 Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Volatile organic compounds (VOCs) in terrestrial extreme environments: implications for life detection beyond Earth
Claire A.
Batty
 *,
Victoria K.
Pearson
,
Karen
Olsson-Francis
and
Geraint
Morgan
*,
Victoria K.
Pearson
,
Karen
Olsson-Francis
and
Geraint
Morgan
The Open University, Walton Hall, Milton Keynes, MK7 6AA, UK. E-mail: claire.batty@open.ac.uk
First published on 21st October 2024
Abstract
Covering: 1961 to 2024
Discovering and identifying unique natural products/biosignatures (signatures that can be used as evidence for past or present life) that are abundant, and complex enough that they indicate robust evidence of life is a multifaceted process. One distinct category of biosignatures being explored is organic compounds. A subdivision of these compounds not yet readily investigated are volatile organic compound (VOCs). When assessing these VOCs as a group (volatilome) a fingerprint of all VOCs within an environment allows the complex patterns in metabolic data to be unravelled. As a technique already successfully applied to many biological and ecological fields, this paper explores how analysis of volatilomes in terrestrial extreme environments could be used to enhance processes (such as metabolomics and metagenomics) already utilised in life detection beyond Earth. By overcoming some of the complexities of collecting VOCs in remote field sites, a variety of lab based analytical equipment and techniques can then be utilised. Researching volatilomics in astrobiology requires time to characterise the patterns of VOCs. They must then be differentiated from abiotic (non-living) signals within extreme environments similar to those found on other planetary bodies (analogue sites) or in lab-based simulated environments or microcosms. Such an effort is critical for understanding data returned from past or upcoming missions, but it requires a step change in approach which explores the volatilome as a vital additional tool to current ‘Omics techniques.
1 Introduction
Finding evidence of life elsewhere in the Solar System is dependent on the discovery of unique biosignatures that are produced by biological activity.1–4 These biosignatures must be sufficiently abundant and/or complex in an environment to display measurable attributes that indicate life and be unlikely to be formed by a non-biological (abiotic) process.5 Understanding the production and modification of potential biosignatures can inform strategies for research pathways, missions, and observations with life detection as a key objective.6Biosignatures have been defined as ‘any phenomenon for which biological processes are a known possible explanation and whose potential abiotic causes have been reasonably explored and ruled out’.7 They can be divided into two distinct categories: (1) inorganic, which include isotope fractionations, morphological fossils, mineral alterations, and sedimentary structures formed from microbial activity;1,8–10 and (2) organic, which includes biomolecules produced as cellular structures and/or metabolic activity.11–14 One sub-division of organic compounds that has yet to be fully explored as viable biosignatures in this context are Volatile Organic Compounds (VOCs).
VOCs are compounds that readily evaporate at room temperature and are low molecular weight15 (below 900 Dalton – but usually in the range 50–200 Daltons).16 They can be further subdivided into 3 main groups as illustrated in Table 1. They have high vapour pressures and low boiling points, that facilitate the evaporative process.18,19
| Description | Abbreviation | Boiling point range (°C) |
|---|---|---|
| Very volatile organic compound | vVOC | <0 to 50–100 |
| Volatile organic compound | VOC | 50–100 to 240–260 |
| Semi-volatile organic compound | sVOC | 240–260 to 380–400 |
For this reason, on Earth, VOCs are intrinsically linked to the atmosphere and play a crucial role in global atmospheric chemistry; they can undergo gas phase oxidation with atmospheric oxidants such as hydroxyl radicals (OH), ozone (O3), and nitrate radicals (NO3) as well as other reactive processes to form volatile organic species such as carbonyls, carboxylic acids, alcohols, esters, organo-sulphates, and organo-nitrates.20 These species can then condense or react with particle phase compounds to form secondary organic aerosols (SOA).21–23 VOCs are necessary for the global cycling of essential elements; for example, oceans release a range of biogenic VOCs (containing carbon, nitrogen, sulfur, and halogens), which are transferred to land, via the atmosphere.24 They are also primary and secondary metabolic by-products of microbial life, used to fulfil a variety of roles in their communication, survival, and persistence,25 along with other volatile gases, for example: methane (CH4) produced via methanogenesis through anaerobic respiration,26,27 ammonia (NH3), which is produced from the metabolism of peptide and amino acids; hydrogen cyanide (HCN), which has been detected in some Pseudomonas, catalysed by the enzyme HCN synthase and forming HCN and carbon dioxide (CO2) from glycine; nitric oxide (NO), which is produced mostly from L-arginine, and hydrogen sulfide (H2S), usually produced from the degradation of cysteine.28 The bioproducts of any biotic activity (including volatile) are therefore, prime targets as biosignatures.28–31
In planetary exploration, it is assumed that life will select chemical compounds that are useful to it (e.g., that allow it to perform chemical processes), creating a disequilibrium; with the acceptance that extra-terrestrial life, could utilise different chemical building blocks and ways to facilitate chemical reactions.32 Indeed, chemical disequilibrium in planetary atmospheres (including exoplanets) has been deemed a plausible biosignature for searching for life elsewhere in the Universe.33,34 The disequilibrium balance and diversity of VOCs produced by phytoplankton has also been explored at the sea–air interface by Halsey & Giovannoni. Here, ecosystem shifts in phytoplankton caused by physical and biological events can lead to a state of surface ocean disequilibrium and VOC accumulation.35 The shifting turnover of VOCs at this interface highlights the importance of investigating VOCs as a group alongside singular compounds as biosignatures.
A broadscale method for group VOC analysis is in the form of the ‘volatilome’. The ‘volatilome’ is a term used to describe all volatile compounds with unique complexity found in an organism, an ecosystem, or a matrix – including those from microbial metabolic processes and exogenously derived compounds.36–38 This method could help to ensure that an unambiguous biosignature is ultimately detected.39 This volatilomic fingerprint of all VOCs within an environment – would allow complex patterns in metabolic data to be unravelled, as has been applied in numerous biological and ecological fields and can be used as a tool for biomonitoring that is non-invasive, non-destructive, and rapid.40 The human volatilome is regularly investigated in health and disease,41–43 for example wound profiling in chronic wounds,44 the gut–brain axis,45,46 clinical medicine,47–49 and diagnostics50 as well as pathogenic microbes such as ampicillin-resistant and -susceptible Escherichia coli,51 for exhaled signs of infection in multiple pathogenic species52 and to help identify bacteria implicated in pneumonia.53 VOCs have been shown to help differentiate groups of subjects54 and identify markers for diseases such as tuberculosis.55
The plant volatilome has been explored by Rhinnan et al. in relation to extreme terrestrial and marine environments.56 It has also been extensively characterised, including the rhizosphere and its interactions with microbes and insects,57–59 and the volatilome has been identified as an essential piece of the soil metabolome.60 As plant diseases caused by phytopathogens cause huge economic loses in agriculture, finding ways to characterise or identify patterns in VOCs as diagnostic markers or to develop strategies for biocontrol could have huge benefits to sustainability in agriculture.61,62 Even research into conditions such as sick building syndrome have shown the value of VOC identification,63 but in many cases each individual VOC chromatogram can contain hundreds, if not thousands of features to identify and classify, making them perfect for chemometric analysis approaches.64 When then combined with metagenomic and metabolomic data, a volatilome could help elicit a holistic perspective on microbial processes30 and offer a new avenue in astrobiology to help enhance current exploration techniques. An ongoing challenge with these techniques however, is that biological variability can be high. For example, variations in VOC concentrations can change with environment, genetics, species, and which communities of microbes exist together.65 There could also be different enzymatic sources for the same volatile signatures, which a chromatogram would not be able to differentiate.66 The complexity of these factors therefore requires the production of multi’Omics data with high precision and robust quality control (QC) protocols to allow for elucidation of multi-level, real-time interactions, that reflect biochemical pathways and highlight any dysregulations.67
Volatilomics can encompass large, untargeted (evaluation of all detectable compounds in the sample)68 or small, targeted (evaluation of compounds that are predetermined by the researcher) analyses of a range of metabolites and explores the characterisation, detection, and quantification of these metabolites in a biological system.37,69 It has also been suggested that microbial VOC data should be integrated into metabolic profiling (metabolomics) to enhance our understanding of microbial systems.30,70,71 In these studies, the overlooked volatile component can lead to incomplete interspecies, and interspecies-to-ecosystem interactions.30 By adding metagenomic analysis, (that characterises the diversity and function of micro-organisms at a genetic level72), we can combine volatilomics, metabolomics and metagenomics to give a holistic view of the dynamics between microbes.73 Although this offers an opportunity for future work, it is not detailed in this review.
The key focus for establishing a volatilome to use as a biosignature is to understand the occurrence of VOCs and organic species in an environmental setting, including how that environment evolves or changes, and how potential atmospheric, geological, and stellar processes/interactions, may suppress, enhance, or mimic a biosignature at different times.74 In this paper we review the production of VOCs from microbial sources (mVOCs) and how they may be produced and interact in extreme environments similar to those on other planetary bodies. We will explore how a volatilome can be used as a putative biosignature for life.
2 Microbial VOCs
Microbial VOCs (mVOCs) are those VOCs produced as a physiological response to environmental conditions.75,76 This can include stresses such as extremes of pH, salinity, desiccation, or temperature, all of which require microorganisms to adapt for survival.77,78 A single organism will emit or utilise a selection of VOCs depending on environmental conditions, for example, nutrient source, pH, growth stage, moisture content, humidity, and aeration.79–81A broad range of compounds can be released by microbes, including alcohols, ketones, nitrogen- and sulfur-containing species, esters, hydrocarbons, carbonyl groups and halogenated compounds.82,83 Further, microbes can utilise VOCs generated anthropogenically,84,85 through intra-and inter-kingdom interactions, e.g., quorum sensing,61,79,86 or biosynthesis, such as via the terpene pathway and fatty acid biosynthesis.87
mVOCs have highly diverse structural variations88 and can move and interact with other volatiles easily because they diffuse well in the gas phase and move readily within the liquid phase (faster than polar compounds) as illustrated in Weisskopf et al. 2021.89 They can interact within many environments, whether within an organism or as part of a complete ecosystem.40,75,90 Subsequently, this affects cell membrane fluidity (by creating membrane expansion through the accumulation of VOC molecules91) or induces disruptions such as leakage of intracellular components. This changes the permeability, allowing VOCs to penetrate interior cell structures and interact with intracellular sites.92 This can allow interaction between microbial groups to promote or reduce communication,93 allow stress alleviation,94 or improve/reduce defence.95 For example, dimethyl disulfide (DMDS) can induce plant growth promotion,96 2-butanone and 2-octanone can affect microbial motility in biofilm formation,97 and 3-carene can affect the production of other metabolites.89 Microbial terpenoids (geosmin, 2-methylisoborneol) have also been shown to promote host health during growth, cell differentiation and rhizoid formation.95,98 Many of these features could create a huge competitive advantage and support micro-organisms to survive large variations in physicochemical conditions within extreme environments.
Microbial metabolism regulation contributes significantly to the complexity and characterisation of their volatilomes, and this varies between species- and strain.99 Tracking species- and strain-level volatilomic diversity across a genus results in a comprehensive understanding of VOCs for specific microbes and microbial groups.100 Strain-level volatilomes have been explored in some pathogenic bacterial species where, at the compound level, the primary metabolites (described in next section) such as alcohols, ketones, and acids, varied between different glucose-dependent volatilomes (e.g., brain heart infusion and tryptone soy broth media). The differences were detected in E. coli and P. aeruginosa, which assisted in the identification of the cellular origin of individual metabolites, and illustrated the complexity in the core (compounds emitted by all strains across all media) and accessory (compounds emitted by at least one strain in at least one medium) volatilomes.99Bacillus subtilis (a bacterium that has consistently demonstrated its resistance to space related extremes101) has been studied for its volatile emissions but mainly in isolates from soil and food sources.82 Interestingly, although many wild-type strains have been analysed for non-volatile metabolites,102 only a few studies have characterised their volatile profile.82,103
At a community level, mVOC profiles differ depending on the community's diversity and the dominant microbial groups.104,105 These volatilomes provide an opportunity to study biochemical pathways, synergetic interrelationships (e.g., quorum sensing), and allow investigation of the unique complexity found either in an organism, an ecosystem, or a matrix – including those from microbial metabolic processes and exogenously derived compounds.37,38,106
2.1 VOCs as metabolic products
Through primary metabolism, biota capture energy from the environment by catalysing complex redox reactions107,108 to build the basic molecules of life, essential for reproduction and growth.30 This process requires no addition of energy109 and produces primary volatile metabolites such as the VOCs acetic acid, acetone, or ethanol.109,110 These are usually produced by microbes in the exponential phase of growth.111 In contrast, secondary metabolism provides another important source of metabolites (called secondary – or specialised – metabolites), which are usually diverse, biologically active, and low molecular weight molecules including mVOCs.112,113In microbes, co-regulation of specialised metabolites has evolved for competitive advantage, where the evolution of traits optimises the retention and production of chemical diversity (at minimal energy cost to the microbe) to allow biomolecular activity.114 Their specialised metabolites also play key roles in metabolic co-regulation between biosynthetic pathways, therefore influencing microbial interactions.115 These enhancements can include, for example, the production and secretion of secondary metabolites as biosurfactants, which can decrease the surface tension of the surrounding liquid.116 Biosurfactants are crucial in cellular communication, and quorum sensing,117 both of which are utilised by microbes using VOCs.87,118,119 The VOC dimethylhexadecylamine has also been shown to affect bacterial growth and swarming motility.120 Utilising VOCs can therefore enhance survival/evolutionary advantages to a population.116 This aids individual microorganisms121 but also benefits the whole community in their quest for survival.116
Specialised metabolites are usually produced during late growth phase122 during transition from active growth to stationary phase.123 They are thought to play no direct part in growth62 and are not essential for at least short-term survival123 but could be a competitive advantage for ongoing microbial survival.124–126 Many specialised metabolites have been identified despite the full biosynthetic pathways being unknown.30 Recent genome sequencing has highlighted that genes involved in specialised metabolite biosynthesis are more abundant than first thought.127 For example, biosynthesis mechanisms for specialised metabolites, are often encoded in biosynthetic gene cluster (BGCs) regions within the genome.128 In the case of most Pseudomonas spp., there are also ‘orphan’ BGCs (loci encoding secondary metabolites) where the products are yet unknown.129 Also, in Streptomyces, there is a higher number of BGCs that expected in relation to production of specialised metabolites, many of which are not expressed under laboratory conditions; hence their products remain unknown.130 As genome sequencing identifies new biosynthetic pathways, more volatilomes that have yet to be fully determined may be discovered. This is an important feature when studying extremophiles.
Specialised VOCs have been the subject of recent interest due to their potential application in biotechnology drug discovery.82,131 Although their diversity has been investigated, however, in Bacillus subtilis isolates,82Acinetobacter johnsonii XY27,131 and in Actinomycete communities,130 they are still relatively unexplored as microbial metabolites.82,130
2.2 Metabolic diversity of microbes
Microorganisms have evolved many different metabolic strategies to obtain energy, as illustrated in Fig. 1. This means the volatilome of differing environments can be complex and combining primary and specialised VOC metabolites from a range of organisms and metabolisms. It is important, therefore, to understand the VOC contributions from the different metabolic pathways before considering the complexity of combining those in an environment. | ||
| Fig. 1 Metabolic diversity of microbes – based on primary source of energy as compared with higher plants and animals (other anaerobic microbial metabolism e.g., fermentation is included in text below) (Adapted from Burgin et al. 2011).107 | ||
Phototrophs are organisms such as photosynthetic bacteria, algae or plants that acquire energy from light.132 Cyanobacteria (blue-green algae) can survive in numerous environments, including cold deserts, oceans, hypersaline waters, and hot springs133,134 and produce VOCs such as aliphatic alcohols, aldehydes, and monoterpene alcohols.135 For example, the cyanobacterium Synechococcus sp. GFB01, isolated from a freshwater lagoon in the Amazon, synthesises five VOCs including 6-pentadecanol and octadecyl acetate which had not previously been described for the phylum.133 Several VOCs such as 3-methyl pyruvate, stearic acid and biuret have been identified to play a critical role in regulation of the composition, and diversity of other prokaryotic communities in hypersaline sediments,136 an environment that could be prevalent on other planetary bodies.
