Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Starvation induces diffusion hindrance at the nanoscale in mammalian cells†
Sakshi
Sareen
,
Alicja
Zgorzelska
,
Karina
Kwapiszewska
 * and
Robert
Hołyst
* and
Robert
Hołyst
 *
*
Institute of Physical Chemistry, Polish Academy of Sciences, Warsaw, Poland. E-mail: kkwapiszewska@ichf.edu.pl; rholyst@ichf.edu.pl
First published on 14th November 2024
Abstract
Prolonged starvation leads to acute stress, inducing a state of cellular dormancy with reduced energy consumption. Our research reveals that nutrient deprivation halts the movement of large ribosomal subunits, trapping them in a gel-like structure within the cytoplasm of surviving cells. This effect is due to water efflux from cells, causing a decrease in cell volume to half the original volume. This simple physical strategy saves, in a dormant state 107 ATP per second, which is needed for normal protein production. We monitored the diffusion of GFP (radius 2.3 nm) and 40S and 60S ribosomes (radii 3.75 and 15 nm, respectively) in the cytoplasm and nucleus during starvation of HeLa cells. GFP and 40S ribosomes slowed their diffusion in the cytoplasm two and five times, respectively. 60S ribosomes exhibited only rotational diffusion. In non-starving cells, biomolecules get stuck in the gel structure of cytoplasm with sizes >100 nm. We show that the gel pore size decreased from 100 nm to 30 nm upon starvation. The diffusive transport in the cell nucleus didn't change during starvation. GFP and ribosomes had the same diffusion coefficients in non-starving and starving cells in the nucleus. This highlights the importance of nuclear transport in cancer cells during extreme stress conditions.
Introduction
Stress affects molecular processes at cell, tissue, and organ levels. In response to stressful conditions, organisms have evolved physiological and biochemical adaptations, leading to a dormant state comparable to hibernation. Warm-blooded hibernators, for example, undergo a decrease in metabolic rates by 10–20%, an increase in energy conservation by up to 90% (especially during protein production), and a reduction in cell division as a response to starvation and temperature fluctuations.1 Analysis of energy consumption in synthetic cellular systems reveals that translation alone requires 34% of total cellular energy, with 45% energy allocated for maintenance and 20% for other processes such as amino acid uptake, transcription, division, and transport.2 Studying energy-intensive cellular processes like ribosomal biogenesis or cytoplasmic maintenance during stress could elucidate important information regarding intracellular processes.Loss of polyribosomes in vital organs, decrease in resting metabolic rate3 (energy conservation reservoir), cessation of mitotic activity, and structural modifications in subcellular compartments – are characteristics universal to a dormant state.4 Entering into a quiescent state during stress is not exclusive to one cell type; it is a function of different body cell types. For instance, tissue-resident stem cells and female egg cells (oocytes) exhibit prolonged dormancy by low metabolic activity5,6 persisting over long periods. Slowed metabolic activity observed ubiquitously throughout different species represents an adaptive response to stressful conditions. While physiological remodelling of internal processes is a highly adaptive strategy in mammalian species, the cellular and molecular mechanisms behind the dormant state remain poorly understood.
One of the stressing factors resulting in dormancy is starvation. Glucose starvation leads to low amounts of ATP, attributed to cellular stress, with harmful effects on the physical characteristics of a cell. Eliminating glucose from the tumour microenvironment by selective starvation is a practical strategy for treating different types of tumors.7 Unlike normal cells exhibiting differential stress resistance8 to ensure viability during starvation, cancer cells are driven by mutations and significant dependence on glucose metabolism for rapid proliferation (Warburg effect), leading to sensitisation towards glucose depletion. Investigations reveal notable impacts of nutrient deprivation on processes such as translation in single-cell organisms like bacteria to intracellular organelles like chloroplast.9 A 2021 study elucidates the effects of starvation through inhibition of de novo ribosome synthesis and degradation of excessive ribosomes, leading to a decrease in translation.10 These findings highlight the role of nutrient deprivation on various cellular processes, establishing it as an essential determinant in cell functioning. Here, we elucidate a simple physical mechanism allowing the entry of a dormancy state in cells during nutrient-deprived conditions by measuring the intracellular diffusion of GFP and ribosomal subunits. We postulate that when diffusion slows down, all metabolic activity slows down. This diffusion – measured as diffusion coefficients of various molecules – is inversely proportional to the effective nanoviscosity inside cells.11
Various biological processes, such as cell migration, motility, and intracellular diffusion of molecules, have been linked to alterations in cellular viscosity.12,13 Changes in internal viscosity can alter the shape and structure of the cytoplasm, with transformative influence on essential processes like cell growth, proliferation, maintenance, and repair.14 The importance of changing intracellular “microviscosity” under stress has significant effects on cell functioning.15 Recent advancements demonstrate additional rheological properties and viscoelasticity governing cellular processes like cell growth, division, movement, and repair in microtubule polymerisation and depolymerization.16 According to this study, an increase in cytoplasmic concentration of macromolecules caused a decreased rate of microtubule polymerisation and depolymerisation and vice versa.16 Intracellular rheological properties seem pivotal in understanding the cell microenvironment during normal and abnormal conditions. For instance, variation in protein concentrations causes increased cytoplasmic viscosity, affecting translation rates in Xenopus oocyte.17 Further research is required to comprehend the impact of these physicochemical properties, particularly concerning changes in the cytoplasm during unfavourable conditions like cell starvation. Our study focuses on the internal nanoviscosity of cell cytoplasm and nucleus, calculated from intracellular probe diffusion coefficients.
Previous studies have demonstrated that inert probes ranging from 0.25–75 nm in radii experience free diffusion in cell nucleus.18 The same applies to probes <50 nm in radii for cell cytoplasm. The cell cytoplasm exhibits liquid-like behaviour at nanoscales (<100 nm) but a gel-like state at larger scales. Objects of sizes >100 nm cannot move by free diffusion in the cytoplasm.11 The cellular nucleus consists of channels of radii ∼75 nm, where diffusion is almost unobstructed and effective nanoviscosity is comparable to water viscosity. These findings were primarily recorded in the HeLa cell cytoplasm and nucleus. Diffusion hindrance experienced by nano-objects like proteins, nucleotides, and other biomolecules within distinct cell compartments (cell nuclei and cytoplasm) can be a limiting factor during various cell processes.
