Open Access Article
Open Access ArticleActive learning high coverage sets of complementary reaction conditions†
Sofia L.
Sivilotti
 ab,
David M.
Friday
ab,
David M.
Friday
 a and
Nicholas E.
Jackson
a and
Nicholas E.
Jackson
 *ac
*ac
aDepartment of Chemistry, University of Illinois at Urbana-Champaign, Urbana, Illinois 61801, USA. E-mail: jacksonn@illinois.edu
bDepartment of Computer Science, University of Illinois at Urbana-Champaign, Urbana, Illinois 61801, USA
cBeckman Institute for Advanced Science and Technology, University of Illinois Urbana-Champaign, Urbana, Illinois 61801, USA
First published on 17th February 2025
Abstract
Chemical reaction conditions capable of producing high yields over diverse reactants are a desired component of nearly all chemical and materials discovery campaigns. While much work has been done to discover individual general reaction conditions, any single conditions are necessarily limited over increasingly diverse chemical spaces. A potential solution to this problem is to identify small sets of complementary reaction conditions that, when combined, cover a larger chemical space than any one general reaction condition. In this work, we analyze experimentally derived datasets to assess the relative performance of individual general reaction conditions vs. sets of complementary reaction conditions. We then propose and benchmark active learning methods to efficiently discover these complimentary sets of conditions. The results show the value of active learning in identifying complementary sets of reaction conditions and provide an avenue for improving synthetic hit rates in high-throughput synthesis campaigns.
The rise in AI methods for chemical optimization has benefited numerous sub-fields including catalysis,1,2 drug discovery,3,4 formulation development,5 material discovery,6–8 optoelectronics,9–16 and energy storage materials.17–22 AI methods have recently been combined with high-throughput synthesis campaigns to rapidly explore chemical space, discovering molecules with improved physical properties,23 leading to more photostable,13,24 more fluorescent,25 and more soluble molecules.13 A common strategy for these campaigns is to define a synthetically accessible chemical space, use machine learning (ML) to select molecules from the chemical space, and leverage high-throughput synthesis and characterization to make and test molecules, the results of which inform the ML algorithm's next selection of molecules. In order to synthesize the molecules selected by the ML algorithm, these campaigns require reaction types and conditions, which may or may not be known, that can cover the predetermined chemical space. The continued desire to explore broader and more diverse chemical spaces makes ensuring synthesizability challenging.
Two common solutions to this problem are (1) to predict specific high-yield reaction conditions tailored to reactant chemistries, and (2) to discover general reaction conditions capable of producing adequate yields across a large reactant space. The former approach has been attempted with some success – while predictions of reaction yields have been unsuccessful due to literature bias and low quality data,26,27 more focused attempts have had some success28,29 with new approaches still under consideration.30 The latter approach has also been explored with varying outcomes.31–35 However, recent works favoring chemical diversity revealed that using a general reaction condition was successful for approximately 40% of the reactants recommended by the ML algorithm,24,25 and when combined with a second general reaction condition, increased to 60%.24 These results highlight the synthetic challenge of campaigns covering diverse chemistries.
A third potential solution to this problem is to develop complementary sets of reaction conditions that together cover larger portions of chemical space than any single reaction condition. This approach could allow individual reaction conditions to specialize in delivering high yields over specific regions of chemical space, allowing the combined set to cover a broader and more diverse chemical space. The task of selecting a minimum set of such reaction conditions that cover a chemical space, when every reaction outcome is known (referred to as the set cover optimization problem in computer science36), is an NP-hard problem with exponential time complexity scaling, but is computationally feasible over limited chemical spaces. For cases where reaction outcomes are unknown, an efficient process for discovering these complementary sets of reaction conditions, to our knowledge, does not exist. Recent publication of a few large-scale synthesis datasets providing reaction yields for a variety of reactants and conditions have made exploring this question possible.31,37–39
In this work we analyze existing experimental datasets covering diverse reactants and reaction conditions to explore the utility of using sets of complementary reaction conditions to provide broader coverage of reactant space. We then develop and test active learning (AL) strategies to rapidly identify these sets, validated on experimental datasets.
