Open Access Article
Open Access ArticleA dual-locked cyclopeptide–siRNA conjugate for tumor-specific gene silencing†
Chen
Li‡
a,
Shuaishuai
Sun‡
b,
Hao
Kong
a,
Xiangqian
Xie
b,
Gaolin
Liang
*c,
Yan
Zhang
 *a,
Huan
Wang
*a,
Huan
Wang
 *b and
Jinbo
Li
*b and
Jinbo
Li
 *a
*a
aState Key Laboratory of Analytical Chemistry for Life Sciences, Jiangsu Key Laboratory of Advanced Organic Materials, School of Chemistry and Chemical Engineering, Chemistry and Biomedicine Innovation Center (ChemBIC), Nanjing University, Nanjing 210023, China. E-mail: njuzy@nju.edu.cn; jinboli@nju.edu.cn
bState Key Laboratory of Coordination Chemistry, Jiangsu Key Laboratory of Advanced Organic Materials, School of Chemistry and Chemical Engineering, Chemistry and Biomedicine Innovation Center (ChemBIC), Nanjing University, Nanjing 210023, China. E-mail: wanghuan@nju.edu.cn
cState Key Laboratory of Digital Medical Engineering, School of Biological Science and Medical Engineering, Southeast University, Nanjing 211189, China. E-mail: gliang@seu.edu.cn
First published on 25th November 2024
Abstract
Strategies allowing tumor-selective siRNA delivery while minimizing off-tumor gene silencing effects are highly demanded to advance cancer gene therapy, which however still remain challenging. We herein report a dual-locking bioconjugation approach to address this challenge. A dual-locked cyclopeptide–siRNA conjugate (DPRC) was designed to simultaneously endow siRNA with tumor-targeting properties and tumor-biomarker/visible-light dually controllable action. The DPRC consisted of a programmed death-ligand 1 (PD-L1)-targeting cyclopeptide as a tumor-homing ligand and B-cell lymphoma-2 (Bcl-2)-targeting siRNA as a payload. They were conjugated via a tandem-responsive cleavable linker containing a photocleavable coumarin moiety quenched by naphthylamide through a disulfide linkage. Owing to the interaction between cell-membrane PD-L1 and the cyclopeptide, the DPRC was efficiently taken up by PD-L1-positive cancer cells. Notably, the internalized DPRC could only release and restore the gene silencing activity of siBcl-2 upon GSH-mediated disulfide bond breakage followed by visible light irradiation on the coumarin moiety to induce photo-cleavage. The released siBcl-2 further silenced the expression of anti-apoptotic Bcl-2 to suppress cancer cell growth. We demonstrated the tumor-targeting and dual-locked action of siRNA by the DPRC in both two-dimensional and three-dimensional cancer cell cultures. This study thus presents a novel strategy for precise tumor-specific gene silencing by siRNA.
Introduction
Small-interfering RNA (siRNA) is a synthetic double-stranded oligonucleotide that theoretically can knock down any gene of interest via the RNA-interference (RNAi) pathway.1 In light of its potential to drug traditionally undruggable oncogenes, siRNA represents a promising anti-cancer modality.2 However, tumor-targeted siRNA delivery still remains a formidable challenge.3 This stems from its lack of both tumor-recognition ability and membrane permeability, with the latter mainly caused by its polyanionic properties and large size.4 To tackle these issues, nanoparticle encapsulation and ligand modification are often employed.5 Lipid nanoparticles and conjugated N-acetylgalactosamine are now the two state-of-the-art delivery vehicles, which have been well validated using clinically approved siRNA drugs.6 Nevertheless, these delivery systems can only transport siRNA into liver, leaving extrahepatic tissues including tumor still out of reach.7 Therefore, strategies allowing tumor-specific siRNA delivery are highly demanded to advance cancer gene therapy.Bioconjugation offers a practical way to achieve cell type-specific siRNA delivery.8,9 Generally, a siRNA bioconjugate is constructed by covalently appending a homing ligand to siRNA through a linker.10 Guided by the ligand, the siRNA bioconjugate binds to a particular membrane receptor on target cells, resulting in cellular internalization and transportation.11 To achieve tumor-targeted delivery, various ligands such as a tumor-homing antibody,12,13 aptamers,14,15 peptides,16,17 and others18 have been demonstrated to be feasible. Notably, it is ideal to restrict RNAi activity in tumor cells while sparing normal cells, as nonspecific cellular uptake is inevitable.19 Therefore, to realize controllable release and activation of siRNA in tumor cells, cleavable linkers responsive to tumor biomarkers (e.g., acidic pH,20 glutathione (GSH),21 reactive oxygen species (ROS),13,22 and enzyme23) or external stimuli (e.g., light12,24 and bioorthogonal chemical25) have been used. In these activatable bioconjugates, tumor-homing ligands are attached to key sites of siRNA to block RNAi activity until linker breakage. However, such a single-lock design may cause undesirable siRNA release in a complex biological milieu because of the moderate expression of biomarkers at noncancerous sites or nonspecific bond-cleavage.26 Hence, tumor-targeted siRNA delivery using the bioconjugation approach still waits to be improved.