Chemotrophs are organisms such as bacteria that obtain energy by oxidising reduced compounds. They are subdivided into chemoorganotrophs (using organic compounds) and chemolithotrophs (using inorganic compounds), as shown in Fig. 1.137,138
Chemoorganotrophy can occur aerobically (e.g., oxygenic respiration) where VOCs such as methanethiol, DMDS and acetoin are produced, and anaerobically (e.g., fermentation) where VOCs such as alcohols, ketones, and fatty acids139 are produced. Chemoorganotrophic microorganisms obtain energy from the oxidation and reduction of carbon/organics, including VOCs; for example, Pelagibacter HTCC1062 a marine bacterium, can consume and metabolise isoprene (C5H8) and acetone (C3H6O), which are prime climate-active (e.g., can alter atmospheric chemistry) VOCs.140 Chemoorganotrophs can also produce smaller organic compounds that can be used for biosynthesis or other assimilatory pathways.141,142 In the case of soil systems, bacterial acetone metabolism occurs via several biochemical pathways, utilising the acetone monooxygenase enzyme to produce different VOCs such as acetaldehyde and formaldehyde (Fig. 2). Methyl acetate, the alternative product of the monooxygenase, can also be further converted to the VOCs methanol and acetic acid.140,143
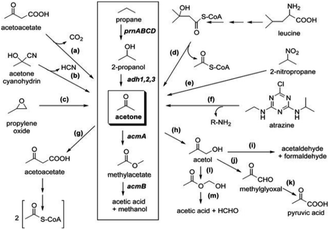 | ||
| Fig. 2 Overview of acetone metabolism.143 | ||
In sub-surface environments on Earth, where photosynthesis is inhibited, e.g., deep-sea hydrothermal vents,144,145 primary production is driven by chemolithotrophic microorganisms. Hence, they can be used as candidates for putative life on other planets, given most extraterrestrial surface environments have detrimental radiation conditions.146–148 Chemolithotrophic microorganisms catalyse inorganic chemical reactions that are in disequilibrium with their environment and drive the reactions towards equilibrium through redox reactions utilising, for example, manganese, iron, nitrogen, or sulfur.107 Chemolithotrophs in hydrothermal vents, for example, utilise volatile H2S as an electron source for growth145 but also produce organic acids as specialised metabolites.149,150 Iron and sulfur oxidising bacteria such as Acidithiobacillus ferrooxidans, Leptospirillum ferriphilum, and Acidithiobacillus caldus – considered to have substantial roles in oxidative dissolution of sulfide minerals in acidic environments – have been shown to produce the VOC glycolic acid in cell free culture liquors.151
This review focuses exclusively on chemolithotrophs since they are the most likely candidates for primary production on other planetary bodies and are common in the more extreme environments on Earth.152,153
3 Detecting VOCs
3.1 Detecting VOCs in extraterrestrial environments
In searching for life beyond the Earth, volatile analysis has predominately been achieved via remote sensing, including characterising exoplanet atmospheres. A range of volatile trace gases have been targeted in this context, such as oxygen (O2), O3, nitrous oxide (N2O) and CH4154–156. CO2 and CH4 have been recently detected remotely by the James Webb Space Telescope (JWST) in the atmosphere of the candidate exoplanet K2-18b,157 where the VOC dimethyl sulfide (DMS) has also been tentatively identified.142 However, at the astronomical distances involved in these contexts, VOCs are unlikely to be harnessed as biosignatures remotely: the concentration within an entire atmosphere would be too low to detect effectively.154 Even if detected over shorter distances, there is a further limitation on using these molecules as biosignatures: owing to the inherent nature of gases, and the extreme environments that exist beyond Earth (e.g., extreme in pH, temperature, salinity, radiation, water availability, and pressure158), VOCs detected on the surface or in the atmosphere of planetary bodies are unlikely to be indicative of any past life. Any VOCs would have rapidly dissipated or been affected on some bodies by UV/ionising radiation, leading to their destruction or transformation to a form that is not readily detectable by current remote or lander instrumentation.159–161 Furthermore, many can be generated abiotically, for example, CH4 can be created by rock–water interactions,156 spallation (e.g., lunar regolith162,163). O2 can be produced by H2O photolysis,164,165 CO2 photolysis, or an extreme hydrogen escape event with subsequent O2 build up.156 DMS has also been identified on comet 67P using data from the Rosetta spacecraft suggesting it may not be a robust indicator for life due to its potential abiotic production.166,167 All these abiotic sources can then lead to a ‘false-positive’ detection or over-impose those that may be produced by life.168VOC analysis is predominantly useful for detecting current life, including life that may exist in the subsurface environment. However, fluids and/or gases can also become entrapped in minerals in rocks or ice (as inclusions), providing a valuable archive of their preserved chemistry.169,170 For example, halite can entrap gases that record ancient chemistry, climate, or evidence of past micro-organisms that may have decomposed to produce CO2, CH4 and VOCs such as aldehydes, alcohols, or ketones.169 Miβbach et al. (2021) showed the presence of volatile compounds within fluid inclusions in 3.5-billion-year-old rocks (e.g., H2S, carbonyl sulfide (COS), methanedithione (CS2), CH4, acetic acid, organic (poly-)sulfanes, and thiols), which could have been important substrates for sulfur and methanogenic metabolisms by the earliest life on Earth. However, thiols are unstable in aqueous medium and tend to oxidise to disulfides171 with the -SH group also causing challenges with chromatographic separation even when the thiol is well preserved172). These inclusions pose an analytical challenge with detection and sensitivity owing to their low abundances173 and are not the focus of this paper.
VOC analysis is a non-invasive, quick, and economical method to detect potential biosignatures and it could, with improvements in analytical capabilities, be deployed in future life detection missions. VOCs and organic compounds on Earth can be identified and quantified using a variety of analytical techniques including ion mobility spectrometry (IMS), gas chromatography mass spectrometry (GC-MS), gas chromatography with flame ionisation detection (GC-FID), fourier transformation infrared spectrometry (FTIR), proton transfer mass spectrometry (PTR-MS), Selective Ion Flow Tube Mass Spectrometry (SIFT-MS), Raman spectroscopy, non-selective gas sensors, photoionisation detectors (PID), and fluorescence spectroscopy.174–181 Some of these instruments have been deployed on past and planned spaceflight missions to search for evidence of life, with GC-based instruments dominating payloads.182–184 For example, fluorescence spectroscopy, Raman spectroscopy and GC-MS are utilised on the Sample Analysis on Mars (SAM) instrument onboard NASA's Mars Science Laboratory (MSL) Curiosity rover,185 and fluorescence and Raman spectroscopy are deployed on the Perseverance rover within its Scanning Habitable Environments with Raman and Luminescence for Organics and Chemicals (SHERLOC) instrument.186 A GC-MS was also deployed on the Mars Viking landers,187 and on the Cassini–Huygens probe, which determined the composition of Titan's atmosphere as the Huygens lander descended.188 The Rosetta orbiter spectrometer for ion and neutral analysis (ROSINA) was deployed on the Ptolemy instrument which was deployed to chase and analyse the comet 67P which included a GC/MS with an ion trap mass spectrometer.189 The Mars Organic Molecule Analyser (MOMA) onboard ESA's ExoMars ‘Rosalind Franklin’ rover (due for launch in the late 2020s) will have a GC instrument with four columns to allow in situ analysis of organic molecules and enantiomers, including VOCs and semi volatiles (sVOCs).190 Similarly, the Europa Clipper mission, launching in October 2024, and will combine spectroscopy and GC-MS techniques to detect organics including VOCs with its ‘MAss Spectrometer for Planetary Exploration’ (MASPEX) instrument191,192 in its search for biosignatures on Jupiter's moon Europa.193 With the presence of subsurface oceans seen and predicted on other planetary bodies in our solar system (icy moons)194 exploration using submersibles in future missions may become a reality.195 Although VOCs are usually detected directly in the gas phase they can be detected in liquids or in the headspace above the liquid. For example, on Earth deep sea ramen spectrometers have been developed to detect variations in H2S, CH4 and CO2,196 a solid phase micro extraction (SPME) sampler has been used on a submersible analysing hydrothermal deep see vents,197 and a hydrothermal organic geochemistry sampler has been developed for deployment on deep sea submersibles.198
It is critical to validate approaches used when seeking VOCs as biosignatures, e.g., gain baseline data, and test instrumentation on Earth. For this, analogue sites are used – sites on Earth that exhibit geological, chemical, or biological similarities to those of the target of a mission. These are habitats that have environmental conditions that are harsh, and beyond the optimal range for humans.199,200 Organisms that have adapted to survive and thrive in these conditions are known as extremophiles (extreme-loving).199,201–203 Identifying and exploring how these extremophile microorganisms survive and interact with their environment through monitoring organic species such as VOCs is therefore vital for determining the potential volatilomes that might be established for equivalent extraterrestrial environments.
3.2 Detecting VOCs in terrestrial environments
There has been limited work relating to mVOC production in terrestrial extreme environments, with most focus instead on mVOCs related to pathogenic species, plant, and soil microbes.204–207 However, in the cold desert environment of the Antarctic, the genera Pseudoalteromonas (in particular the Antarctic bacterium P. haloplanktis TAC125 – a model example of cold-adapted bacteria) has been shown to produce bioactive specialised metabolites including anti-biofilm molecules, compounds that promote antiproliferative action, and antimicrobials.206 These functional responses in the bacterium can then give a competitive advantage to some community members208 or create disadvantages for competing communities,209 since cells in biofilms survive harsh growth conditions210 and are more resistant to UV, toxicity from metals, acid exposure, and desiccation.211 pH has also been shown to influence the mVOCs produced and their utilisation. For example, microalgae, when exposed to acid conditions in a marine environment, produced different VOCs compared to diatoms, and these changes in VOCs altered the behavioural responses of benthic invertebrates.212 Further, Streptomyces venezuelae produces the VOC trimethylamine (TMA), which increases the pH to a more alkaline state (pH 9.5), subsequently acting as a ‘Streptomyces communication cue’ (causing the microbes to initiate exploration into separate Streptomyces colonies) as well as acting as a weapon to reduce the survival of other soil bacteria such as Bacillus subtilis.213The analytical methods for characterising VOCs from natural sources have been well documented by Rowan (2011)16 and Li (2023).214 To increase the breadth of environments characterised for their volatilomes, there has been steady progress in the technology of portable field instruments and their applications, offering a wide range of organic and inorganic analysis in multiple matrices.215 In addition, techniques that can give a snapshot of the volatiles present at a timepoint, or a real-time/near real-time representation over time have been developed.216–218 Due to the huge diversity of these volatile compounds, no ‘one single’ analytical method can detect them all, which then leads to challenges in the identification or characterisation of certain compounds of interest214 and therefore any determination of an entire volatilome. Especially within dynamic processes such as metabolism. Working in extreme field sites also requires staying in remote locations far away from a regular source of power, from lab-based analytical equipment, and laboratory supplies. This somewhat limits the type of equipment that can be used in the field, especially if it is also heavy or bulky. Some areas of exploration have access only by foot with challenging hiking in very high or low temperatures, or with variable terrain to navigate. With local pH or salinity also in the extreme, some equipment may become compromised or dysfunctional.
To mitigate this, sampling techniques have been developed to entrap or isolate volatile gases. This also ensures targeted sampling, and not just the ‘air’ around it. Several types of containers – gas-tight syringes, stainless steel canisters, or borosilicate glass bulbs/containers – can be utilised219,220 but these can be fragile, with limited volume. Pre-evacuated canisters are robust and provide good sample stability but are bulky, heavy and require rigorous cleaning between samples.219 A solution to this is food grade polymer bags (for example made of Nalophan® (polyethylene terephthalate), or more expensive bags made from Teflon® (polytetrafluoroethylene) or Tedlar® (polyvinyl fluoride) materials). These are chemically inert, have low permeability to VOCs,221 are economical and more convenient for field analysis220 being foldable, low weight, resistant to breakage, and available in various sizes.222
These bagging methods are well established222 and are extremely useful in many contexts, including enclosing stems and leaves on living plants,223 in breath analysis,219,224 for the enclosure of a huge variety of biological samples,225–227 and biofilms97,228,229 or sediment samples.230–232 These approaches allow for a more volatile rich, and specific sample to be collected, which helps equilibrate VOCs between sample and headspace, and create a larger dynamic headspace to sample from ref. 233 and 234. Dynamic enclosure techniques are also being utilised to enclose larger areas such as to study fluxes in sub-artic tundra plants235 and permafrost affected peatland.236 By applying an enclosure over an item of interest, a snapshot of VOCs can be collected. This can also be applied over a gas emanation source (Fig. 3a), biofilms (Fig. 3b), or by placing sediment samples or biofilms into a fully enclosed bag/container (Fig. 3c).
There are some disadvantages to bagging methods: limited time to keep the sample in the bag – suggested up to 6 hours before loss of volatiles via diffusion through the material,237 loss of sample through wall adsorption, leaking, or potential loss of organics via partitioning into water vapour.222 Some gas sampling bags (e.g., reusable ones such as Tedlar®) may also need to be cleaned or conditioned extensively prior to use as they can produce and adsorb VOCs, such as phenol.238 To counteract this in the field, samples can be analysed either directly from the bags or by concentrating the VOCs onto a solid adsorbent, such as those in a thermal desorption (TD) tube or trap, and then purging and/or heating within the 6 hours window to remove excess water vapour.234
Analytical tools such as TD tubes,239,240 or pocket diffusive (POD) samplers,241,242 are useful as they are portable, small, light, and contain sorbent material such as Tenax™ or Carbotrap™. These sorbents are hydrophobic and preconcentrate VOCs onto them243,244 making them easy to carry to and use in a remote site. Electronic nose245 has a multi sensor array that can respond to numerous chemical classes that are then identified by artificial neural network (ANN) software.246 The type of sensor used in portable versions can be more specific to measuring low-concentration gases but struggle with high concentration environments.247 However, recent developments have helped create a more stable system, with reduced interference from humidity and temperature, that has been deployed successfully in the VOC analysis of whitefly infestations on tomato plants.248 Portable commercial gas sensors (e.g., electrochemical, infra-red, photo ionisation detectors249) provide specific detection for individual or small groups of gases,250 and are often tailored to gas leak detection, fire detection, or control of ventilation.251 They have recently become smaller, more affordable, and easier to use250 making them a useful tool when in the field.