Several techniques, such as microfluidics, atomic force microscopy, optical tweezers, and magnetic twisting cytometry, have been applied to investigate the extracellular physicochemical properties of specific cell lines. These techniques provided information on the viscoelastic properties of the whole cell and its environment, focusing on qualitative and quantitative measurements. In contrast to the above-mentioned techniques, we study intracellular viscosity or molecular diffusion by precisely focusing on a very small volume inside the cell cytoplasm. For this study, a fluorescent protein with minimal internal interactions exerting minimal influence on the outcome in fluorescence intensity was chosen. Fluorescence correlation spectroscopy (FCS) facilitates measurements of intracellular diffusion coefficients without influencing the cell environment.19 Fluorescence lifetime imaging microscopy coupled with fluorescence correlation spectroscopy has the potential to quantify viscosity in living systems in a concentration-independent manner.20
Our previous works on complex fluids and polymers, using various nano- and microrheological measurement techniques, have contributed to developing a length-scale dependent viscosity (LSDV) model.21,22 The model elucidates that an increase in probe size in the nanometre range encounters an increase in nanoviscosity (eqn (1)). This model was used to analyse data collected from FCS measurements during this study.
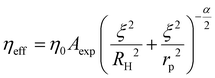 | (1) |
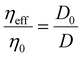 | (2) |
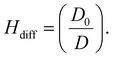 | (3) |
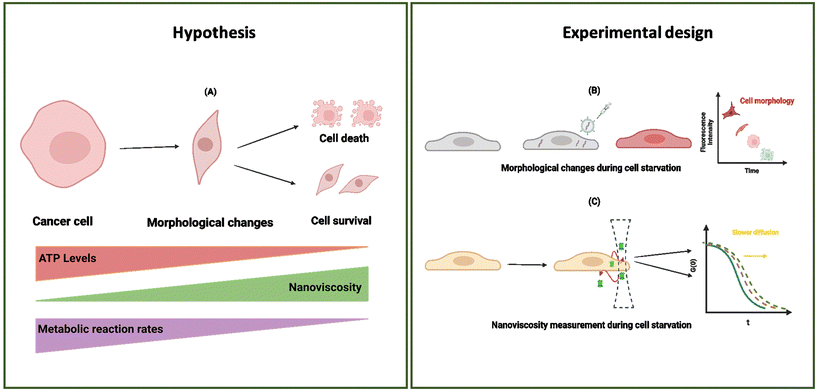
In this study, we present our results regarding diffusion hindrance, Hdiff, which describes how much diffusion is hindered with respect to water. Here, D0 is the diffusion coefficient of GFP in water (calculated using the Stokes–Sutherland–Einstein relation, ESI eqn (S2)†), and D is the diffusion coefficient of GFP measured in cell cytoplasm. Using FCS, we measure diffusion coefficients of moving GFP particles and ribosomes in and out of confocal volume. Hdiff was calculated from these measured values. This technique uses fluorescent probes to analyse the correlation of temporal fluorescence intensity fluctuations, which in turn was used to calculate diffusion of GFP in cytoplasm. Along with quantifying this physical parameter, morphological changes during starvation were studied. Refer to the schematic representation of the study in Fig. 1.
Materials and methods
Cells and cell culture
769-P cells (ATCC) were cultured in RPMI-1640 media (ATCC) supplemented with 10% fetal bovine serum (Sigma-Aldrich) and 1% penicillin (100 mg ml−1), streptomycin (100 mg ml−1) solution purchased from (Gibco). A GFP/HeLa stable cell line from amsbio (AMS14-904ACL) was used for all diffusion-related experiments. It was cultured in DMEM media (Sigma-Aldrich) supplemented with 10%v/v Fetal bovine serum (Sigma), 1%v/v solution of penicillin (100 mg ml−1), streptomycin (100 mg ml−1) and 1%v/v solution of L-glutamine (2 mM stock; Sigma-Aldrich). Cells were maintained at 5% CO2 in an incubator (Biogenet) at 37 °C under humidified filter conditions. Cells were grown in T75 corning flasks (Sarstedt AG & Co.) till passage five before seeding them on a 35 nm Glass bottom cell culture dish (GBO) for starvation experiments.Transfection and ribosome staining
769-P cells were seeded on a 35 mm thin Glass bottom cell culture dish (Eppendorf). Transfection with a pcDNA3.1(–) plasmid vector containing MALIONR gene insert of 1100 bp (Addgene plasmid #113908) was used containing ε subunit of the ATP-binding region of the bacterial F0F1-ATP synthase labelled with the red fluorescent mApple protein, which increased its fluorescence in the presence of ATP. Transfection was performed using a Lipofectamine 3000 transfection kit (Sigma). After 5 hours of transfection, the cell medium was changed. Transfected cells were left in the incubator for 48 hours before measurements.Ribosomes were stained using Yo-Pro-1 dye (Invitrogen), as described previously.28 Briefly, cells were washed and incubated in 50 nM Yo-Pro-1 in PBS for 30 minutes before the measurement. The Yo-Pro-1 stained ribosomes exhibited fluorescence at λex = 485 nm, λem > 500 nm.
Glucose cell starvation
Cell culture medium was replaced from transfected cells with phosphate saline buffer (PBS; Sigma) containing calcium and magnesium ions to starve cells for multiple days.Confocal microscopy
Images were collected on a Nikon A1 confocal setup. Melle's griot 488 nm ion laser, model 35-IMA-410-019, was used for imaging. t0 measurements of transfected cells were recorded in cell culture media using a confocal microscope. Subsequent readings on days 0, 1, 2, 3 and 4 were taken with cells in PBS containing calcium and magnesium ions.Fluorescence correlation spectroscopy
HeLa GFP was seeded on glass bottom dishes overnight before measurements. Cell measurements were taken in a cell culture medium as a control, and then the medium was immediately changed to PBS for recording starvation measurements. FCS measurements were performed using a commercially available setup based on a confocal microscope (Nikon Eclipse TE2000U) coupled with Pico Harp 300 TCSPC module (PicoQuant). A climate chamber (Okolab) was used to perform all cell measurements, providing temperature control (36 ± 0.5 °C) and the required composition and humidity atmosphere. A pulse diode laser (PicoQuant LDH-D-C-485; 485 nm) driven by a Sepia II module (both from PicoQuant) with a long-pass filter of 488 nm in the detector was used. The power of the laser was kept in the range of 5–10 μW. Minimal potential photobleaching of GFP molecules was observed during the short measurement time of 30 seconds/measurement. Fluorescent GFP particles were observed and measured through the 60× (N.A. 1.2) objective with water immersion. The fluorescent signal for FCS was collected by single photon avalanche diodes (MPD and PerkinElmer).A calibration step preceded each day of measurements. Rhodamine 110 (Sigma-Aldrich) in 2.5% weight glucose in PBS1 was used for calibration and correction collar adjustment. The diffusion coefficient of Rhodamine 110 in the calibration conditions was 560 μm2 s−1, which was used to calculate the size of the confocal volume. On average, the short axis of the confocal volume (ω0) was equal to 0.196 ± 0.009 μm, structure parameter (κ) to 6.4 ± 0.8, and effective volume (Veff) to 0.27 ± 0.04 fL. The position of a cell was adjusted using a confocal microscope, as described in ESI 8.† The confocal focus was positioned in a cytoplasmic area of a cell away from the nucleus. SymphoTime 64 software (Pico Quant) was used to acquire time trace data and autocorrelation curves. Time trace of fluorescence intensity I(t) collected from the effective volume, Veff is recorded and the autocorrelation function (eqn (4)) is calculated in FCS measurements.