1 Methods
1.1 Reactants–condition datasets
This work uses four experimentally derived datasets that measure reaction yield for a set of reactants (reactant space, made of 1–2 reactant types: ra, rb) using all possible combinations of reaction conditions (condition space, made of 1–3 condition parameters: ca, cb, cc, e.g. catalyst, solvent, base) described in Table 1. Three datasets are purely experimental data,31,37,38 and the fourth dataset uses a ML model trained on 3300 reactions to predict yields for a space of 450![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 possible reactions.39 Each of these datasets completely enumerate the reaction yields for every combination of reactant(s) and condition(s) (reactant–condition space).
000 possible reactions.39 Each of these datasets completely enumerate the reaction yields for every combination of reactant(s) and condition(s) (reactant–condition space).
| Reaction type (dataset abbreviation) | Reactants | Conditions | Total reactions | OHE vector length |
|---|---|---|---|---|
| Deoxyfluorination (DeoxyF)37 | 37ra | 4ca × 5 cb | 740 | 37 + 4 + 5 |
| Palladium-catalysed C–H arylation31 (Pd-aryl) | 8ra × 8rb | 24ca | 1536 | 8 + 8 + 24 |
| Ni-catalyzed aryl-halide borylation38 (Ni-boryl) | 33ra | 23ca × 2 cb | 1518 | 33 + 23 + 2 |
| Buchwald–Hartwig39 (B–H) | 50ra × 50rb | 3ca × 3 cb × 20cc | 450![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 (3300 exp.) 000 (3300 exp.) |
50 + 50 + 3 + 3 + 20 |
The success of each reaction in a dataset is determined by whether the yield is at or above a yield cutoff (described below). Using this binary classification of reaction success, the coverage of a reaction condition γ{c} or set of conditions γ{ci,cj,…} is defined as the fraction of reactant space with a condition in the set producing a yield greater than the yield cutoff. This binary classification has the advantage of simplifying the definition of success when analyzing datasets and employing AL methods, and is also more amenable to an experimental campaign where measuring precise yields might be costly or time consuming. When comparing two sets with the same coverage, the smaller set is ranked higher, as it requires fewer conditions to cover the same amount of reactant space and can be combined with any other condition not in the set to produce an equal or better performing set.
1.2 Active learning
Discovering a set of complementary reaction conditions, when reaction outcomes are not known, is a challenging task; therefore we have employed an AL strategy to guide the process. The AL strategy involves (1) selecting an initial batch of reactions, (2) experimentally determining the success of the batch of reactions, (3) training a ML classifier on all experimental data, (4) predicting the expected probability of reaction success for all reactant–condition space, (5) selecting the next batch of reactions using an acquisition function, and returning to (2) and iterating until ending the campaign. Central to this approach, the ML classifier predicts the probability of reaction success (ϕr,c) for reactant(s) r and condition(s) c, where 0 is certain to fail, 1 is certain to succeed, and 0.5 is completely unknown. Reaction sets are compared using ϕr,c rounded to 0 or 1 to identify the best set of reaction conditions via combinatorial enumeration of all possible sets up to a maximum set size. In this work, a Gaussian Process Classifier (GPC) and a Random Forest Classifier (RFC) were compared for predicting ϕr,c. GPC is a standard method for classifying combinatorial spaces and have been used for similar AL classifier tasks40,41 with good performance. RFC has recently been shown to have superior performance in classification tasks in chemistry.42 Here, individual reactions are described by concatenated One Hot Encoded (OHE) vectors for each type of reactant and condition parameter in the dataset (e.g. ra, …, and ca, …, see Table 1). This encoding contains no physical or chemical information about the reactions, making it the simplest and most naive reaction representation.To select batches of reactions that will maximally improve the classifier's ability to identify the best set of complementary reaction conditions, a combination of explore and exploit acquisition functions were proposed. For each acquisition function, the selected reactions (r, c combinations) maximize the function's value. The explore function (eqn (1)) computes the uncertainty for a given reaction, with a probability of success of 0.5 maximizing the function. Several exploitative acquisition functions were tested (see ESI†), and the most effective is presented here. This exploit function (eqn (2)) favors reactions that use conditions (c) which complement other conditions (ci) for high predicted coverage (γ{c,ci}). Additionally, it favors reactant(s) (r) where the other conditions are unlikely to be successful (low ϕr,ci). For explanatory purposes, an example exploit calculation and AL campaign on a toy dataset are provided in the ESI.† All functions use the GPC or RFC model's predicted probability of success ϕr,c.