In many extreme environments, the variety and concentration of gases emitted are likely unknown, so broad VOC detection is a necessity. However, many of these analyses are complicated to achieve in the field, as they require sensitive non-portable equipment. Physical samples such as sediments or biofilms can be collected on site and transferred back to a laboratory. However, metabolic changes may occur within the time frame of transportation, or the samples may be subjected to contamination. To minimise this, cooling and freezing samples as soon as possible is essential252 so they can be returned to laboratory conditions for further analysis and potential culturing. Using microcosms which reproduce conditions as close to those in analogue environments, it is possible to measure VOCs under controlled conditions. This technique has been proven effective with terrestrial samples from boreal peatland253 and in reproducing the behaviour of VOCs in subsurface materials.254
There is also analytical equipment that is becoming more portable for in-field analysis. Ion mobility spectrometry (IMS) is an established technique for VOC detection in military security (detection of chemical warfare agents and explosives) and air quality control and monitoring in industrial processes.255–257 IMS does not require the addition of bulky vacuum pumps and can detect down to ppb levels;257,258 it is robust and has a rapid response and wide application.255 However, IMS can have limited selectivity alone (e.g., to separate and characterise compounds) and requires coupling with a GC, MS or with liquid chromatography (LC).257,259 For example, it has been deployed alongside GC (GC-IMS) by NASA to function as the volatile organic analyser (VOA) on the International Space Station (ISS) to monitor air quality for outgassing material, contaminants from human excretion, containment breaches of utility chemicals (fuels, coolants etc), and thermal degradation by products.260
An ‘in the field’ portable GC-MS has also been developed but in practice was bulky, fragile, and required a significant power supply.261 More recently, military units have utilised portable GC-MS to rapidly confirm chemical warfare agents (CWAs) at very low concentrations, but these systems proved complex from both a hardware and software perspective making them difficult to use in harsh and dangerous environments262 and subject to continued testing.263 For example, a needle trap system has been incorporated into one portable GC-MS design to allow dynamic or static headspace analyses, but this could only detect 31 volatile compounds, with a sensitivity too low for full characterisation. In addition, deploying such a system in the field would require key processes such as calibration curves and method development to also be carried out, increasing the demand and duration of use.264 For example, an internal ion trap mass library must be generated using standards to allow for proper compound identification. Compound resolution can also be an issue in complex samples as column lengths in field-based kits are usually short, making it more likely to need deconvolution due to more complicated overlapping resolutions.265 These issues continue to be problematic for portable equipment, however the reduction in storage or transportation issues can outweigh the need for ultimate sensitivity.215
A further compromise needed with vVOCs and other trace gas species, as many cannot be collected or stored on a sorbent or in containers unless under cryogenic conditions. This requires either large heavy cylinders/canisters to collect the sample in or liquid nitrogen which is difficult to transport to remote field sites. For example, CH4 cannot be retained on traditional sorbents in a TD tube, so this method is not viable for CH4 detection.266 Recent developments in photoacoustic spectroscopy gas sensors for trace gases such as CH4 and C2H6 have led to a range of highly sensitive, fast sensors that are reliable and chemically sensitive.267 Optical fibre sensors based on absorption, which incorporate LED, hollow core waveguides (HCW) and photodiodes, have also been developed.268 They can also be coated with layers of nano-assembled ultrathin films to allow for specificity to certain VOCs269,270 and are small, portable, resistant to harsh environments and corrosion, and immune to electromagnetic interference.270,271 These have not yet been tested in extreme environments, but the hope is that a broad range of portable, affordable devices will allow the fullest analysis of volatiles from trace gases to sVOCs and aerosols.
4 Simulations, data analytics and beyond
4.1 Simulated environments
With the high costs of directly sampling planetary environments and their access limited to specific missions, a wide variety of simulation chambers have been developed to mimic conditions that have either previously been measured by spacecraft or are predicted to exist on certain planetary bodies such as different radiation environments, varying temperature/pressure conditions, or water/rock ratios. These can provide insights into conditions expected on rocky planets (e.g., Mars272,273 or Venus274–276), icy worlds,152,277 or early-Earth conditions.278 The stability of organics and interactions with organisms can be observed in such simulated environments.275,279 These data type will be invaluable to elaborate on for future missions.280 For example, by understanding multiple biochemical pathways and potential transformational processes, it may be possible to predict the behaviours of organics in certain environments, as well as understanding how they are affected by different parameter changes, or the addition of microbial life.Identifying VOCs or other trace gases over a wide simulated parameter space could provide insights for understanding data returned from future subsurface exploration or lander missions.278 By coupling VOC analysis to simulation experiments we can understand how VOCs behave within these environments, how they chemically change under differing conditions, and how they could be used to help make decisions about future mission targets. If organisms are active or can survive under the simulated planetary simulation conditions, including the subsurface environment,281 this could inform habitability studies immensely.280
4.2 Maximising lab-based studies
In current laboratory settings, techniques to analyse VOCs such as TD, GC-MS, SIFT-MS, and PTR-MS are gold standard or are becoming a gold standard.174,282,283 These techniques are being successfully applied to planetary science and will be essential to lab-based experiments in astrobiology. For example, the recent Winchcombe meteorite analysis in the UK included SIFT-MS analysis, which yielded a number of volatile species, including alcohols (C1–C6), carboxylic acids, aldehydes, and ketones.284 While organic species have been identified in meteorites for several decades, the application of SIFT-MS was novel and yielded a greater understanding of the volatile component of this type of meteorite, which has been difficult to elucidate.285 Evolved gas analysis (linear heating of material to release volatile gases that are then detected by gas chromatography, infrared spectroscopy, or mass spectrometry) is a well explored technique that has been successfully applied to studies of meteorites, including Winchcombe286,287 and lunar samples from the Apollo missions. Other methods such as smartphone based, SPME, and adsorbent traps are detailed in Tholl et al. (2021).265 HiSorb® is also a recently developed technique which is compatible with aqueous samples and requires no solvent extraction. It also allows for a larger volume of sorbent than SPME (HiSorb® 65 μL vs. SPME 0.5 μL), improving the extraction capabilities.Samples collected in field sites or from return missions will include many complex matrices (for example a combination of sediments, microbial mats, and gases in hydrothermal fluids288). This added complexity can be hard to completely unravel with the previously mentioned analytical techniques. A more expensive but advanced tool for analysis of these complex signatures is using two-dimensional gas chromatography (GCxGC) where compounds are separated in two dimensions, allowing overlapping peaks to be separated.289 GCxGC can not only be coupled with multiple detectors (FID290/sulfur chemoluminescence detector (SCD)291/Time of flight MS292) but also handle complex samples at low limits of detection (LOD).293 It can also be coupled with TD systems allowing it to be utilised in the lab after field sampling of VOCs.294 2D GCxGC-ToFMS has already been applied to the analysis of extraterrestrial samples.295
4.3 Analysing VOCs as a whole unit
Volatilomics targets a small or large range of metabolites and explores the characterisation, detection, and quantification of these metabolites in a biological system.37,69 It has been highlighted that VOCs should be integrated into metabolic profiling to enhance our understanding of microbial systems. In metabolomic studies, overlooked volatile components can lead to an incomplete understanding of the interspecies- and intraspecies-to ecosystem interactions.30 To get a greater insight into the function and diversity of volatile compounds as, for example, secondary metabolites, it is vital to follow workflows that are well documented, as well as look at production of VOCs at a single cell level, to potentially attribute metabolites to certain developmental stages or cell forms.82 As the volatilome has not readily been explored within astrobiology, this is a key area to focus on in future.An advantage to studying the whole volatilome is that you can perform targeted and untargeted analyses, as is undertaken in traditional metabolomics.30 In targeted analyses, compounds that are predetermined by the researcher are selected for analysis. The untargeted analysis involves the evaluation of all the detectable compounds in the sample68 including chemical unknowns and is the current preference for analysis.296 The untargeted method can be particularly useful when studying unknown environments or micro-organisms, such as those that could be present on other planetary bodies, as many different VOCs could be identified. Untargeted volatilomics focuses on the dynamic adjustments of small molecules in response to subtle disruption made by organisms.297 These methods can also be performed in ‘real-time or near real-time’, which allows non-invasive monitoring, particularly on microbial communities.30
In astrobiology, being able to analyse a group of compounds and their relationships to each other, rather than relying on a single gas, could provide a wealth of information as has been shown successfully with other ‘omics techniques within astrobiology such as exploring the diversity of microbial mats in the Makgadikgadi Salt Pans,298 and genomic modelling of extremophiles.299,300 Alone, gene-based tools are currently insufficient to explore the full plethora of chemical reactions and small molecules that compose a living cell.301 Different ‘omics techniques have provided insights into the survivability of microbes living in terrestrial environments that resemble potentially habitable environments on other worlds.302 By analysing a variety of small metabolic products of living organisms including combining with volatilomics, we can help target the search for biomarkers and their interactions.301
4.4 Data analytics
To explore the volatilome, complex robust statistical analysis is becoming more commonplace303 but it is a complex technique that has numerous dimensions (compounds), and different magnitudes of each dimension.304 Consequently, evaluating these data can be challenging and requires specialist expertise. Chemometric methods are used to retrieve greater information from the chemical information. Primary goals often include differentiation of compounds between different groups.305 With advances in computer technology, statistical software, and analytical techniques, the chemometric approach has supported analytical chemists to obtain more robust, quicker results for analyses.306 Typically, a full spectrum of data is imported, pre-processed, and then subjected to numerous different data analysis techniques or machine learning to tease out the interesting information.64 For extensive reviews of these processes see Eisen et al.,305 and Lubes & Goodarzi,307 and references therein.Prevalent methods in chemometrics include unsupervised pattern recognition (UPR), supervised pattern recognition (SPR), and exploratory data analysis (EDA). Unsupervised methods include principal component analysis (PCA), and cluster analysis (CA) and focus on the interconnectedness of the data and the intrinsic structure and relationship of the different features (e.g. peak area).307 Supervised methods (often termed predictive models) for example classification models, partial least squares (PLS), discriminant analysis (DA), and neural networks267,307 require training the data. This allows the development of classification models that are built on prior information about the samples. These models are then tested and validated on a known independent sample set, before then being applied to unknown samples.307
Classification models have been widely used in VOC research within many medical and food-based studies. For example, for differentiating air–liquid interface cultures after Staphylococcus aureus infection,308 separating healthy and infected mushrooms via microbial VOCs,309 differentiating VOCs associated with security issues (e.g., drug trafficking, explosives, or the presence of humans in forbidden areas),310 identifying microorganisms in pulmonary bacterial infections,311 assessing microbial and mite contamination in cereal grains and coffee beans,312 or profiling the faecal metabolome in horses with colic.313 They have also been used in many metabolomic studies on individual or groups of microbes,314 including NMR metabolomics of bacterial extracts,315 in combination with proteomics, to analyse the effect of spaceflight on rice progeny,316 and the identification of microbes using their metabolomic profiles.317 These studies all highlight that by applying multivariate techniques to both untargeted environmental volatile profiles, and microbial volatile profiles, large amounts of complex data can be integrated and compared. Subsequently, analysis time is also shortened by the reduction in manual processing, human errors are minimised and there is less need for analytically trained experts.318
By using these classification techniques, data analysis can be expanded within astrobiology to distinguish differences or similarities between VOCs in environments, microbial communities, or can even assist in searching for contamination. Exploring volatile compounds and their interactions within environments that could hold biological systems, rather than just searching for individual compounds of interest produced by abiotic processes, could allow subtle changes in VOC dynamics produced by life to be tracked. The amount, ratio, or diversity of emitted VOCs are part of a microbe's phenotype,319,320 so when their volatilomic data are then also combined with metabolomic, transcriptomic and genomic data, a more complete picture of metabolic pathways, and the potential routes VOCs are produced and utilised through this. This is useful to astrobiology because it helps to elucidate a clear picture of a whole system. If only parts of it are investigated, subtle elements could be missed in the relationship between rock, environment, and potential life. By understanding how they fit into complex systems could also help predict how they may evolve or be detectable in places where the immediate surface environment is too harsh to sustain life.
4.5 Future missions
Real-time measurements, miniaturisation, automation, reliability, portability, accuracy and sensitivity and low power consumption are all pre-requisites for analytical techniques that may be deployed on a spacecraft.321 This provides a great challenge to analysts without even considering the finer points of landing a mission or running equipment that is millions of miles away.321,322 Technical innovation has allowed technology such the mass spectrometer to be mobilised and miniaturised to allow in situ and field deployment.323 In the context of VOCs, an OrbitrapTM cell-based mass spectrometer termed OLYMPIA (Orbitrap anaLYser MultiPle IonisAtion) is being developed for spaceflight, as well as a laboratory instrument for high-resolution studies of space-relevant chemical processes.324Application of high throughput ‘omics’ analysis and deployment of omics instrumentation into space is also coming to the fore in relation to in situ biological analysis in locations such as the International Space Station (facilitating life science research and enabling dynamic biological studies in low gravity environments).325 However, when samples from planetary bodies, such as Mars or the Moon, are returned to Earth, VOC analysis, including high throughput ‘omics’, would be beneficial in the search for life and for planetary protection.326–328 For example, in the planned NASA/ESA Mars sample return programme, a sample receiving facility (SRF) will be in place for curation, where sample tubes will be opened and initially processed.329 Preliminary stages of this processing will include safety protocols to avoid backward contamination (release of extraterrestrial material into Earth's biosphere).330 All material collected (which includes samples of gas, rock, and sediments329,331) needs to be carefully stored, handled, and analysed with minimal alteration or contamination from the Earth, while also ensuring the samples are non-hazardous and sterilised before distribution.329 Initial volatile analysis of the collected gases and rocks offers a way to minimally interact with the samples prior to sterilisation332 since harsh chemicals are not required for compound extraction, and the gas can be gently removed from sample containers to establish its indigenous volatilome. This allows the collection of valuable initial data with minimal interference, acting as a robust precursor to the wealth of other analyses likely to be conducted on extraterrestrial samples where sample alteration or destruction will be required.
For now, researching volatilomics in astrobiology requires time spent characterising patterns of VOCs and differentiating these from abiotic signals within analogue or simulated environments. Such an effort is critical for understanding the data returned from past or upcoming missions, but it requires a step change in approach. Exploring the volatilome in astrobiology offers this step change.
5 Author contributions
Claire Batty: conceptualization, writing – original draft, writing – review & editing. Victoria Pearson: conceptualization, writing – review & editing. Karen Olsson-Francis: conceptualization, writing – review & editing. Geraint Morgan: writing – review & editing.6 Conflicts of interest
There are no conflicts to declare.7 Acknowledgements
The authors would like to thank team project officers for their ongoing support including Camilla Wilkinson and Simona Nicoara.8 References
- J. F. Banfield, J. W. Moreau, C. S. Chan, S. A. Welch and B. Little, Astrobiology, 2001, 1, 447–465 CrossRef CAS PubMed.
- A. Price, M. C. Macey, V. K. Pearson, S. P. Schwenzer, N. K. Ramkissoon and K. Olsson-Francis, Front. Microbiol., 2022, 13, 800219, DOI:10.3389/fmicb.2022.800219.
- J. Tan and M. A. Sephton, Astrobiology, 2020, 20, 53–72 CrossRef CAS.
- P. Vandevivere, S. A. Welch, W. J. Ullman and D. L. Kirchman, Microb. Ecol., 1994, 27, 241–251, DOI:10.1007/BF00182408.
- D. J. Des Marais, J. A. Nuth, L. J. Allamandola, A. P. Boss, J. D. Farmer, T. M. Hoehler, B. M. Jakosky, V. S. Meadows, A. Pohorille, B. Runnegar and A. M. Spormann, Astrobiology, 2008, 8, 715–730 CrossRef.
- A. Pohorille and J. Sokolowska, Astrobiology, 2020, 20, 1236–1250 CrossRef PubMed.
- C. Gillen, C. Jeancolas, S. McMahon and P. Vickers, Astrobiology, 2023, 23, 1228–1237 CrossRef CAS PubMed.
- F. Westall, Space Sci. Rev., 2008, 135, 95–114 CrossRef.
- A. K. Garcia, C. M. Cavanaugh and B. Kacar, ISME J., 2021, 15, 2183–2194 CrossRef CAS PubMed.
- S. Cogliati, E. Wolsey, N. K. Ramkissoon, S. P. Schwenzer, V. K. Pearson and K. Olsson-Francis, Front. astron. space sci., 2022, 9, 1062007, DOI:10.3389/fspas.2022.1062007.
- R. E. Summons, J. P. Amend, D. Bish, R. Buick, G. D. Cody, D. J. Des Marais, G. Dromart, J. L. Eigenbrode, A. H. Knoll and D. Y. Sumner, Astrobiology, 2011, 11, 157–181 CrossRef PubMed.
- W. F. M. Röling, J. W. Aerts, C. H. L. Patty, I. L. Ten Kate, P. Ehrenfreund and S. O. L. Direito, Astrobiology, 2015, 15, 492–507 CrossRef.
- L. E. Hays, H. V. Graham, D. J. Des Marais, E. M. Hausrath, B. Horgan, T. M. McCollom, M. N. Parenteau, S. L. Potter-McIntyre, A. J. Williams and K. L. Lynch, Astrobiology, 2017, 17, 363–400 CrossRef PubMed.