| G(τ) = 〈δI(t)δI(t + τ)〉/〈I(t)〉2 | (4) |
Data acquisition and statistical analysis
Data were acquired from 3–5 cells, with five measurements per cell lasting 30 seconds each. Three sets of experiments were performed with similar recording conditions over five days per experiment. FCS analysis was performed in two steps: (a) the autocorrelation function G(t) was calculated from the measurements of fluorescence intensity time trace, and (b) the diffusion coefficient was calculated using one component free diffusion model. For each cell, the average value of the diffusion coefficient of EGFP was obtained from 5 consecutive measurements. Each point on the graph obtained at a different time reflects the average diffusion time value obtained from different cells on that day from three independent experiments (Fig. 2). Data were fitted using eqn (5), where N is the number of particles, κ is the structural parameter obtained during calibration, and τD is the time of flight across the focal volume.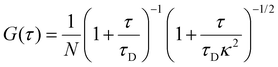 | (5) |
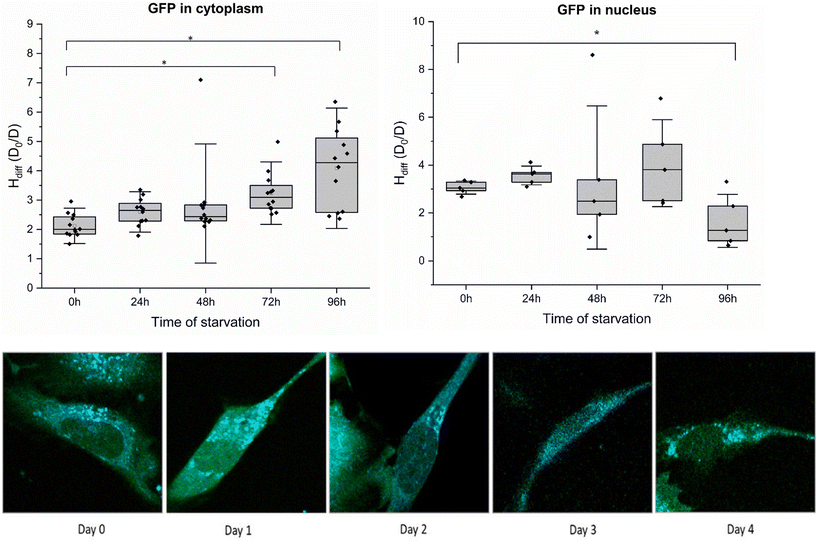 | ||
| Fig. 2 H diff of starved HeLa-GFP cells in the cytoplasm (A) and nucleus (B) measured using fluorescence correlation spectroscopy. Results are presented as box plots. Each box represents average data collected from 4–5 cells in PBS daily. Error bars represent the standard error of the mean. Using time of flight as a fitting parameter, the diffusion coefficient of the GFP molecule was calculated using eqn (5), ESI eqn (S1) and (S2).† Diffusion hindrance was calculated from the diffusion time of GFP particles in the cell cytoplasm and nucleus (ESI, eqn (S1) and (S2)†). FLIM images of HeLa-GFP cells during starvation over 4 days are shown at the bottom (from left to right). Two sample variance t-tests were performed (*p values >0.05). Magnification: 40×. | ||
At τ = 0, G(0) = 1 (for N = 1), where fluorescence intensity is perfectly correlated with itself for a probe. As τ increases, G(τ) decreases, capturing subsequent decay of autocorrelation. The average residence time of fluorescent molecule in confocal volume corresponds to the lag time at which G(τ) decays to half its maximal value. τD and N are found by analysing experimentally obtained autocorrelation function. The rotational diffusion of ribosomes was measured using the method described in,28 with the details (including the fitted model) presented in ESI 5.†
Data analysis is usually performed in two steps: the first part includes the calculation of autocorrelation function {G(τ), eqn (4)} obtained from measured fluorescence intensity time trace I(t) and the second part focuses on fitting the autocorrelation function curves. Interpretation of the calculated G(τ) via an appropriate theoretical model depends on the nature of diffusion in the sample. These models define the shape of autocorrelation function G(τ), which is further derived by approximating effective detection volume by a 3-D Gaussian profile describing the probability of detecting the emitted photon by fluorophore at a given position. These models are used to fit characteristics of the processes occurring during fluorescence intensity fluctuations with the shape of the autocorrelation function. For our study, we use a one-component diffusion model defining freely diffusing single particles in all 3 dimensions.23
Image and data plotting
Using MathWorks's software MATLAB, Hdiff graphs were plotted. Images acquired from FCS experiments were analysed using an in-built software called SymphoTime.Results
Changes in cytoplasmic diffusion of GFP in HeLa cells during starvation
To determine Hdiff within cell cytoplasm, we selected a HeLa cell line with stable GFP expression. Fluctuations in fluorescence, resulting from diffusion of GFP (∼2.3 nm)24 molecules in and out of confocal volume were quantified using time-correlated single photon counting. Autocorrelation curves obtained from measurements were subjected to fitting employing a single-component diffusion model. Results obtained after subjecting the cells to starvation are presented in Fig. 2A. The experimental procedure involved exchanging media with phosphate saline buffer (PBS) and recording diffusion coefficients immediately after on day 0. Cells were incubated in PBS for four consecutive days. A slight increase in Hdiff was observed on days 1 and 2 compared to day 0 measurements. Throughout the four days of starvation, measurements were performed on cells attached to the slide until day 4. However, from day 2 onwards, cell detachment was observed in all the wells of a glass bottom plate, and cell death increased from 10% to 60%. Day 4 constituted ∼80% cell death in all the wells. By the end of the fourth day, cells succumbed ultimately to death, and notable amounts of cellular debris remained in the PBS. All FCS measurements were performed on cells sticking to the surface and having spindle shape morphology.Diffusion coefficient measurements were averaged from 4–5 cells during starvation in every experiment. The diffusion coefficient (D) measured in the cytoplasm from the non-starving state until the last day of starvation (100 