| Explorer,c = 1 − 2(|ϕr,c − 0.5|) | (1) |
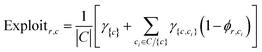 | (2) |
To merge these strategies into a single function, the explore and exploit functions were linearly combined using a weighting value, α (eqn (3)).
| Combinedr,c = (α)explorer,c + (1 − α)exploitr,c | (3) |
Following each iteration of the AL algorithm, the performance was measured by enumerating the predicted coverage of all possible reaction condition sets up to a specified size, selecting the highest-coverage set, and reporting the true coverage of that set.
When selecting reactions to test, batched reaction recommendations are useful for accelerating discovery with fewer iterations by testing multiple reactions in parallel. Simulations with batch sizes ranging from 1 to 160 were tested. For simulations using the combined explore–exploit strategy, each batch's alpha values were evenly spaced from 0 to 1 to select a range of exploratory and exploitative reactions.43 The initial batches of reactions were selected with Latin hypercube sampling.
2 Results and discussion
2.1 Coverage of reactant space
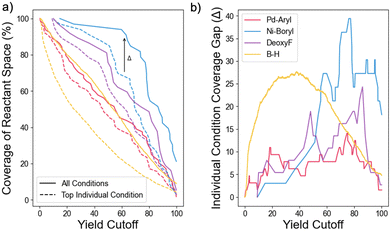
Fig. 1a compares how the coverage of reactant space changes with yield cutoff when using either all reaction conditions or only the single best reaction condition for that yield cutoff. The difference between the coverage of the best individual condition and all conditions (Δ) is plotted in Fig. 1b, showing how it varies with dataset and cutoff. This difference consistently shows an additional coverage of 10% of the total reactant space for cutoffs > 50%, and reaches as high as 40%. Using this data, a yield cutoff corresponding to 75% coverage by all reactions was chosen for all datasets as it maximized the coverage gap Δ and was used in previous optimization work.31 This large difference in coverage provides the most opportunistic case for testing reaction condition set AL algorithms. Furthermore, the 25% of reactant space not covered by any reaction will test the algorithm's ability to explore challenging reactants efficiently.In all four datasets, optimizing a set of reaction conditions allows for broader coverage of reactant space than the single most general reaction condition, as illustrated in Fig. 2. In this dataset, an optimal set of three reaction conditions improves the coverage of reactant space from 62% for the most general single reaction condition, to 75% of the reactant space. Furthermore, even if an optimal reaction condition set is not found, suboptimal sets of reaction conditions regularly outperform the single most general reaction condition. For the smaller three datasets, the smallest reaction set with maximal coverage contains 3–6 reaction conditions (Fig. S1†). The analysis of the B–H dataset was limited to sets of size 4 due to computational cost, yielding a maximum coverage of 73%, which is close to the true maximum possible coverage of 75% (Fig. S2†).
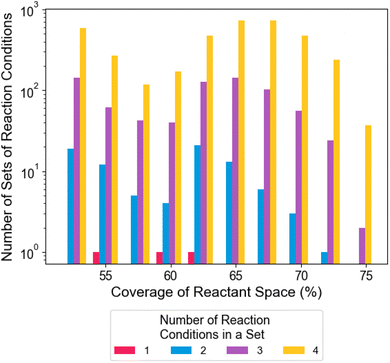 | ||
| Fig. 2 Histogram of the coverage of reactant space by high-coverage reaction condition sets in the DeoxyF dataset. The bars are colored by the number of conditions in a set. A DeoxyF reaction is considered successful if the reaction yield exceeds 50%, corresponding to a maximum coverage of 75% of reactant space as described in the text. Coverage is maximized at three conditions. See Fig. S1 and S2† for histograms for the other datasets. | ||
2.2 Optimal reaction condition sets