- J. L. Vago, F. Westall, A. J. Coates, R. Jaumann, O. Korablev, V. Ciarletti, I. Mitrofanov, J. L. Josset, M. C. De Sanctis, J. P. Bibring, F. Rull, F. Goesmann, H. Steininger, W. Goetz, W. Brinckerhoff, C. Szopa, F. Raulin, H. G. M. Edwards, L. G. Whyte, A. G. Fairén, J. Bridges, E. Hauber, G. G. Ori, S. Werner, D. Loizeau, R. O. Kuzmin, R. M. E. Williams, J. Flahaut, F. Forget, D. Rodionov, H. Svedhem, E. Sefton-Nash, G. Kminek, L. Lorenzoni, L. Joudrier, V. Mikhailov, A. Zashchirinskiy, S. Alexashkin, F. Calantropio, A. Merlo, P. Poulakis, O. Witasse, O. Bayle, S. Bayón, U. Meierhenrich, J. Carter, J. M. García-Ruiz, P. Baglioni, A. Haldemann, A. J. Ball, A. Debus, R. Lindner, F. Haessig, D. Monteiro, R. Trautner, C. Voland, P. Rebeyre, D. Goulty, F. Didot, S. Durrant, E. Zekri, D. Koschny, A. Toni, G. Visentin, M. Zwick, M. Van Winnendael, M. Azkarate and C. Carreau, Astrobiology, 2017, 17, 471–510 CrossRef PubMed.
- F. Hadacek and G. Bachmann, Front. Environ. Sci., 2015, 3(12), 1–21 Search PubMed.
- D. D. Rowan, Metabolites, 2011, 1, 41–63 CrossRef CAS PubMed.
- EURO Rep Stud, 1989, pp. , pp. 1–70 Search PubMed.
- E. Hong-Geller and S. Adikari, in Biosensing Technologies for the Detection of Pathogens – A Prospective Way for Rapid Analysis, InTech, 2018 Search PubMed.
- T. Salthammer, Indoor Air, 2016, 26, 25–38 CrossRef CAS PubMed.
- S. Gagan, K. Sarang, K. J. Rudzinski, R. Liu, R. Szmigielski and Y. Zhang, Atmos. Environ., 2023, 312, 120017 CrossRef CAS.
- F. Vichi, A. Ianniello, M. Frattoni, A. Imperiali, G. Esposito, M. C. Tomasi Scianò, M. Perilli and A. Cecinato, Atmosphere, 2021, 12, 1609 CrossRef CAS.
- L. K. Sahu, R. Yadav and N. Tripathi, Atmos. Res., 2020, 246, 105114 CrossRef CAS.
- G. Capes, J. G. Murphy, C. E. Reeves, J. B. McQuaid, J. F. Hamilton, J. R. Hopkins, J. Crosier, P. I. Williams and H. Coe, Atmos. Chem. Phys., 2009, 9, 3841–3850 CrossRef CAS.
- F. E. Hopkins, P. D. Nightingale, J. A. Stephens, C. Mark Moore, S. Richier, G. L. Cripps and S. D. Archer, Biogeosciences, 2020, 17, 163–186 CrossRef CAS.
- T. Netzker, E. M. F. Shepherdson, M. P. Zambri and M. A. Elliot, Annu. Rev. Microbiol., 2020, 74, 409–430 CrossRef CAS.
- R. Seeburger, P. M. Higgins, N. P. Whiteford and C. S. Cockell, Astrobiology, 2023, 23, 415–430 CrossRef CAS PubMed.
- M. Jebbar, K. Hickman-Lewis, B. Cavalazzi, R. S. Taubner, S. K. M. R. Rittmann and A. Antunes, Space Sci. Rev., 2020, 216, 10 CrossRef.
- B. Audrain, M. A. Farag, C. M. Ryu and J. M. Ghigo, FEMS Microbiol. Rev., 2015, 39, 222–233 CrossRef CAS.
- B. Singh and E. Christina, in Volatiles and Metabolites of Microbes, Elsevier, 2021, pp. 205–234 Search PubMed.
- L. K. Meredith and M. M. Tfaily, Trends Microbiol., 2022, 30, 622–631 CrossRef CAS PubMed.
- S. M. Marshall, C. Mathis, E. Carrick, G. Keenan, G. J. T. Cooper, H. Graham, M. Craven, P. S. Gromski, D. G. Moore, S. I. Walker and L. Cronin, Nat. Commun., 2021, 12, 3033, DOI:10.1038/s41467-021-23258-x.
- K. M. Seaton, M. L. Cable and A. M. Stockton, Anal. Chem., 2021, 93, 5981–5997 CrossRef CAS.
- N. F. Wogan and D. C. Catling, Astrophys. J., 2020, 892, 127 CrossRef CAS.
- S. Seager, W. Bains and J. J. Petkowski, Astrobiology, 2016, 16, 465–485 CrossRef CAS PubMed.
- K. H. Halsey and S. J. Giovannoni, Earth Sci. Rev., 2023, 240, 104360, DOI:10.1016/j.earscirev.2023.104360.
- R. Cumeras, Curr. Metabolomics, 2017, 5(2), 79–89, DOI:10.2174/2213235x05666170502103408.
- A. E. Lytou, E. Z. Panagou and G. J. E. Nychas, Curr. Opin. Food Sci., 2019, 28, 88–95 CrossRef.
- M. Llambrich, J. Brezmes and R. Cumeras, Biol. Proced. Online, 2022, 24, 20 CrossRef.
- P. Poinot and C. Geffroy-Rodier, TrAC, Trends Anal. Chem., 2015, 65, 1–12 CrossRef CAS.
- M. Steinke, L. Randell, A. J. Dumbrell and M. Saha, in Advances in Ecological Research, Academic Press, 2018, pp. 75–92 Search PubMed.
- N. Drabińska, C. Flynn, N. Ratcliffe, I. Belluomo, A. Myridakis, O. Gould, M. Fois, A. Smart, T. Devine and B. D. L. Costello, J. Breath Res., 2021, 15, 034001 Search PubMed.
- A. S. Bannaga, H. Tyagi, E. Daulton, J. A. Covington and R. P. Arasaradnam, Molecules, 2021, 2447, DOI:10.3390/molecules26092447.
- N. M. Sagar, I. A. Cree, J. A. Covington and R. P. Arasaradnam, Gastroenterol Res. Pract., 2015, 2015 Search PubMed.
- A. N. Thomas, S. Riazanskaia, W. Cheung, Y. Xu, R. Goodacre, C. L. P. Thomas, M. S. Baguneid and A. Bayat, Wound Repair and Regeneration, 2010, 18, 391–400 Search PubMed.
- M. de Angelis, R. Francavilla, M. Piccolo, A. De Giacomo and M. Gobbetti, Gut Microbes, 2015, 6, 207–213 Search PubMed.
- S. Ghaisas, J. Maher and A. Kanthasamy, Pharmacol. Ther., 2016, 158, 52–62 CrossRef CAS.
- A. M. Casas-Ferreira, M. del Nogal-Sánchez, J. L. Pérez-Pavón and B. Moreno-Cordero, Anal. Chim. Acta, 2019, 1045, 10–22 CrossRef CAS.
- K. Ioannidis, C. Batty, C. Turner, D. Smith, F. Mannocci and S. Deb, Int. Endod. J., 2021, 54, 601–615 CrossRef CAS PubMed.
- C. Walton, D. P. Fowler, C. Turner, W. Jia, R. N. Whitehead, L. Griffiths, C. Dawson, R. H. Waring, D. B. Ramsden, J. A. Cole, M. Cauchi, C. Bessant and J. O. Hunter, Inflammatory Bowel Dis., 2013, 19, 2069–2078 CrossRef.
- C. Lourenço and C. Turner, Metabolites, 2014, 4, 465–498 CrossRef PubMed.
- B. Dixon, W. M. Ahmed, A. A. Mohamed, T. Felton and S. J. Fowler, J. Appl. Microbiol., 2022, 133, 2445–2456 CrossRef CAS PubMed.
- W. M. Ahmed, O. Lawal, T. M. Nijsen, R. Goodacre and S. J. Fowler, ACS Infect. Dis., 2017, 3, 695–710 CrossRef CAS PubMed.
- O. Lawal, H. Muhamadali, W. M. Ahmed, I. R. White, T. M. E. Nijsen, R. Goodacre and S. J. Fowler, J. Breath Res., 2018, 12(2), 026002, DOI:10.1088/1752-7163/aa8efc.
- C. A. Batty, M. Cauchi, J. O. Hunter, J. Woolner, T. Baglin and C. Turner, Clin. Chim. Acta, 2016, 461, 61–68 CrossRef CAS PubMed.
- R. Mcnerney, K. Mallard, P. I. Okolo and C. Turner, FEMS Microbiol. Lett., 2012, 328, 150–156 CrossRef CAS PubMed.
- R. Rinnan, M. Steinke, T. Mcgenity and F. Loreto, Plant, Cell Environ., 2014, 37, 1776–1789 CrossRef CAS.
- D. Mateirć, D. Blenkhorn, R. González-Méndez, D. Bruhn, C. Turner, G. Morgan, N. Mason and V. Gauci, Appl. Ecol. Environ. Res., 2016, 14, 667–681 CrossRef.
- A. Mulero-Aparicio, T. Cernava, D. Turrà, A. Schaefer, A. Di Pietro, F. J. López-Escudero, A. Trapero and G. Berg, Front. Microbiol., 2019, 10, 1808, DOI:10.3389/fmicb.2019.01808.
- H. Yuan, G. Cao, X. Hou, M. Huang, P. Du, T. Tan, Y. Zhang, H. Zhou, X. Liu, L. Liu, Y. Jiangfang, Y. Li, Z. Liu, C. Fang, L. Zhao, A. R. Fernie and J. Luo, Mol. Plant, 2022, 15, 189–202 CrossRef CAS.
- L. K. Honeker, K. R. Graves, M. M. Tfaily, J. E. Krechmer and L. K. Meredith, Front. Environ. Sci., 2021, 9, 649905, DOI:10.3389/fenvs.2021.649905.
- O. A. C. Almeida, N. O. de Araujo, A. T. N. Mulato, G. F. Persinoti, M. L. Sforça, M. J. Calderan-Rodrigues and J. V. C. de Oliveira, Front. Plant Sci., 2023, 13, 1056082, DOI:10.3389/fpls.2022.1056082.
- U. Salam, S. Ullah, Z. H. Tang, A. A. Elateeq, Y. Khan, J. Khan, A. Khan and S. Ali, Life, 2023, 13 Search PubMed.
- B. Sahlberg, M. Gunnbjörnsdottir, A. Soon, R. Jogi, T. Gislason, G. Wieslander, C. Janson and D. Norback, Sci. Total Environ., 2013, 444, 433–440 CrossRef CAS.
- J. Christmann, S. Rohn and P. Weller, Food Chem., 2022, 394, 133476, DOI:10.1016/j.foodchem.2022.133476.
- S. Li, N. Looby, V. Chandran and V. Kulasingam, Metabolites, 2024, 14 CAS.
- S. H. Chu, M. Huang, R. S. Kelly, E. Benedetti, J. K. Siddiqui, O. A. Zeleznik, A. Pereira, D. Herrington, C. E. Wheelock, J. Krumsiek, M. Mc Geachie, S. C. Moore, R. G. Snell and J. L. S. Lasky-Su, Metabolites, 2019, 9, 117, DOI:10.3390/metabo9060117.
- E. P. Resurreccion and K. W. Fong, Metabolites, 2022, 12 Search PubMed.
- J. Morimoto, M. C. Rosso, N. Kfoury, C. Bicchi, C. Cordero and A. Robbat, Molecules, 2019, 24, 3757, DOI:10.3390/molecules24203757.
- C. V. Berenguer, F. Pereira, J. A. M. Pereira and J. S. Câmara, Cancers, 2022, 14, 3982, DOI:10.3390/cancers14163982.
- F. Monedeiro, V. Railean-Plugaru, M. Monedeiro-Milanowski, P. Pomastowski and B. Buszewski, Int. J. Mol. Sci., 2021, 22, 4696, DOI:10.3390/ijms22094696.
- K. L. Reese, A. Rasley, J. R. Avila, A. D. Jones and M. Frank, Sci. Rep., 2020, 10, 9333, DOI:10.1038/s41598-020-66136-0.
- B. D. Green and M. Keller, Curr. Opin. Biotechnol., 2006, 17, 236–240 CrossRef CAS.
- A. Vaccalluzzo, G. Celano, A. Pino, F. M. Calabrese, P. Foti, C. Caggia and C. Randazzo, Front. Microbiol., 2022, 12, 771636, DOI:10.3389/fmicb.2021.771636.
- SS Board, An Astrobiology Strategy for the Search for Life in the Universe, National Academies Press, Washington, D.C., 2019 Search PubMed.
- L. Abis, B. Loubet, R. Ciuraru, F. Lafouge, S. Houot, V. Nowak, J. Tripied, S. Dequiedt, P. A. Maron and S. Sadet-Bourgeteau, Sci. Rep., 2020, 10, 6104, DOI:10.1038/s41598-020-63091-8.
- E. Parrilli, P. Tedesco, M. Fondi, M. L. Tutino, A. Lo Giudice, D. de Pascale and R. Fani, Phys. Life Rev., 2021, 36, 137–161 CrossRef CAS PubMed.
- C. Fagorzi, S. Del Duca, S. Venturi, C. Chiellini, G. Bacci, R. Fani and F. Tassi, Diversity, 2019, 11, 140, DOI:10.3390/d11080140.
- R. Jiménez-Mejía, R. I. Medina-Estrada, S. Carballar-Hernández, M. D. C. Orozco-Mosqueda, G. Santoyo and P. D. Loeza-Lara, Microorganisms, 2022, 10 Search PubMed.
- D. E. Sidorova, V. A. Plyuta, D. A. Padiy, E. V. Kupriyanova, N. V. Roshina, O. A. Koksharova and I. A. Khmel, Microorganisms, 2022, 10, 69, DOI:10.3390/microorganisms10010069.
- S. R. Haines, E. C. Hall, K. Marciniak, P. K. Misztal, A. H. Goldstein, R. I. Adams and K. C. Dannemiller, Microbiome, 2021, 9, 209, DOI:10.1186/s40168-021-01158-y.
- D. Blom, C. Fabbri, E. C. Connor, F. P. Schiestl, D. R. Klauser, T. Boller, L. Eberl and L. Weisskopf, Environ. Microbiol., 2011, 13, 3047–3058 CrossRef CAS PubMed.
- M. Kai, Front. Microbiol., 2020, 11, 559 CrossRef.
- R. M. S. Thorn and J. Greenman, J. Breath Res., 2012, 6, 024001, DOI:10.1088/1752-7155/6/2/024001.
- P. K. Misztal, D. S. Lymperopoulou, R. I. Adams, R. A. Scott, S. E. Lindow, T. Bruns, J. W. Taylor, J. Uehling, G. Bonitp, R. Vilgalys and A. H. Goldstein, Environ. Sci. Technol., 2018, 52, 8272–8282 CrossRef CAS.
- M. Yoshikawa, M. Zhang and K. Toyota, Microbes Environ., 2017, 32, 188–200 CrossRef PubMed.
- O. A. Koksharova and N. A. Safronov, Biophys. Rev., 2022, 14, 843–856 CrossRef CAS PubMed.
- R. Schmidt, V. Cordovez, W. De Boer, J. Raaijmakers and P. Garbeva, ISME J., 2015, 9, 2329–2335 CrossRef CAS.
- S. Schulz, C. Schlawis, D. Koteska, T. Harig and P. Biwer, in Bacterial Volatile Compounds as Mediators of Airborne Interactions, Springer Singapore, Singapore, 2020, pp. 93–121 Search PubMed.