hours) was compared to the diffusion coefficient of GFP molecules in water. Minimal fluctuations in diffusion coefficients were observed for the first two days of starvation. The highest decrease in diffusion coefficients was observed on the last day of starvation from cells still attached to the plate. Decreased diffusion of GFP or a subsequent increase in diffusion hindrance in cell cytoplasm indicates a halt in intracellular metabolism. We postulate that diffusion hindrance is a physical way that hinders metabolic reactions, decreasing the probability of molecule collision. Cells attached to the surface (for day 0 – day 4 starvation periods) were considered for all the diffusion coefficient measurements. Temperature conditions were kept constant for both measurements. Hdiff is plotted against the time of starvation (in hours) for HeLa-GFP cells. Refer to ESI 1, 2 (Fig. S1†) for results obtained in Fig. S2.†
No change in diffusion of GFP in HeLa cell nucleus during starvation
The observed increase in diffusion hindrance of GFP in cytoplasm prompted another question regarding internal spatial arrangement: whether this increase is uniform throughout the cell or compartmentalised. To investigate, we measured Hdiff in the cell nucleus, where the genetic code resides and regulates cell function. The results from Hdiff measurements in the HeLa cell nucleus are presented (Fig. 2B), where the focal volume was placed precisely inside the cell nucleus18 and the diffusion coefficient of GFP was measured. Cells were subjected to four consecutive days of starvation, and the diffusion coefficients were measured daily. Each box plot consolidates data from 5 measurements spanning 4–5 different cells, with an exposure time of 30 seconds. Our measurements conclude that diffusion hindrance to GFP remains approximately constant at 2–4 (D0/D) in the cell nucleus throughout the transition from a nutrient-abundant to a nutrient-deprived state. Fluctuations recorded in Hdiff from day 0 (cells in media) till day 4 (cells in PBS, 90% cell death) are not significantly different from those measured in cell cytoplasm under similar starvation conditions. Considering the pivotal role of the nucleus as the cell's command centre, these results suggest that the diffusion hindrance in the nuclear compartment is maintained even under stressful conditions of starvation. Hdiff values obtained from these experiments correspond numerically to the same order of magnitude in both cell cytoplasm and nucleus. This suggests a particular range of Hdiff in cellular compartments, specifically for starvation conditions in HeLa GFP cells. Our observation of physical changes in cancer cells highlights subtle distinctions in cell response to similar stressful conditions within different compartments. These results are congruent with the cellular response in organisms such as yeast and C. elegans25,26 towards stressful conditions like nutrient deprivation.Examination of rotational and translational diffusion of 60S and 40S ribosomal subunit
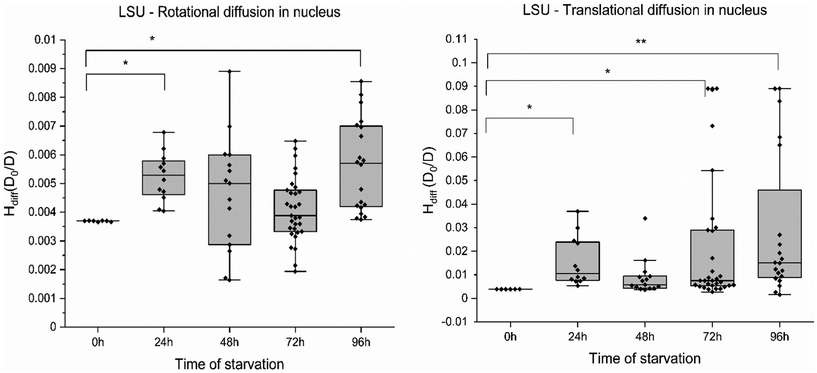
In agreement with these reports, we note similar compartmentalisation when measuring the diffusion of ribosomal subunits in cancer cells. To examine the impact across different length scales in different compartments, we measured translational and rotational diffusion of 60S ribosomes (LSU) in the nucleus and cytoplasm of HeLa cells (without GFP) using YO-PRO-1 dye.27 YO-PRO-1 is a DNA intercalator. Surprisingly, a recent study demonstrated the binding interaction of YO-PRO-1 with dsRNA.28 The large ribosomal subunit in eukaryotic cells consists of 3 RNAs – 5S, 5.8S and 23S RNA, 46 proteins, and 3 binding sites for tRNA.29 Refer to Yo-Pro-1 staining protocol and uptake in ESI 5, Fig. S5.† YO-PRO-1 stained cells were utilised for four consecutive days to obtain rotational and translational diffusion coefficients in the nucleus and cytoplasm in glucose-depleted conditions. FCS curves registered in the cells were fitted with a two-component free diffusion model, incorporating a rotational diffusion term (following Aragon and Pecora30). The model and an exemplary fit are presented in ESI 5, Fig. S6–S8.†The rotational (Fig. 3A) and translational motion (Fig. 3B) of the 60S ribosome exhibited no discernible changes with increasing stress in nutrient-deprived conditions in the nucleus. Consequently, we report the absence of any significant alteration in the rotational and translational diffusion of the ribosome at the nanoscale. Similar results were observed for GFP (∼2.5 nm) and ribosome LSU (∼15 nm). Across the probe range of 2–20 nm, the nucleus maintains a constant diffusion profile under starvation conditions in HeLa cells. Diffusion coefficients from this study indicate that ribosomes move unaffected in the cell nucleus but not in the cytoplasm during starvation. The Hdiff measured in the nucleus has proven relatively stable on various length scales. This could indicate a significant cellular survival strategy during harsh conditions where a cell protects its command centre compartment (the chromatin and associated proteins) as an act of survival.