Here, we report a dual-locked cyclopeptide–siRNA conjugate (DPRC) for tumor-specific siRNA delivery and gene silencing (Scheme 1). The DPRC consists of a PD-L1 (programmed cell death ligand 1)-targeting cyclopeptide, a Bcl-2 (B-cell lymphoma-2)-targeting siRNA (siBcl-2), and a glutathione (GSH)/visible-light tandem-responsive cleavable linker. PD-L1 is a membrane receptor overexpressed on cancer cells,27 and has been adopted for tumor-targeted drug delivery.12,28 The DPRC thus can target cancer cells through the interaction between the cyclopeptide and PD-L1. Following endocytosis, tumor biomarker GSH partially cleaves the linker to switch on its photosensitivity, which subsequently leads to siRNA release after exposure to visible light irradiation. The released free siBcl-2 then translocates into the cytoplasm to knock down Bcl-2 that is a pro-survival oncogene responsible for cancer development.29 As a result, the DPRC affords a tumor-specific gene silencing effect to suppress cancer cell growth. To our knowledge, this study demonstrates the first dual-locking bioconjugation approach for tumor-selective siRNA delivery and activation.
 | ||
| Scheme 1 Schematic illustration of the design of the DPRC and its application for tumor-biomarker/visible-light dual-locked control of siRNA release and activation. Created with BioRender. | ||
Results and discussion
We designed a DPRC as follows (Scheme 1). Since light is a non-invasive external stimulus permitting spatiotemporal regulator of biomolecular events,30 it thus holds great potential to activate a siRNA bioconjugate. We previously developed a photoactivatable antibody-siRNA conjugate using a photocleavable o-nitrophenyl linker.12 However, such a linker only responds to ultraviolet (UV) light, restricting the biological applications due to limited penetration capability and phototoxicity of UV light.30 In this work, to overcome this drawback, we used a visible light-responsive coumarin moiety as a photocleavable linker.31 To avoid nonspecific photoactivation under ambient light and in a complex biological environment, we caged the coumarin moiety with a naphthylamide group through a disulfide bond. The naphthylamide serves as a fluorescence resonance energy transfer (FRET) acceptor for coumarin, thereby disabling both the fluorescence and photocleavage sensitivity of coumarin.32 Only after GSH-mediated breakage of the disulfide linker to liberate naphthylamide can coumarin recover its fluorescence and undergo photocleavage when exposure to visible light illumination. On the other hand, to target PD-L1, we selected a cyclopeptide instead of an antibody because of its small size, stability, biocompatibility, and site-specific chemical labelling properties.33 Kp101 is a validated cyclopeptide ligand against PD-L1,34 which is thus used for tumor-targeting in our study. To achieve conditional siRNA activation, the 3′-end of the siBcl-2 guide strand was covalently modified with Kp101 through the GSH/visible light dual-responsive moiety, because this site has been demonstrated to be essential for RNAi activity in our previous studies.12,13,35,36 Therefore, this dual-lock design can assure a “AND” logic gated control on the DPRC to enable precise release and activation of siRNA in cancer cells.The synthetic routes for the DPRC are shown in Scheme 2. Briefly, we first synthesized the GSH/visible light tandem-responsive linker C11, which encompassed coumarin linked with naphthylamide through a disulfide bond as well as terminal carboxylic acid/alkyne groups (Fig. S1, ESI†). Kp101 bearing a free amine group was synthesized by cyclizing the linear peptide following solid-phase peptide synthesis (Fig. S2, ESI†). siBcl-2 was synthesized using a standard solid-phase phosphoramidite method, and was further functionalized with azide at the 3′-end of the guide strand (Fig. S3, ESI†). C11 was then conjugated with Kp101 through a condensation reaction between carboxylic acid and amine moieties, followed by coupling with the guide strand of siBcl-2 through copper-catalyzed azide–alkyne cycloaddition (CuAAC). All the key intermediates and final products were confirmed by nuclear magnetic resonance (NMR) spectroscopy and mass spectrometry (MS) analyses (ESI†). The DPRC was further obtained through annealing the final product with the passenger strand of siBcl-2.