- L. Weisskopf, S. Schulz and P. Garbeva, Nat. Rev. Microbiol., 2021, 19, 391–404 CrossRef CAS PubMed.
- J. Lasne, ACS Earth Space Chem., 2021, 5, 149–162 CrossRef CAS.
- J. Sikkema, J. A. M. De Bont and B. Poolman, J. Biol. Chem., 1994, 269, 8022–8028 CrossRef CAS.
- D. Trombetta, F. Castelli, M. G. Sarpietro, V. Venuti, M. Cristani, C. Daniele, A. Saija, G. Mazzanti and G. Bisignano, Antimicrob. Agents Chemother., 2005, 49, 2474–2478 CrossRef CAS PubMed.
- J. Niu, X. Li, S. Zhang, Y. Yao, Y. Zhang, Y. Liu, X. Peng, J. Huang and F. Peng, Front. Plant Sci., 2023, 13, 941929, DOI:10.3389/fpls.2022.941929.
- M. Chandrasekaran, M. Paramasivan and J. J. Sahayarayan, Microorganisms, 2023, 11 Search PubMed.
- M. Avalos, P. Garbeva, L. Vader, G. P. Van Wezel, J. S. Dickschat and D. Ulanova, Nat. Prod. Rep., 2022, 39, 249–272 RSC.
- D. G. Meldau, S. Meldau, L. H. Hoang, S. Underberg, H. Wünsche and I. T. Baldwin, Plant Cell, 2013, 25, 2731–2747 CrossRef CAS.
- D. E. Sidorova, M. I. Skripka, I. A. Khmel, O. A. Koksharova and V. A. Plyuta, Microorganisms, 2022, 10, 1512 CrossRef CAS PubMed.
- F. Goecke, A. Labes, J. Wiese and J. F. Imhoff, Mar. Ecol.: Prog. Ser. , 2010, 409, 267–300 CrossRef CAS.
- S. Fitzgerald, L. Holland and A. Morrin, Front. Microbiol., 2021, 12, 693075, DOI:10.3389/fmicb.2021.693075.
- S. Fitzgerald, C. Furlong, L. Holland and A. Morrin, Metabolites, 2022, 12, 432, DOI:10.3390/metabo12050432.
- M. Cortesão, F. M. Fuchs, F. M. Commichau, P. Eichenberger, A. C. Schuerger, W. L. Nicholson, P. Setlow and R. Moeller, Front. Microbiol., 2019, 10, 333, DOI:10.3389/fmicb.2019.00333.
- T. Stein, Mol. Microbiol., 2005, 56, 845–857 CrossRef CAS PubMed.
- S. Iqbal, F. Begum, A. A. Rabaan, M. Aljeldah, B. R. Al Shammari, A. Alawfi, A. Alshengeti, T. Sulaiman and A. Khan, Molecules, 2023, 28, 927, DOI:10.3390/molecules28030927.
- W. Raza and Q. Shen, in Molecular Aspects of Plant Beneficial Microbes in Agriculture, Elsevier, 2020, pp. 209–219 Search PubMed.
- J. Wang, X. Mei, Z. Wei, W. Raza and Q. Shen, Soil Ecol. Lett. , 2021, 3, 32–41 CrossRef CAS.
- R. Cumeras, Curr. Metabolomics, 2017, 5, 79–89, DOI:10.2174/2213235x05666170502103408.
- A. J. Burgin, W. H. Yang, S. K. Hamilton and W. L. Silver, Front. Ecol. Environ., 2011, 9, 44–52 CrossRef.
- X. N. Wang, G. X. Sun and Y. G. Zhu, J. Soils Sediments, 2017, 17, 2831–2846 CrossRef CAS.
- J. Rabha and D. K. Jha, in New and Future Developments in Microbial Biotechnology and Bioengineering, Elsevier, 2018, pp. 217–234 Search PubMed.
- H. S. Elshafie, I. Camele and A. A. Mohamed, Int. J. Mol. Sci., 2023, 24, 3266, DOI:10.3390/ijms24043266.
- S. Sanchez and A. L. Demain, Microb. Biotechnol., 2008, 1, 283–319 CrossRef CAS PubMed.
- D. Thirumurugan, A. Cholarajan, S. S. S. Raja and R. Vijayakumar, in Secondary Metabolites – Sources and Applications, InTech, 2018 Search PubMed.
- O. Mosunova, J. C. Navarro-Muñoz and J. Collemare, in Encyclopedia of Mycology, Elsevier, 2021, pp. 458–476 Search PubMed.
- G. L. Challis and D. A. Hopwood, Synergy and contingency as driving forces for the evolution of multiple secondary metabolite production by Streptomyces species, 2003 Search PubMed.
- Q. Yan, B. Philmus, J. H. Chang and J. E. Loper, Elife, 2017, 6, e22835, DOI:10.7554/eLife.22835.
- G. Santamaria, C. Liao, C. Lindberg, Y. Chen, Z. Wang, K. Rhee, F. R. Pinto, J. Yan and J. B. Xavier, Elife, 2022, 11, e76119, DOI:10.7554/eLife.76119.
- C. Ibacache-Quiroga, J. Ojeda, G. Espinoza-Vergara, P. Olivero, M. Cuellar and M. A. Dinamarca, Microb. Biotechnol., 2013, 6, 394–405 CrossRef CAS.
- A. Samanta, D. Roy, D. Lahiri, R. R. Ray, D. Bhattacharya, and M. Nag, in Natural Products, CRC Press, Boca Raton, 2023, pp. 200–222 Search PubMed.
- C. De Clerck, L. Josselin, V. Vangoethem, L. Lassois, M. L. Fauconnier and H. Jijakli, Microorganisms, 2022, 10, 2459, DOI:10.3390/microorganisms10122459.
- R. Martínez-Cámara, V. Montejano-Ramírez, G. Moreno-Hagelsieb, G. Santoyo and E. Valencia-Cantero, Folia Microbiol., 2020, 65, 523–532 CrossRef.
- J. Sharma, D. Sundar and P. Srivastava, Front. Mol. Biosci., 2021, 8, 727070, DOI:10.3389/fmolb.2021.727070.
- S. Sanchez and A. L. Demain, in Comprehensive Biotechnology, Elsevier, 2011, pp. 155–167 Search PubMed.
- L. V. Roze, A. Chanda and J. E. Linz, Fungal Genet. Biol., 2011, 48, 35–48 CrossRef CAS PubMed.
- P. K. Babele, A. Srivastava and J. D. Young, Trends Microbiol., 2023, 31, 1118–1130 CrossRef CAS.
- F. Modolon, A. R. Barno, H. D. M. Villela and R. S. Peixoto, J. Appl. Microbiol., 2020, 129, 1441–1457 CrossRef CAS PubMed.
- A. M. Calvo and J. W. Cary, Front. Microbiol., 2015, 6(62), 1–16, DOI:10.3389/fmicb.2015.00062.
- Y.-M. Chiang, K.-H. Lee, J. F. Sanchez, N. P. Keller and C. C. C. Wang, Unlocking Fungal Cryptic Natural Products NIH Public Access, 2009 Search PubMed.
- N. Lee, S. Hwang, W. Kim, Y. Lee, J. H. Kim, S. Cho, H. U. Kim, Y. J. Yoon, M. K. Oh, B. O. Palsson and B. K. Cho, Nat. Prod. Rep., 2021, 38, 1330–1361 RSC.
- H. Gross and J. E. Loper, Nat. Prod. Rep., 2009, 26, 1408–1446 RSC.
- L. Cuervo, C. Méndez, J. A. Salas, C. Olano and M. G. Malmierca, Cells, 2022, 11, 3510, DOI:10.3390/cells11213510.
- X. Y. Wang and J. Xie, LWT, 2022, 162, 113453, DOI:10.1016/j.lwt.2022.113453.
- R. Y. Stanier, Bacteriol Rev., 1961, 25, 1–17 CrossRef CAS PubMed.
- S. C. do Amaral, A. V. Santos, M. P. C. da Schneider, J. K. R. da Silva and L. P. Xavier, Molecules, 2020, 25, 4744, DOI:10.3390/molecules25204744.
- L. Sanchez-Garcia, M. A. Fernandez-Martinez, M. García-Villadangos, Y. Blanco, S. L. Cady, N. Hinman, M. E. Bowden, S. B. Pointing, K. C. Lee, K. Warren-Rhodes, D. Lacap-Bugler, N. A. Cabrol, V. Parro and D. Carrizo, Front. Microbiol., 2019, 9, 3350, DOI:10.3389/fmicb.2018.03350.
- O. A. Koksharova and N. A. Safronov, Biophys. Rev., 2022, 14, 843–856 CrossRef CAS PubMed.
- X. Ding, K. Liu, G. Gong, L. Tian and J. Ma, Extremophiles, 2020, 24, 307–318 CrossRef CAS.
- K. R. Choi, Y. J. Ahn and S. Y. Lee, J. CO2 Util., 2022, 58, 101929, DOI:10.1016/j.jcou.2022.101929.
- R. Amils, in Encyclopedia of Astrobiology, Springer Berlin Heidelberg, Berlin, Heidelberg, 2014, p. 1 Search PubMed.
- Bacterial Volatile Compounds as Mediators of Airborne Interactions, ed. C.-M. Ryu, L. Weisskopf and B. Piechulla, Springer Singapore, Singapore, 2020 Search PubMed.
- E. R. Moore, A. J. Weaver, E. W. Davis, S. J. Giovannoni and K. H. Halsey, Environ. Microbiol., 2022, 24, 212–222 CrossRef CAS PubMed.
- P. Jurtshuk, in Medical Microbiology, 4th edn, 1996 Search PubMed.
- M. Müller, M. Mentel, J. J. van Hellemond, K. Henze, C. Woehle, S. B. Gould, R.-Y. Yu, M. van der Giezen, A. G. M. Tielens and W. F. Martin, Microbiol. Mol. Biol. Rev., 2012, 76, 444–495 CrossRef.
- R. P. Hausinger, J. Bacteriol., 2007, 189, 671–673 CrossRef CAS PubMed.
- D. V Meier, P. Pjevac, W. Bach, S. Hourdez, P. R. Girguis, C. Vidoudez, R. Amann and A. Meyerdierks, ISME J., 2017, 11, 1545–1558 CrossRef.
- G. J. Dick, Nat. Rev. Microbiol., 2019, 17, 271–283 CrossRef CAS.
- D. Schulze-Makuch, A. G. Fairén and A. F. Davila, Int. J. Astrobiol., 2008, 7, 117–141 CrossRef CAS.
- F. Westall, F. Foucher, N. Bost, M. Bertrand, D. Loizeau, J. L. Vago, G. Kminek, F. Gaboyer, K. A. Campbell, J. G. Bréhéret, P. Gautret and C. S. Cockell, Astrobiology, 2015, 15, 998–1029 CrossRef PubMed.
- S. A. Napieralski, H. L. Buss, S. L. Brantley, S. Lee, H. Xu, E. E. Roden and D. E. Canfield, Proc. Natl. Acad. Sci. U.S.A., 2019, 116, 26394–26401, DOI:10.1073/pnas.1909970117.
- X. Liu, R. J. Koestler, T. Warscheid, Y. Katayama and J.-D. Gu, Nat. Sustain., 2020, 3, 991–1004 CrossRef.
- F. Nas, N. Aissaoui, M. Mahjoubi, A. Mosbah, M. Arab, S. Abdelwahed, R. Khrouf, A. S. Masmoudi, A. Cherif and N. Klouche-Khelil, Int. J. Microbiol., 2021, 24, 455–470 CrossRef CAS.
- I. Ñancucheo and D. B. Johnson, Appl. Environ. Microbiol., 2010, 76, 461–467 CrossRef PubMed.
- K. Olsson-Francis, N. K. Ramkissoon, M. C. Macey, V. K. Pearson, S. P. Schwenzer and D. N. Johnson, J. Microbiol. Methods, 2020, 172, 105883, DOI:10.1016/j.mimet.2020.105883.
- M. C. Macey, M. Fox-Powell, N. K. Ramkissoon, B. P. Stephens, T. Barton, S. P. Schwenzer, V. K. Pearson, C. R. Cousins and K. Olsson-Francis, Sci. Rep., 2020, 10, 10941, DOI:10.1038/s41598-020-67815-8.
- S. Seager, M. Schrenk and W. Bains, Astrobiology, 2012, 12, 61–82 CrossRef CAS.
- S. Seager, W. Bains and J. J. Petkowski, Astrobiology, 2016, 16, 465–485 CrossRef CAS PubMed.
- E. W. Schwieterman, N. Y. Kiang, M. N. Parenteau, C. E. Harman, S. Dassarma, T. M. Fisher, G. N. Arney, H. E. Hartnett, C. T. Reinhard, S. L. Olson, V. S. Meadows, C. S. Cockell, S. I. Walker, J. L. Grenfell, S. Hegde, S. Rugheimer, R. Hu and T. W. Lyons, Astrobiology, 2018, 18, 663–708 CrossRef PubMed.
- N. Madhusudhan, S. Sarkar, S. Constantinou, M. Holmberg, A. A. A. Piette and J. I. Moses, Astrophys. J. Lett., 2023, 956, L13 CrossRef CAS.
- L. J. Preston and L. R. Dartnell, Int. J. Astrobiol., 2014, 13, 81–98 CrossRef.
- A. Brack, in From Habitability to Life on Mars, Elsevier, 2018, pp. 13–35 Search PubMed.
- A. C. Schuerger, J. E. Moores, C. A. Clausen, N. G. Barlow and D. T. Britt, J. Geophys. Res. Planets, 2012, 117, E08007, DOI:10.1029/2011JE004023.
- I. L. Ten Kate, R. Ruiterkamp, O. Botta, B. Lehmann, C. G. Hernandez, N. Boudin, B. H. Foing and P. Ehrenfreund, Int. J. Astrobiol., 2002, 1, 387–399 CrossRef CAS.
- C. T. Pillinger and G. Eglinton, The Chemistry of Carbon in the Lunar Regolith, 1977, vol. 285 Search PubMed.
- Y. Langevin and J. R. Arnold, Annu. Rev. Earth Planet. Sci., 1977, 5, 449–489 CrossRef CAS.
- D. J. Des Marais, M. O. Harwit, K. W. Jucks, J. F. Kasting, D. N. C. Lin, J. I. Lunine, J. Schneider, S. Seager, W. A. Traub and N. J. Woolf, Remote Sensing of Planetary Properties and Biosignatures on Extrasolar Terrestrial Planets, 2002, 2, pp. 153–181 Search PubMed.
- S. Seager and W. Bains, Sci. Adv., 2015, 1, e1500047, DOI:10.1126/sciadv.1500047.
- N. Hänni, K. Altwegg, D. Müller, M. Rubin, and S. Wampfler, in EGU General Assembly 2024, EGU24-16695, Vienna, 2024 Search PubMed.
- M. Rubin, K. Altwegg, H. Balsiger, J. J. Berthelier, M. R. Combi, J. de Keyser, M. Drozdovskaya, B. Fiethe, S. A. Fuselier, S. Gasc, T. I. Gombosi, N. Hänni, K. C. Hansen, U. Mall, H. Rème, I. R. H. G. Schroeder, M. Schuhmann, T. Sémon, J. H. Waite, S. F. Wampfler and P. Wurz, Mon. Not. R. Astron. Soc., 2019, 489, 594–607 CrossRef CAS.
- R. Claudi and E. Alei, Galaxies, 2019, 7, 82, DOI:10.3390/galaxies7040082.
- N. J. F. Blamey and U. Brand, Gondwana Res., 2019, 69, 163–176 CrossRef CAS.
- H. Mißbach, J. P. Duda, A. M. van den Kerkhof, V. Lüders, A. Pack, J. Reitner and V. Thiel, Nat. Commun., 2021, 12, 1101, DOI:10.1038/s41467-021-21323-z.
- T. Toyo’oka, J. Chromatogr. B: Anal. Technol. Biomed. Life Sci., 2009, 877, 3318–3330 CrossRef.