As shown in Fig. 3, the highest coverage set of reaction conditions is typically not a simple combination of the most general reaction conditions. While the most general conditions are frequently included in the best sets, specific less general complimentary conditions covering difficult reactants not commonly covered by the general reaction conditions are often necessary. For producing high coverage sets in the DeoxyF dataset (Fig. 3), the 4th and 5th ranked conditions (out of 20) are important complementary reaction conditions for the 1st or 2nd ranked conditions. In contrast, the 3rd ranked condition is nearly absent in high coverage sets, and the 1st and 2nd ranked conditions are never combined in high coverage sets despite their broader individual coverages. Similar trends were observed in the Pd-aryl dataset, where the 2nd and 4th ranked conditions formed a complementary pair, and in the B–H dataset, where 11th ranked condition is present in 39/45 top sets of size 3 (see Fig. S3†). Lastly, the Ni-boryl dataset (Fig. S3b†) showed that the 8th ranked condition was necessary for the highest coverage set of conditions, and present in half of the sets with the next highest coverage. In all cases, the presence of important non-general reaction conditions motivates us to find ways to rapidly identify these complementary sets of reaction conditions.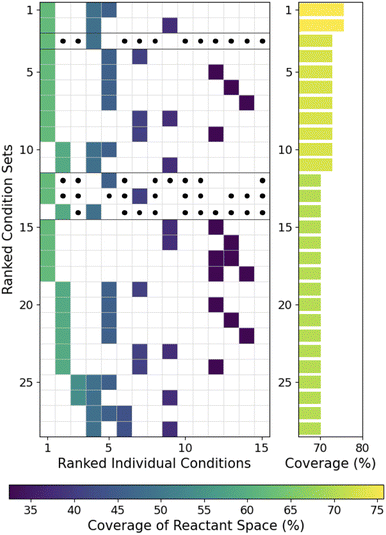 | ||
| Fig. 3 Grid showing the specific reaction conditions (ranked by coverage) within the highest coverage sets of reaction conditions for the DeoxyF dataset, with the coverage of each set on the right. The color of each grid point corresponds to the coverage of the individual reaction condition. For sets of two conditions, each black dot represents a third condition that could be added to the set without changing the coverage, highlighting the value of those pairs of reactants. Black lines divide sets of reaction conditions with different performance: either different coverage or different numbers of conditions required. Equivalent results for the other datasets are provided in Fig. S3.† | ||
2.3 Discovering high-coverage sets
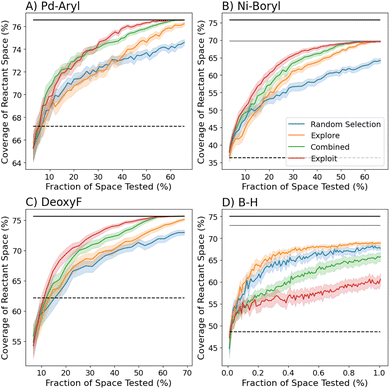
While the prior analyses of optimal sets of reaction conditions required full knowledge of all reaction outcomes, the most efficient process for selecting high-coverage sets of reaction conditions, when reaction outcomes are unknown, is unclear. Therefore, we approached this task using the AL strategy described previously. In general, the exploit acquisition function in eqn (2) performed better than other proposed exploitative acquisition functions (Fig. S4†). The RFC outperformed the GPC, possibly due to the RFC's ability to respond to sharp changes in success when varying reactants or condition parameters (Fig. S5†). Batch sizes of up to 40 reactions showed minimal performance degradation (see Fig. S6†), making the algorithms robust to parallelization and accelerating the optimization in real time. The following results used a batch size of 20 for the three smaller datasets (DeoxyF, Pd-aryl, Ni-boryl), and 40 for the B–H dataset. Fig. 4 shows true coverage of the classifier's predicted best set of conditions after gathering successive batches of reaction results as recommended by the acquisition functions.These results show that the exploit strategy is generally effective for the three smaller datasets, with the explore strategy typically showing reduced performance, and the combined explore–exploit strategy's performance being intermediate between the two strategies. For the sake of comparison, previous experimental work using ML to discover a general reaction condition (dashed lines in Fig. 4) required exploring 57% of space (300 reactions).32 The exploit acquisition function's superior performance in these three datasets is attributed to it prioritizing challenging reactants in potentially complementary sets. This strategy either confirms that challenging reactants are covered or quickly corrects overly optimistic predictions, allowing it to refine its sets quickly. This approach was especially successful for the DeoxyF and Ni-boryl datasets, which both contained a 1-dimensional reactant space (see Table 1). This observation indicates that reaction yields may not correlate well with individual reactants in bi-molecular reactions, motivating exploration of more informative reactant representations.