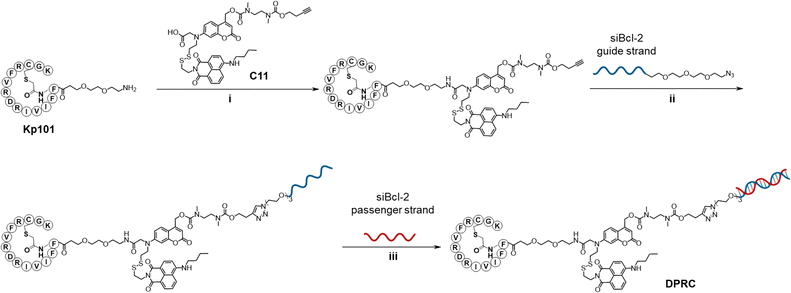
We first studied the optical and stimuli-responsive properties of the DPRC. Its UV-vis absorption spectra showed two characteristic peaks at 380 and 455 nm, which could be ascribed to coumarin and naphthylamide, respectively (Fig. 1a). However, the fluorescence emission of coumarin at 470 nm was quenched in the DPRC as compared to free coumarin, whereas the emission of naphthylamide at 525 nm was unaffected (Fig. 1b). This result suggests an anticipated FRET effect between coumarin and naphthylamide. We then irradiated the DRPC under 420 nm visible light to trigger the photocleavage effect of coumarin.37 Nevertheless, the DPRC remained intact after photoirradiation as indicated by native polyacrylamide gel electrophoresis (PAGE) analysis (Fig. 1c). These data thus demonstrate that the FRET effect could disable both the fluorescence and photocleavage sensitivity of coumarin in the DPRC. We therefore treated the DPRC with GSH to break the disulfide bond. As shown in Fig. 1d and Fig. S4 (ESI†), fluorescence emission of coumarin at 470 nm gradually recovered, reaching a plateau at 50 min after incubation. We further irradiated the GSH-treated DPRC under 420 nm light. Formation of a new band migrating faster than the DPRC accompanied by the disappearance of the DPRC was clearly observed according to PAGE analysis (Fig. 1c), implying photocleavage of the coumarin moiety to liberate siBcl-2. This photo-induced siBcl-2 liberation is dependent on the photoirradiation time, and completed after 15 min irradiation (Fig. S5, ESI†). We confirmed the structure of released siBcl-2 via MS (Fig. 1e, calculated 9540, found 9543), verifying the expected photocleavage of the coumarin moiety in the DPRC. In contrast, co-treatment of the DPRC with GSH and N-ethylmaleimide (NEM), a GSH scavenger,38 abolished both the recovery of coumarin fluorescence (Fig. 1d and Fig. S4, ESI†) and photo-induced siBcl-2 release (Fig. 1c), indicative of a GSH-initiated photo-cleavage effect. These results thus jointly demonstrate a GSH/visible-light tandem-responsive release of siBcl-2 by the DPRC.