- L. Chen, D. L. Capone and D. W. Jeffery, Molecules, 2019, 24, 2472, DOI:10.3390/molecules24132472.
- I. Mulder, S. G. Huber, T. Krause, C. Zetzsch, K. Kotte, S. Dultz and H. F. Schöler, Chem. Geol., 2013, 358, 148–155 CrossRef CAS.
- D. Smith and P. Španěl, Mass Spectrom. Rev., 2005, 24, 661–700 CrossRef CAS.
- R. E. Summons, J. P. Amend, D. Bish, R. Buick, G. D. Cody, D. J. Des Marais, G. Dromart, J. L. Eigenbrode, A. H. Knoll and D. Y. Sumner, Astrobiology, 2011, 11, 157–181 CrossRef.
- M. Jünger, W. Vautz, M. Kuhns, L. Hofmann, S. Ulbricht, J. I. Baumbach, M. Quintel and T. Perl, Appl. Microbiol. Biotechnol., 2012, 93, 2603–2614 CrossRef.
- A. Romano, V. Capozzi, G. Spano and F. Biasioli, Appl. Microbiol. Biotechnol., 2015, 99, 3787–3795 CrossRef CAS.
- V. Abrahamsson and I. Kanik, Front. astron. space sci., 2022, 9, 959670, DOI:10.3389/fpas.2022.959670.
- R. Epping and M. Koch, Molecules, 2023, 28, 1598, DOI:10.3390/molecules28041598.
- K. Elke, J. Begerow, H. Oppermann, U. Krämer, E. Jermanna and L. Dunemanna, J. Environ. Monit., 1999, 1, 445–452 RSC.
- P. R. Gordon and M. A. Sephton, Astrobiology, 2016, 16, 831–845 CrossRef CAS.
- P. Poinot and C. Geffroy-Rodier, TrAC, Trends Anal. Chem., 2015, 65, 1–12 CrossRef CAS.
- K. M. Seaton, M. L. Cable and A. M. Stockton, Anal. Chem., 2021, 93, 5981–5997 CrossRef CAS.
- K. M. Seaton, M. L. Cable and A. M. Stockton, Annu. Rev. Anal. Chem., 2022, 15, 197–219 CrossRef CAS.
- P. R. Mahaffy, C. R. Webster, M. Cabane, P. G. Conrad, P. Coll, S. K. Atreya, R. Arvey, M. Barciniak, M. Benna, L. Bleacher, W. B. Brinckerhoff, J. L. Eigenbrode, D. Carignan, M. Cascia, R. A. Chalmers, J. P. Dworkin, T. Errigo, P. Everson, H. Franz, R. Farley, S. Feng, G. Frazier, C. Freissinet, D. P. Glavin, D. N. Harpold, D. Hawk, V. Holmes, C. S. Johnson, A. Jones, P. Jordan, J. Kellogg, J. Lewis, E. Lyness, C. A. Malespin, D. K. Martin, J. Maurer, A. C. McAdam, D. McLennan, T. J. Nolan, M. Noriega, A. A. Pavlov, B. Prats, E. Raaen, O. Sheinman, D. Sheppard, J. Smith, J. C. Stern, F. Tan, M. Trainer, D. W. Ming, R. V. Morris, J. Jones, C. Gundersen, A. Steele, J. Wray, O. Botta, L. A. Leshin, T. Owen, S. Battel, B. M. Jakosky, H. Manning, S. Squyres, R. Navarro-González, C. P. McKay, F. Raulin, R. Sternberg, A. Buch, P. Sorensen, R. Kline-Schoder, D. Coscia, C. Szopa, S. Teinturier, C. Baffes, J. Feldman, G. Flesch, S. Forouhar, R. Garcia, D. Keymeulen, S. Woodward, B. P. Block, K. Arnett, R. Miller, C. Edmonson, S. Gorevan and E. Mumm, Space Sci. Rev., 2012, 170, 401–478 CrossRef.
- R. Bhartia, L. W. Beegle, L. DeFlores, W. Abbey, J. Razzell Hollis, K. Uckert, B. Monacelli, K. S. Edgett, M. R. Kennedy, M. Sylvia, D. Aldrich, M. Anderson, S. A. Asher, Z. Bailey, K. Boyd, A. S. Burton, M. Caffrey, M. J. Calaway, R. Calvet, B. Cameron, M. A. Caplinger, B. L. Carrier, N. Chen, A. Chen, M. J. Clark, S. Clegg, P. G. Conrad, M. Cooper, K. N. Davis, B. Ehlmann, L. Facto, M. D. Fries, D. H. Garrison, D. Gasway, F. T. Ghaemi, T. G. Graff, K. P. Hand, C. Harris, J. D. Hein, N. Heinz, H. Herzog, E. Hochberg, A. Houck, W. F. Hug, E. H. Jensen, L. C. Kah, J. Kennedy, R. Krylo, J. Lam, M. Lindeman, J. McGlown, J. Michel, E. Miller, Z. Mills, M. E. Minitti, F. Mok, J. Moore, K. H. Nealson, A. Nelson, R. Newell, B. E. Nixon, D. A. Nordman, D. Nuding, S. Orellana, M. Pauken, G. Peterson, R. Pollock, H. Quinn, C. Quinto, M. A. Ravine, R. D. Reid, J. Riendeau, A. J. Ross, J. Sackos, J. A. Schaffner, M. Schwochert, M. O Shelton, R. Simon, C. L. Smith, P. Sobron, K. Steadman, A. Steele, D. Thiessen, V. D. Tran, T. Tsai, M. Tuite, E. Tung, R. Wehbe, R. Weinberg, R. H. Weiner, R. C. Wiens, K. Williford, C. Wollonciej, Y. H. Wu, R. A. Yingst and J. Zan, Space Sci. Rev., 2021, 217, 58, DOI:10.1007/s11214-021-00812-z.
- K. Biemann, Proc. Natl. Acad. Sci. U.S.A., 2007, 104(25), 10310–10313, DOI:10.1073/pnas.0703732104.
- H. B. Niemann, S. K. Atreya, J. E. Demick, D. Gautier, J. A. Haberman, D. N. Harpold, W. T. Kasprzak, J. I. Lunine, T. C. Owen and F. Raulin, J. Geophys. Res. Planets, 2010, 115, E12006, DOI:10.1029/2010JE003659.
- J. F. J. Todd, S. J. Barber, I. P. Wright, G. H. Morgan, A. D. Morse, S. Sheridan, M. R. Leese, J. Maynard, S. T. Evans, C. T. Pillinger, D. L. Drummond, S. C. Heys, S. E. Huq, B. J. Kent, E. C. Sawyer, M. S. Whalley and N. R. Waltham, J. Mass Spectrom., 2007, 42, 1–10 CrossRef CAS.
- M. Guzman, C. Szopa, C. Freissinet, A. Buch, F. Stalport, D. Kaplan and F. Raulin, Planet Space Sci., 2020, 186, 104903, DOI:10.1016/j.pss.2020.104903.
- J. H. Waite, J. L. Burch, T. G. Brockwell, D. T. Young, G. P. Miller, S. C. Persyn, J. M. Stone, P. Wilson, K. E. Miller, C. R. Glein, R. S. Perryman, M. A. McGrath, S. J. Bolton, W. B. McKinnon, O. Mousis, M. A. Sephton, E. L. Shock, M. Choukroun, B. D. Teolis, D. Y. Wyrick, M. Y. Zolotov, C. Ray, A. L. Magoncelli, R. R. Raffanti, R. L. Thorpe, A. Bouquet, T. L. Salter, K. J. Robinson, C. Urdiales, Y. D. Tyler, G. J. Dirks, C. R. Beebe, D. A. Fugett, J. A. Alexander, J. J. Hanley, Z. A. Moorhead-Rosenberg, K. A. Franke, K. S. Pickens, R. J. Focia, B. A. Magee, P. J. Hoeper, D. P. Aaron, S. L. Thompson, K. B. Persson, R. C. Blase, G. F. Dunn, R. L. Killough, A. De Los Santos, R. J. Rickerson and O. H. W. Siegmund, Space Sci. Rev., 2024, 220, 30 CrossRef.
- S. D. Vance, K. L. Craft, E. Shock, B. E. Schmidt, J. Lunine, K. P. Hand, W. B. McKinnon, E. M. Spiers, C. Chivers, J. D. Lawrence, N. Wolfenbarger, E. J. Leonard, K. J. Robinson, M. J. Styczinski, D. M. Persaud, G. Steinbrügge, M. Y. Zolotov, L. C. Quick, J. E. C. Scully, T. M. Becker, S. M. Howell, R. N. Clark, A. J. Dombard, C. R. Glein, O. Mousis, M. A. Sephton, J. Castillo-Rogez, F. Nimmo, A. S. McEwen, M. S. Gudipati, I. Jun, X. Jia, F. Postberg, K. M. Soderlund and C. M. Elder, Space Sci. Rev., 2023, 219, 81, DOI:10.1007/s11214-023-01025-2.
- K. P. Hand, C. B. Phillips, A. Murray, J. B. Garvin, E. H. Maize, R. G. Gibbs, G. Reeves, A. M. San Martin, G. H. Tan-Wang, J. Krajewski, K. Hurst, R. Crum, B. A. Kennedy, T. P. McElrath, J. C. Gallon, D. Sabahi, S. W. Thurman, B. Goldstein, P. Estabrook, S. W. Lee, J. A. Dooley, W. B. Brinckerhoff, K. S. Edgett, C. R. German, T. M. Hoehler, S. M. Hörst, J. I. Lunine, C. Paranicas, K. Nealson, D. E. Smith, A. S. Templeton, M. J. Russell, B. Schmidt, B. Christner, B. Ehlmann, A. Hayes, A. Rhoden, P. Willis, R. A. Yingst, K. Craft, M. E. Cameron, T. Nordheim, J. Pitesky, J. Scully, J. Hofgartner, S. W. Sell, K. J. Barltrop, J. Izraelevitz, E. J. Brandon, J. Seong, J. P. Jones, J. Pasalic, K. J. Billings, J. P. Ruiz, R. V. Bugga, D. Graham, L. A. Arenas, D. Takeyama, M. Drummond, H. Aghazarian, A. J. Andersen, K. B. Andersen, E. W. Anderson, A. Babuscia, P. G. Backes, E. S. Bailey, D. Balentine, C. G. Ballard, D. F. Berisford, P. Bhandari, K. Blackwood, G. S. Bolotin, E. A. Bovre, J. Bowkett, K. T. Boykins, M. S. Bramble, T. M. Brice, P. Briggs, A. P. Brinkman, S. M. Brooks, B. B. Buffington, B. Burns, M. L. Cable, S. Campagnola, L. A. Cangahuala, G. A. Carr, J. R. Casani, N. E. Chahat, B. K. Chamberlain-Simon, Y. Cheng, S. A. Chien, B. T. Cook, M. Cooper, M. DiNicola, B. Clement, Z. Dean, E. A. Cullimore, A. G. Curtis, J. P. de la Croix, P. Di Pasquale, E. M. Dodd, L. A. Dubord, J. A. Edlund, R. Ellyin, B. Emanuel, J. T. Foster, A. J. Ganino, G. J. Garner, M. T. Gibson, M. Gildner, K. J. Glazebrook, M. E. Greco, W. M. Green, S. J. Hatch, M. M. Hetzel, W. A. Hoey, A. E. Hofmann, R. Ionasescu, A. Jain, J. D. Jasper, J. R. Johannesen, G. K. Johnson, I. Jun, A. B. Katake, S. Y. Kim-Castet, D. I. Kim, W. Kim, E. F. Klonicki, B. Kobeissi, B. D. Kobie, J. Kochocki, M. Kokorowski, J. A. Kosberg, K. Kriechbaum, T. P. Kulkarni, R. L. Lam, D. F. Landau, M. A. Lattimore, S. L. Laubach, C. R. Lawler, G. Lim, J. Y. Lin, T. E. Litwin, M. W. Lo, C. A. Logan, E. Maghasoudi, L. Mandrake, Y. Marchetti, E. Marteau, K. A. Maxwell, J. B. Mc Namee, O. McIntyre, M. Meacham, J. P. Melko, J. Mueller, D. A. Muliere, A. Mysore, J. Nash, H. Ono, J. M. Parker, R. C. Perkins, A. E. Petropoulos, A. Gaut, M. Y. Piette Gomez, R. P. Casillas, M. Preudhomme, G. Pyrzak, J. Rapinchuk, J. M. Ratliff, T. L. Ray, E. T. Roberts, K. Roffo, D. C. Roth, J. A. Russino, T. M. Schmidt, M. J. Schoppers, J. S. Senent, F. Serricchio, D. J. Sheldon, L. R. Shiraishi, J. Shirvanian, K. J. Siegel, G. Singh, A. R. Sirota, E. D. Skulsky, J. S. Stehly, N. J. Strange, S. U. Stevens, E. T. Sunada, S. P. Tepsuporn, L. P. C. Tosi, N. Trawny, I. Uchenik, V. Verma, R. A. Volpe, C. T. Wagner, D. Wang, R. G. Willson, J. L. Wolff, A. T. Wong, A. K. Zimmer, K. G. Sukhatme, K. A. Bago, Y. Chen, A. M. Deardorff, R. S. Kuch, C. Lim, M. L. Syvertson, G. A. Arakaki, A. Avila, K. J. DeBruin, A. Frick, J. R. Harris, M. C. Heverly, J. M. Kawata, S. K. Kim, D. M. Kipp, J. Murphy, M. W. Smith, M. D. Spaulding, R. Thakker, N. Z. Warner, C. R. Yahnker, M. E. Young, T. Magner, D. Adams, P. Bedini, L. Mehr, C. Sheldon, S. Vernon, V. Bailey, M. Briere, M. Butler, A. Davis, S. Ensor, M. Gannon, A. Haapala-Chalk, T. Hartka, M. Holdridge, A. Hong, J. Hunt, J. Iskow, F. Kahler, K. Murray, D. Napolillo, M. Norkus, R. Pfisterer, J. Porter, D. Roth, P. Schwartz, L. Wolfarth, E. H. Cardiff, A. Davis, E. W. Grob, J. R. Adam, E. Betts, J. Norwood, M. M. Heller, T. Voskuilen, P. Sakievich, L. Gray, D. J. Hansen, K. W. Irick, J. C. Hewson, J. Lamb, S. C. Stacy, C. M. Brotherton, A. S. Tappan, D. Benally, H. Thigpen, E. Ortiz, D. Sandoval, A. M. Ison, M. Warren, P. G. Stromberg, P. M. Thelen, B. Blasy, P. Nandy, A. W. Haddad, L. B. Trujillo, T. H. Wiseley, S. A. Bell, N. P. Teske, C. Post, L. Torres-Castro, C. Grosso and M. Wasiolek, Planet. Sci. J., 2022, 3(31pp), 22, DOI:10.3847/PSJ/ac4493.
- F. Nimmo and R. T. Pappalardo, J. Geophys. Res. Planets, 2016, 121, 1378–1399 CrossRef.
- J. D. Lawrence, A. D. Mullen, F. E. Bryson, C. J. Chivers, A. M. Hanna, T. Plattner, E. M. Spiers, J. S. Bowman, J. J. Buffo, J. L. Burnett, C. E. Carr, D. J. Dichek, K. H. G. Hughson, W. King, E. Glenn Lightsey, E. Ingall, J. McKaig, M. R. Meister, S. Pierson, Y. Tomar and B. E. Schmidt, Planet. Sci. J., 2023, 4(27pp), 22, DOI:10.3847/PSJ/aca6ed.
- J. Liang, J.-C. Feng, S. Zhang, Y. Cai, Z. Yang, T. Ni and H.-Y. Yang, iScience, 2021, 24, 103299 CrossRef.
- J. J. Grandy, B. Onat, V. Tunnicliffe, D. A. Butterfield and J. Pawliszyn, Sci. Rep., 2020, 10, 1360, DOI:10.1038/s41598-020-58418-4.