The performance of the exploit strategy on the B–H dataset was notably diminished, with the explore strategy and even random selection significantly out-performing it. To better understand the performance of the different acquisition functions on the B–H dataset, we conducted additional AL campaigns (Fig. S9†) as a function of variations in data size, reactant space, and condition parameters. Over the large, multidimensional spaces, explore is initially more effective than exploit because it prioritizes reducing RFC model uncertainty, helping it rapidly identify global trends across the reactant–condition space. However, explore does not favor understanding high coverage reaction conditions, regularly converging to a reasonably good set, and only optimizing slowly with additional data. In contrast, the exploit algorithm only focuses on the highest coverage conditions that have been identified. This yields poor performance when the space is large and highest coverage conditions are unknown, but provides more tailored recommendations as the campaign continues. The combined algorithm benefits from the advantages of explore and exploit, allowing for exploration at earlier times and more focused exploration at later times, evidenced by the combined algorithm frequently surpassing explore by the time 10% of the reaction-condition space has been tested.
An important consideration for the effectiveness of complementary reaction conditions involves the selection of descriptors used to characterize the reactant and catalyst spaces in the AL campaigns. The primary results of this work utilize OHE due to the dissimilar nature of the chemical spaces in the four studied datasets. However, an obvious avenue for improvement of the method would be to use more informative molecular featurizations, such as molecular fingerprints, that could help further navigate a broad chemical space. To explore this direction, we repeated our AL campaigns using Daylight molecular fingerprinting (Fig. S10 and S11†) to see if this added chemical information within the descriptors would improve the discovery of complementary reaction sets. Across all reaction datasets, we did not see any significant benefit in the use of molecular fingerprints over OHE. Subsequent dataset analysis (Fig. S12†) showed that this was likely due to the dilution of the fingerprint by molecular features far from the reactive site. However, these results do not suggest that OHE is the best featurization strategy. Previous work has shown that molecular fingerprints, DFT calculated properties (i.e. bond orders, charge distributions), and geometrical descriptors (i.e. steric effects, atomic arrangements) have improved prediction of reaction yields, and could enhance the AL strategy's ability to predict reaction success.28,29,39 However, such metrics would likely be unique to each dataset.
One caveat for the present study involves the combinatorial nature of the data in the optimization campaigns. Real chemical data is not necessarily combinatorial, and when considering all chemical entities that could undergo a given reaction, the different hypothetical reactions could be non-uniform and not possess a combinatorial structure. An interesting future direction for the present work pertains to examining AL across larger, unstructured datasets such as USPTO or ORD; for such a study, using a featurization other than OHE would be essential.
3 Conclusions
By analyzing experimentally derived chemical reaction datasets covering a variety of reactants and conditions, we have shown that sets of complementary reaction conditions consistently outperform the most general single reaction conditions. For all datasets, it was found that the highest-coverage sets of reaction conditions frequently contained certain lower-coverage conditions capable of complementing the more general conditions. Furthermore, AL algorithms were tested, demonstrating accelerated discovery of high-coverage sets of complementary conditions at comparable cost to discovering single general reaction conditions.The most effective AL strategy for discovering high coverage sets of reaction conditions was the exploit acquisition function. The exploit function prioritized testing reactions of highly complementary conditions on reactant(s) where other conditions are not expected to be successful. More efficient searches for sets of conditions could be realized by describing the reactants or conditions with features relevant to the synthesis (e.g. sterics, bond strengths, electrostatic potentials, stability of intermediates, or solubilities).44,45
These results present an opportunity for synthetic chemists to approach the challenge of high throughput synthesis more systematically with improved coverage of reaction space by exploring new reaction conditions that can cover more challenging reactants. We hope that these high coverage sets of reaction conditions will accelerate high-throughput synthesis campaigns by providing the highest probability of successful synthesis with the fewest required attempted reactions. More broadly, this AL method for set optimization can be applied to any situation where it would be advantageous to have a small set of ‘default’ options that are likely to produce at least one successful hit over a wide variety of scenarios. Specifically, we believe that material processing conditions, drug and agriculture formulation compositions,46 assay optimization, and ensemble model hyper-parameter optimization could benefit from having sets of complementary conditions.