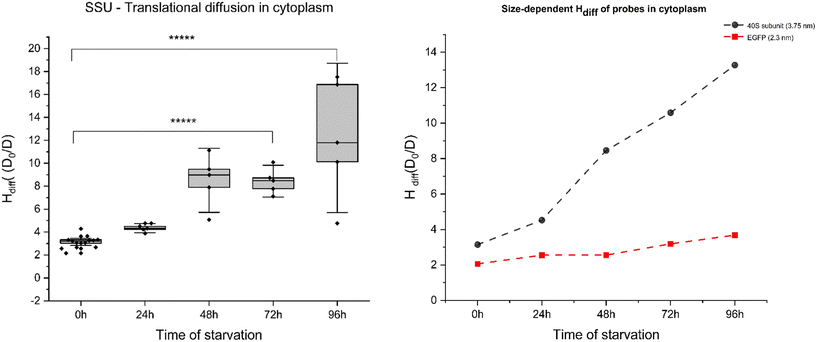
On the other hand, FCS curves obtained from YO-PRO-1 experiments for cytoplasm revealed significant diffusion hindrance of ribosomes in the cytoplasm. Single spot measurements of YO-PRO-1 treated starved cells resulted in extensive photobleaching before signal stabilising, suggesting that two populations were present in the sample – freely diffusing and immobile (see ESI 5, Fig. S6–S8†). Analysis of FCS curves of mobile fraction led to the conclusion that LSU was immobilised in the pores of physical gel and either got photobleached due to complete immobilisation or exhibited only rotations with characteristic times ∼10 μs. The resisting mobile fraction (diffusion times >100 μs) were small subunits (SSU) – not detectable in the presence of mobile LSU due to a much lower probability of staining.27 With these results, we postulate that cytoplasm undergoes pore size reduction in its “mesh/gel-like state” from <100 nm (native cells) to <30 nm in glucose-depleted cells. Moreover, diffusion hindrance (Hdiff) for the length-scale of 3.75 nm (radius of SSU) increased 5-fold during 4 days of starvation – from 3.1 D0/D (day 0: reference probe – dextran 20 kDa of the same hydrodynamic radius) – to ∼13.5 D0/D (day 4; refer to Fig. 4(A)). Diffusion hindrance of ribosomes in cytoplasm noted in our experiments could be due to their preservation in liquid-like condensates. Similar results have been observed in yeast during heat shock, where ribosome production is the first process to get shut down to conserve cellular energy.31 Our results also show that length-scale dependency of the apparent viscosity is maintained in starved cells cytoplasm.
Cell volume changes during starvation
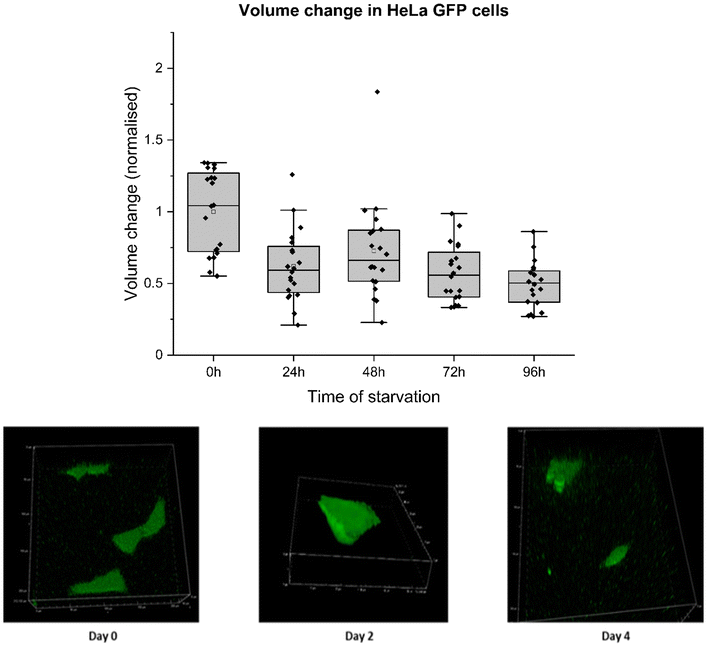
Along with using fluorescence correlation spectroscopy to understand the physical properties of cells, confocal microscopy was implemented to document morphological changes during starvation (ESI 6, Fig. S9†). Visual interpretation revealed a reduction in cell size for those subjected to starvation for 4 consecutive days compared to the cells on day 0 or in complete media. We measured volume changes during starvation in HeLa cells to quantify these alterations. 25%–40% of cytoplasmic volume mainly consists of proteins, nucleic acids, and lipids, and the water content of cell32 covers the rest. Water movement in and out of a cell is highly regulated and critical in proper cell functioning and response to pathological and physical stress stimuli.33 Cell volume maintenance is essential for cellular processes, including cell growth, cell division, cell cycle progression at different checkpoints, lipid synthesis, and other critical translational processes inside a cell cytoplasm.34Alterations in cell volume have been extensively documented, with a 1986 study highlighting that cells can detect volume changes as low as 3%.35 Volume perturbations leading to cell shrinkage or swelling cause activation of active membrane transport and various metabolic processes within the cell to restore it to a normal resting state.36 This phenomenon could explain the stability of molecular diffusion during the cell cycle.37 Changes in cell volume (osmotic compression) have been recorded to disrupt cell signalling in yeast.38 A decrease in cell volume hinders the cell signalling processes, although the ability of a cell to activate cell signalling returns as soon as the cell volume is restored. Cell signalling is highly affected by reducing the rate of reactions and diffusion coefficients. Nuclear transport of multiple proteins, as well as a transcription factor, showed similar results. This signifies the importance of cell volume changes in determining the molecular crowding in cell cytoplasm, causing alterations in cell signalling due to varied protein–protein interactions. Therefore, comprehensive quantification of these changes becomes imperative for understanding intracellular dynamics.
For this experiment, HeLa GFP cells were cultured in DMEM media and allowed to grow for 24 hours before the medium was replaced with PBS. Subsequently, the cells underwent four consecutive days of starvation, during which their volumes were assessed through z-stack imaging on a confocal microscope. Cell volume was measured by masking fluorescent cell surface area in a 3D plane in a NIS confocal setup. These values were used to calculate cell volume and surface area change. Cell volume decreased to 50% from its initial volume by the fourth day of starvation. The reduction in cell volume could be attributed to intramolecular crowders coming close to each other due to water efflux, reducing pore size between them. This decrease directly affects the movement of molecules involved in cellular metabolism. Therefore, increased diffusion hindrance noted on the fourth day of starvation can be due to water efflux, as shown by decreased cell volume (Fig. 5) and surface area (ESI 3, Fig. S4†). Cell volume decrease throughout starvation was directly related to increased Hdiff near cell death. The relationship between Hdiff and cell volume changes was analysed during the starvation of HeLa cells. Measured Hdiff in starved HeLa cells and changes in the volume of starved HeLa cells perfectly correlated as depicted in ESI 3, Fig. S2 and S3.†
Using ESI, eqn (S1)–(S3),† the Hdiff of cells was calculated, which differed from the measured values obtained. Although water efflux was linked to changes in measured Hdiff, the measured and calculated values from the LSDV model could not be solely explained by changes in ξ (intermolecular space) after water efflux. To account for the observed discrepancy, we postulate that the difference in calculated and measured Hdiff values could result from the changes in interactions between diffusing molecules within cells.