The binding profiles of the DPRC towards PD-L1 were also investigated. Micro-scale thermophoresis (MST) assay revealed that the DPRC interacts with the recombinant PD-L1 protein with a dissociation constant (Kd) of 130.1 ± 15 nM (Fig. 1f), comparable to that of Kp101 (Kd = 35.1 ± 8.3 nM). Flow cytometry analysis also showed that the DPRC binds to PD-L1-positive breast cancer MDA-MB-231 cells with a Kd of 153.3 ± 41.3 nM, whereas diminished binding was observed in PD-L1-knockdown cells (Fig. S6, ESI†). Therefore, the DPRC maintains the PD-L1 binding activity of Kp101 with an ignorable impact after siRNA bioconjugation.
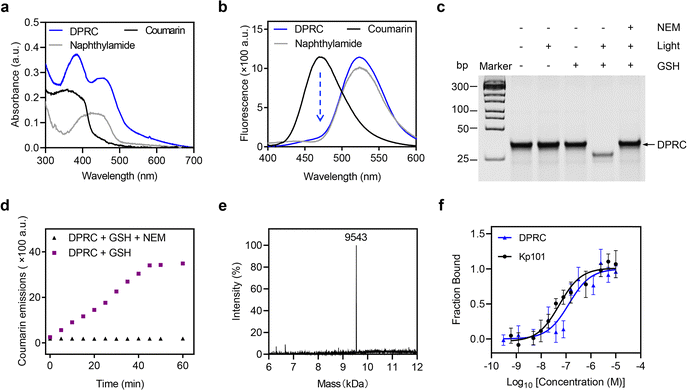
We next explored cellular uptake of the DPRC. To visualize the siRNA location, we used Cy3-tagged siBcl-2 to prepare the DPRC (Fig. 2a). After treating MDA-MB-231 cells with Cy3-DPRC, we observed a time-dependent increase of Cy3 fluorescence signals inside the cells as revealed by flow cytometry analysis, peaking at 4 h post incubation (Fig. 2b and Fig. S7, ESI†). This result indicated an efficient cellular internalization of the DPRC. However, largely diminished cellular uptake was found when simultaneously adding excess Kp101 to MDA-MB-231 cells to block PD-L1-binding (Fig. 2b) or using Cy3-DPRC to treat PD-L1-knockdown MDA-MB-231 cells (Fig. S8, ESI†). Moreover, negligible uptake of Cy3-DPRC was detected in PD-L1-negative normal liver cell line LO-2 either (Fig. S9, ESI†). These results thus confirm PD-L1-targeted siRNA delivery mediated by the Kp101 cyclopeptide in the DPRC. We then checked the subcellular location of siRNA via confocal fluorescence imaging. After co-staining lysosomes, we observed well-overlapped signals between Cy3 and Lyso-Tracker with a Pearson's coefficient of 0.986 (Fig. 2c, light (−) group). This suggests lysosomal trapping of siRNA post PD-L1-targeted cellular internalization, which is also a common obstacle in the field of nucleic acid drug delivery.39 Meanwhile, we confirmed GSH-mediated fluorescence recovery of coumarin after cellular uptake of the DPRC, as only weak signals were detected in NEM-pre-treated cells (Fig. 2d). Therefore, at 4 h post treating MDA-MB-231 cells with Cy3-DPRC, we irradiated cells with a 420 nm light for 15 min. After this, we found uniformly distributed Cy3 signals throughout the cytoplasm under a confocal fluorescence microscope (Fig. 2c, light (+) group). The Pearson's coefficients between Cy3 and Lyso-Tracker decreased to 0.371, validating a successful lysosomal escape of siBcl-2. However, using NEM to block GSH-mediated recovery of photosensitivity of coumarin prevented light-promoted siRNA translocation from the lysosome to cytoplasm (Fig. S10, ESI†). While mechanistic reasons still remain to be explored, these results suggest that timely release of siRNA from the targeting ligand may facilitate its lysosomal escape after cellular uptake, which also agrees with previous reports.12