Data availability
The datasets used for this article can be found at: https://doi.org/10.1021/jacs.8b01523 (DeoxyF), https://doi.org/10.5281/zenodo.8181283 (Pd-aryl), https://doi.org/10.1021/acs.organomet.2c00089 (Ni-boryl), https://doi.org/10.5281/zenodo.8185014 (B–H). The code repository can be found at the following link: https://doi.org/10.5281/zenodo.14861625.Author contributions
Sofia Sivilotti: conceptualization, methodology, formal analysis, investigation, writing – original draft. David Friday: conceptualization, methodology, data curation, writing – review & editing, supervision. Nicholas Jackson: conceptualization, methodology, writing – review & editing, supervision, project administration, funding acquisition.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the Molecule Maker Lab Institute, an AI Research Institutes program supported by the US National Science Foundation under grant no. 2019897. We would like to thank Scott Denmark, N. Ian Rinehart, and Blake Ocampo for their assistance in generating the B–H dataset.Notes and references
- Q. Zhu, Y. Huang, D. Zhou, L. Zhao, L. Guo, R. Yang, Z. Sun, M. Luo, F. Zhang and H. Xiao, et al. , Nat. Synth., 2024, 3, 319–328 CrossRef CAS.
- A. Ramirez, E. Lam, D. P. Gutierrez, Y. Hou, H. Tribukait, L. M. Roch, C. Copéret and P. Laveille, Chem Catal., 2024, 4, 100888 CrossRef CAS.
- S. M. Pant, A. Mukonoweshuro, B. Desai, M. K. Ramjee, C. N. Selway, G. J. Tarver, A. G. Wright, K. Birchall, T. M. Chapman and T. A. Tervonen, et al. , J. Med. Chem., 2018, 61, 4335–4347 CrossRef CAS PubMed.
- A. Ortiz-Perez, D. van Tilborg, R. van der Meel, F. Grisoni and L. Albertazzi, Digital Discovery, 2024, 3, 1280–1291 RSC.
- L. Cao, D. Russo, K. Felton, D. Salley, A. Sharma, G. Keenan, W. Mauer, H. Gao, L. Cronin and A. A. Lapkin, Cell Rep. Phys. Sci., 2021, 2, 100295 CrossRef.
- B. DeCost, H. Joress, S. Sarker, A. Mehta and J. Hattrick-Simpers, J. Met., 2022, 74, 2941–2950 Search PubMed.
- M. B. Rooney, B. P. MacLeod, R. Oldford, Z. J. Thompson, K. L. White, J. Tungjunyatham, B. J. Stankiewicz and C. P. Berlinguette, Digital Discovery, 2022, 1, 382–389 RSC.
- T. Erps, M. Foshey, M. K. Luković, W. Shou, H. H. Goetzke, H. Dietsch, K. Stoll, B. von Vacano and W. Matusik, Sci. Adv., 2021, 7, eabf7435 CrossRef CAS PubMed.
- K. Higgins, S. M. Valleti, M. Ziatdinov, S. V. Kalinin and M. Ahmadi, ACS Energy Lett., 2020, 5, 3426–3436 CrossRef CAS.
- A. K. Low, F. Mekki-Berrada, A. Gupta, A. Ostudin, J. Xie, E. Vissol-Gaudin, Y.-F. Lim, Q. Li, Y. S. Ong and S. A. Khan, et al. , npj Comput. Mater., 2024, 10, 104 CrossRef CAS.
- R. W. Epps, M. S. Bowen, A. A. Volk, K. Abdel-Latif, S. Han, K. G. Reyes, A. Amassian and M. Abolhasani, Adv. Mater., 2020, 32, 2001626 CrossRef CAS PubMed.
- A. Vikram, K. Brudnak, A. Zahid, M. Shim and P. J. Kenis, Nanoscale, 2021, 13, 17028–17039 RSC.
- B. A. Koscher, R. B. Canty, M. A. McDonald, K. P. Greenman, C. J. McGill, C. L. Bilodeau, W. Jin, H. Wu, F. H. Vermeire and B. Jin, et al. , Science, 2023, 382, eadi1407 CrossRef CAS PubMed.