- S. Q. Lang and B. Benitez-Nelson, Deep Sea Res. Part 1 Oceanogr. Res. Pap., 2021, 173, 103529, DOI:10.1016/j.dsr.2021.103529.
- N. Merino, H. S. Aronson, D. P. Bojanova, J. Feyhl-Buska, M. L. Wong, S. Zhang and D. Giovannelli, Front. Microbiol., 2019, 10, 780, DOI:10.3389/fmicb.2019.00780.
- F. Gomez, in Encyclopedia of Astrobiology, Springer Berlin Heidelberg, Berlin, Heidelberg, 2015, pp. 2473–2479 Search PubMed.
- D. A. Wharton, in Cellular Origin, Life in Extreme Habitats and Astrobiology, Kluwer Academic Publishers, Dordrecht, 2004, vol. 6, pp. 607–620 Search PubMed.
- D. Prieur, in Encyclopedia of Astrobiology, Springer Berlin Heidelberg, Berlin, Heidelberg, 2015, pp. 830–835 Search PubMed.
- A. Matthews, J. Lima-Zaloumis, R. V. Debes, G. Boyer and E. Trembath-Reichert, Astrobiology, 2023, 23, 446–459 CrossRef CAS.
- S. Fitzgerald, L. Holland and A. Morrin, Front. Microbiol., 2021, 12, 693075, DOI:10.3389/fmicb.2021.693075.
- H. Cherif-silini, A. Silini, A. C. Bouket, F. N. Alenezi, L. Luptakova, N. Bouremani, J. A. Nowakowska, T. Oszako and L. Belbahri, Sustainability, 2021, 13, 4422, DOI:10.3390/su13084422.
- E. Parrilli, P. Tedesco, M. Fondi, M. L. Tutino, A. Lo Giudice, D. de Pascale and R. Fani, Phys. Life Rev., 2021, 36, 137–161 CrossRef CAS PubMed.
- M. Avalos, P. Garbeva, L. Vader, G. P. Van Wezel, J. S. Dickschat and D. Ulanova, Nat. Prod. Rep., 2022, 39, 249–272 RSC.
- A. Penesyan, I. T. Paulsen, S. Kjelleberg and M. R. Gillings, NPJ Biofilms Microbiomes, 2021, 7, 80, DOI:10.1038/s41522-021-00251-2.
- R. E. Wheatley, Antonie van Leeuwenhoek, 2002, 81, 357–364 CrossRef CAS PubMed.
- S. S. Branda, Å. Vik, L. Friedman and R. Kolter, Trends Microbiol., 2005, 13, 20–26 CrossRef CAS PubMed.
- J. W. Costerton, P. S. Stewart and E. P. Greenberg, Science, 1999, 284, 1318–1322 CrossRef CAS PubMed.
- M. Mutalipassi, V. Mazzella, M. Schott, P. Fink, F. Glaviano, L. Porzio, M. Lorenti, M. C. Buia, E. von Elert and V. Zupo, Front. Mar. Sci., 2022, 9, 809702, DOI:10.3389/fmars.2022.809702.
- S. E. Jones, C. A. Pham, M. P. Zambri, J. Mckillip, E. E. Carlson and M. A. Elliot, mBio, 2019, 10(2), e219, DOI:10.1128/mBio.00171-19.
- Y. Li, Nat. Prod. Rep., 2023, 40, 922–956 RSC.
- A. Gałuszka, Z. M. Migaszewski and J. Namieśnik, Environ. Res., 2015, 140, 593–603 CrossRef.
- V. S. Langford, Chemosensors, 2023, 11, 111, DOI:10.3390/chemosensors11020111.
- H. Chu, K.-S. Jang, B. Choi, J. W. Kang, C. E. Son and Y. G. Ahn, Instrum. Sci. Technol., 2023, 51, 260–272 CrossRef CAS.
- X. Ding, H. Lu, J. Jiang, A. Tasoglou, A. D. Shah and N. Jung, Build Environ., 2023, 246, 110953, DOI:10.1016/j.buildenv.2023.110953.
- S. Ghimenti, T. Lomonaco, F. G. Bellagambi, S. Tabucchi, M. Onor, M. G. Trivella, A. Ceccarini, R. Fuoco and F. Di Francesco, J. Breath Res., 2015, 9, 047110, DOI:10.1088/1752-7155/9/4/047110.
- A. Stewart-Jones and G. M. Poppy, J. Chem. Ecol., 2006, 32, 845–864 CrossRef CAS PubMed.
- M. M. Sampson, D. M. Chambers, D. Y. Pazo, F. Moliere, B. C. Blount and C. H. Watson, Anal. Chem., 2014, 86, 7088–7095 CrossRef CAS PubMed.
- W. A. Groves and E. T. Zellers, Am. Ind. Hyg. Assoc. J., 1996, 57, 257 CrossRef CAS PubMed.
- M. A. Ibrahim, M. C. Egigu, A. Kasurinen, A. Yahya and J. K. Holopainen, Flavour Fragr. J., 2010, 25, 83–92 CrossRef CAS.
- E. Schulz, M. Woollam, P. Grocki, M. D. Davis and M. Agarwal, Molecules, 2023, 28, 4533, DOI:10.3390/molecules28114533.
- C. Bax, R. Bernasconi, F. Massironi, L. Magagnin, F. Grizzi, L. Capelli and G. Taverna, J. Electrochem. Soc., 2021, 168, 047513 CrossRef CAS.
- C. A. Batty, M. Cauchi, C. Lourenço, J. O. Hunter and C. Turner, PLoS One, 2015, 10(6), e0130301, DOI:10.1371/journal.pone.0130301.
- I. Belluomo, S. E. Whitlock, A. Myridakis, A. G. Parker, V. Converso, M. J. Perkins, V. S. Langford, P. Španěl and G. B. Hanna, Anal. Chem., 2024, 96, 1397–1401 CrossRef CAS PubMed.
- F. J. Gilchrist, H. Sims, A. Alcock, J. Belcher, A. M. Jones, D. Smith, P. Španěl, A. K. Webb and W. Lenney, Anal. Methods, 2012, 4, 3661–3665 RSC.
- V. Plyuta, V. Lipasova, A. Popova, O. Koksharova, A. Kuznetsov, E. Szegedi, L. Chernin and I. Khmel, APMIS, 2016, 124, 586–594 CrossRef CAS.
- P. Gschwend, J. Macfarlane, D. Jensen, J. Soo, G. Saparbaiuly, R. Borrelli, F. Vago, A. Oldani, L. Zaninetta, I. Verginelli and R. Baciocchi, Environ. Sci. Technol., 2022, 56, 7810–7819 CrossRef CAS PubMed.
- B. V. Lancellotti, D. A. Hensley and R. Stryker, Sci. Rep., 2023, 13, 17238, DOI:10.1038/s41598-023-44455-2.
- X. H. Lu, Y. S. Zhang, L. Yi, W. G. Su, Z. Ma and F. Q. Han, Appl. Ecol. Environ. Res., 2022, 20, 447–464 CrossRef.
- C. Turner, Bioanalysis, 2016, 8, 677–690 CrossRef CAS.
- S. Beghi and J. M. Guillot, J. Chromatogr. A, 2008, 1183, 1–5 CrossRef CAS.
- T. Li, N. Baggesen, R. Seco and R. Rinnan, Atmos. Environ., 2023, 292, 119430, DOI:10.1016/j.atmosenv.2022.119430.
- Y. Jiao, C. L. Davie-Martin, M. Kramshøj, C. T. Christiansen, H. Lee, I. H. J. Althuizen and R. Rinnan, Geoderma, 2023, 430, 116355, DOI:10.1016/j.geoderma.2023.116355.
- P. Mochalski, J. King, K. Unterkofler and A. Amann, Analyst, 2013, 138, 1405–1418 RSC.
- S. Ghosh, K. H. Kim and J. R. Sohn, ScientificWorldJournal, 2011, 11, 2160–2177 CrossRef CAS.
- X. M. Cai, X. X. Xu, L. Bian, Z. X. Luo and Z. M. Chen, Anal. Bioanal. Chem., 2015, 407, 9105–9114 CrossRef CAS PubMed.
- S. L. Trabue, K. D. Scoggin, H. Li, R. Burns and H. Xin, Environ. Sci. Technol., 2008, 42, 3745–3750 CrossRef CAS PubMed.
- P. Pérez Ballesta, A. Baù, R. A. Field and E. Woolfenden, Environ. Int., 2023, 179, 108119, DOI:10.1016/j.envint.2023.108119.
- P. Pérez Ballesta, E. Grandesso, R. A. Field and A. Cabrerizo, Anal. Chim. Acta, 2016, 908, 102–112 CrossRef.
- M. R. Ras, F. Borrull and R. M. Marcé, TrAC, Trends Anal. Chem., 2009, 28, 347–361 CrossRef CAS.
- E. Woolfenden, J. Air Waste Manage. Assoc., 1997, 47, 20–36 CrossRef CAS.
- L. Conner, S. Chin and K. G. Furton, Sens. Actuators, B, 2006, 116, 121–129 CrossRef CAS.
- A. D. Wilson and M. Baietto, Sensors, 2009, 9, 5099–5148 CrossRef CAS PubMed.
- A. Özmen and E. Do
![[g with combining tilde]](https://www.rsc.org/images/entities/char_0067_0303.gif) an, IEEE Trans. Instrum. Meas., 2009, 58, 3609–3618 Search PubMed.
an, IEEE Trans. Instrum. Meas., 2009, 58, 3609–3618 Search PubMed. - S. Cui, L. Cao, N. Acosta, H. Zhu and P. P. Ling, Chemosensors, 2021, 9, 297, DOI:10.3390/chemosensors9110297.
- M. Aleixandre and M. Gerboles, Chem. Eng. Trans., 2012, 30, 169–174, DOI:10.3303/CET1230029.
- W. I. Lai, Y. Y. Chen and J. H. Sun, Sensors, 2022, 22, 4393, DOI:10.3390/s22124393.
- S. Capone, A. Forleo, L. Francioso, R. Rella, P. Siciliano, J. Spadavecchia, D. S. Presicce and A. M. Taurino, Solid State Gas Sensors: State of the Art and Future Activities, 2003, vol. 5 Search PubMed.
- R. M. Shah, K. E. Hillyer, S. Stephenson, J. Crosswell, A. V. Karpe, E. A. Palombo, O. A. H. Jones, D. Gorman, L. Bodrossy, J. Van De Kamp, A. Bissett, A. W. Whiteley, A. D. L. Steven and D. J. Beale, Sci. Total Environ., 2021, 781, 146526, DOI:10.1016/j.scitotenv.2021.146526.
- R. Rinnan, Å. Rinnan, T. Holopainen, J. K. Holopainen and P. Pasanen, Atmos. Environ., 2005, 39, 921–930 CrossRef CAS.
- J. T. Wilson, G. B. Smith, J. W. Cochran, J. F. Barker and P. V. Roberts, Water Resour. Res., 1987, 23, 1547–1553 CrossRef CAS.
- Y. Guo, M. Q. Lu and Y. T. Long, Field Anal. Chem. Technol., 1997, 1, 195–211 CrossRef CAS.
- J. Puton and J. Namieśnik, TrAC, Trends Anal. Chem., 2016, 85, 10–20 CrossRef CAS.
- P. Mochalski, H. Wiesenhofer, M. Allers, S. Zimmermann, A. T. Güntner, N. J. Pineau, W. Lederer, A. Agapiou, C. A. Mayhew and V. Ruzsanyi, J. Chromatogr. B: Anal. Technol. Biomed. Life Sci., 2018, 1076, 29–34 CrossRef CAS PubMed.
- C. P. Shih, K. C. Yu, H. T. Ou and P. L. Urban, Anal. Chem., 2021, 93, 2424–2432 CrossRef CAS PubMed.
- V. Ruzsanyi, J. I. Baumbach, S. Sielemann, P. Litterst, M. Westhoff and L. Freitag, J. Chromatogr. A, 2005, 1084, 145–151 CrossRef CAS PubMed.
- P. V. Johnson, L. W. Beegle, H. I. Kim, G. A. Eiceman and I. Kanik, Int. J. Mass Spectrom., 2007, 262, 1–15 CrossRef CAS.
- B. A. Eckenrode, FOCUS: FIELD-PORTABLE AND MINIATURE MS Environmental and Forensic Applications of Field-Portable GC-MS: An Overview, 2001 Search PubMed.
- P. E. Leary, B. W. Kammrath, K. J. Lattman and G. L. Beals, Appl. Spectrosc., 2019, 73, 841–858 CrossRef CAS.
- T. V Truong, E. Lee and R. Thomas, Rapid Screening of Volatile and Semi-Volatile Organic Components in Cocoa Beans and Chocolate Products Using a Portable GC/MS System, 2017 Search PubMed.
- J. J. Beck, N. Porter, D. Cook, W. S. Gee, C. M. Griffith, A. D. Rands, T. V. Truong, L. Smith and I. San Román, Phytochem. Anal., 2015, 26, 395–403 CrossRef CAS PubMed.
- D. Tholl, O. Hossain, A. Weinhold, U. S. R. Röse and Q. Wei, Plant J., 2021, 106, 314–325 CrossRef CAS PubMed.
- E. Woolfenden, J. Chromatogr. A, 2010, 1217, 2674–2684 CrossRef CAS.
- X. Zhang, L. Liu, L. Zhang, X. Yin, H. Huan, L. Zhang and X. Shao, J. Appl. Phys., 2022, 131, 174501, DOI:10.1063/5.0088257.
- S. Khan, D. Newport and S. Le Calvé, Sensors, 2019, 19, 5210, DOI:10.3390/s19235210.
- S. Korposh and S. W. Lee, Chemosensors, 2021, 9, 269, DOI:10.3390/chemosensors9090269.
- J. Hromadka, B. Tokay, R. Correia, S. P. Morgan and S. Korposh, Sens. Actuators, B, 2018, 260, 685–692 CrossRef CAS.
- H. Ding, J. Guan, P. Lu, S. J. Mihailov, C. T. Kingston and B. Simard, IEEE Sens. Lett., 4(10), 5000804, DOI:10.1109/LSENS.2020.3023702.
- P. Brož, O. Krýza, L. Wilson, S. J. Conway, E. Hauber, A. Mazzini, J. Raack, M. R. Balme, M. E. Sylvest and M. R. Patel, Nat. Geosci., 2020, 13, 403–407 CrossRef.
- N. K. Ramkissoon, M. C. Macey, S. P. Schwenzer, V. K. Pearson and K. Olsson-Francis, in Microbial growth in simulated martian environments, European Planetary Science Congress, 2018, p. 248 Search PubMed.
- A. Santos, J. Balcerski, D. M. Burr, N. Johnson, P. Byrne, J. Cordova, U.-M. Darby Dyar, M. Gilmore, J. Hall, J. Kandis-Lea Jessup, B. A. Jonathan Lewis, N. O. JSC Joe, C. Ostberg, C. Tsang, S. Michael Way, N. GISS Thomas Widemann, L. E. Kathleen Vander Kaaden and J.-N. Jsc, Title of the white paper: The Importance of Venus Experimental Facilities Co-authors with their respective institutions: Co-signers with their respective institutions, 2021 Search PubMed.
- N. K. Ramkissoon and M. Baqué, in Encyclopedia of Astrobiology, Springer Berlin Heidelberg, 2022, pp. 1–4 Search PubMed.
- A. R. Santos, M. S. Gilmore, J. P. Greenwood, L. M. Nakley, K. Phillips, T. Kremic and X. Lopez, J. Geophys. Res. Planets, 2023, 128, e2022JE007423, DOI:10.1029/2022JE007423.
- Rachael E Hamp, The Open University, 2022 Search PubMed.