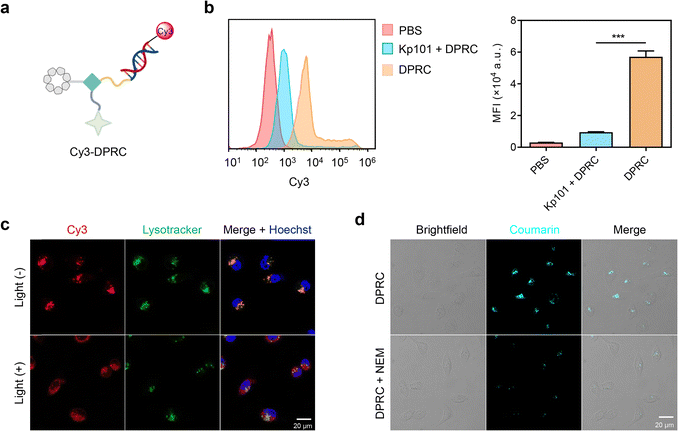
The biological activity of siBcl-2 delivered by the DPRC was further evaluated in MDA-MB-231 cells. Real-time quantitative polymer chain reaction (RT-qPCR) assay showed that the DPRC upon 15 min photoirradiation dose dependently reduced Bcl-2 mRNA levels (Fig. S11a, ESI†). At a concentration of 500 nM, the DPRC with photoirradiation induced ∼81% decrease of Bcl-2 mRNA, whereas no changes were detected by the DRPC in the dark (Fig. 3a). Western blot analysis of Bcl-2 protein levels also revealed similar outcomes (Fig. 3b and Fig. S11b, ESI†). Moreover, both excess Kp101 and NEM could block the gene silencing activity of the DRPC under light (Fig. S12, ESI†). These data are consistent with the cellular uptake results shown in Fig. 2, also demonstrating that siBcl-2 delivered by the DPRC exerts its gene silencing activity under a GSH/visible-light tandem control. As Bcl-2 is an anti-apoptotic biomolecule contributing to cancer cell growth,40 we further measured viabilities of MDA-MB-231 cells post DPRC treatment. The half-maximal inhibitory concentration (IC50) values of the DPRC under photoirradiation were 343.7 ± 16.9 nM, whereas only slight cytotoxicity was detectable for the DPRC in the dark at a concentration up to 1000 nM (Fig. 3c and Fig. S13, ESI†). Notably, excess Kp101 and NEM were also able to reverse the cytotoxicity of the DPRC under photoirradiation (Fig. S14, ESI†). Moreover, flow cytometry with annexin V-FITC/PI (propidium iodide) double staining analysis showed that the DPRC with photoirradiation elicited ∼58% cellular apoptosis, whereas insignificant apoptosis was triggered by the DPRC in the dark (Fig. 3d and e). These results further verify a GSH/visible-light tandemly controlled cytotoxicity by the DRPC.
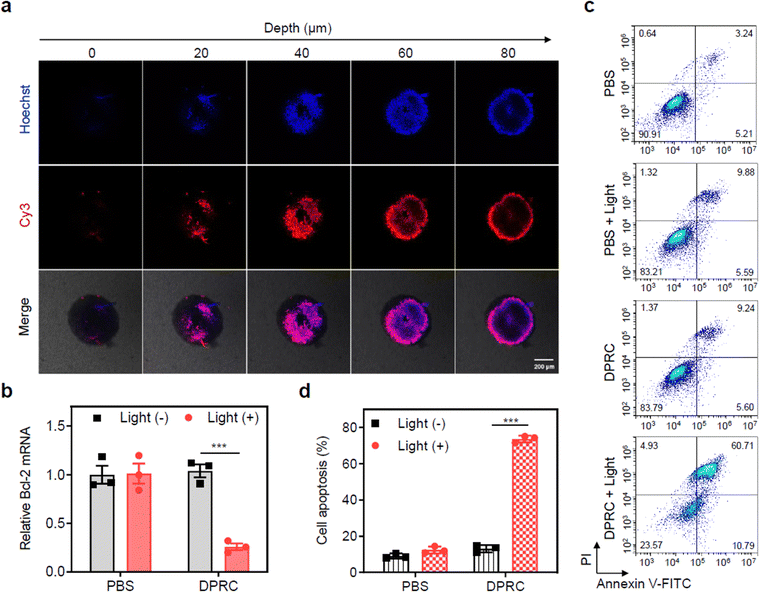
Finally, we evaluated the potential of the DPRC using three-dimensional tumor spheroids (3DTSs) to mimic tumor tissues. The penetration of siBcl-2 into MDA-MB-231 3DTSs was first evaluated. After incubation with Cy3-DPRC, the 3DTSs presented clear Cy3 fluorescence signals, which had an intense fluorescence signal from the outside in, suggesting a penetration of the DPRC (Fig. 4a). RT-qPCR assay also showed that only under photoirradiation could the DPRC decrease Bcl-2 mRNA levels in the 3DTSs by 75% (Fig. 4b). The pro-apoptosis capability of the DPRC toward MDA-MB-231 3DTSs was further quantified. Flow cytometry analysis showed that indiscernible cellular apoptosis was caused by the DPRC in the dark, whereas the apoptotic rate was raised up to 71% by the DPRC under photoirradiation (Fig. 4c). Calcein-AM/PI double staining assay also indicated that the number of PI-positive dead cells in the DPRC group with photoirradiation was significantly higher compared to that in the dark (Fig. S15, ESI†).