- B. P. MacLeod, F. G. Parlane, T. D. Morrissey, F. Häse, L. M. Roch, K. E. Dettelbach, R. Moreira, L. P. Yunker, M. B. Rooney and J. R. Deeth, et al. , Sci. Adv., 2020, 6, eaaz8867 CrossRef CAS PubMed.
- S. Langner, F. Häse, J. D. Perea, T. Stubhan, J. Hauch, L. M. Roch, T. Heumueller, A. Aspuru-Guzik and C. J. Brabec, Adv. Mater., 2020, 32, 1907801 CrossRef CAS PubMed.
- Z. Liu, N. Rolston, A. C. Flick, T. W. Colburn, Z. Ren, R. H. Dauskardt and T. Buonassisi, Joule, 2022, 6, 834–849 CrossRef CAS.
- I. Oh, M. A. Pence, N. G. Lukhanin, O. Rodríguez, C. M. Schroeder and J. Rodríguez-López, Device, 2023, 1, 100103 CrossRef.
- A. Dave, J. Mitchell, S. Burke, H. Lin, J. Whitacre and V. Viswanathan, Nat. Commun., 2022, 13, 5454 CrossRef CAS PubMed.
- S. Matsuda, G. Lambard and K. Sodeyama, Cell Rep. Phys. Sci., 2022, 3, 100832 CrossRef CAS.
- L. Porwol, D. J. Kowalski, A. Henson, D.-L. Long, N. L. Bell and L. Cronin, Angew. Chem., 2020, 132, 11352–11357 CrossRef.
- J. Noh, H. A. Doan, H. Job, L. A. Robertson, L. Zhang, R. S. Assary, K. Mueller, V. Murugesan and Y. Liang, Nat. Commun., 2024, 15, 2757 CrossRef CAS PubMed.
- E. Fatehi, M. Thadani, G. Birsan and R. W. Black, arXiv, 2023, preprint, arXiv:2305.12541, DOI:10.48550/arXiv.2305.12541.
- G. Tom, S. P. Schmid, S. G. Baird, Y. Cao, K. Darvish, H. Hao, S. Lo, S. Pablo-García, E. M. Rajaonson and M. Skreta, et al. , Chem. Rev., 2024, 124, 9633–9732 CrossRef CAS PubMed.
- N. H. Angello, D. M. Friday, C. Hwang, S. Yi, A. H. Cheng, T. C. Torres-Flores, E. R. Jira, W. Wang, A. Aspuru-Guzik, M. D. Burke, C. M. Schroeder, Y. Diao and N. E. Jackson, Nature, 2024, 1–8 Search PubMed.
- F. Strieth-Kalthoff, H. Hao, V. Rathore, J. Derasp, T. Gaudin, N. H. Angello, M. Seifrid, E. Trushina, M. Guy, J. Liu, X. Tang, M. Mamada, W. Wang, T. Tsagaantsooj, C. Lavigne, R. Pollice, T. C. Wu, K. Hotta, L. Bodo, S. Li, M. Haddadnia, A. Wolos, R. Roszak, C.-T. Ser, C. Bozal-Ginesta, R. J. Hickman, J. Vestfrid, A. Aguilar-Gránda, E. L. Klimareva, R. C. Sigerson, W. Hou, D. Gahler, S. Lach, A. Warzybok, O. Borodin, S. Rohrbach, B. Sanchez-Lengeling, C. Adachi, B. A. Grzybowski, L. Cronin, J. E. Hein, M. D. Burke and A. Aspuru-Guzik, Science, 2024, 384, eadk9227 CrossRef CAS PubMed.
- W. Beker, R. Roszak, A. Wołos, N. H. Angello, V. Rathore, M. D. Burke and B. A. Grzybowski, J. Am. Chem. Soc., 2022, 144, 4819–4827 CrossRef CAS PubMed.
- P. Schwaller, A. C. Vaucher, T. Laino and J.-L. Reymond, Mach. Learn.: Sci. Technol., 2021, 2, 015016 Search PubMed.
- Z. Fu, X. Li, Z. Wang, Z. Li, X. Liu, X. Wu, J. Zhao, X. Ding, X. Wan, F. Zhong, D. Wang, X. Luo, K. Chen, H. Liu, J. Wang, H. Jiang and M. Zheng, Org. Chem. Front., 2020, 7, 2269–2277 RSC.