Interaction parameter for diffusion in starved cells
The interaction parameter γ refers to interactions of the probe with molecular crowders during diffusion in a fluid (see ESI 4†).40 As we consider cytoplasm a complex fluid, we calculated γ for the GFP (2.3 nm) motion at different times of starvation in cell cytoplasm. γ is the energy required to overcome the friction between the probe and intracellular molecules, resulting in the overall flow of the fluid.39 The energy of viscous flow in cell cytoplasm depends mainly on these internal interactions. For systems where the probe radius (rp) is significantly larger than the hydrodynamic radius of molecular crowders (RH), changes in γ reach a limit similar to those measured in macroscopic flow.40 For small proteins (rp ∼ 1–5 nm), the diffusivities were orders of magnitude faster than the ones predicted for SSE.41 Thus, we deploy LSDV model equations to calculate changes in γ in a nanoscale system, considering that the probe size in our studies is 2.3 nm. Upon water efflux during starvation (discussed in the previous segment, Fig. 5), the cell undergoes shrinkage, causing molecules to move close to each other, consequently reducing the intermolecular space from ξ1 to ξ2.We assume that this decrease in distance between major crowders results in a change in interaction parameter (γ) between the probe and the crowders. Changes in γ were calculated to obtain energy values to correlate with viscous flows in the cytoplasm during starvation (ESI, eqn (S6)–(S8)†). An increase in γ for GFP (from 0.7 kJ mol−1 to 1.3 kJ mol−1) was observed with decreasing intermolecular crowder space (from 3.2 nm to 2.5 nm) over four days of starvation (Table 1).
| Sample | ξ [nm] | γ [kJ mol−1] |
|---|---|---|
| Control cells | 3.2 | 0.7 |
| 24 h starved cells | 2.0 | 0.7 |
| 48 h starved cells | 2.8 | 1.1 |
| 72 h starved cells | 2.6 | 1.2 |
| 96 h starved cells | 2.5 | 1.2 |
We calculated diffusion coefficients corresponding to changes in ξ, which were higher than those measured during experiments; Hdiff was higher for measured values than calculated values (ESI 4, Table S1†). By factoring in the interaction parameter (γ), an increase in Hdiff's measured values can be easily explained. Increasing Hdiff corresponds to a higher interaction regime, with increased friction between probe and crowders, over an intense period of cellular starvation. With decreasing intermolecular crowder space, particles require more energy for unhindered diffusion, accounting for higher measured Hdiff than calculated Hdiff. These results highlight that as the cytoplasm shrinks, the interactions between the molecules increase.
Growth and regrowth of cells under stress
HeLa GFP cells were again subjected to a four-day starvation period to further understand cell behaviour during and after starvation. Bright-field images of pre-seeded cells were captured from three regions of interest in two glass slide wells before initiating starvation, and similar procedures were followed on day 0, day 2, and day 4 of starvation. Following the four-day starvation period, cell media was replaced from PBS to complete DMEM media containing growth factors (Fig. 6). Images were recorded on day 5, day 7, and day 9, following the same capturing procedure. By day 4, a significant cell population had detached from the surface, floating as debris in PBS (Fig. 6). The PBS containing cell debris was spun at 1100g for 5 min. The supernatant was removed, and the circular debris was resuspended in complete growth media (data not shown). This resuspended media containing cells was plated in another 35 mm glass slide. A similar media change was given to the previous glass slide containing the remaining 1% cell population still attached to the glass surface. The small cell population, still sticking to the glass slide, revived to 60% confluency three days after complete media was supplied. Cell growth was also observed in the cell debris wells, freshly supplied with complete growth media. These observations suggest that starvation imposes significant stress on mammalian cells, causing cellular shrinkage due to increased water loss, which could be an energy conservation mechanism to induce cell dormancy. Some cancer cells are assumed to remain dormant until they find a suitable nourishment source. Therefore, we suspect that water efflux is employed by cells as a defence mechanism to survive under stressful conditions like starvation. Cells undergo a state of dormancy and subsequently revive when a source of nourishment is reintroduced. These findings align with observations in other single-cell organisms like yeast and bacteria (refer to discussion).Discussion
This study presents that cells enter metabolic dormancy characterised by restricted macromolecular motion under stress conditions. Here, we propose a physical mechanism of cell survival under stress conditions by studying changes in diffusion coefficients. We propose (1) macromolecular motion is hindered during low ATP levels in cells, causing cytoplasmic processes to halt (2) this intracellular partial freezing is the result of reduced pore size of cytoplasmic mesh from 100 nm to 30 nm due to water efflux (3) diffusion rates are affected in the cytoplasm at two different length scales but macromolecular motion remains unaffected in the nucleus, pointing towards a state of cell dormancy (4) interaction energy parameter of molecules plays an important role in diffusion (5) the proposed model suggests that changes in diffusion coefficients dictate the process of cellular hibernation.Bacterial cytoplasmic behaviour was reported to shift from glass-like to liquid-like for particles below 30 nm.42 This effect is influenced by the water molecule's hydration effect on the cytoplasm's volume fraction. Our study found that water efflux reduces the intermolecular spaces in mammalian cell cytoplasm. Macromolecular movement in the cytoplasm is highly dependent on the space available, and this movement is the basis of all intracellular processes. Intensive starvation leads to water expulsion, decreasing cell volume by up to 50%. This reduction is directly linked with the increase in viscosity or hindered diffusion experienced by probes in the nanometre range, according to the LSDV model described in this study.43 Diffusion hindrance increased from 2 (D0/D) to 4 (D0/D) for the length scale of EGFP (2.3 nm) and from 3 D0/D to 15 D0/D for the length scale of 40S ribosomes (3.75 nm) in the cell cytoplasm, while the ratios remained relatively constant in the cell nucleus. The translational diffusion was completely halted for the 60S ribosomal subunit (15 nm), leaving only the subunits to rotate in the cytoplasm and decreasing pore size from 100 nm to 30 nm or below.