Conclusions
In summary, we have developed a novel dual-locking bioconjugation approach for tumor-specific siRNA delivery and on-tumor gene silencing. A DPRC was synthesized via connecting a PD-L1 targeting cyclopeptide and Bcl-2-targeting siRNA through a GSH/visible-light tandem-responsive cleavable linker. This design enables precise tumor targeting and controlled siRNA activation. We demonstrated that the DPRC can be effectively taken up by PD-L1-positive cancer cells via the interaction between the cyclopeptide and membrane PD-L1. However, only after GSH-initiated recovery of both the fluorescence and photosensitivity of coumarin could it undergo photocleavage to release siBcl-2. Moreover, we found that timely release of siRNA from the bioconjugates could facilitate its lysosomal escape. We validated the dual-locked controllable gene silencing and cytotoxicity of the DPRC in both 2D and 3D tumor cell models. Overall, we provide a promising avenue for precise siRNA delivery and activation in tumor cells only, benefiting from the dual-locking design. In addition to cancer treatment, this tumor-specific genetic tool should also be broadly applicable to various chemical biology studies.Data availability
Data are available upon request to the authors.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the Natural Science Foundation of Jiangsu Province (BK20232020, BE2022835, and BK20232007) and the National Natural Science Foundation of China (22322703, 22077063, 21877058, 22225703, and 22137003). Jinbo Li also acknowledges the support from Xiaomi Foundation.References
- V. Jadhav, A. Vaishnaw, K. Fitzgerald and M. A. Maier, Nat. Biotechnol., 2024, 42, 394–405 CrossRef CAS.
- S. G. Huayamares, D. Loughrey, H. Kim, J. E. Dahlman and E. J. Sorscher, Nat. Rev. Clin. Oncol., 2024, 21, 407–427 CrossRef CAS.
- M. Winkle, S. M. El-Daly, M. Fabbri and G. A. Calin, Nat. Rev. Drug Discovery, 2021, 20, 629–651 CrossRef CAS.
- H. Yin, R. L. Kanasty, A. A. Eltoukhy, A. J. Vegas, J. R. Dorkin and D. G. Anderson, Nat. Rev. Genet., 2014, 15, 541–555 CrossRef CAS PubMed.
- K. Paunovska, D. Loughrey and J. E. Dahlman, Nat. Rev. Genet., 2022, 23, 265–280 CrossRef CAS.
- Q. Tang and A. Khvorova, Nat. Rev. Drug Discovery, 2024, 23, 341–364 CrossRef CAS.
- J. A. Kulkarni, D. Witzigmann, S. B. Thomson, S. Chen, B. R. Leavitt, P. R. Cullis and R. van der Meel, Nat. Nanotechnol., 2021, 16, 630–643 CrossRef CAS PubMed.