- P. Raghavan, A. J. Rago, P. Verma, M. M. Hassan, G. M. Goshu, A. W. Dombrowski, A. Pandey, C. W. Coley and Y. Wang, J. Am. Chem. Soc., 2024, 146, 15070–15084 CrossRef CAS PubMed.
- E. Shim, A. Tewari, T. Cernak and P. M. Zimmerman, J. Chem. Inf. Model., 2023, 63, 3659–3668 CrossRef CAS PubMed.
- J. Y. Wang, J. M. Stevens, S. K. Kariofillis, M.-J. Tom, D. L. Golden, J. Li, J. E. Tabora, M. Parasram, B. J. Shields, D. N. Primer, B. Hao, D. Del Valle, S. DiSomma, A. Furman, G. G. Zipp, S. Melnikov, J. Paulson and A. G. Doyle, Nature, 2024, 626, 1025–1033 CrossRef CAS PubMed.
- N. H. Angello, V. Rathore, W. Beker, A. Wołos, E. R. Jira, R. Roszak, T. C. Wu, C. M. Schroeder, A. Aspuru-Guzik, B. A. Grzybowski and M. D. Burke, Science, 2022, 378, 399–405 CrossRef CAS PubMed.
- R. V. Jagadeesh, K. Murugesan, A. S. Alshammari, H. Neumann, M.-M. Pohl, J. Radnik and M. Beller, Science, 2017, 358, 326–332 CrossRef CAS PubMed.
- Z. Feng, Q.-Q. Min, H.-Y. Zhao, J.-W. Gu and X. Zhang, Angew. Chem., Int. Ed., 2015, 54, 1270–1274 CrossRef CAS PubMed.
- S. P. Schmid, E. M. Rajaonson, C. T. Ser, M. Haddadnia, S. X. Leong, A. Aspuru-Guzik, A. Kristiadi, K. Jorner and F. Strieth-Kalthoff, AI for Accelerated Materials Design - NeurIPS 2024, 2024 Search PubMed.
- U. Feige, J. Assoc. Comput. Mach., 1998, 45, 634–652 CrossRef.
- M. K. Nielsen, D. T. Ahneman, O. Riera and A. G. Doyle, J. Am. Chem. Soc., 2018, 140, 5004–5008 CrossRef CAS PubMed.
- J. M. Stevens, J. Li, E. M. Simmons, S. R. Wisniewski, S. DiSomma, K. J. Fraunhoffer, P. Geng, B. Hao and E. W. Jackson, Organometallics, 2022, 41, 1847–1864 CrossRef CAS.
- N. I. Rinehart, R. K. Saunthwal, J. Wellauer, A. F. Zahrt, L. Schlemper, A. S. Shved, R. Bigler, S. Fantasia and S. E. Denmark, Science, 2023, 381, 965–972 CrossRef CAS PubMed.
- R.-R. Griffiths, L. Klarner, H. Moss, A. Ravuri, S. Truong, Y. Du, S. Stanton, G. Tom, B. Rankovic and A. Jamasb, et al. , Adv. Neural Inf. Process. Syst., 2023, 36, 76923–76946 Search PubMed.
- H. Tao, T. Wu, M. Aldeghi, T. C. Wu, A. Aspuru-Guzik and E. Kumacheva, Nat. Rev. Mater., 2021, 6, 701–716 CrossRef.
- Q. Gallagher and M. Webb, Digital Discovery, 2025, 4(1), 135–148 RSC.
- F. Hase, L. M. Roch, C. Kreisbeck and A. Aspuru-Guzik, ACS Cent. Sci., 2018, 4, 1134–1145 CrossRef CAS PubMed.
- J. Lu, S. Donnecke, I. Paci and D. C. Leitch, Chem. Sci., 2022, 13, 3477–3488 RSC.
- P. A. Cox, M. Reid, A. G. Leach, A. D. Campbell, E. J. King and G. C. Lloyd-Jones, J. Am. Chem. Soc., 2017, 139, 13156–13165 CrossRef CAS PubMed.
- P. Bannigan, R. J. Hickman, A. Aspuru-Guzik and C. Allen, Adv. Healthcare Mater., 2024, 2401312 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4dd00365a |
| This journal is © The Royal Society of Chemistry 2025 |