Cell dormancy is widely prevalent as a survival mode and can be reversed to metabolically active, fully functional states with changing environments.44 Cells require 107 ATP per s to maintain daily protein turnover. Stress inhibits these energetically costly processes, causing reduced metabolic reaction rates, thus inducing dormancy45 The rates of macromolecular diffusion play a crucial role in determining various cellular processes, and their regulation has been demonstrated as a tuneable property in yeast cells responding to altering ATP levels.46 Our results suggest that cancer cells experience increased cytoplasmic viscosity during prolonged starvation, thus hindering diffusion in stressful conditions. These cells may undergo alterations in one of their cellular compartments (cytoplasm) while keeping a more steady state in another compartment (nucleus), protecting their genetic material. The study emphasises the importance of considering a physical perspective beyond changes in DNA-related processes (translation, transcription) to explain alterations in macromolecular movement.
Constant values obtained for diffusion hindrance in the nucleus, probed with GFP (Fig. 2B) and YO-PRO-1-stained ribosomes (Fig. 3A, B and 4A), suggest that the nucleus is structurally protected at the nanoscale during water efflux. Meanwhile, in the cytoplasm, the decrease in molecular diffusion is influenced by factors beyond water efflux. Decreasing cytoplasmic space (cell shrinkage due to water efflux) compels major macromolecules to approach each other. The closer the molecules are to each other, the higher energies will be required to overcome friction to carry out new processes. An increase in interaction parameter from 0.7 kJ mol−1 to 1.3 kJ mol−1 was calculated for GFP in cytoplasm. We show how starvation induces cytoplasmic condensation during a shortage of glucose in the surroundings. Starvation-induced cell death may be reversible, as observed for cell revival from a near-death stage back to 60% confluency in these experiments (Fig. 6). The modulation of intracellular molecular crowding, proposed in this study as a survival response, adds to the complexity of factors contributing towards tumour progression and could influence and inform about potential therapeutic strategies in the future.
Conclusion
Our study has revealed that starved cancer cells employ a physical mechanism of changing the fluidity of cytoplasm by removing water, consequentially affecting the internal diffusion of biomolecules. This is observed in cells surviving the harsh conditions of prolonged starvation. This phenomenon helps reduce energy utilisation for protein synthesis, inducing a “dormancy” “-like state. Cytoplasm experiences a reduction in mesh size from 100 to 30 nm, hindering translational diffusion of 60S ribosomal subunit in smaller pore size. Cellular response to starvation in cancer cells is compartmentalised. While cell cytoplasm registers changes in macromolecular crowding during starvation, nuclear transport in cells remains unaffected. Molecular diffusion of small probes could be employed as a quantitative indicator of changes happening in the cell during starvation to evaluate targeted therapies in tumours.Data availability
The supporting data for this article has been prepared as part of ESI.†Conflicts of interest
There are no competing interests.Acknowledgements
This work was supported by the National Science Centre, Poland, within the grant OPUS UMO-2019/33/B/ST4/00557.References
- S. M. Mohr, S. N. Bagriantsev and E. O. Gracheva, Cellular, Molecular, and Physiological Adaptations of Hibernation: The Solution to Environmental Challenges, Annu. Rev. Cell Dev. Biol., 2020, 36, 315–338 CrossRef CAS PubMed
.
- H. R. Sikkema, B. F. Gaastra, T. Pols and B. Poolman, Cell Fuelling and Metabolic Energy Conservation in Synthetic Cells, ChemBioChem, 2019, 20, 2581–2592 CrossRef CAS
.
- B. Olson, D. L. Marks and A. J. Grossberg, Diverging metabolic programmes and behaviours during states of starvation, protein malnutrition, and cachexia, J. Cachexia Sarcopenia Muscle, 2020, 11, 1429–1446 CrossRef PubMed
.
- H. V. Carey, M. T. Andrews and S. L. Martin, Mammalian Hibernation: Cellular and Molecular Responses to Depressed Metabolism and Low Temperature, Physiol. Rev., 2003, 83, 1153–1181 CrossRef CAS PubMed
.
- I. B. Dias, H. R. Bouma and R. H. Henning, Unraveling the Big Sleep: Molecular Aspects of Stem Cell Dormancy and Hibernation, Front. Physiol., 2021, 12, 624950 CrossRef PubMed
.
- S. Z. Swartz,
et al., Quiescent Cells Actively Replenish CENP-A Nucleosomes to Maintain Centromere Identity and Proliferative Potential, Dev. Cell, 2019, 51, 35–48 CrossRef CAS
.
- E. M. Selwan, B. T. Finicle, S. M. Kim and A. L. Edinger, Attacking the supply wagons to starve cancer cells to death, FEBS Lett., 2016, 590, 885–907 CrossRef CAS PubMed
.
- S. Naveed, M. Aslam and A. Ahmad, Starvation based differential chemotherapy: A novel approach for cancer treatment, Oman Med. J., 2014, 29, 391–398 CrossRef PubMed
.
- R. Trösch and F. Willmund, The conserved theme of ribosome hibernation: from bacteria to chloroplasts of plants, Biol. Chem., 2019, 400, 879–893 Search PubMed
.
- T. Prossliner, K. Gerdes, M. A. Sørensen and K. S. Winther, Hibernation factors directly block ribonucleases from entering the ribosome in response to starvation, Nucleic Acids Res., 2021, 49, 2226–2239 CrossRef CAS PubMed
.
- K. Kwapiszewska,
et al., Nanoscale Viscosity of Cytoplasm Is Conserved in Human Cell Lines, J. Phys. Chem. Lett., 2020, 11, 6914–6920 CrossRef CAS
.
- J. Gonzalez-Molina,
et al., Extracellular fluid viscosity enhances liver cancer cell mechanosensing and migration, Biomaterials, 2018, 177, 113–124 CrossRef CAS
.
- P. Rodríguez-Sevilla,
et al., Upconverting Nanorockers for Intracellular Viscosity Measurements During Chemotherapy, Adv. Biosyst., 2019, 3, 1–9 Search PubMed
.
- A. Sohrabi Kashani and M. Packirisamy, Cancer-nano-interaction: From cellular uptake to mechanobiological responses, Int. J. Mol. Sci., 2021, 22, 9587 Search PubMed
.
- M. K. Kuimova,
et al., Imaging intracellular viscosity of a single cell during photoinduced cell death, Nat. Chem., 2019, 1, 69–73 CrossRef
.