- K. M. Brown, J. K. Nair, M. M. Janas, Y. I. Anglero-Rodriguez, L. T. H. Dang, H. Peng, C. S. Theile, E. Castellanos-Rizaldos, C. Brown, D. Foster, J. Kurz, J. Allen, R. Maganti, J. Li, S. Matsuda, M. Stricos, T. Chickering, M. Jung, K. Wassarman, J. Rollins, L. Woods, A. Kelin, D. C. Guenther, M. W. Mobley, J. Petrulis, R. McDougall, T. Racie, J. Bombardier, D. Cha, S. Agarwal, L. Johnson, Y. Jiang, S. Lentini, J. Gilbert, T. Nguyen, S. Chigas, S. LeBlanc, U. Poreci, A. Kasper, A. B. Rogers, S. Chong, W. Davis, J. E. Sutherland, A. Castoreno, S. Milstein, M. K. Schlegel, I. Zlatev, K. Charisse, M. Keating, M. Manoharan, K. Fitzgerald, J. T. Wu, M. A. Maier and V. Jadhav, Nat. Biotechnol., 2022, 40, 1500–1508 CrossRef CAS PubMed.
- A. Mullard, Nat. Rev. Drug Discovery, 2022, 21, 6–8 CrossRef CAS.
- J. W. Lee, J. Choi, Y. Choi, K. Kim, Y. Yang, S. H. Kim, H. Y. Yoon and I. C. Kwon, J. Controlled Release, 2022, 351, 713–726 CrossRef CAS.
- W. Tai, Molecules, 2019, 24, 2211–2235 CrossRef CAS PubMed.
- X. Wang, X. Xiao, Y. Feng, J. Li and Y. Zhang, Chem. Sci., 2022, 13, 5345–5352 RSC.
- X. Wang, Y. Wang, J. Li, T. Tian, J. Li, Z. Guo and Y. Zhang, Adv. Ther., 2022, 5, 2100161 CrossRef.
- S. Xie, W. Sun, T. Fu, X. Liu, P. Chen, L. Qiu, F. Qu and W. Tan, J. Am. Chem. Soc., 2023, 145, 7677–7691 CrossRef PubMed.
- J. O. McNamara, 2nd, E. R. Andrechek, Y. Wang, K. D. Viles, R. E. Rempel, E. Gilboa, B. A. Sullenger and P. H. Giangrande, Nat. Biotechnol., 2006, 24, 1005–1015 CrossRef PubMed.
- X. Liu, W. Wang, D. Samarsky, L. Liu, Q. Xu, W. Zhang, G. Zhu, P. Wu, X. Zuo, H. Deng, J. Zhang, Z. Wu, X. Chen, L. Zhao, Z. Qiu, Z. Zhang, Q. Zeng, W. Yang, B. Zhang and A. Ji, Nucleic Acids Res., 2014, 42, 11805–11817 CrossRef PubMed.
- A. Gandioso, A. Massaguer, N. Villegas, C. Salvans, D. Sanchez, I. Brun-Heath, V. Marchan, M. Orozco and M. Terrazas, Chem. Commun., 2017, 53, 2870–2873 RSC.
- S. Benizri, A. Gissot, A. Martin, B. Vialet, M. W. Grinstaff and P. Barthelemy, Bioconjugate Chem., 2019, 30, 366–383 CrossRef PubMed.
- S. F. Dowdy, Nat. Biotechnol., 2017, 35, 222–229 CrossRef PubMed.
- M. Oishi, Y. Nagasaki, K. Itaka, N. Nishiyama and K. Kataoka, J. Am. Chem. Soc., 2005, 127, 1624–1625 CrossRef.
- M. Cochran, D. Arias, R. Burke, D. Chu, G. Erdogan, M. Hood, P. Kovach, H. W. Kwon, Y. Chen, M. Moon, C. D. Miller, H. Huang, A. Levin and V. R. Doppalapudi, J. Med. Chem., 2024, 67, 14852–14867 CrossRef.
- J. Ruhle, I. Klemt, T. Abakumova, O. Sergeeva, P. Vetosheva, T. Zatsepin and A. Mokhir, Chem. Commun., 2022, 58, 4388–4391 RSC.
