oncol. Clin., 2022, 31(2), 127–141 CrossRef.
- H. Yan, R. Talty and C. H. Johnson, Targeting ferroptosis to treat colorectal cancer, Trends Cell Biol., 2023, 33(3), 185–188 CrossRef PubMed.
- M. I. Del Principe, E. De Bellis, C. Gurnari, E. Buzzati, A. Savi, M. A. I. Consalvo and A. Venditti, Applications and efficiency of flow cytometry for leukemia diagnostics, Expert Rev. Mol. Diagn, 2019, 19(12), 1089–1097 CrossRef PubMed.
- L. M. Channon, V. M. Tyma, Z. Xu, D. W. Greening, J. S. Wilson, C. J. Perera and M. V. Apte, Small extracellular vesicles (exosomes) and their cargo in pancreatic cancer: Key roles in the hallmarks of cancer, Biochim. Biophys. Acta, Rev. Cancer, 2022, 1877(3), 188728 CrossRef PubMed.
- X. Guo, R. Sui and H. Piao, Tumor-derived small extracellular vesicles: potential roles and mechanism in glioma, J. Nanobiotechnol., 2022, 20(1), 383 CrossRef PubMed.
- E. Gargiulo, E. Viry, E. Moussay and J. Paggetti, Small extracellular vesicles: multi-faceted tools for leukemia immune evasion in vivo, Oncoimmunology, 2022, 11(1), 2127507 CrossRef.
- M. Li, G. Sun, J. Zhao, S. Pu, Y. Lv, Y. Wang, Y. Li, X. Zhao, Y. Wang, S. Yang, T. Cheng and H. Cheng, Small extracellular vesicles derived from acute myeloid leukemia cells promote leukemogenesis by transferring miR-221-3p, Haematologica, 2024, 109(10), 3209–3221 CrossRef.
- Y. Jia, L. Yu, T. Ma, W. Xu, H. Qian, Y. Sun and H. Shi, Small extracellular vesicles isolation and separation: Current techniques, pending questions and clinical applications, Theranostics, 2022, 12(15), 6548–6575 CrossRef PubMed.
- F. Urabe, N. Kosaka, K. Ito, T. Kimura, S. Egawa and T. Ochiya, Extracellular vesicles as biomarkers and therapeutic targets for cancer, Am. J. Physiol. Cell Physiol., 2020, 318(1), C29–C39 CrossRef PubMed.
- D. K. Jeppesen, A. M. Fenix, J. L. Franklin, J. N. Higginbotham, Q. Zhang, L. J. Zimmerman, D. C. Liebler, J. Ping, Q. Liu, R. Evans, W. H. Fissell, J. G. Patton, L. H. Rome, D. T. Burnette and R. J. Coffey, Reassessment of Exosome Composition, Cell, 2019, 177(2), 428–445 CrossRef PubMed.
- S. Araujo-Abad, M. Saceda and C. de Juan Romero, Biomedical application of small extracellular vesicles in cancer treatment, Adv. Drug Deliv. Rev., 2022, 182, 114117 CrossRef.
- C. Zhang, C. Qin, S. Dewanjee, H. Bhattacharya, P. Chakraborty, N. K. Jha, M. Gangopadhyay, S. K. Jha and Q. Liu, Tumor-derived small extracellular vesicles in cancer invasion and metastasis: molecular mechanisms, and clinical significance, Mol. Cancer, 2024, 23(1), 18 CrossRef PubMed.
- A. Kurata, S. Kiyohara, T. Imai, S. Yamasaki-Yashiki, N. Zaima, T. Moriyama, N. Kishimoto and K. Uegaki, Characterization of extracellular vesicles from Lactiplantibacillus plantarum, Sci. Rep., 2022, 12(1), 13330 CrossRef.
- H. Im, K. Lee, R. Weissleder, H. Lee and C. M. Castro, Novel nanosensing technologies for exosome detection and profiling, Lab Chip, 2017, 17(17), 2892–2898 RSC.
- J. Wang, P. Ma, D. H. Kim, B. F. Liu and U. Demirci, Towards Microfluidic-Based Exosome Isolation and Detection for Tumor Therapy, Nano Today, 2021, 37, 101066 CrossRef PubMed.
- W. Chen, Z. Li, W. Cheng, T. Wu, J. Li, X. Li, L. Liu, H. Bai, S. Ding, X. Li and X. Yu, Surface plasmon resonance biosensor for exosome detection based on reformative tyramine signal amplification activated by molecular aptamer beacon, J. Nanobiotechnol., 2021, 19(1), 450 CrossRef.
- F. Liu, N. Li, Y. Shang, Y. Wang, Q. Liu, Z. Ma, Q. Jiang and B. Ding, A DNA-Based Plasmonic Nanodevice for Cascade Signal Amplification, Angew Chem. Int. Ed. Engl., 2022, 61(22), e202114706 CrossRef CAS PubMed.
- A. Bagheri Hashkavayi, B. S. Cha, E. S. Lee, S. Kim and K. S. Park, Advances in Exosome Analysis Methods with an Emphasis on Electrochemistry, Anal. Chem., 2020, 92(19), 12733–12740 CrossRef CAS PubMed.
- V. Gajdosova, L. Lorencova, P. Kasak and J. Tkac, Electrochemical Nanobiosensors for Detection of Breast Cancer Biomarkers, Sensors, 2020, 20(14), 4022 CrossRef.
- X. Zhao, C. Luo, Q. Mei, H. Zhang, W. Zhang, D. Su, W. Fu and Y. Luo, Aptamer-Cholesterol-Mediated Proximity Ligation Assay for Accurate Identification of Exosomes, Anal. Chem., 2020, 92(7), 5411–5418 CrossRef.
- X. Zhao, L. Zeng, Q. Mei and Y. Luo, Allosteric Probe-Initiated Wash-Free Method for Sensitive Extracellular Vesicle Detection through Dual Cycle-Assisted CRISPR-Cas12a, ACS Sens., 2020, 5(7), 2239–2246 CrossRef PubMed.
- X. Zhao, W. Zhang, X. Qiu, Q. Mei, Y. Luo and W. Fu, Rapid and sensitive exosome detection with CRISPR/Cas12a, Anal. Bioanal. Chem., 2020, 412(3), 601–609 CrossRef.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4ay01845a |
| ‡ Contributed equally to this article. |
| This journal is © The Royal Society of Chemistry 2025 |