- A. T. Molines,
et al., Physical properties of the cytoplasm modulate the rates of microtubule polymerisation and depolymerisation, Dev. Cell, 2022, 57, 466–479 CrossRef CAS PubMed
.
- Y. Chen, J.-H. Huang, C. Phong and J. E. Ferrell, Viscosity-dependent control of protein synthesis and degradation, Nat. Commun., 2014, 15, 2149 CrossRef PubMed
.
- G. Bubak,
et al., Quantifying Nanoscale Viscosity and Structures of Living Cells Nucleus from Mobility Measurements, J. Phys. Chem. Lett., 2021, 12, 294–301 CrossRef CAS PubMed
.
- I. Emilie,
et al., Targeted fluorescence lifetime probes reveal responsive organelle viscosity and membrane fluidity, PLoS One, 2019, 1–20 Search PubMed
.
- E. M. Woodcock,
et al., Measuring Intracellular Viscosity in Conditions of Hypergravity, Biophys. J., 2019, 116, 1984–1993 CrossRef CAS PubMed
.
- R. Holyst,
et al., Scaling form of viscosity at all length-scales in poly(ethylene glycol) solutions studied by fluorescence correlation spectroscopy and capillary electrophoresis, Phys. Chem. Chem. Phys., 2009, 11, 9025–9032 RSC
.
- T. Kalwarczyk,
et al., Motion of nanoprobes in complex liquids within the framework of the length-scale dependent viscosity model, Adv. Colloid Interface Sci., 2015, 223, 55–63 CrossRef CAS PubMed
.
- T. Kalwarczyk,
et al., Apparent Anomalous Diffusion in the Cytoplasm of Human Cells: The Effect of Probes’ Polydispersity, J. Phys. Chem. B, 2017, 121, 9831–9837 CrossRef CAS PubMed
.
- K. Kwapiszewska,
et al., Determination of oligomerisation state
of Drp1 protein in living cells at nanomolar concentrations, Sci. Rep., 2019, 9, 5906 CrossRef
.
- C. P. Brangwynne,
et al., Germline P Granules Are Liquid Droplets That Localize by Controlled Dissolution/Condensation, Science, 2009, 324, 1729–1732 CrossRef CAS PubMed
.
- R. Narayanaswamy,
et al., Widespread reorganisation of metabolic enzymes into reversible assemblies upon nutrient starvation, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 10147–10152 CrossRef CAS PubMed
.
- A. Magiera, K. Kucharska, T. Kalwarczyk, P. Haniewicz, K. Kwapiszewska and R. Hołyst, Measurement of large ribosomal subunit size in cytoplasm and nucleus of living human cells, 2024 Search PubMed.
- J. Michalski,
et al., Rotational and translational diffusion of biomolecules in complex liquids and HeLa cells, Soft Matter, 2024, 20, 5810–5821 RSC
.
-
T. M. Schmeing, Ribosome Structure, in Encyclopedia of Biological Chemistry, 2013, 2nd edn, pp. 128–135 Search PubMed
.
- S. R. Aragón and R. Pecora, Fluorescence correlation spectroscopy and Brownian rotational diffusion, Biopolymers, 1975, 14, 119–137 CrossRef
.
- A. Ali,
et al., Adaptive preservation of orphan ribosomal proteins in chaperone-dispersed condensates, Nat. Cell Biol., 2023, 25, 1691–1703 CrossRef CAS PubMed
.
- I. Yu,
et al., Biomolecular interactions modulate macromolecular structure and dynamics in atomistic model of a bacterial cytoplasm, eLife, 2016, 5, 1–22 Search PubMed
.
- E. H. Zhou,
et al., Universal behavior of the osmotically compressed cell and its analogy to the colloidal glass transition, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 10632–10637 CrossRef CAS
.
- V. Mhatre,
et al., Cell Volume Regulation in Immune Cell Function, Activation and Survival, Cell. Physiol. Biochem., 2021, 55, 71–88 Search PubMed
.
- J. W. Lohr and J. J. Grantham, Isovolumetric regulation of isolated S2 proximal tubules in anisotonic media, J. Clin. Invest., 1986, 78, 1165–1172 CrossRef CAS PubMed
.
- K. Strange, Cellular volume homeostasis, Am. J. Physiol., 2004, 28, 155–159 Search PubMed
.
- K. Szczepański, K. Kwapiszewska and R. Hołyst, Stability of cytoplasmic nanoviscosity during cell cycle of HeLa cells synchronised with Aphidicolin, Sci. Rep., 2019, 9, 16486 CrossRef
.
- A. Miermont,
et al., Severe osmotic compression triggers a slowdown of intracellular signaling, which can be explained by molecular crowding, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 5725–5730 CrossRef CAS PubMed
.
- A. Agasty, A. Wisniewska, T. Kalwarczyk, K. Koynov and R. Holyst, Macroscopic Viscosity of Polymer Solutions from the Nanoscale Analysis, ACS Appl. Polym. Mater., 2021, 3, 2813–2822 CrossRef CAS PubMed
.
- K. Sozański, A. Wiśniewska, T. Kalwarczyk and R. Hołyst, Activation Energy for Mobility of Dyes and Proteins in Polymer Solutions: From Diffusion of Single Particles to Macroscale Flow, Phys. Rev. Lett., 2013, 111, 228301 CrossRef
.
- T. Kalwarczyk,
et al., Comparative Analysis of Viscosity of Complex Liquids and Cytoplasm of Mammalian Cells at the Nanoscale, Nano Lett., 2011, 11, 2157–2163 CrossRef CAS
.
- B. R. Parry,
et al., The Bacterial Cytoplasm Has Glass-like Properties and Is Fluidised by Metabolic Activity, Cell, 2014, 156, 183–194 CrossRef CAS PubMed
.
- M. Guo,
et al., Cell volume change through water efflux impacts cell stiffness and stem cell fate, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, E8618–E8627 CAS
.
- J. T. Lennon and S. E. Jones, Microbial seed banks: the ecological and evolutionary implications of dormancy, Nat. Rev. Microbiol., 2011, 9, 119–130 CrossRef CAS PubMed
.
- A. Flamholz, R. Phillips and R. Milo, The quantified cell, Mol. Biol. Cell, 2014, 25, 3497–3500 CrossRef PubMed
.
- D. Ranava,
et al., Bidirectional sequestration between a bacterial hibernation factor and a glutamate metabolising protein, Proc. Natl. Acad. Sci. U. S. A., 2022, 119, 257119 CrossRef PubMed
.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d4nr03620d |
| This journal is © The Royal Society of Chemistry 2025 |