- B. R. Meade, K. Gogoi, A. S. Hamil, C. Palm-Apergi, A. van den Berg, J. C. Hagopian, A. D. Springer, A. Eguchi, A. D. Kacsinta, C. F. Dowdy, A. Presente, P. Lonn, M. Kaulich, N. Yoshioka, E. Gros, X. S. Cui and S. F. Dowdy, Nat. Biotechnol., 2014, 32, 1256–1261 CrossRef PubMed.
- Q. Wang, X. Fan, N. Jing, H. Zhao, L. Yu and X. Tang, ChemBioChem, 2021, 22, 1901–1907 CrossRef.
- X. Zhang, A. Gubu, J. Xu, N. Yan, W. Su, D. Feng, Q. Wang and X. Tang, Molecules, 2022, 27, 4377–4385 CrossRef PubMed.
- L. Wu, J. Huang, K. Pu and T. D. James, Nat. Rev. Chem., 2021, 5, 406–421 CrossRef PubMed.
- H. Yamaguchi, J. M. Hsu, W. H. Yang and M. C. Hung, Nat. Rev. Clin. Oncol., 2022, 19, 287–305 CrossRef.
- C. W. Li, S. O. Lim, E. M. Chung, Y. S. Kim, A. H. Park, J. Yao, J. H. Cha, W. Xia, L. C. Chan, T. Kim, S. S. Chang, H. H. Lee, C. K. Chou, Y. L. Liu, H. C. Yeh, E. P. Perillo, A. K. Dunn, C. W. Kuo, K. H. Khoo, J. L. Hsu, Y. Wu, J. M. Hsu, H. Yamaguchi, T. H. Huang, A. A. Sahin, G. N. Hortobagyi, S. S. Yoo and M. C. Hung, Cancer Cell, 2018, 33, 187–201 CrossRef .e110.
- A. Ashkenazi, W. J. Fairbrother, J. D. Leverson and A. J. Souers, Nat. Rev. Drug Discovery, 2017, 16, 273–284 CrossRef.
- J. Li and K. Pu, Chem. Soc. Rev., 2019, 48, 38–71 RSC.
- Y. Xue, H. Bai, B. Peng, B. Fang, J. Baell, L. Li, W. Huang and N. H. Voelcker, Chem. Soc. Rev., 2021, 50, 4872–4931 RSC.
- Y. Jiao, B. Zhu, J. Chen and X. Duan, Theranostics, 2015, 5, 173–187 CrossRef PubMed.
- X. Ji, A. L. Nielsen and C. Heinis, Angew. Chem., Int. Ed., 2024, 63, e202308251 CrossRef PubMed.
- K. Magiera-Mularz, K. Kuska, L. Skalniak, P. Grudnik, B. Musielak, J. Plewka, J. Kocik, M. Stec, K. Weglarczyk, D. Sala, B. Wladyka, M. Siedlar, T. A. Holak and G. Dubin, Adv. Ther., 2021, 4, 2000195 CrossRef.
- J. Li, L. Huang, X. Xiao, Y. Chen, X. Wang, Z. Zhou, C. Zhang and Y. Zhang, J. Am. Chem. Soc., 2016, 138, 15943–15949 CrossRef PubMed.
- L. Chen, Y. Sun, J. Li and Y. Zhang, Chem. Commun., 2020, 56, 627–630 RSC.
- Q. Lin, Z. Du, Y. Yang, Q. Fang, C. Bao, Y. Yang and L. Zhu, Chemistry, 2014, 20, 16314–16319 CrossRef PubMed.
- T. Schirmeister, J. Med. Chem., 1999, 42, 560–572 CrossRef PubMed.
- T. Tieu, Y. K. Wei, A. Cifuentes-Rius and N. H. Voelcker, Adv. Ther., 2021, 4, 2100108 CrossRef.
- D. J. Fowler-Shorten, C. Hellmich, M. Markham, K. M. Bowles and S. A. Rushworth, Blood Rev., 2024, 65, 101195 CrossRef PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4cb00247d |
| ‡ These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2025 |