- A. Elsaesser, D. J. Burr, P. Mabey, R. G. Urso, D. Billi, C. Cockell, H. Cottin, A. Kish, N. Leys, J. J. W. A. van Loon, E. Mateo-Marti, C. Moissl-Eichinger, S. Onofri, R. C. Quinn, E. Rabbow, P. Rettberg, R. de la Torre Noetzel, K. Slenzka, A. J. Ricco, J. P. de Vera and F. Westall, NPJ Microgravity, 2023, 9(1), 43, DOI:10.1038/s41526-023-00292-1.
- K. Hamp, in 49th Lunar and Planetary Science Conference, Lunar and Planetary Institute, Houston, 2018 Search PubMed.
- R. S. Taubner, K. Olsson-Francis, S. D. Vance, N. K. Ramkissoon, F. Postberg, J. P. de Vera, A. Antunes, E. Camprubi Casas, Y. Sekine, L. Noack, L. Barge, J. Goodman, M. Jebbar, B. Journaux, Ö. Karatekin, F. Klenner, E. Rabbow, P. Rettberg, T. Rückriemen-Bez, J. Saur, T. Shibuya and K. M. Soderlund, Space Sci. Rev., 2020, 216, 9, DOI:10.1007/s11214-020-0635-5.
- A. C. Schuerger and W. L. Nicholson, Astrobiology, 2016, 16, 964–976 CrossRef CAS PubMed.
- A. Agapiou, A. Amann, P. Mochalski, M. Statheropoulos and C. L. P. Thomas, TrAC, Trends Anal. Chem., 2015, 66, 158–175 CrossRef CAS.
- C. Lourenço, R. González-Méndez, F. Reich, N. Mason and C. Turner, Int. J. Mass Spectrom., 2017, 419, 1–10 CrossRef.
- A. J. King, L. Daly, J. Rowe, K. H. Joy, R. C. Greenwood, H. A. R. Devillepoix, M. D. Suttle, Q. H. S. Chan, S. S. Russell, H. C. Bates, J. F. J. Bryson, P. L. Clay, D. Vida, M. R. Lee, Á. O’brien, L. J. Hallis, N. R. Stephen, R. Tartèse, E. K. Sansom, M. C. Towner, M. Cupak, P. M. Shober, P. A. Bland, R. Findlay, I. A. Franchi, A. B. Verchovsky, F. A. J. Abernethy, M. M. Grady, C. J. Floyd, M. Van Ginneken, J. Bridges, L. J. Hicks, R. H. Jones, J. T. Mitchell, M. J. Genge, L. Jenkins, P.-E. Martin, M. A. Sephton, J. S. Watson, T. Salge, K. A. Shirley, R. J. Curtis, T. J. Warren, N. E. Bowles, F. M. Stuart, L. Di Nicola, D. Györe, A. J. Boyce, K. M. M. Shaw, T. Elliott, R. C. J. Steele, P. Povinec, M. Laubenstein, D. Sanderson, A. Cresswell, A. J. T. Jull, I. Sýkora, S. Sridhar, R. J. Harrison, F. M. Willcocks, C. S. Harrison, D. Hallatt, P. J. Wozniakiewicz, M. J. Burchell, L. S. Alesbrook, A. Dignam, N. V Almeida, C. L. Smith, B. Clark, E. R. Humphreys-Williams, P. F. Schofield, L. T. Cornwell, V. Spathis, G. H. Morgan, M. J. Perkins, R. Kacerek, P. Campbell, J. Horak, D. Jones, N. James, S. Bosley, A. Shuttleworth, P. Dickinson, I. Mcmullan, D. Robson, A. R. D. Smedley, B. Stanley, R. Bassom, M. Mcintyre, A. A. Suttle, R. Fleet, L. Bastiaens, M. B. Ihász, S. Mcmullan, S. J. Boazman, Z. I. Dickeson, P. M. Grindrod, A. E. Pickersgill, C. J. Weir, F. M. Suttle, S. Farrelly, I. Spencer, S. Naqvi, B. Mayne, D. Skilton, D. Kirk, A. Mounsey, S. E. Mounsey, S. Mounsey, P. Godfrey, L. Bond, V. Bond, C. Wilcock, H. Wilcock and R. Wilcock, Sci. Adv., 2022, 8, eabq3925, DOI:10.1126/sciadv.abq3925.
- G. R. Osinski, C. S. Cockell, A. Pontefract and H. M. Sapers, Astrobiology, 2020, 20, 1121–1149 CrossRef CAS.
- A. Verchovsky, F. A. J. Abernethy, M. Anand, S. J. Barber, R. Findlay, I. A. Franchi, R. C. Greenwood and M. M. Grady, Meteorit. Planet. Sci., 2023, 59(5), 1145–1169, DOI:10.1111/maps.13983.
- A. B. Verchovsky, M. Anand, S. J. Barber, S. Sheridan and G. H. Morgan, Planet Space Sci., 2020, 181, 104830, DOI:10.1016/j.pss.2019.104830.
- P. Mara, R. K. Nelson, C. M. Reddy, A. Teske and V. P. Edgcomb, Commun. Earth Environ., 2022, 3, 250, DOI:10.1038/s43247-022-00582-8.
- J. Dallüge, J. Beens and U. A. T. Brinkman, J. Chromatogr. A, 2003, 1000, 69–108 CrossRef.
- R. L. Cordell, R. Panchal, E. Bernard, M. Gatari, E. Waiguru, M. Ng’ang’a, J. Nyang’aya, M. Ogot, M. J. Wilde, K. P. Wyche, A. A. Abayomi, R. Alani, P. S. Monks and J. D. Vande Hey, Atmosphere, 2021, 12, 1329, DOI:10.3390/atmos12101329.
- E. Colleoni, V. G. Samaras, P. Guida, A. Frassoldati, T. Faravelli and W. L. Roberts, J. Anal. Appl. Pyrolysis, 2023, 175, 106204, DOI:10.1016/j.jaap.2023.106204.
- P. Zhang, S. Carlin, C. Lotti, F. Mattivi and U. Vrhovsek, Metabolomics, 2020, 16, 102, DOI:10.1007/s11306-020-01718-7.
- M. Libardoni and C. Fix, Utilizing GCxGC for Advanced Analytical Analysis of Volatile and Semi-Volatile Organic Compounds, 2010 Search PubMed.
- E. B. Franklin, M. R. Alves, A. N. Moore, D. B. Kilgour, G. A. Novak, K. Mayer, J. S. Sauer, R. J. Weber, D. Dang, M. Winter, C. Lee, C. D. Cappa, T. H. Bertram, K. A. Prather, V. H. Grassian and A. H. Goldstein, Environ. Sci. Technol., 2021, 55, 15705–15714 CrossRef CAS.
- J. S. Watson, in Mass Spectrometry Handbook, Wiley, 2012, pp. 407–415 Search PubMed.
- Y. Zhang, R. Su, H. Yuan, H. Zhou, Y. Jiangfang, X. Liu and J. Luo, Metabolites, 2023, 13, 700, DOI:10.3390/metabo13060700.
- A. T. Brinca, O. Anjos, M. M. C. Alves, Â. Sousa, A. H. Oliani, L. Breitenfeld, L. A. Passarinha, A. C. Ramalhinho and E. Gallardo, Biomedicines, 2022, 10, 2852, DOI:10.3390/biomedicines10112852.
- S. Filippidou, A. Price, C. Spencer-Jones, A. Scales, M. C. Macey, F. Franchi, L. Lebogang, B. Cavalazzi, S. P. Schwenzer and K. Olsson-Francis, Microorganisms, 2024, 12, 147, DOI:10.3390/microorganisms12010147.
- N. Noirungsee, S. Changkhong, K. Phinyo, C. Suwannajak, N. Tanakul and S. Inwongwan, Environ. Microbiol. Rep., 2024, 16(1), e13231 CrossRef CAS PubMed.
- S. DasSarma, P. DasSarma, V. J. Laye and E. W. Schwieterman, Extremophiles, 2020, 24, 31–41 CrossRef CAS PubMed.
- L. Seyler, E. B. Kujawinski, A. Azua-Bustos, M. D. Lee, J. Marlow, S. M. Perl and H. J. Cleaves, Astrobiology, 2020, 20, 1251–1261 CrossRef CAS.
- L. J. Rothschild and R. L. Mancinelli, Nature, 2001, 409, 1092–1101 CrossRef CAS.
- G. Stavropoulos, D. Salman, Y. Alkhalifah, F.-J. van Schooten and A. Smolinska, in Breathborne Biomarkers and the Human Volatilome, Elsevier, 2020, pp. 633–647 Search PubMed.
- A. O. Brown, P. J. Green, G. J. Frankham, B. H. Stuart and M. Ueland, ACS Omega, 2023, 8, 22042–22054 CrossRef CAS.
- K. E. Eisen, J. M. Powers, R. A. Raguso and D. R. Campbell, Front. Ecol. Evol., 2022, 10, 1006416, DOI:10.3389/fevo.2022.1006416.
- M. Noshad, B. A. Behbahani and I. K. Karabagias, Eur. Food Res. Technol., 2023, 249, 2215–2226 CrossRef CAS.
- G. Lubes and M. Goodarzi, Chem. Rev., 2017, 117, 6399–6422 CrossRef CAS.
- W. Ahmed, E. Bardin, M. D. Davis, I. Sermet-Gaudelus, S. Grassin Delyle and S. J. Fowler, Analyst, 2023, 148, 618–627 RSC.
- D. Radványi, Heliyon, 2023, 9, e12703, DOI:10.1016/j.heliyon.2022.e12703.
- A. Argyris, J. J. Filippi and D. Syvridis, J. Chemom., 2015, 29, 38–48 CrossRef CAS.
- A. W. Boots, A. Smolinska, J. J. B. N. Van Berkel, R. R. R. Fijten, E. E. Stobberingh, M. L. L. Boumans, E. J. Moonen, E. F. M. Wouters, J. W. Dallinga and F. J. Van Schooten, J. Breath Res., 2014, 8, 027106, DOI:10.1088/1752-7155/8/2/027106.
- Â. C. Salvador, I. Baptista, A. S. Barros, N. C. M. Gomes, Â. Cunha, A. Almeida and S. M. Rocha, PLoS One, 2013, 8(4), e59338, DOI:10.1371/journal.pone.0059338.
- C. Turner, C. Batty, E. Escalona, J. Hunter and C. Proudman, Curr. Anal. Chem., 2013, 9, 614–621 CrossRef CAS.
- D. Ye, X. Li, J. Shen and X. Xia, TrAC, Trends Anal. Chem., 2022, 148, 116540, DOI:10.1016/j.trac.2022.116540.
- S. Urquiza-Zurich, V. A. Garcia-Angulo, P. Burdisso, M. Fernanda Palominos, L. Fernandez-Hubeid, P. A. Harcha, J. P. Castillo and A. Calixto, mBio, 2023, 14(2), e0340222, DOI:10.1128/mbio.03402-22.
- D. Zeng, J. Cui, Y. Yin, C. Dai, H. Zhao, C. Song, S. Guan, D. Cheng, Y. Sun and W. Lu, Front. Plant Sci., 2022, 13, 900143, DOI:10.3389/fpls.2022.900143.
- D. Wang, P. Greenwood and M. S. Klein, Metabolites, 2021, 11, 863, DOI:10.3390/metabo11120863.
- I. Nordström, P. Sherwood, B. Bohman, S. Woodward, D. L. Peterson, J. Niño-Sánchez, T. Sánchez-Gómez, J. J. Díez and M. Cleary, Sci. Rep., 2022, 12, 21661, DOI:10.1038/s41598-022-26078-1.
- T. J. Davis, A. V. Karanjia, C. N. Bhebhe, S. B. West, M. Richardson and H. D. Bean, mSphere, 2020, 5(5), e920, DOI:10.1128/mSphere.00834-20.
- W. Jud, J. B. Winkler, B. Niederbacher, S. Niederbacher and J. P. Schnitzler, Plant Methods, 2018, 14, 109, DOI:10.1186/s13007-018-0378-4.
- J. Huidobro, J. Aramendia, G. Arana and J. M. Madariaga, Anal. Chim. Acta, 2022, 1197, 339499, DOI:10.1016/j.aca.2022.339499.
- J. Starek, B. Açıkmes, I. A. Nesnas and M. Pavone, Advances in Control System Technology for Aerospace Applications, ed. E. Feron, Springer, Berlin, 2016, vol. 460, pp. 1–48 Search PubMed.
- R. Arevalo, Z. Ni and R. M. Danell, J. Mass Spectrom., 2020, 55, e4454, DOI:10.1002/jms.4454.
- I. Zymak, J. Žabka, M. Polášek, A. Sanderink, J. P. Lebreton, B. Gaubicher, B. Cherville, A. Zymaková and C. Briois, Aerospace, 2023, 10, 522, DOI:10.3390/aerospace10060522.
- F. Karouia, K. Peyvan and A. Pohorille, Biotechnol. Adv., 2017, 35, 905–932 CrossRef PubMed.
- A. Coustenis, N. Hedman, P. T. Doran, O. Al Shehhi, E. Ammannito, M. Fujimoto, O. Grasset, F. Groen, A. G. Hayes, V. Ilyin, K. P. Kumar, C. E. Morisset, C. Mustin, K. Olsson-Francis, J. Peng, O. Prieto-Ballesteros, F. Raulin, P. Rettberg, S. Sinibaldi, Y. Suzuki, K. Xu and M. Zaitsev, Front. astron. space sci., 2023, 10, 1172546, DOI:10.3389/fspas.2023.1172546.
- T. Cheney, C. Newman, K. Olsson-Francis, S. Steele, V. Pearson and S. Lee, Front. astron. space sci., 2020, 7, 589817, DOI:10.3389/fspas.2020.589817.
- K. Olsson-Francis, P. T. Doran, V. Ilyin, F. Raulin, P. Rettberg, G. Kminek, M. P. Z. Mier, A. Coustenis, N. Hedman, O. Al Shehhi, E. Ammannito, J. Bernardini, M. Fujimoto, O. Grasset, F. Groen, A. Hayes, S. Gallagher, P. Kumar K, C. Mustin, A. Nakamura, E. Seasly, Y. Suzuki, J. Peng, O. Prieto-Ballesteros, S. Sinibaldi, K. Xu and M. Zaitsev, Life. Sci. Space Res., 2023, 36, 27–35 CrossRef PubMed.
- K. T. Tait, F. M. McCubbin, C. L. Smith, C. B. Agee, H. Busemann, B. Cavalazzi, V. Debaille, A. Hutzler, T. Usui, G. Kminek, M. A. Meyer, D. W. Beaty, B. L. Carrier, T. Haltigin, L. E. Hays, C. S. Cockell, D. P. Glavin, M. M. Grady, E. Hauber, B. Marty, L. M. Pratt, A. B. Regberg, A. L. Smith, R. E. Summons, T. D. Swindle, N. J. Tosca, A. Udry, M. A. Velbel, M. Wadhwa, F. Westall and M.-P. Zorzano, Astrobiology, 2022, 22, S57–S80 CrossRef.
- Fondation européenne de la science. and European space sciences committee., Mars Sample Return backward contamination - Strategic advice and requirements: report from the ESF-ESSC Study Group on MSR Planetary Protection Requirements., European Science Foundation, 2012 Search PubMed.
- European Space Agency, ExoMars - Has life ever existed on Mars? Search PubMed.
- N. J. Tosca, C. B. Agee, C. S. Cockell, D. P. Glavin, A. Hutzler, B. Marty, F. M. McCubbin, A. B. Regberg, M. A. Velbel, G. Kminek, M. A. Meyer, D. W. Beaty, B. L. Carrier, T. Haltigin, L. E. Hays, H. Busemann, B. Cavalazzi, V. Debaille, M. M. Grady, E. Hauber, L. M. Pratt, A. L. Smith, C. L. Smith, R. E. Summons, T. D. Swindle, K. T. Tait, A. Udry, T. Usui, M. Wadhwa, F. Westall and M.-P. Zorzano, Astrobiology, 2022, 22, S81–S111, DOI:10.1089/ast.2021.0115.
| This journal is © The Royal Society of Chemistry 2025 |





